The absence of a chloroplast protein kinase leads to remodeling of photosynthetic protein complexes, uncovering an intricate protein phosphorylation network.
Abstract
In plants and algae, the serine/threonine kinase STN7/STT7, orthologous protein kinases in Chlamydomonas reinhardtii and Arabidopsis (Arabidopsis thaliana), respectively, is an important regulator in acclimation to changing light environments. In this work, we assessed STT7-dependent protein phosphorylation under high light in C. reinhardtii, known to fully induce the expression of LIGHT-HARVESTING COMPLEX STRESS-RELATED PROTEIN3 (LHCSR3) and a nonphotochemical quenching mechanism, in relationship to anoxia where the activity of cyclic electron flow is stimulated. Our quantitative proteomics data revealed numerous unique STT7 protein substrates and STT7-dependent protein phosphorylation variations that were reliant on the environmental condition. These results indicate that STT7-dependent phosphorylation is modulated by the environment and point to an intricate chloroplast phosphorylation network responding in a highly sensitive and dynamic manner to environmental cues and alterations in kinase function. Functionally, the absence of the STT7 kinase triggered changes in protein expression and photoinhibition of photosystem I (PSI) and resulted in the remodeling of photosynthetic complexes. This remodeling initiated a pronounced association of LHCSR3 with PSI-LIGHT HARVESTING COMPLEX I (LHCI)-ferredoxin-NADPH oxidoreductase supercomplexes. Lack of STT7 kinase strongly diminished PSII-LHCII supercomplexes, while PSII core complex phosphorylation and accumulation were significantly enhanced. In conclusion, our study provides strong evidence that the regulation of protein phosphorylation is critical for driving successful acclimation to high light and anoxic growth environments and gives new insights into acclimation strategies to these environmental conditions.
Oxygenic photosynthesis converts solar energy into chemical energy. This energy is utilized for carbon dioxide assimilation, allowing the formation of complex organic material. Plant photosynthesis is performed by a series of reactions in and at the thylakoid membrane, resulting in light-dependent water oxidation, NADP reduction, and ATP formation (Whatley et al., 1963). These light reactions are catalyzed by two photosystems (PSI and PSII). A third multiprotein complex, also embedded in the thylakoid membrane, is the cytochrome b6f (cyt b6f) complex that links photosynthetic electron transfer processes between the two photosystems and functions in proton translocation. The ATP synthase takes advantage of the proton-motive force that is generated by the light reactions (Mitchell, 1961) to produce ATP. ATP and NADPH, generated through linear electron flow from PSII to PSI, drive the Calvin-Benson-Bassham cycle (Bassham et al., 1950) to fix CO2. Alternatively, cyclic electron flow (CEF) between PSI and the cyt b6f complex solely produces ATP (Arnon, 1959).
Under normal growth conditions, CEF provides additionally required ATP for CO2 fixation (Lucker and Kramer, 2013), counteracts overreduction of the PSI acceptor side under stressful environmental cues, and readjusts the ATP poise, leading to increased lumen acidification important for photoprotection (Alric, 2010; Peltier et al., 2010; Leister and Shikanai, 2013; Shikanai, 2014). In microalgae and vascular plants, CEF relies on the NAD(P)H dehydrogenase-dependent and/or PROTON GRADIENT REGULATION5 (PGR5)-related pathways (Munekage et al., 2002, 2004; Petroutsos et al., 2009; Tolleter et al., 2011; Johnson et al., 2014). For both pathways, supercomplexes consisting of PSI-LIGHT HARVESTING COMPLEX I (LHCI) and components of the respective electron transfer routes have been identified. In Arabidopsis (Arabidopsis thaliana), a unique NAD(P)H dehydrogenase-PSI supercomplex with a molecular mass of more than 1,000 kD was discovered (Peng et al., 2008). From Chlamydomonas reinhardtii, Iwai et al. (2010) isolated a protein supercomplex composed of PSI-LHCI, LHCII, the cyt b6/f complex, ferredoxin-NADPH oxidoreductase (FNR), and PROTON GRADIENT REGULATION-LIKE1 (PGRL1).
PGRL1 and PGR5 interact physically in Arabidopsis and associate with PSI to allow the operation of CEF (DalCorso et al., 2008). Functional data suggest that PGRL1 might operate as a ferredoxin-plastoquinone reductase (Hertle et al., 2013). The PGRL1-containing CEF supercomplex isolated from C. reinhardtii is capable of CEF under in vitro conditions in the presence of exogenously added soluble plastocyanin and ferredoxin (Iwai et al., 2010). Terashima et al. (2012) isolated a CEF supercomplex of similar composition from anaerobic growth conditions that was active in vitro and contained proteins such as the chloroplast-localized Ca2+ sensor CAS and ANAEROBIC RESPONSE1 (ANR1), which were also shown to be functionally important for efficient CEF in the alga. Notably, it was suggested that the onset of CEF in C. reinhardtii is redox controlled (Takahashi et al., 2013).
It has been demonstrated that efficient CEF is crucial for successful acclimation to excess light (Munekage et al., 2004; Dang et al., 2014; Johnson et al., 2014; Kukuczka et al., 2014). The most rapid response to excess light, however, relies on a mechanism called nonphotochemical quenching (NPQ). The fastest constituent of NPQ is energy-dependent (qE) quenching, which operates at a time scale of seconds to minutes and regulates the thermal dissipation of excess absorbed light energy, thereby providing effective photoprotection. In vascular plants, the PSII protein PSII SUBUNIT S is essential for qE (Li et al., 2000), whereas qE induction in the green alga C. reinhardtii is mediated by LIGHT-HARVESTING COMPLEX STRESS-RELATED PROTEIN3 (LHCSR3), an ancient light-harvesting protein that is missing in vascular plants (Peers et al., 2009). CEF and qE are complementary for acclimation to excess light, as double mutants deficient in both mechanisms possess additive phenotypes and are highly sensitive to light (Kukuczka et al., 2014). Another constituent of NPQ is the quenching by state transitions. State transitions are important to balance the excitation energy between PSI and PSII (Bonaventura and Myers, 1969; Murata, 1969). Under light conditions where PSII is preferentially excited, both PSII core and LHCII proteins become phosphorylated (Lemeille and Rochaix, 2010). As a consequence, phosphorylated LHCII proteins detach from PSII and partly connect to PSI (state 2). Under conditions where PSI excitation is predominant, this process is reversed. LHCII proteins are dephosphorylated and associate with PSII (state 1). The extent of state transition between vascular plants such as Arabidopsis and C. reinhardtii differs significantly. The proportion of mobile LHCII antenna is about 80% in the alga, whereas in Arabidopsis, only 15% to 20% of LHCII is transferred to PSI under state 2 conditions (Lemeille and Rochaix, 2010). However, the large increase in PSI antenna size in C. reinhardtii has recently been challenged (Nagy et al., 2014; Ünlü et al., 2014): while 70% to 80% of mobile LHCII detached from PSII in response to transition to state 2 conditions, only a fraction of about 20% functionally attached to PSI.
Phosphorylation of LHC proteins requires the function of the STT7 kinase or its ortholog STN7 in C. reinhardtii or Arabidopsis, respectively. In the absence of the STT7/STN7 kinase, the initiation of state transitions is blocked (Depège et al., 2003; Bellafiore et al., 2005). The mobile LHCII fraction of C. reinhardtii includes the two monomeric minor LHCII antenna proteins, CP26 and CP29 (encoded by lhcb5 and lhcb4 genes), and the major chlorophyll a/b binding protein of LHCII, LHCBM5 (Takahashi et al., 2006), but also the LHCSR3 protein was suggested to migrate during state transitions (Allorent et al., 2013). Takahashi et al. (2014) suggested that only CP29 and LHCBM5 directly associate with PSI to form the PSI-LHCI-LHCII supercomplex, while the binding of CP26 could occur indirectly or via the other two proteins. However, it is not yet known whether STT7 directly phosphorylates the LHCII proteins or if this takes place as part of a kinase cascade (Rochaix, 2007). Nevertheless, the direct interaction between STT7 and the LHCII proteins is quite likely, since none of the other chloroplast kinases was found to be specifically required for LHCII phosphorylation (Rochaix, 2014). The activity of the STT7 kinase is mainly determined by the redox status of the plastoquinone pool (Vener et al., 1997; Zito et al., 1999). The identification of a PROTEIN PHOSPHATASE 2C (PP2C)-type phosphatase responsible for the dephosphorylation of the LHCII proteins in Arabidopsis has been described by two studies in parallel pointing to the fact that this enzyme, called PROTEIN PHOSPHATASE1/THYLAKOID-ASSOCIATED PHOSPHATASE38, acts directly on phosphorylated LHCII proteins, in particular when they are associated with the PSI-LHCI supercomplex (Pribil et al., 2010; Shapiguzov et al., 2010). Moreover, it is not known whether these phosphatases are constitutively active or if they are regulated by other means, for example through the redox state of the plastoquinone pool. Nonetheless, both enzymes are conserved in land plants and exhibit orthologous proteins in C. reinhardtii (Rochaix et al., 2012).
Another kinase related to STN7/STT7 is encoded in the Arabidopsis and C. reinhardtii genomes and named STN8 and STATE TRANSITION-LIKE1 (STL1), respectively. STN8 is involved in PSII core subunit phosphorylation and influences the repair of PSII after photodamage (Bonardi et al., 2005; Vainonen et al., 2005). Remarkably, the disassembly of the PSII holocomplex is inhibited in STN7/STN8 double mutants (Tikkanen et al., 2008; Fristedt et al., 2009; Dietzel et al., 2011; Nath et al., 2013), suggesting that the phosphorylation of core subunits is required for PSII disassembly. It was further suggested that STN8 controls the transition between linear electron flow and CEF by the phosphorylation of PGRL1 in Arabidopsis (Reiland et al., 2011). As described for STN7, the activity of STN8 is probably regulated via the redox state of the plastoquinone pool (Bennett, 1991; Fristedt et al., 2009). Notably, the action of STN8 is counteracted by a chloroplast PP2C phosphatase (Samol et al., 2012), allowing for the fast reversibility of STN8-mediated acclimation responses. Thus, it appears that an intricate regulatory network of chloroplast protein kinases and phosphatases evolved in vascular plants and algae that drives the acclimation response to various environmental cues, including excess and changing light settings (Rochaix et al., 2012). As STN7/STT7 and STN8/STL1 kinase activities appear to be controlled by the redox poise of the plastoquinone pool, the plastoquinone pool would be a central player in these acclimation responses. On the other hand, the kinases themselves are subjected to phosphorylation (Reiland et al., 2009, 2011; Lemeille et al., 2010; Wang et al., 2013). However, the functional consequences of this phosphorylation are unknown.
Recent comparative analyses revealed the presence of at least 15 distinct chloroplast protein kinases, suggesting an intricate kinase phosphorylation network in the chloroplast (Bayer et al., 2012). Generally, the phosphorylation of proteins is one of the most abundant posttranslational modifications. In complex eukaryotic systems, protein phosphorylation occurs most frequently on Ser followed by Thr residues, whereas protein phosphorylation of Tyr residues (1,800:200:1) is comparatively rare (Hunter, 1998; Mann et al., 2002). Protein phosphorylation is a general phenomenon in vivo; it is assumed that about one-third of all proteins are phosphorylated at a given time (Cohen, 2000; Ahn and Resing, 2001; Venter et al., 2001; Manning et al., 2002; Knight et al., 2003). A recent large-scale quantitative evaluation of human proteomic data strengthened the importance of protein phosphorylation for cellular function and human biology (Wilhelm et al., 2014). The C. reinhardtii and Arabidopsis genomes encode large kinase families (Arabidopsis Genome Initiative, 2000; Kerk et al., 2002; Merchant et al., 2007), supporting the view that protein phosphorylation also plays an important role in a plant’s life cycle. It is thus evident that the understanding of protein phosphorylation, including the specificity of residues phosphorylated or dephosphorylated in response to cellular as well as environmental factors, is one key to understanding the complex functional biological networks at the whole-system level. Likewise, it is crucial to design experimental setups allowing the linkage between phosphorylation events and particular physiological consequences to be elucidated.
In this regard, we designed experiments to investigate STT7 kinase-dependent phosphorylation dynamics in C. reinhardtii in response to high light and anoxia, employing quantitative proteomics in conjunction with in-depth physiological characterization. These conditions are particularly interesting, as high light conditions are known to fully induce LHCSR3 protein expression and qE, while anoxia promotes CEF activity. Recently, it was demonstrated that qE and CEF are complementary and crucial in acclimation to these environmental cues (Kukuczka et al., 2014). Notably, LHCSR3 phosphorylation was suggested to depend on STT7 function (Bonente et al., 2011), while CEF supercomplex formation was found to be independent of STT7 kinase function (Takahashi et al., 2013), indicating that STT7 function might impact the acclimation to high light and anoxia in different ways. However, our quantitative proteomics and physiological data reveal that STT7-dependent variations in protein phosphorylation profiles have similar dramatic phenotypic consequences in both conditions, strongly suggesting that the regulation of protein phosphorylation is critical for driving successful acclimation to high light and anoxic growth environments.
RESULTS
The acclimation responses of plants toward adverse environmental conditions require and correlate with the phosphorylation of chloroplast proteins, implying the functional importance of this posttranslational modification for a successful survival strategy. The STT7 kinase is an important chloroplast kinase involved in protein phosphorylation events driving acclimation processes. To gain deeper insights into STT7-dependent chloroplast protein phosphorylation in acclimation to high light and anaerobic growth conditions, we analyzed two STT7 mutants, stt7-1 (Depège et al., 2003) and stt7-9 (Cardol et al., 2009), and the wild type for protein phosphorylation by employing phosphoproteomics and quantitative mass spectrometry (MS).
To this end, thylakoid membranes were isolated from the three strains under the respective conditions and digested with trypsin (Figs. 1–3; Supplemental Tables S1–S3; Turkina et al., 2006). The resulting peptides were subjected to sequential elution from immobilized metal affinity chromatography (SIMAC) and using titanium dioxide (TiO2) chromatography for the enrichment of phosphopeptides (Thingholm et al., 2008). In parallel, thylakoid membranes from the wild type (14N labeled) and stt7-1 (15N labeled) were isolated from cells grown for 24 h under high light. These membranes were solubilized with detergent and fractionated by Suc density centrifugation according to previously published work (Tokutsu and Minagawa, 2013). In total, six distinct Suc fractions were collected (see Fig. 4B below). These fractions contained LHCII monomers and trimers, PSII core complexes, PSI-LHCI and PSII-LHCII complexes, as well as putative CEF supercomplexes. Suc gradient fractions were tryptically digested according to the filter-aided sample preparation (FASP) method (Wiśniewski et al., 2009) followed by TiO2 phosphopeptide enrichment. Subsequently, peptides were analyzed by tandem mass spectrometry (MS/MS). For peptide and phosphopeptide identification, the OMSSA and X!TANDEM algorithms were used to evaluate the MS/MS data incorporated as pipeline in Proteomatic (Specht et al., 2011). In total, 884 phosphoproteins and 2,434 distinct phosphopeptides were identified from the thylakoid membranes (Supplemental Tables S1–S3). To enhance identification and quantification, liquid chromatography (LC)-MS/MS runs of enriched (after SIMAC enrichment) and nonenriched peptide samples (after tryptic digestion) were aligned in intensity and retention time domains using piqDB (Höhner et al., 2013; Barth et al., 2014). This database/platform integrated information on peptide identification and quantification qualities into the definition of accurate isotopolog retention time windows, thereby allowing high-quality alignments to be performed. As a result, confident quantification of all peptides in all LC-MS/MS runs is possible, an approach very similar to that of SuperHirn (Mueller et al., 2007). Subsequent label-free quantitation was performed using pyQms, a tool for MS1-based quantitation (Höhner et al., 2013; Barth et al., 2014). Protein and peptide quantitation and peptide phosphorylation events are listed in Supplemental Table S2. For quantitative arguments, these data will be solely considered in the following. Although these criteria are very stringent, hundreds of peptide phosphorylations can be described, to our knowledge for the first time, in a quantitative way and can be partially related to STT7 kinase-dependent action. Correspondingly, the focus of this work was to dissect the phosphorylation events dependent on STT7 kinase activity.
Figure 1.
(Continues on following page.)
Figure 3.
MS-based label-free quantitation (pyQms) of D2/PSBD peptides (numbers) released from trypsin-digested thylakoid membranes. For detailed description, see Figure 1. Results stemming from nonenriched and phosphopeptide-enriched samples of two independent experiments are depicted in log10 scale.
Figure 4.
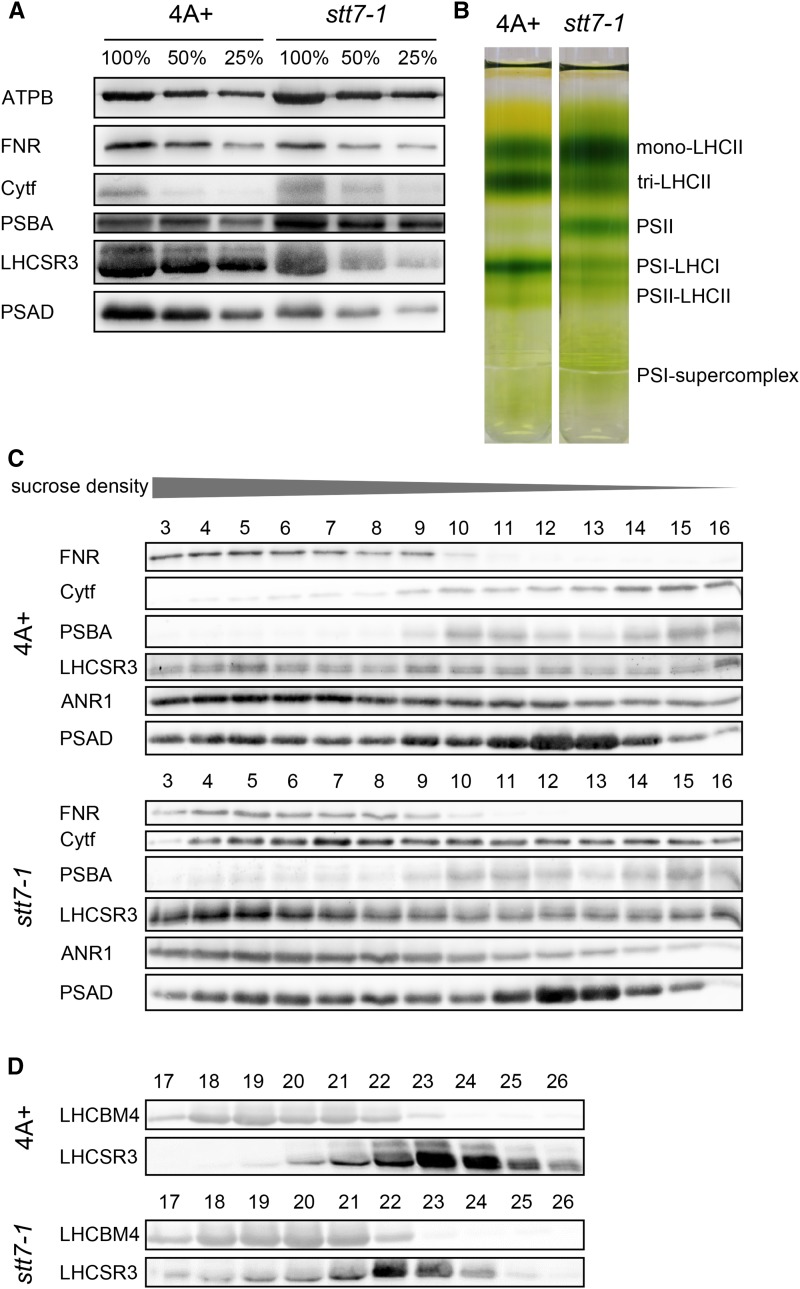
STT7 deficiency alters the organization and protein association of photosynthetic complexes. A, Immunoblot analyses of thylakoid membrane proteins separated by SDS-PAGE (100% = 40 µg of total protein) from high light-acclimated wild-type 4A+ and stt7-1 cells (grown photoautotrophically under 200 µE m−2 s−1 for 24 h) reveal increased amounts of PSBA and cytochrome f and decreased abundances of LHCSR3, PSAD, and FNR in stt7-1. Protein levels of the PSI subunit PSAD, PSII subunit PSBA, cytochrome f, FNR, and LHCSR3 were determined using specific antibodies against these proteins. Detection of the ATP synthase subunit B (ATPB) was used as a loading control. B, Separation of photosynthetic complexes from wild-type 4A+ and stt7-1 by SDG centrifugation after solubilization of the thylakoid membranes with n-dodecyl-α-d-maltoside (α-DM). stt7-1 shows a more pronounced LHCII monomer and PSII core band, while PSI-LHCI and PSII-LHCII supercomplexes were diminished in stt7-1 compared with the wild type. C and D, SDGs were fractionated, and an 80-µL volume of SDG fractions 3 to 16 (C) and 17 to 26 (D) were used for immunoblot detection with the indicated antibodies. The intensities of cytochrome f and LHCSR3 signals were stronger in high molecular-weight fractions 3 to 8 in stt7-1 versus wild-type 4A+, while LHCSR3 levels were decreased in monomeric LHCII fractions 21 to 24.
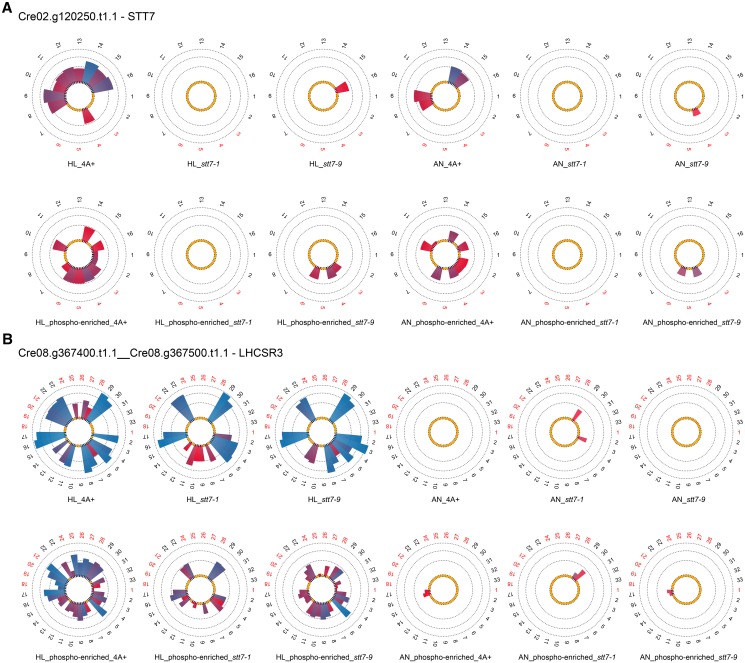
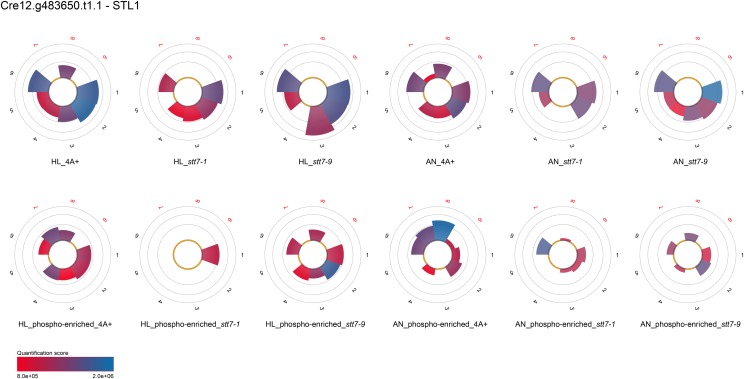
Surprisingly, the quantitative data presented in Figure 1A demonstrated that the STT7 kinase is present in strain stt7-9, since an STT7 phosphopeptide, corresponding to a previously recognized STT7 phosphopeptide (Lemeille et al., 2010), could be identified from stt7-9 thylakoid membranes. The presence of STT7 in stt7-9 is verified by SDS-PAGE and immunoblot analyses using STT7-specific antibodies, showing that stt7-9 accumulates as an approximately 100-kD fusion protein of STT7 and, probably, ARG7, which can be explained by the in-frame insertion in the C-terminal domain of the arg7 cassette used for genetic disruptions of the stt7 gene (Depège et al., 2003; Supplemental Fig. S1). Thus, stt7-9 can be considered a leaky STT7 mutant. Due to the presence of the phosphopeptide DAGLA(p)S533MEEAILK and peptide VNQGALTEAQLMEELGLQEPAPVAPR, we conclude that the kinase domain in stt7-9 is possibly intact while the C terminus with yet unknown function might be absent. In contrast, no STT7 peptides could be identified from stt7-1 either from thylakoids (Fig. 1A) or in Suc gradient fractions (Supplemental Table S2), in line with the fact that no STT7 protein could be observed in the immunoblot assay (Supplemental Fig. S1). Taking advantage of the label-free quantitation event of peptide 16 (VNQGALTEAQLMEELGLQEPAPVAPR) present in wild-type and stt7-9 thylakoids from high light conditions (Supplemental Table S2), we estimate that the STT7 kinase is about 6-fold more abundant in the wild type as compared with stt7-9. Notably, several proteins in this data set show STT7-dependent phosphorylation (Supplemental Tables S1–S3). In many cases, proteins that lacked phosphorylation in stt7-1 were phosphorylated in stt7-9. In more rare cases, protein phosphorylation was present in the wild type but absent in both stt7-1 and stt7-9.
Here, we identified and quantified, to our knowledge for the first time, LHCSR3 peptides that were phosphorylated in an STT7-dependent fashion. Quantification results for LHCSR3 demonstrated the lack of phosphorylation of Ser-26, Ser-28, Thr-32, Thr-33, and Thr-39 in the stt7-1 mutant (Fig. 1B), whereas phosphorylation was observed in stt7-9 (phosphopeptide-enriched fraction) and the wild type (nonenriched and SIMAC-enriched fractions). The absence of phosphorylation in LHCSR3 from stt7-1 was confirmed by analyses of the Suc gradient fractions (Supplemental Table S2). More importantly, it became evident that nonphosphorylated peptides harboring Ser-26, Ser-28, Thr-32, Thr-33, and Thr-39 were more abundant in the stt7-1 mutant compared with the wild type (see peptides 22, 23, 29, and 30). At the same time, the average abundance of peptides, which do not contain known phosphorylation sites and which were quantified in both strains, is slightly higher in the wild type (see peptides 4, 5, 11, and 16). Consequently, the absence of detectable phosphopeptides in the mutant is not due to a lack of LHCSR3 expression. Moreover, the differences between the wild type and the mutant with respect to the abundance of the nonphosphorylated peptides allowed the estimation of the degree of phosphorylation. Accordingly, we could detect 60% more nonphosphorylated peptides harboring the phosphorylatable residues Ser-26, Ser-28, Thr-32, Thr-33, and Thr-39 for the mutant compared with the wild type, likely reflecting the fraction of wild-type-specific phosphorylated peptides. While the STT7-dependent phosphorylation was observed in the N-terminal part of the protein, there was an STT7-independent phosphorylation found in a peptide closer to the C terminus that is not proteotypic and shared between LHCSR1 and LHCSR3. Thus, it remains unclear whether this phosphorylation is present in only one LHCSR protein or in both, but it demonstrates that a second chloroplast kinase, other than STT7, is phosphorylating LHCSR protein(s) at their C-terminal part. Besides the above-mentioned nonproteotypic phosphorylation site, LHCSR1 was not found to be phosphorylated at other proteotypic sites.
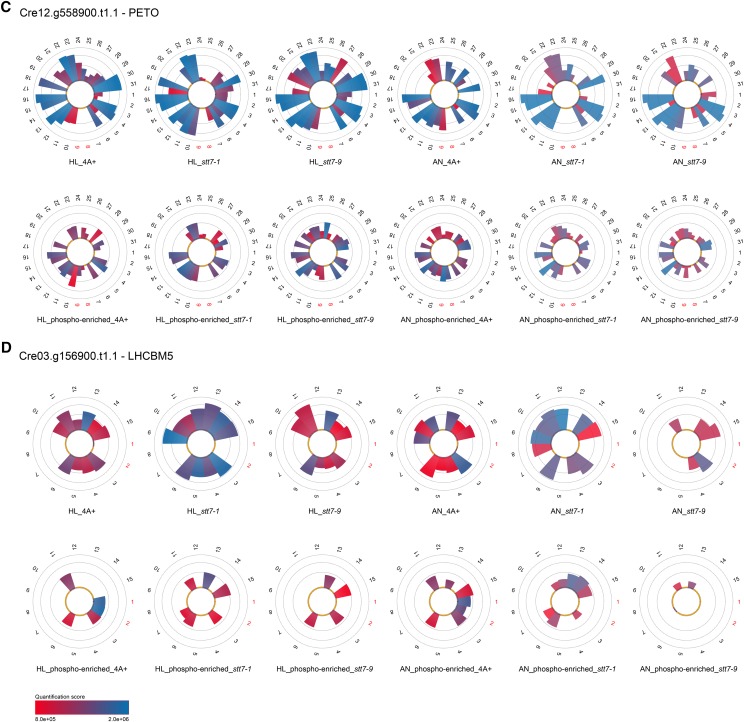
Other noteworthy proteins that showed STT7-dependent phosphorylation as revealed by MS/MS analyses of both thylakoid and Suc density fractions were the cytochrome b6f complex subunit V (PETO), LHCBM5 (Fig. 1, C and D), and THYLAKOID-ENRICHED FRACTION23 (TEF23; Supplemental Tables S2 and S3). Phosphorylation of LHCBM5 has been reported before (Lemeille et al., 2010). Interestingly, phosphorylation events of PETO and TEF23 were also detected in stt7-9, whereas LHCBM5 phosphorylation could not be detected in stt7-1 or stt7-9. For Cre12.g540700.t1.2, a protein predicted to localize to the chloroplast by PredAlgo (Tardif et al., 2012), phosphorylated peptides could be detected in the wild type and stt7-9 but not in stt7-1 in nonenriched high light thylakoid samples, while phosphorylated peptides could be observed in stt7-1 after SIMAC-based enrichment, indicating that the amount of phosphorylation of Cre12.g540700.t1.2 was drastically reduced in the stt7-1 mutant, particularly under high light growth conditions. This is supported by the fact that only one phosphorylation quantitation event (peptide 5) could be detected for Cre12.g540700.t1.2 after TiO2-based phosphopeptide enrichment in Suc density fractions stemming from high light thylakoids of stt7-1, in contrast to the wild type, where four phosphorylation events were recognized. In the case of STL1 (Fig. 2), LHCA6, and PGRL1 (Supplemental Tables S2 and S3), no phosphorylation was seen under high light conditions in stt7-1 (thylakoids and Suc density fractions) in contrast to the wild type. However, phosphorylation was present in stt7-1 under anaerobic conditions as observed in the wild type, implying that phosphorylation of these proteins is independent of STT7 under anoxia. On the other hand, STT7 is involved in phosphorylation under high light settings. Also, PSAF was not phosphorylated in high light thylakoids from stt7-1, while it was in the wild type. However, under anaerobic conditions, phosphorylation of PSAF was enhanced in stt7-1 with respect to the wild type. The absence of phosphorylation for STL1 and PGRL1 under high light is confirmed by the MS/MS analyses of the Suc fractions (Fig. 2; Supplemental Tables S2 and S3). In contrast, PSAF was found to be phosphorylated in Suc density fractions of stt7-1. Subunit IV of the cyt b6f complex appeared to be phosphorylated in an STT7-dependent manner. In thylakoids from high light conditions, its phosphorylation was 3 times less abundant in stt7-1 as compared with the wild type, while it was missing in the mutant under anaerobic conditions. After Suc density centrifugation of detergent-solubilized thylakoids treated with high light, no phosphorylation of subunit IV was detectable in stt7-1 but was present in the wild type, confirming the STT7-dependent phosphorylation. Other proteins, which were phosphorylated in an STT7-dependent way (Supplemental Tables S2 and S3) and only found in phosphopeptide-enriched and/or nonenriched fractions from isolated thylakoids, are the one-half-size ATP-binding cassette (ABC) transporter CADMIUM SENSITIVE1 (CDS1; Cre12.g561550.t1.1), HEAT SHOCK PROTEIN 70B (HSP70B; Cre06.g250100.t1.1), Cre17.g737450.t1.2, Cre06.g284100.t1.1, Cre09.g398400.t1.1, and Cre17.g724150.t1.1. Cre01.g071450.t1.2 shares similarity with STN8 and STN7 kinases from Arabidopsis. This protein was found to be phosphorylated in enriched fractions of high light thylakoids from the wild type and stt7-9 but not in stt7-1. As observed for STL1, this putative kinase was phosphorylated in stt7-1 under anaerobic conditions. However, according to PredAlgo (Tardif et al., 2012) prediction, it is not chloroplast localized.
Figure 2.
MS-based label-free quantitation (pyQms) of STL1 peptides (numbers) released from trypsin-digested thylakoid membranes. For detailed description, see Figure 1. Results stemming from nonenriched and phosphopeptide-enriched samples of two independent experiments are depicted in log10 scale.
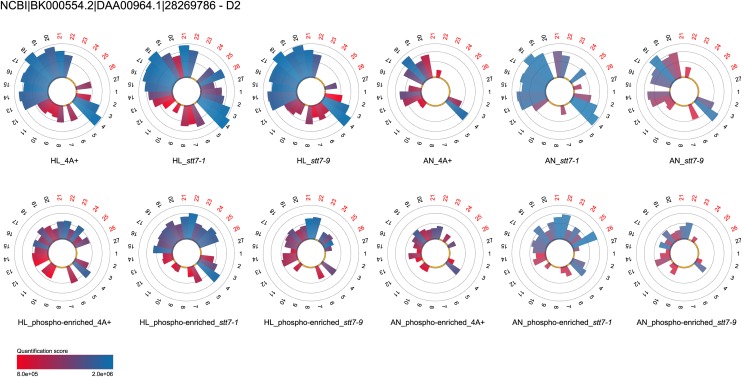
In contrast to proteins showing a lack of phosphorylation in the absence of the STT7 kinase, the reaction center-building PSII subunit PSBD (D2) displayed increased phosphorylation in the absence of the kinase (Fig. 3; Supplemental Table S2). The amounts of phosphorylated PSBD peptides stemming from thylakoids isolated from high light conditions increased in stt7-1 5.6- and 4.4-fold as compared with the wild type and stt7-9, respectively (peptides 21, 22, 25, and 26). At the same time, the overall amount of PSBD nonphosphorylated peptides was only slightly augmented, 1.08- and 1.03-fold in stt7-1 in comparison with the wild type and stt7-9, respectively. In thylakoids from anaerobic conditions, the differences in amounts of phosphorylated PSBD peptides between stt7-1 versus the wild type and stt7-9 were even more pronounced. In this regard, 73- and 44-fold induction of phosphorylation were observed in stt7-1, while the quantities of nonphosphorylated PSBD increased 1.6-fold in relation to the wild type and stt7-9. A similar picture was observed for the phosphorylation of PSBC (CP43; Supplemental Tables S2 and S3). Phosphopeptides 38 and 39 were 2.3- and 1.7-fold more abundant in stt7-1 thylakoids isolated from high light conditions in comparison with the wild type and stt7-9, respectively. These differences further increased to 10.4- and 2.4-fold for the wild type and stt7-9, respectively, when thylakoids stemming from anaerobic conditions were analyzed. Comparing the amounts of nonphosphorylated peptides of stt7-1 under high light and anaerobic conditions revealed that PSBC was slightly enriched, with ratios of 1.2 and 1.07 and 1.1 and 1.4 for the wild type and stt7-9, respectively. In conclusion, deletion of the STT7 kinase caused a significant increase in PSBD and PSBC phosphorylation, whereas depletion of the kinase in stt7-9 led to only slightly higher PSBD and PSBC phosphorylation as compared with the wild type, suggesting that the amount of PSII core phosphorylation is not linked to STT7 in a strict concentration-dependent manner but rather to a threshold in STT7 activity. Notably, other core subunits of PSII, such as PSBA (D1) and PSBB (CP47), were not phosphorylated.
Besides differences in protein phosphorylation in stt7-1 versus the wild type and stt7-9, many proteins were not altered in their phosphorylation status. As examples, it can be affirmed that photosynthetic proteins such as PSAH, PSBR, and PSBO showed a similar protein phosphorylation profile in the three strains as reported previously for PSAH and PSBR in stt7-1 and the wild type (Lemeille et al., 2010). In the case of LHCB4, no qualitative changes in phosphorylation events could be detected between the three strains. However, quantitative changes in LHCB4 phosphorylation were observed. Phosphopeptides 20 and 21 (NNKGS103VEAIVQATPDEVSSENR; charge 2 and 3, respectively) of LHCB4 in stt7-1 were diminished 3-fold with regard to the wild type and stt7-9, whereas phosphopeptides 5 and 6 and the overall amount of nonphosphorylated peptides in stt7-1 were slightly more abundant as compared with the wild type and stt7-9. MS/MS analysis of nonenriched samples derived from FASP-digested sucrose density gradient (SDG) fractions showed that the same phosphopeptides (18 and 19, corresponding to phosphopeptides 20 and 21 found in thylakoids) were present in the wild type (monomeric LHCII) but not in stt7-1, underpinning the notion that this phosphopeptide is diminished in stt7-1 under high light growth conditions (Supplemental Table S2).
In the quantitative analyses of STT7-dependent phosphorylation, the comparison of phosphopeptides from thylakoids and Suc density fractionated thylakoids determined differences independently. However, the Suc density fractionation of detergent-solubilized thylakoids offered another advantage, namely the possibility to analyze the comigration of proteins with known multiprotein complexes such as PSII and PSI, the cyt b6f complex, the ATPase, or the CEF supercomplex. Thus, the aim of these experiments was to determine whether STT7 deficiency alters the dynamic association of proteins in the thylakoid membrane. To define amounts of PSII, PSI, the cyt b6f complex, and the ATPase in thylakoids isolated after 24 h of high light treatment, SDS-PAGE fractionation of wild-type and stt7-1 thylakoids and immunoblotting experiments were performed (Fig. 4A). Using anti-ATPB antibodies, we demonstrated that ATPB, a subunit of the chloroplast ATPase, was similar in amounts in the wild type and stt7-1, while the use of anti-PSBA and anti-CYTF antibodies showed that the amounts of PSBA and CYTF are increased in stt7-1. In contrast, LHCSR3, PSAD, and FNR were slightly decreased in stt7-1, as revealed with specific antibodies directed against these proteins. Overall, these differences were also reflected in quantitative mass spectrometric data stemming from isotopic labeling experiments (Fig. 5). In the next step, thylakoid membranes isolated from wild-type and stt7-1 cells grown in high light were solubilized with detergent and fractionated by Suc density centrifugation according to previously published work (Tokutsu and Minagawa, 2013). Figure 4B shows the corresponding Suc gradients after ultracentrifugation. Six green bands could be distinguished. These green bands corresponded, as revealed by SDS-PAGE fractionation and immunoblotting experiments (Fig. 4, C and D), to LHCII monomers, LHC trimers, PSII core complexes, PSI-LHCI and PSII-LHCII complexes, and putative CEF supercomplexes. The data clearly showed that the intensity of the LHCII monomer and the PSII core band increased in stt7-1 versus the wild type. On the other hand, PSI-LHCI and PSII-LHCII supercomplexes were diminished in stt7-1.
Figure 5.
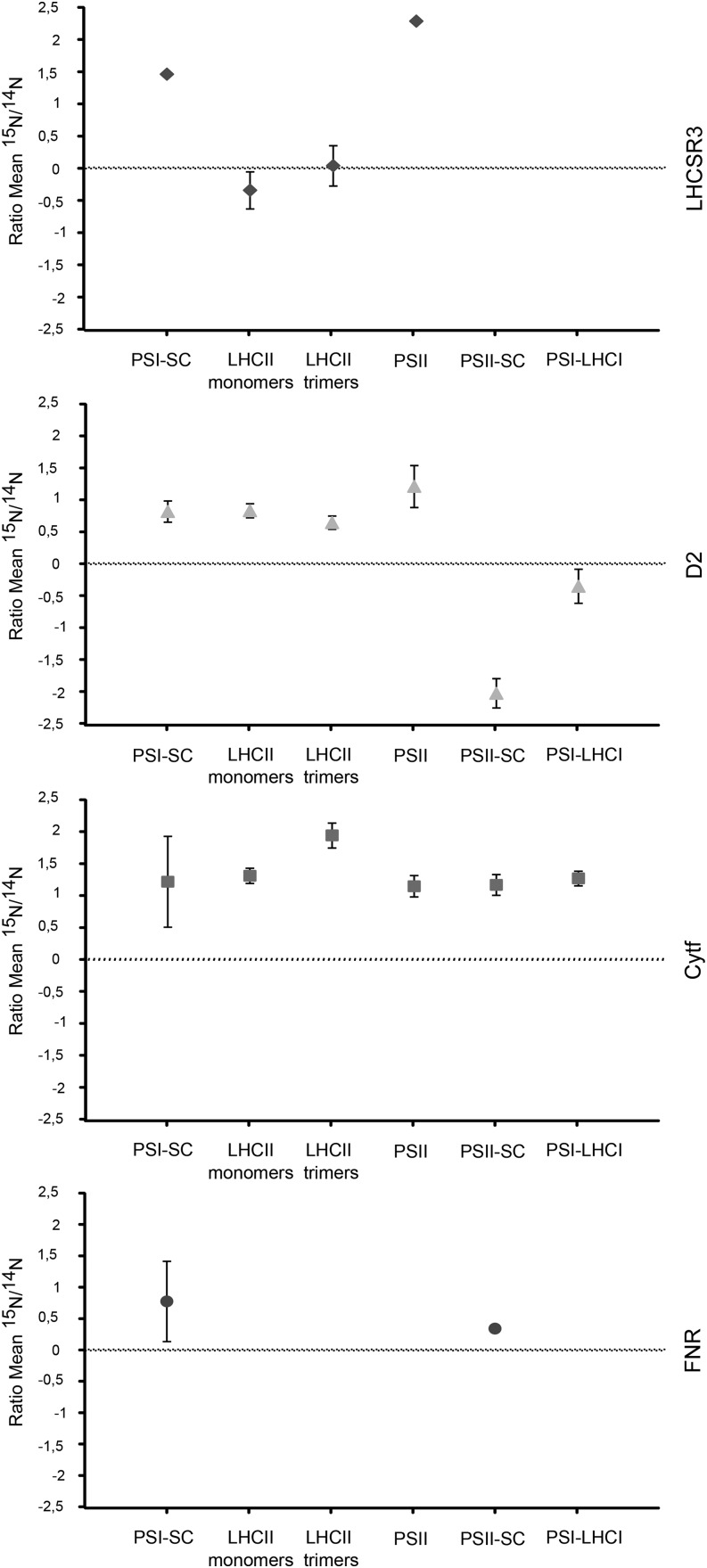
Comparative quantitative proteomic analyses of LHCSR3, D2, cytochrome f, and FNR peptides stemming from chlorophyll-protein complexes of the wild type (14N) and stt7-1 (15N). Volumes corresponding to 100 µg of chlorophyll of isolated thylakoids from 14N-labeled (wild-type 4A+) and 15N-labeled (stt7-1) high light-acclimated cells were mixed, solubilized with α-DM, and separated by SDG centrifugation. The protein complexes obtained were fractionated and analyzed by MS. Log2 ratios of 15N and 14N polypeptides are presented (mean values and their respective sd). PSI-SC, PSI-LHCI-FNR supercomplex; PSII-SC, PSII-LHCII supercomplex.
The immunoblotting experiments after SDS-PAGE fractionation of the Suc gradient fractions using various specific antibodies revealed some very interesting results. The amounts of PSBA in PSII-LHCII (fractions 9–11) and PSAD in PSI-LHCI (fractions 11–13) supercomplexes were slightly diminished in stt7-1 compared with the wild type, as seen in the band pattern of the Suc gradients. Fraction 5 in the wild type and stt7-1 corresponded to the peak fraction of a putative CEF supercomplex, where PSAD, ANR1, CYTF, and FNR comigrated as reported previously (Iwai et al., 2010; Terashima et al., 2012; Takahashi et al., 2013). However, the CYTF signal peaked in stt7-1 in fraction 7 and was not detectable for the wild type. Thus, we designated the putative protein complex in fraction 5, present in the wild type and stt7-1, as the PSI-LHCI-FNR supercomplex as described previously (Takahashi et al., 2014). Moreover, the intensity of the LHCSR3 signal in fraction 5 was also stronger in stt7-1 than in the wild type. To our knowledge, the fact that LHCSR3 comigrated with this PSI- and FNR-containing supercomplex has not been described before. The association of LHCSR3 with PSI-LHCI in state 2 and high light had been described instead (Allorent et al., 2013; Xue et al., 2015). Notably, the amounts of LHCSR3 in the LHCII monomeric and trimeric fractions in stt7-1 decreased in comparison with the wild type (Fig. 4D). Thus, the increase of LHCSR3 in the PSI-LHCI-FNR supercomplex fraction of stt7-1 is likely due to a change in association caused by a lack in STT7 rather than an overall increase in LHCSR3 abundance. This result is further underpinned when considering the LHCSR3 amount in stt7-1 thylakoids (Fig. 4A). On the other hand, the amounts of CYTF increased significantly in fractions 4 to 16 in stt7-1 as compared with the corresponding fractions in the wild type.
To independently assess the amounts of thylakoid membrane proteins in the distinct protein complexes in stt7-1 and the wild type, we performed quantitative mass spectrometric analyses based on isotopic labeling. To this end, thylakoid membranes from 14N-labeled wild-type and 15N-labeled stt7-1 cells grown for 24 h in high light were isolated and subjected to detergent-mediated solubilization and SDG centrifugation and investigated by MS. The data confirmed that CYTF was more abundant in stt7-1 than in the wild type in all Suc fractions analyzed (Fig. 5; Supplemental Table S2). Moreover, FNR was enriched in stt7-1 PSI-LHCI-FNR supercomplex fractions. PSAD amounts were found to be diminished in stt7-1, in agreement with the immunoblot data shown in Figure 4C. Furthermore, these data showed that LHCSR3 was not only significantly more abundant in PSI-LHCI-FNR supercomplexes in stt7-1 but also in PSII core complexes. At the same time, the abundance of PSBD in stt7-1 was increased considerably in PSII core complexes and decreased in PSII-LHCII supercomplexes. The latter notion is in line with the fact that PSII-LHCII supercomplexes were decreased in the stt7-1 Suc density band pattern (Fig. 4B), in agreement with the observation that PSBD and PSBC phosphorylation was higher in stt7-1 PSII core complexes (Supplemental Table S2).
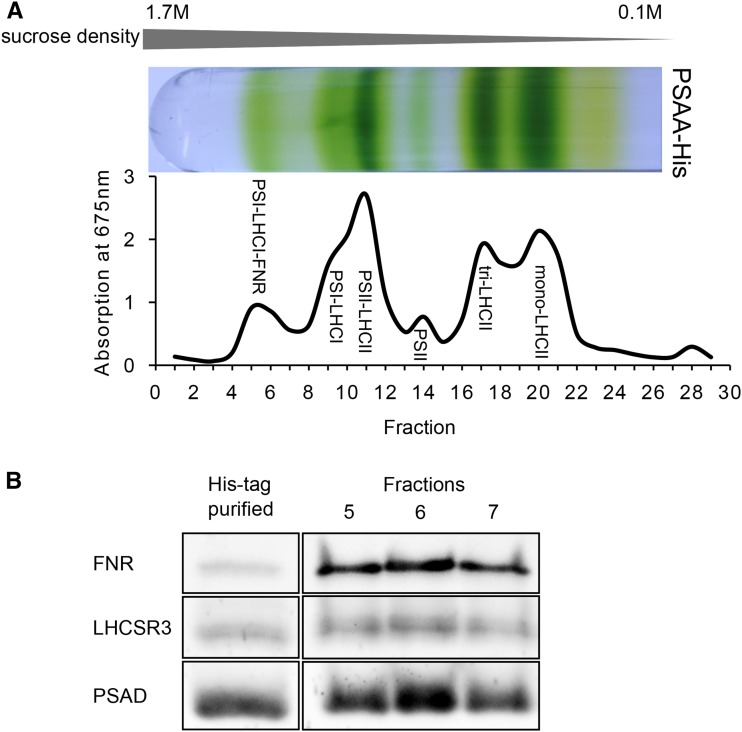
To provide an experimental proof for a link between LHCSR3 and the PSI-LHCI-FNR supercomplex, we took advantage of a C. reinhardtii His6-tagged PSAA strain (Gulis et al., 2008) and purified the supercomplex from cells grown for 24 h in high light (Fig. 6A) using detergent solubilization of isolated thylakoids and Suc density ultracentrifugation. In the next step, the PSI supercomplex from Suc density fractions 5 to 7 were further purified by the tagged PSI subunit (PSAA) via nickel-nitrilotriacetic acid agarose (Ni-NTA) chromatography. Fractions 5 to 7 before and after Ni-NTA purification were separated by SDS-PAGE and analyzed by immunoblotting, employing specific antibodies directed against LHCSR3, PSAD, and FNR (Fig. 6B). The immunoblotting data revealed that PSAD and LHCSR3 were similarly copurified, while FNR was lost via Ni-NTA separation. Therefore, we can conclude that LHCSR3 is associated with PSI in the supercomplex.
Figure 6.
LHCSR3 is attached to PSI-LHCI-FNR supercomplexes. A, Thylakoid membranes from high light-acclimated PSAA-His strain (PSI-His6 tagged) were isolated, α-DM solubilized, and separated by SDG centrifugation. Bottom-to-top fractionation was performed prior to measuring absorption at 675 nm. Fractions 5 to 7 (corresponding to the PSI-LHCI-FNR supercomplex) were subjected to nickel-affinity purification. B, Polypeptides in fractions 5 to 7 and the His-purified eluate (PSI-LHCI-FNR supercomplex) were analyzed by immunoblotting with antibodies directed against PSAD, FNR, and LHCSR3.
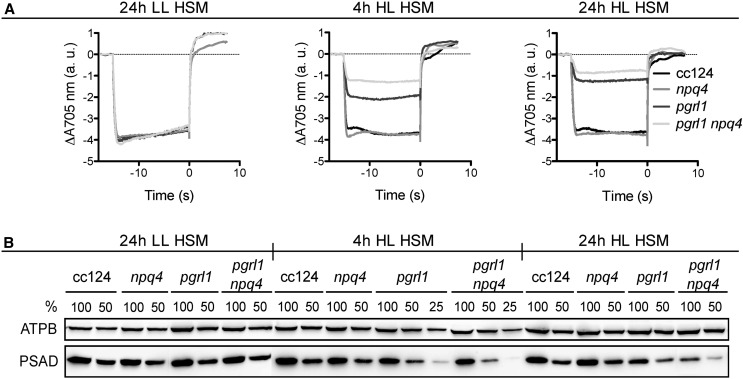
What is the function of LHCSR3 associated with PSI within the PSI-LHCI-FNR supercomplex? A possible scenario might be that LHCSR3 is involved in the photoprotection of PSI within such supercomplexes. As recently reported, PSI from pgr5, pgrl1, and pgrl1 npq4 mutant strains (npq4 is lacking LHCSR3; Peers et al., 2009) was susceptible to photoinhibition in high light (Johnson et al., 2014; Kukuczka et al., 2014). To investigate whether the PSI photoinhibition occurred faster in pgrl1 npq4 due to a lack of LHCSR3, we performed a time-course experiment under high light conditions. As reported earlier (Kukuczka et al., 2014), the amount of oxidizable P700 (special chlorophyll pair in PSI), representing the quantity of efficient PSI, was comparable in the wild type, pgrl1, npq4, and pgrl1 npq4 at low light intensities (Fig. 7A). Interestingly, after 4 h at high light, the extent of oxidizable P700 was already further diminished in pgrl1 npq4 as compared with pgrl1. The same was true after 24 h of high light, while the amounts of oxidizable P700 in the wild type and npq4 remained comparable to the low-light control. Fractionation of wild-type, pgrl1, npq4, and pgrl1 npq4 cells by SDS-PAGE and immunoblotting using anti-PSAD and anti-ATPB antibodies revealed that the amount of PSAD diminished in pgrl1 and pgrl1 npq4 after 4 h and even more after 24 h of high light, whereas the amount of ATPB remained similar over the time course (Fig. 7B). In line with the observation made for the decrease in amounts of oxidizable P700, the immunoblotting data confirmed that the decrease in PSAD was more pronounced in pgrl1 npq4 than in pgrl1. This supports the notion that LHCSR3 has a functional impact, either direct or indirect, on the photoprotection of PSI under conditions where PGRL1 function is absent. The deficiency of PGRL1 affects CEF and will alter the redox poise and lumen ΔpH of the chloroplast, implying that PSI is particularly prone to photoinhibition under conditions when CEF is impaired, as suggested before (Johnson et al., 2014; Kukuczka et al., 2014).
Figure 7.
Lack of LHCSR3 results in a pronounced photoinhibition of PSI in pgrl1 npq4 compared with pgrl1 cells under high light conditions. A, Maximal P700 oxidation amplitude is severely compromised in pgrl1 and even more in pgrl1 npq4. P700 oxidoreduction kinetics were measured under aerobic conditions with actinic light intensity of 3,300 μmol photons m−2 s−1 in the presence of 40 µm 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) to block PSII, giving access to the full amount of oxidizable P700. Cells were acclimated to low-light conditions (Tris-acetate phosphate [TAP] medium) before incubation (4 μg chlorophyll mL−1) in minimal medium (high salt medium [HSM]) at low light (LL) for 24 h or at high light (HL) for 4 and 24 h. Data represent three biological replicates ± sd. B, PSI is degraded faster in pgrl1 npq4 than in pgrl1 due to high light exposure, as shown by western-blot analysis of the PSAD subunit. ATPB served as a loading control. Whole-cell extracts were loaded per chlorophyll basis (100% = 2.5 µg).
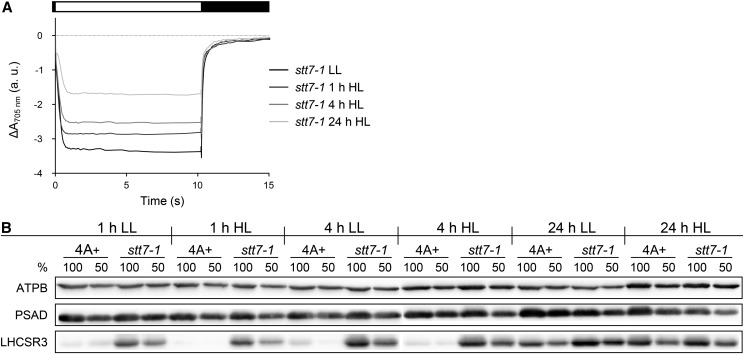
We next investigated PSI photoinhibition under high light conditions in stt7-1. Wild-type and stt7-1 cells were shifted from low-light to high light conditions, and the amount of oxidizable P700 was measured after 1, 4, and 24 h of high light and 24 h of low light. The data shown in Figure 8A revealed that the amounts of oxidizable P700 decreased for stt7-1 gradually to about 50% oxidizable P700 after 24 h in high light in comparison with cells grown for 24 h in low light. SDS-PAGE fractionation of wild-type and stt7-1 cells followed by immunoblotting using anti-PSAD, anti-LHCSR3, and anti-ATPB antibodies revealed that amounts of PSAD diminished in stt7-1 after 24 h of high light to about 50% of the low-light control cells (Fig. 8B). In addition, the comparison to PSAD amounts between stt7-1 cells harvested after 1 and 24 h of high light revealed a 50% reduction of PSAD after 24 h, while no change in abundance was observed for PSAD when comparing wild-type cells after 1 and 24 h of high light. Thus, the conclusion that PSI in stt7-1 is also prone to photoinhibition, although not to the same extent as observed for pgrl1 and pgrl1 npq4, was further supported. Moreover, the immunoblot clearly revealed that in stt7-1, LHCSR3 expression is already present in low light and induced faster in high light, in contrast to the wild type (Fig. 8B), underpinning the assumption that stt7-1 is already stressed under conditions where the wild type is not and possibly explaining why the PSI-LHCI-FNR complex contains more LHCSR3 in stt7-1.
Figure 8.
The stt7-1 mutant shows PSI photoinhibition upon prolonged high light treatment. Wild-type 4A+ and stt7-1 cells grown photoheterotrophically under low-light intensity (LL; 60 µE m−2 s−1) were shifted to high light (HL; 200 µE m−2 s−1)/photoautotrophic conditions for 24 h. A control culture was kept in low light/photoautotrophic conditions. A, PSI oxidation amplitude in samples containing an equal amount of chlorophyll (20 µg mL−1). DCMU was added prior to the measurements to exclude PSI reduction via linear electron flow. The diminishment in the steady-state-level amplitude indicates decreasing amounts of active, photooxidizable PSI. Curves represent averages of three independent samples. Steady-state oxidation levels in relative absorption units were as follows: stt7-1 LL, −3.4 ± 0.2; stt7-1 1 h HL, −2.8 ± 0.1; stt7-1 4 h HL, −2.5 ± 0.2; and stt7-1 24 h HL, −1.7 ± 0.2. B, Changes in protein abundance of the PSI subunit PSAD and the stress-induced light-harvesting protein LHCSR3 detected by immunoblotting. Detection of ATPB was utilized as a loading control. Samples from wild-type 4A+ and stt7-1 strains were taken at the indicated time points from cultures cultivated in low-light and high light conditions. Cell cultures were adjusted to a total chlorophyll amount of 5 µg (100%) or 2.5 µg (50%) prior to protein separation by SDS-PAGE.
DISCUSSION
In this work, we investigated STT7-related protein phosphorylation dynamics in C. reinhardtii in response to high light and anoxia. We intended to assess STT7-dependent protein phosphorylation under conditions where LHCSR3 and qE were fully induced in comparison with conditions known to promote CEF activity. Our quantitative proteomics data revealed that the protein phosphorylation status in STT7-deficient cells was dependent on the environmental condition examined. Furthermore, the phosphoproteome differed between STT7-deficient cells (stt7-1) and cells with diminished expression of a mutant form of STT7 kinase (stt7-9). These are indications that the intricate chloroplast phosphorylation network is highly sensitive and dynamic toward environmental cues and alterations in STT7 kinase function. Variations in chloroplast protein phosphorylation profiles due to the absence of STT7 kinase caused changes in protein expression and photoinhibition of PSI, resulted in the remodeling of photosynthetic complexes, and triggered a pronounced association of LHCSR3 with PSI-LHCI-FNR supercomplexes. Moreover, the absence of STT7 kinase strongly diminished PSII-LHCII supercomplexes, while PSII core complex phosphorylation and formation were enhanced. In conclusion, our study provides strong evidence that the regulation of protein phosphorylation is critical for driving successful acclimation to high light and anoxic growth environments. Furthermore, numerous new potential STT7 kinase substrates were identified and quantified.
Expression of cyt b6f Complex Subunits and Association of LHCSR3 with PSI-LHCI-FNR Are Enhanced in the Absence of STT7 Function
The formation of CEF supercomplexes has been described for state 2 and anoxic conditions (Iwai et al., 2010; Terashima et al., 2012; Takahashi et al., 2013). Here, we provide evidence that such supercomplexes might be formed in high light under aerobic phototrophic conditions, as CYTF comigrates with PSI, FNR, and ANR1 in stt7-1 (Fig. 4C). However, the peak fractions of CYTF in comparison with ANR1, PSAD, and FNR are different, and it is currently unknown whether the putative CEF supercomplex is functional under high light conditions. The comigration of PSI, FNR, and ANR1 in the wild type and stt7-1 points to the formation of a PSI-LHCI-FNR supercomplex in high light, as described previously (Takahashi et al., 2014). Notably, the expression of cyt b6f complex subunits in stt7-1 was enhanced (Fig. 4, A and C; Supplemental Tables S2 and S3). This increased expression was also observed under anoxic conditions (Supplemental Table S2; Supplemental Fig. S3). Interestingly, as observed for high light, under anoxic conditions more CYTF and PETO comigrated with the CEF supercomplex peak fraction (Fig. 4C; Supplemental Table S3; Supplemental Fig. S2). It is currently unclear how this increase of expression is achieved. As the STT7 kinase is associated with the cyt b6f complex (Lemeille et al., 2009), its absence might impact the assembly-controlled biosynthesis of the complex (Choquet et al., 2001). On the other hand, the increase in complex abundance could be indirectly modulated (e.g. via changes in the physiological status of the cells). Depletion in CEF supercomplex-associated proteins such as ANR1 or PGRL1 combined with a reduced CEF capacity caused a severe growth defect under high light and/or anoxic conditions (Terashima et al., 2012; Kukuczka et al., 2014), indicating a requirement of efficient CEF for acclimation to these conditions.
At the same time, a remodeling of PSI-LHCI-FNR supercomplexes occurred by increasing the amount of LHCSR3 associated with PSI within the supercomplex, as revealed by affinity purification using the His6-tagged PSAA strain (Fig. 6). The function of LHCSR3 bound to PSI is likely to protect the complex from photoinhibition (Figs. 7 and 8). The support for this idea is that a pgrl1 npq4 mutant strain displayed a more pronounced PSI photoinhibition in high light as compared with pgrl1 (Kukuczka et al., 2014; this study). However, these data cannot exclude an indirect protective function of LHCSR3 with regard to shielding PSI from photoinhibition under high light stress.
Nonetheless, these data suggest that nonphosphorylated LHCSR3 was bound preferentially to PSI-LHCI-FNR in stt7-1. In agreement, only a small amount of phosphorylated LHCSR3 was found to be associated with the respective supercomplex in the wild type. In conclusion, the binding of LHCSR3 to PSI-LHCI-FNR is negatively correlated with the activity of the STT7 kinase. In this scenario, the inactivation of STT7-dependent phosphorylation via reduction of the luminal STT7 disulfide bridge due to prolonged high light, as outlined by Lemeille et al. (2009), would increase the amount of nonphosphorylated LHCSR3, which in turn would increase its binding to PSI-LHCI-FNR, thereby protecting PSI and resembling the situation in stt7-1.
Absence of STT7 Function Causes High Light-Induced Photoinhibition of PSI and Increased PSII Core Phosphorylation and Formation
Despite the binding of LHCSR3 to PSI, a sensitivity of PSI to photoinhibition was observed in stt7-1. The extent of photoinhibition was not as pronounced as seen for the mutant strains pgrl1 and pgr5 (Figs. 7 and 8; Johnson et al., 2014; Kukuczka et al., 2014), but it was significant. It has been reported that STT7/STN7 function is essential for state transitions in algae and vascular plants, respectively (Depège et al., 2003; Bellafiore et al., 2005). State transitions are important for an efficient balancing of excitation energy between PSI and PSII (Bonaventura and Myers, 1969; Murata, 1969). They involve the phosphorylation of LHCII proteins by STT7/STN7, which then detach from PSII and in part migrate to PSI (state 2). As reported before, LHCBM5 is phosphorylated in state 2 settings under anaerobic and high light conditions in C. reinhardtii. Moreover, LHCBM5 phosphorylation was shown to be STT7 dependent (Lemeille et al., 2010). Notably, as found for stt7-1 in our study, no LHCBM5 phosphorylation was detectable in stt7-9. Like stt7-1, stt7-9 is unable to perform state transitions (Cardol et al., 2009). So far, phosphorylation of LHCBM5 has only been detected when the protein was associated with PSI-LHCI complexes (Takahashi et al., 2006). Hence, the inability to phosphorylate LHCBM5 might explain the fact that both mutants are locked in state 1. On the other hand, no increased expression of cyt b6f complex subunits, augmented PSII core formation, or enhanced binding of LHCSR3 to PSI-LHCI-FNR was observed in stt7-9 (Supplemental Table S2; Supplemental Fig. S2). Moreover, phosphorylation profiles of stt7-9 and stt7-1 showed clear differences, likely explaining these phenotypic differences. These findings indicate that triggering the association of LHCII with PSI requires far more active STT7 kinase than triggering the acclimation process, which permits full PSII-LHCII supercomplex formation and circumvents PSI photoinhibition in high light conditions. We cannot exclude that STT7 from stt7-9 is differently regulated due to its altered protein sequence (see below). However, the lack of a functional interconnection between PSI and LHCII cannot explain the phenotypic differences between stt7-1 and stt7-9. The requirement of STN7 for effective retrograde signaling has been also described for Arabidopsis, as growth phenotypes and diminished relative amounts of PSI had been found in stn7 Arabidopsis plants grown in fluctuating light conditions (Tikkanen et al., 2010). Along the same line, growth phenotypes were also observed in plants treated with fluctuating PSII and PSI lights (Bellafiore et al., 2005).
Recently, the process of state transitions was revisited in C. reinhardtii (Nagy et al., 2014; Ünlü et al., 2014). Interestingly, these new data indicated that although 70% to 80% of LHCII detached from PSII in state 2 conditions, only a fraction of about 20% was attached to PSI. It was postulated that the disconnected antenna complexes form energetically quenched complexes that are protected against photodamage and exhibit a shortened excited-state fluorescence lifetime (Ünlü et al., 2014). In addition, it was suggested that phosphorylated LHCII may also remain with PSII but become quenched (Nagy et al., 2014). Importantly, these studies were conducted under low light in photoheterotrophic conditions where the expression of LHCSR3 is not expected. Thus, the observed quenching should be independent of LHCSR3 and qE. In this regard, it is important to note that the phosphorylation of LHCB4 phosphopeptides 20 and 21 (NNKGS103VEAIVQATPDEVSSENR; charge 2 and 3, respectively) were diminished 2- to 3-fold in stt7-1 thylakoids isolated from high light conditions in comparison with stt7-9 and the wild type (Supplemental Table S2), respectively. However, here, a postulated STT7-dependent LHCB4 phosphorylation (Lemeille et al., 2010) was found to be present in the absence of STT7. In total, 12 different phosphorylation sites were detected, five more than described previously (Turkina et al., 2006). Our data indicate that under high light and photoautotrophic conditions, the phosphorylation of LHCB4 is changed merely quantitatively by the absence of STT7. LHCB4 is a minor core antenna protein of PSII that is suggested to be a linker between PSII core and LHCBM proteins and likely involved in excitation energy transfer between the trimeric LHCBM proteins and the PSII core (Dainese and Bassi, 1991; Dekker and Boekema, 2005; van Amerongen and Croce, 2013) as well as in state transitions in C. reinhardtii (Tokutsu et al., 2009). Moreover, it has been suggested that the phosphorylation of LHCB4 and LHCB5 is responsible for the detachment of all LHCBM polypeptides from PSII in the transition from state 1 to state 2. Notably, we were unable to detect LHCB5 phosphorylation in the quantitative data. Since changes in LHCB4 phosphorylation status between the wild type and stt7-9 were similar, it is possible that the larger reduction in LHCB4 phosphorylation in stt7-1 directly affected the unbinding of LHCBM proteins from PSII. Additionally, phosphorylated LHCB4 was not detected to be bound to the PSII core and was suggested to be associated with other free LHCBMs (Iwai et al., 2008), thereby decreasing the amount and possibly the organization of unbound LHCII polypeptides. It is tempting to speculate that this altered phosphorylation status might affect the formation of energetically quenched LHCII complexes, since differences in LHCII complex formation became apparent as more monomeric LHCII and less trimeric LHCII was observed in stt7-1 as compared with the wild type (Fig. 4B).
Moreover, the amount of LHCSR3, which comigrated with the other free LHCII polypeptides, was severely diminished in stt7-1 (Fig. 4D) due to enhanced comigration with PSI in the PSI-LHCI-FNR fraction, which in turn would certainly have an impact on the formation of LHCII complexes that are energetically quenched by LHCSR3, as it has been shown to quench excited LHC protein-bound chlorophyll molecules (Bonente et al., 2011). Therefore, a lack in LHCSR3 association with aggregated LHCII complexes would likely promote photooxidative stress, because quenching capacity should be lower.
It had been suggested that PSII core subunit phosphorylation functions as a mechanism to protect photodamaged PSII complexes under high light conditions from proteolytic degradation (Koivuniemi et al., 1995; Kruse et al., 1997; Turkina et al., 2006). Additionally, PSII core phosphorylation has been implicated in the regulation of the repair cycle of photodamaged PSII (Aro et al., 1993; Tikkanen et al., 2008; Fristedt et al., 2009; Dietzel et al., 2011; Nath et al., 2013) by facilitating the migration of damaged PSII centers from grana to stroma thylakoid, where damaged PSBA is replaced with a newly synthesized copy. Phosphorylation of PSBD and PSBC had been also proposed to facilitate the dissociation of peripheral light-harvesting antenna (Turkina et al., 2006; Iwai et al., 2008). Our quantitative data clearly showed that PSBD and PSBC were significantly phosphorylated to a higher degree in stt7-1 thylakoids and particularly in PSII core complexes compared with the wild type. This is in line with recent anti-phospho-Thr immunoblot data on isolated stt7-1 and stn7 thylakoids (Fristedt et al., 2010; Lemeille and Rochaix, 2010). Our quantitative data also demonstrated that the formation of PSII core complexes was highly enhanced in stt7-1 and that at the same time the amount of PSII-LHCII supercomplexes was significantly decreased in comparison with the wild type, thus indicating that the absence of STT7 was responsible for this transition and suggesting that PSII-LHCII complexes were prone to photoinhibition in high light-acclimated stt7-1.
It is conceivable that strong PSII core phosphorylation in stt7-1, possibly due to a higher requirement in the PSII repair cycle, is linked to enhanced PSII core formation, as it is suggested to trigger a dissociation of LHCII proteins from PSII. As in Arabidopsis (Bonardi et al., 2005; Vainonen et al., 2005), it is proposed that STL1, the C. reinhardtii STN8 ortholog, is required for the phosphorylation of PSII core proteins PSBD and PSBC (Rochaix et al., 2012). Notably, STL1 phosphorylation was absent in stt7-1 under high light but present in anoxic conditions. Yet, PSBD and PSBC phosphorylation was detected under high light and anoxic conditions. Thus, STL1 phosphorylation is not required for PSII core phosphorylation in C. reinhardtii, and STL1 phosphorylation is not strictly dependent on STT7.
The Intricate Chloroplast Phosphorylation Network Is Highly Dynamic and Sensitive toward Environmental Cues and Alterations in Kinase Function
Phosphorylation of STL1, LHCSR3, and PETO, for the latter in particular under anoxia, was present in stt7-9 but reduced with regard to the wild type, suggesting that the level of decline corresponds and correlates to the diminishment in amounts of STT7 kinase in stt7-9. On the other hand, no phosphorylation was observed for LHCBM5, another abundant substrate, in stt7-1 and stt7-9. Here, it is tempting to speculate that besides the depletion in kinase abundance, the proposed regulatory C-terminal part of STT7, which is missing in stt7-9, is also involved in substrate recognition and phosphorylation efficiency and could also be responsible for the changes in phosphorylation detected between stt7-1 and stt7-9.
As revealed for STL1, the phosphorylation of PGRL1 was absent under high light but present under anoxic conditions in stt7-1, thus pointing to highly dynamic chloroplast protein phosphorylation that is dependent on environmental cues and alterations in kinase function. It has been suggested that STN8 is responsible for PGRL1 phosphorylation (Reiland et al., 2011). The phosphorylation of PGRL1 by STN8 slowed the transition from CEF to linear electron flow in Arabidopsis, thereby providing a link between phosphorylation and the control of CEF activity (Reiland et al., 2011). In C. reinhardtii, the phosphorylation of PGRL1 was observed under anoxia in stt7-1, but it was not increased compared with the wild type or stt7-9. In contrast, under the same conditions, the capacity of CEF was not diminished in stt7-1 with regard to the two other strains (Supplemental Fig. S3), implying that the CEF activity per se is not dependent on PGRL1 phosphorylation, as CEF rates and PGRL1 phosphorylation are not proportional. In the same line, CEF activity is also not dependent on PETO phosphorylation, as it is absent in stt7-1 under anoxia. It is tempting to speculate that there is a link between PGRL1 and STL1 phosphorylation, since their phosphorylation patterns coincide. Currently, it is unknown which other chloroplast kinase could phosphorylate STL1 or STN8. The protein Cre01.g071450.t1.1 shares similarity with STN7/STN8 from Arabidopsis and showed the same phosphorylation dynamics as STL1. However, this protein is probably not chloroplast localized (Tardif et al., 2012). Interestingly, the phosphorylation of subunit IV of the cyt b6f complex was strongly diminished in stt7-1 under high light but completely missing in anoxia. This is another example showing that protein phosphorylation is not only kinase dependent but also related to the respective environmental conditions. TEF23 was the other STT7-dependent substrate that was not phosphorylated under high light or anoxia in stt7-1. TEF23 is an ortholog of ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN1 (EGY1), a zinc metalloprotease required in Arabidopsis and tomato (Solanum lycopersicum) for proper chloroplast development (Chen et al., 2005; Barry et al., 2012). Plants that are deficient in the protease possessed reduced grana stacks, poorly developed thylakoid lamella networks, and low amounts of chlorophyll a/b-binding proteins. Furthermore, it has been suggested that EGY1 participates in abscisic acid/reactive oxygen species (ROS)-dependent plastid retrograde signaling networks, in particular with regard to the regulation of ammonium stress (Li et al., 2012, 2013). Mutant plants deficient in EGY1 are characterized by diminished hydrogen peroxide accumulation in guard cell chloroplasts, likely caused by an impaired turnover and assembly of membrane-associated components of PSI and PSII (Li et al., 2012). In Arabidopsis, STN7 was proposed to be involved in ROS homoeostasis and ROS-based retrograde signaling through controlling LHCII distribution and the redox balance in the electron transfer chain (Tikkanen et al., 2012). Hence, the STT7 dependency of TEF23 phosphorylation is striking and may reveal that STT7 affects ROS-based retrograde signaling on multiple levels.
Besides the control of redox balance, STT7 might also be involved in the regulation of transport processes in the chloroplast and/or the cell. Accordingly, another STT7 substrate is the protein Cre13.g571500.t1.1, which is phosphorylated in the wild type and stt7-9 but not in stt7-1. This protein shows similarity to various transporters and permeases (Merchant et al., 2007), although its location is unknown. Other transport proteins that were found to be phosphorylated in the wild type and stt7-9 are CDS1 (Cre12.g561550.t1.1), the major Rhesus protein RHP1 (Cre06.g284100.t1.1), and TRANSIENT RECEPTOR POTENTIAL5 (TRP5; Cre09.g398400.t1.1). In stt7-1, however, there was no evidence for protein phosphorylation nor the presence of nonphosphorylated peptides, indicating that either phosphorylation was absent and/or protein expression vastly diminished. CDS1, a one-half-size ABC transporter with similarity to ABC TRANSPORTER OF THE MITOCHONDRION3, suggested to be involved in cadmium resistance (Kim et al., 2006) and iron homeostasis (Chen et al., 2007), is supposedly mitochondrion localized, although proteomic data also suggest chloroplast localization in C. reinhardtii (Terashima et al., 2011). RHP1, on the other hand, is a CO2 channel (Soupene et al., 2002). TRP5 is a member of the TRP multigene superfamily encoding integral membrane proteins that function as ion channels. Notably, this multigene family of channels is absent in vascular plants and represented by 14 potential TRP genes in C. reinhardtii (Fujiu et al., 2011). Currently, it is unknown whether TRP5 is chloroplast localized. However, the absence of STT7-dependent phosphorylation suggests direct or indirect control of TRP5 function and/or presence via STT7. The same holds for the two other discussed transport proteins, since indirect impact cannot be excluded. As TRP channels are frequently involved in tuning Ca2+-dependent responses (Philipp et al., 1998), it is tempting to speculate that STT7-mediated phosphorylation might impact Ca2+-modulated photosynthetic processes such as NPQ and CEF (Petroutsos et al., 2011; Terashima et al., 2012), although a link remains to be shown. In this regard, it is remarkable that the phosphorylation and abundance of chloroplast HSP70B (Cre06.g250100.t1.1) were clearly diminished in stt7-1. Phosphorylation of HSP70B was shown to be Ca2+ stimulated (Amir-Shapira et al., 1990), and more generally, the heat shock response in C. reinhardtii was recently revealed to be sensitive to the kinase inhibitor staurosporine and the removal of extracellular Ca2+ (Schmollinger et al., 2013). Thus, it is conceivable that an increase in HSP70B phosphorylation is either directly or indirectly meditated by STT7 and is part of the acclimation process to high light, and it remains elusive whether changes in chloroplast Ca2+ due to the loss of STT7 function contribute to the process.
Thus, deletion of STT7 caused manifold phenotypic consequences, which manifested in alterations in the phosphoproteome and the abundance of individual proteins. The differences observed under high light and anoxic conditions clearly point to an intricate kinase network in the chloroplast that is more complex than anticipated. Moreover, our data indicate that changes in the chloroplast phosphorylation network extend to alterations in phosphorylation events outside the chloroplast, where direct and indirect relations are challenging to differentiate, yet where phenotypic consequences are very evident, demonstrating the multifaceted consequences of STT7 deletions. Thus, further work is required to dissect the chloroplast kinase-signaling network.
MATERIALS AND METHODS
Strains
The Chlamydomonas reinhardtii 4A+ wild-type line (Peers et al., 2009) was used to compare phosphorylation pattern with STT7 mutant strains stt7-1 (obtained from J.-D. Rochaix; Depège et al., 2003) and stt7-9 (Cardol et al., 2009). Wild-type strain CC124 (nit2−, mt−) served as a control for the comparison of photoinhibition sensitivity with C. reinhardtii pgrl1 (Tolleter et al., 2011), npq4 (Peers et al., 2009), and the pgrl1 npq4 double mutant (Kukuczka et al., 2014). The C. reinhardtii PSAA-His strain (Gulis et al., 2008) was employed to purify His-tagged PSI from supercomplex fractions.
Culture Conditions
Strains were precultured in standard TAP medium at 25°C under continuous light of 20 to 50 μE m−2 s−1 on a rotary shaker (120 rpm). Anaerobic conditions were induced by 4 h of argon bubbling in medium containing TAP at 20 μE m−2 s−1, and cultures were harvested in the exponential growing phase (3–4 × 106 cells mL−1).
Cells were acclimated to low-light conditions (20 μE m−2 s−1) before shifting (4 µg chlorophyll mL−1) to phototrophic growth in minimal medium at 20 μE m−2 s−1 (low light) or 200 μE m−2 s−1 (high light).
For quantitative MS measurements, stt7-1 cells were metabolically labeled by maintaining on 15N-containing TAP agar plates for several generations, then were grown in TAP medium with 15NH4Cl accordingly.
Isolation of Thylakoid Membranes
Thylakoid membranes from C. reinhardtii were isolated as described previously (Terashima et al., 2012; Trompelt et al., 2014). Buffers H2 to H6 additionally contained 10 mm NaF, an inhibitor of endogenous protein phosphatases, preserving in vivo protein phosphorylation of thylakoid membranes.
SDG Ultracentrifugation
Dodecyl β-maltoside-solubilized SDG centrifugation of isolated thylakoids from anaerobic conditions was conducted as described previously (Trompelt et al., 2014).
α-DM-solubilized SDG centrifugation was performed as described (Tokutsu et al., 2012) with the following modifications. Isolated thylakoid membranes from high light-acclimated cells were suspended in H6 buffer (5 mm HEPES, pH 7.5, and 10 mm EDTA) at 0.4 mg mL−1 and solubilized with 1% (v/v) α-DM for 5 min on ice in the dark. Separation of photosynthetic complexes was obtained by loading solubilized membranes (200 µg of chlorophyll) on discontinuous SDGs with Suc concentrations from 1.3 to 0.1 m (0.02% [v/v] α-DM and 10 mm NaF) and centrifugation at 33,000 rpm for 14 to 16 h using an SW41Ti rotor (Beckmann) at 4°C.
SDGs were fractionated from bottom to top, and chlorophyll absorption of the fractions was measured at 675 nm.
Protein Separation and Immunoblot Analyses
Protein samples were equalized to the same volume of each fraction (SDGs), the same amount of chlorophyll (whole cells and thylakoids), and the same amount of total protein (thylakoids) and separated by 13% (w/v) SDS-PAGE. SDS-PAGE and immunoblot analyses were conducted as described (Hippler et al., 2001; Naumann et al., 2005) using the following antibodies: ANR1 (Terashima et al., 2010) ATPB (Agrisera), LHCSR3 (Naumann et al., 2007), PSAD (Naumann et al., 2005), PSBA (Agrisera), FNR (a kind gift from Y. Takahashi, Okayama University), cytochrome f (Agrisera), and PETO (a kind gift from F.-A. Wollman, Institut de Biologie Physico-Chimique). Secondary antibody was anti-rabbit (Invitrogen).
Nickel Affinity Chromatography
To determine whether LHCSR3 was specifically bound to the PSI-LHCI-FNR supercomplex, Ni-NTA chromatography of PSI-containing supercomplexes was carried out using the PSAA-His strain carrying a His6-tagged PSAA protein (Gulis et al., 2008).
Thylakoid membranes of 24-h high light-acclimated PSAA-His cultures were isolated as described. Membranes were then solubilized with α-DM, and photosynthetic complexes were separated by SDG centrifugation as described except for the following. An SDG with Suc concentrations from 1.7 to 0.1 m was used. After SDG ultracentrifugation and fractionation of the gradient, a PSI supercomplex containing fractions 5 to 7 was pooled, diluted with a 4× volume of column buffer (containing 25 mm HEPES-KOH (pH 7.5), 100 mm NaCl, 5 mm MgSO4, 10% [v/v] glycerin, and 0.02% α-DM), and loaded onto an Ni-NTA column that had been preequilibrated with column buffer. The column was washed with column buffer containing 2 mm imidazole. PSI was finally eluted with column buffer containing 200 mm imidazole and 40 mm MES-NaOH (pH 6).
Tryptic Digestion of Thylakoid Membrane Proteins
For the identification of phosphorylated proteins in the 4A+ wild type and stt7 mutants, isolated thylakoid membranes corresponding to 300 µg of chlorophyll were tryptically shaved as described (Turkina et al., 2006). Thylakoid membranes were washed twice with 25 mm NH4HCO3 (containing 10 mm NaF) and subsequently digested with sequencing-grade modified trypsin (Promega).
FASP of SDG Fractions
Isotopic 14N and 15N labeling was performed for the 4A+ wild type and stt7-1, respectively. Thylakoid membranes were isolated from high light-acclimated (24-h) cell cultures. For quantitative MS analysis, thylakoid membranes from 14N-labeled wild type and 15N-labeled stt7-1 were mixed (based on equal amounts of chlorophyll), solubilized with α-DM, and separated by SDG centrifugation. The Suc gradient was fractionated, and protein concentrations of peak fractions of LHCII monomers, LHCII trimers, PSII core complexes, PSI-LHCI, PSII-LHCII, and supercomplexes were determined using the Pierce BCA Protein Assay Kit (Thermo Scientific) according to the manufacturer’s instructions. Samples were tryptically digested in 0.5-mL Amicon Ultra ultrafiltration devices (30-kD cutoff; Millipore) according to the FASP method (Wiśniewski et al., 2009, 2011) with minor modifications. Proteins were reduced with 100 mm dithiothreitol in 100 mm Tris-HCl (pH 8.5) and 8 m urea (UA) at room temperature for 30 min. Subsequently, excess dithiothreitol was removed by buffer exchange using UA. Alkylation of Cys residues was performed by adding 50 mm iodoacetamide in UA followed by incubation for 20 min in the dark. Subsequently, samples were washed three times each with UA and 50 mm NH4HCO3.
Proteins were tryptically digested overnight on the filter devices with an enzyme-to-protein sample ratio of 1:50. After the digestion, peptides were eluted from the column by centrifugation, acidified with 20 μL of 2% (v/v) formic acid, and dried by vacuum centrifugation. Phosphopeptides were subsequently enriched by TiO2 affinity chromatography prior to MS analysis.
Enrichment of Phosphopeptides
Peptides released from the thylakoid membranes by trypsin were dried in a vacuum centrifuge and subjected to a combined phosphopeptide enrichment involving immobilized metal affinity chromatography (IMAC) followed by TiO2 chromatography (Thingholm et al., 2008).
SIMAC
Eighty microliters of PhosSelect Iron Affinity Gel (Sigma) was used per sample. Beads were washed five times by centrifugation (6.7g, 30 s) with 250 μL of loading buffer (0.25% [v/v] trifluoroacetic acid [TFA] and 50% [v/v] acetonitrile). Lyophilized peptide samples were resuspended in 300 μL of loading buffer and incubated with the beads at room temperature for 30 to 60 min with end-to-end rotation. The mixture was transferred to a SnapCap column (Pierce), the flow through was collected, and the column was washed twice with 500 μL of loading buffer. Flow-through and wash solutions were pooled and submitted to TiO2-based phosphopeptide enrichment (see below). Monophosphorylated peptides were eluted from the IMAC column by adding 300 μL of 1% TFA and 20% acetonitrile. Subsequently, multiply phosphorylated peptides were eluted by applying 300 μL of 0.4 m ammonia and immediately acidified with 15 μL of formic acid. The IMAC flow through/wash and IMAC eluates were dried in a vacuum centrifuge.
TiO2 Chromatography
The dry flow-through/wash fraction and eluate 1 (containing the monophosphorylated and contaminating nonphosphorylated peptides) from the IMAC procedure were enriched in a second step using NuTips (NT3TIO). For every sample, a NuTip was washed twice with 50 µL of acetonitrile and equilibrated with loading buffer (1 m glycolic acid in 80% acetonitrile and 2.5% TFA). Sample was suspended in 50 µL of loading buffer and loaded to the tip by slowly aspirating and expelling 50 times. The NuTip was washed with 25 µL of loading buffer followed by 25 µL of wash buffer (80% acetonitrile and 1% TFA). Phosphopeptides bound to the tip were eluted by slowly pipetting 50 µL of elution buffer (0.4 m ammonia water) 10 times up and down. The eluate was acidified by adding 3 µL of 20% formic acid and dried by vacuum centrifugation prior to MS analysis.
LC-MS/MS and Data Analysis
Peptides were separated using the Ultimate 3000 Nanoflow HPLC system (Dionex). The mobile phases were composed of 2% (v/v) acetonitrile and 0.1% (v/v) formic acid in ultrapure water (A) and 80% acetonitrile and 0.1% formic acid in ultrapure water (B). The sample (1 µL) was loaded on a trap column (C18 PepMap 100; 300 µm × 5 mm, 5-µm particle size, 100-Å pore size; Thermo Scientific) and desalted for 4 min using eluent A at a flow rate of 25 µL min−1. Then chromatographic separation was carried out on an RP18 capillary column (Acclaim PepMap100 C18; 75 µm × 15 cm, 3-µm particle size, 100-Å pore size; Thermo Scientific). Peptides were eluted at a flow rate of 300 nL min−1 with the following gradient: 0% to 35% B (0–90 min), 35% to 100% B (90–97 min), and 100% B (97–102 min). The column was reequilibrated with 100% A for 10 min.
The LC system was coupled via a nanospray source to an LTQ Orbitrap XL mass spectrometer (Thermo Finnigan). MS full scans (mass-to-charge ratio 375–1,600) were acquired by Fourier transform-MS in the Orbitrap at a resolution of 60,000 (full width at half maximum) with internal lock mass calibration on mass-to-charge ratio 445.12003. The five most intense ions were fragmented in the linear ion trap by collision-induced dissociation (35% normalized collision energy) with multistage activation of the neutral losses of phosphoric acid (−24.5, −32.7, −49, and −98 D). Automatic gain control was enabled with target values of 5 × 105 and 5 × 103 for MS full scans and MS/MS, respectively. Each sample was analyzed twice, with 100-ms (three microscans) and 150-ms (two microscans) maximum ion trap fill time for MS/MS. Dynamic exclusion was enabled with an exclusion duration of 90s, repeat count of one, list size of 500, and exclusion mass width of ±5 ppm. Unassigned charge states and charged state 1 were rejected.
Peak lists were generated from raw files using msconvert 3.0.5047, developed by the ProteoWizard project (Chambers et al., 2012). For the identification of peptides, multistage activation spectra were matched against the JGI4.3 Augustus 10.2 protein sequence database using OMSSA 2.1.9 (Geer et al., 2004) and X!Tandem (version 2013.09.01; Craig and Beavis, 2004). The maximum number of missed cleavages allowed was two. Mass accuracy was set to 20 ppm for MS1 precursor ions and 0.8 D for product ions. Ser, Thr, and Tyr phosphorylation and oxidation of Met were used as variable parameters. In addition, carbamidomethylation of Cys was set as a fixed modification for samples derived from Suc gradients. Phosphorylation site validation was carried out using PhosphoRS 3.1 (fragment ion tolerance of 0.5 D; Taus et al., 2011) incorporated into Proteomatic (Specht et al., 2011). Retention time alignment and peptide quantification were performed as described previously (Höhner et al., 2013; Barth et al., 2014).
Raw MS data and tables containing all peptide identifications have been deposited at PeptideAtlas with identifier PASS00618.
Chlorophyll Fluorescence Measurements
Fluorescence was measured at room temperature using a Maxi-Imaging PAM chlorophyll fluorometer (Heinz Walz). C. reinhardtii strains were dark adapted for 20 min to allow full oxidation of the plastoquinone pool. The effective photochemical quantum yield of PSII was measured as (Fm′ − F)/Fm′, where Fm′ is maximum PSII fluorescence in the light-adapted state, and total NPQ was calculated as (Fm − Fm′)/Fm′), where Fm is maximum PSII fluorescence in the dark-adapted state.
Spectrophotometric Measurements
P700 absorption measurements of C. reinhardtii cells were performed as described previously (Alric et al., 2010) using a pump and probe light-emitting diode-based JTS 10 spectrophotometer (BioLogic). Prior to the measurements, cells were resuspended in 20 mm HEPES-KOH (pH 7.2) containing 20% (w/v) Ficoll to avoid sedimentation and dark incubated for 20 min. A concentration of 40 μm of the PSII inhibitor DCMU was added to reach conditions in which the sustained steady-state electron transfer occurring in the light can be solely attributed to CEF around PSI. Continuous red actinic light illumination at 630 nm with an intensity of 3,300 μE m−2 s−1 was applied for multiple seconds, followed by a short (30-ms) flash of strong actinic light giving access to the full amount of oxidizable P700.
CEF rates were measured as described (Takahashi et al., 2013) in the presence of 10 µm DCMU. The rate of CEF turnover equals the product of the CEF rate constant with the fraction of reduced P700 under continuous illumination (130 µE m−2 s−1) in steady state. The CEF rate constant was calculated by normalizing the change in the electrochromic shift signal measured at 520 nm at the onset of continuous illumination to the electrochromic shift signal caused by charge separation in PSI due to a saturating, single-turnover flash. The fraction of reduced P700 was assessed as the difference between the P700 oxidation signal measured at 705 nm, under continuous illumination at steady state, and the 705-nm signal caused by a saturating flash leading to full oxidation of PSI in the sample, normalized to the full P700 amplitude. Anaerobic conditions were induced by the addition of 100 mm Glc and 2 mg mL−1 Glc oxidase from Aspergillus niger (Sigma-Aldrich).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Immunoblot detection of STT7 protein from C. reinhardtii thylakoid extracts.
Supplemental Figure S2. STT7 deficiency alters organization and protein association of photosynthetic complexes.
Supplemental Figure S3. CEF rates are significantly increased under anaerobic conditions in wild-type 4A+ and stt7-1.
Supplemental Table S1. Identified phosphoproteins from the wild type, stt7-9, and stt7-1.
Supplemental Table S2. Quantitative representation of peptides and phosphopeptides identified from isolated thylakoid membranes and protein samples obtained after Suc density fractionation.
Supplemental Table S3. Representative phosphopeptides derived from photosynthetic complexes and other proteins relevant to the article.
Supplementary Material
Glossary
- cyt b6f
cytochrome b6f
- CEF
cyclic electron flow
- NPQ
nonphotochemical quenching
- SIMAC
sequential elution from immobilized metal affinity chromatography
- TiO2
titanium dioxide
- FASP
filter-aided sample preparation
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- LC
liquid chromatography
- Ni-NTA
nickel-nitrilotriacetic acid agarose
- ROS
reactive oxygen species
- TAP
Tris-acetate phosphate
- SDG
sucrose density gradient
- α-DM
n-dodecyl-α-d-maltoside
- UA
100 mm Tris-HCl (pH 8.5) and 8 m urea
- IMAC
immobilized metal affinity chromatography
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
Footnotes
This work was supported by the Swiss National Foundation (grant no. 31003A_146300 to M.G.-C.) and by the Deutsche Forschungsgemeinschaft (grant nos. HI 739/7.2 in FOR964 and HI 739/8.1 to M.H.).
References
- Ahn NG, Resing KA (2001) Toward the phosphoproteome. Nat Biotechnol 19: 317–318 [DOI] [PubMed] [Google Scholar]
- Allorent G, Tokutsu R, Roach T, Peers G, Cardol P, Girard-Bascou J, Seigneurin-Berny D, Petroutsos D, Kuntz M, Breyton C, et al. (2013) A dual strategy to cope with high light in Chlamydomonas reinhardtii. Plant Cell 25: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alric J. (2010) Cyclic electron flow around photosystem I in unicellular green algae. Photosynth Res 106: 47–56 [DOI] [PubMed] [Google Scholar]
- Alric J, Lavergne J, Rappaport F (2010) Redox and ATP control of photosynthetic cyclic electron flow in Chlamydomonas reinhardtii. I. Aerobic conditions. Biochim Biophys Acta 1797: 44–51 [DOI] [PubMed] [Google Scholar]
- Amir-Shapira D, Leustek T, Dalie B, Weissbach H, Brot N (1990) Hsp70 proteins, similar to Escherichia coli DnaK, in chloroplasts and mitochondria of Euglena gracilis. Proc Natl Acad Sci USA 87: 1749–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arnon DI. (1959) Conversion of light into chemical energy in photosynthesis. Nature 184: 10–21 [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B (1993) Photoinhibition of photosystem II: Inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Barry CS, Aldridge GM, Herzog G, Ma Q, McQuinn RP, Hirschberg J, Giovannoni JJ (2012) Altered chloroplast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent2 locus of tomato. Plant Physiol 159: 1086–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth J, Bergner SV, Jaeger D, Niehues A, Schulze S, Scholz M, Fufezan C (2014) The interplay of light and oxygen in the reactive oxygen stress response of C. reinhardtii dissected by quantitative mass spectrometry. Mol Cell Proteomics 13: 969–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham JA, Benson AA, Calvin M (1950) The path of carbon in photosynthesis. J Biol Chem 185: 781–787 [PubMed] [Google Scholar]
- Bayer RG, Stael S, Rocha AG, Mair A, Vothknecht UC, Teige M (2012) Chloroplast-localized protein kinases: a step forward towards a complete inventory. J Exp Bot 63: 1713–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellafiore S, Barneche F, Peltier G, Rochaix JD (2005) State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Bennett J. (1991) Protein phosphorylation in green plant chloroplasts. Annu Rev Plant Physiol Plant Mol Biol 42: 281–311 [Google Scholar]
- Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D (2005) Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437: 1179–1182 [DOI] [PubMed] [Google Scholar]
- Bonaventura C, Myers J (1969) Fluorescence and oxygen evolution from Chlorella pyrenoidosa. Biochim Biophys Acta 189: 366–383 [DOI] [PubMed] [Google Scholar]
- Bonente G, Ballottari M, Truong TB, Morosinotto T, Ahn TK, Fleming GR, Niyogi KK, Bassi R (2011) Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol 9: e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardol P, Alric J, Girard-Bascou J, Franck F, Wollman FA, Finazzi G (2009) Impaired respiration discloses the physiological significance of state transitions in Chlamydomonas. Proc Natl Acad Sci USA 106: 15979–15984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, et al. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30: 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Bi YR, Li N (2005) EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J 41: 364–375 [DOI] [PubMed] [Google Scholar]
- Chen S, Sánchez-Fernández R, Lyver ER, Dancis A, Rea PA (2007) Functional characterization of AtATM1, AtATM2, and AtATM3, a subfamily of Arabidopsis half-molecule ATP-binding cassette transporters implicated in iron homeostasis. J Biol Chem 282: 21561–21571 [DOI] [PubMed] [Google Scholar]
- Choquet Y, Wostrikoff K, Rimbault B, Zito F, Girard-Bascou J, Drapier D, Wollman FA (2001) Assembly-controlled regulation of chloroplast gene translation. Biochem Soc Trans 29: 421–426 [DOI] [PubMed] [Google Scholar]
- Cohen P. (2000) The regulation of protein function by multisite phosphorylation: a 25 year update. Trends Biochem Sci 25: 596–601 [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467 [DOI] [PubMed] [Google Scholar]
- Dainese P, Bassi R (1991) Subunit stoichiometry of the chloroplast photosystem II antenna system and aggregation state of the component chlorophyll a/b binding proteins. J Biol Chem 266: 8136–8142 [PubMed] [Google Scholar]
- DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schünemann D, Finazzi G, Joliot P, Barbato B, Leister D (2008) A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285 [DOI] [PubMed] [Google Scholar]
- Dang KV, Plet J, Tolleter D, Jokel M, Cuiné S, Carrier P, Auroy P, Richaud P, Johnson X, Alric J, et al. (2014) Combined increases in mitochondrial cooperation and oxygen photoreduction compensate for deficiency in cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 26: 3036–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker JP, Boekema EJ (2005) Supramolecular organization of thylakoid membrane proteins in green plants. Biochim Biophys Acta 1706: 12–39 [DOI] [PubMed] [Google Scholar]
- Depège N, Bellafiore S, Rochaix JD (2003) Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299: 1572–1575 [DOI] [PubMed] [Google Scholar]
- Dietzel L, Bräutigam K, Steiner S, Schüffler K, Lepetit B, Grimm B, Schöttler MA, Pfannschmidt T (2011) Photosystem II supercomplex remodeling serves as an entry mechanism for state transitions in Arabidopsis. Plant Cell 23: 2964–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Granath P, Vener AV (2010) A protein phosphorylation threshold for functional stacking of plant photosynthetic membranes. PLoS ONE 5: e10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Willig A, Granath P, Crèvecoeur M, Rochaix JD, Vener AV (2009) Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. Plant Cell 21: 3950–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiu K, Nakayama Y, Iida H, Sokabe M, Yoshimura K (2011) Mechanoreception in motile flagella of Chlamydomonas. Nat Cell Biol 13: 630–632 [DOI] [PubMed] [Google Scholar]
- Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH (2004) Open mass spectrometry search algorithm. J Proteome Res 3: 958–964 [DOI] [PubMed] [Google Scholar]
- Gulis G, Narasimhulu KV, Fox LN, Redding KE (2008) Purification of His6-tagged photosystem I from Chlamydomonas reinhardtii. Photosynth Res 96: 51–60 [DOI] [PubMed] [Google Scholar]
- Hertle AP, Blunder T, Wunder T, Pesaresi P, Pribil M, Armbruster U, Leister D (2013) PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol Cell 49: 511–523 [DOI] [PubMed] [Google Scholar]
- Hippler M, Klein J, Fink A, Allinger T, Hoerth P (2001) Towards functional proteomics of membrane protein complexes: analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J 28: 595–606 [DOI] [PubMed] [Google Scholar]
- Höhner R, Barth J, Magneschi L, Jaeger D, Niehues A, Bald T, Grossman A, Fufezan C, Hippler M (2013) The metabolic status drives acclimation of iron deficiency responses in Chlamydomonas reinhardtii as revealed by proteomics based hierarchical clustering and reverse genetics. Mol Cell Proteomics 12: 2774–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (1998) The Croonian Lecture 1997. The phosphorylation of proteins on tyrosine: its role in cell growth and disease. Philos Trans R Soc Lond B Biol Sci 353: 583–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai M, Takahashi Y, Minagawa J (2008) Molecular remodeling of photosystem II during state transitions in Chlamydomonas reinhardtii. Plant Cell 20: 2177–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai M, Takizawa K, Tokutsu R, Okamuro A, Takahashi Y, Minagawa J (2010) Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Johnson X, Steinbeck J, Dent RM, Takahashi H, Richaud P, Ozawa S, Houille-Vernes L, Petroutsos D, Rappaport F, Grossman AR, et al. (2014) Proton gradient regulation 5-mediated cyclic electron flow under ATP- or redox-limited conditions: a study of ΔATpase pgr5 and ΔrbcL pgr5 mutants in the green alga Chlamydomonas reinhardtii. Plant Physiol 165: 438–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk D, Bulgrien J, Smith DW, Barsam B, Veretnik S, Gribskov M (2002) The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol 129: 908–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Bovet L, Kushnir S, Noh EW, Martinoia E, Lee Y (2006) AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol 140: 922–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, Schilling B, Row RH, Kenski DM, Gibson BW, Shokat KM (2003) Phosphospecific proteolysis for mapping sites of protein phosphorylation. Nat Biotechnol 21: 1047–1054 [DOI] [PubMed] [Google Scholar]
- Koivuniemi A, Aro EM, Andersson B (1995) Degradation of the D1- and D2-proteins of photosystem II in higher plants is regulated by reversible phosphorylation. Biochemistry 34: 16022–16029 [DOI] [PubMed] [Google Scholar]
- Kruse O, Zheleva D, Barber J (1997) Stabilization of photosystem two dimers by phosphorylation: implication for the regulation of the turnover of D1 protein. FEBS Lett 408: 276–280 [DOI] [PubMed] [Google Scholar]
- Kukuczka B, Magneschi L, Petroutsos D, Steinbeck J, Bald T, Powikrowska M, Fufezan C, Finazzi G, Hippler M (2014) Proton Gradient Regulation5-Like1-mediated cyclic electron flow is crucial for acclimation to anoxia and complementary to nonphotochemical quenching in stress adaptation. Plant Physiol 165: 1604–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D, Shikanai T (2013) Complexities and protein complexes in the antimycin A-sensitive pathway of cyclic electron flow in plants. Front Plant Sci 4: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeille S, Rochaix JD (2010) State transitions at the crossroad of thylakoid signalling pathways. Photosynth Res 106: 33–46 [DOI] [PubMed] [Google Scholar]
- Lemeille S, Turkina MV, Vener AV, Rochaix JD (2010) Stt7-dependent phosphorylation during state transitions in the green alga Chlamydomonas reinhardtii. Mol Cell Proteomics 9: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeille S, Willig A, Depège-Fargeix N, Delessert C, Bassi R, Rochaix JD (2009) Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol 7: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Kronzucker HJ, Shi W (2013) Molecular components of stress-responsive plastid retrograde signaling networks and their involvement in ammonium stress. Plant Signal Behav 8: e23107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Li Q, Xiong L, Kronzucker HJ, Krämer U, Shi W (2012) Arabidopsis plastid AMOS1/EGY1 integrates abscisic acid signaling to regulate global gene expression response to ammonium stress. Plant Physiol 160: 2040–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Lucker B, Kramer DM (2013) Regulation of cyclic electron flow in Chlamydomonas reinhardtii under fluctuating carbon availability. Photosynth Res 117: 449–459 [DOI] [PubMed] [Google Scholar]
- Mann M, Ong SE, Grønborg M, Steen H, Jensen ON, Pandey A (2002) Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol 20: 261–268 [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298: 1912–1934 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. (1961) Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191: 144–148 [DOI] [PubMed] [Google Scholar]
- Mueller LN, Rinner O, Schmidt A, Letarte S, Bodenmiller B, Brusniak MY, Vitek O, Aebersold R, Müller M (2007) SuperHirn: a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 7: 3470–3480 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hashimoto M, Miyake C, Tomizawa K, Endo T, Tasaka M, Shikanai T (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429: 579–582 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Murata N. (1969) Control of excitation transfer in photosynthesis. I. Light-induced change of chlorophyll a fluorescence in Porphyridium cruentum. Biochim Biophys Acta 172: 242–251 [DOI] [PubMed] [Google Scholar]
- Nagy G, Ünnep R, Zsiros O, Tokutsu R, Takizawa K, Porcar L, Moyet L, Petroutsos D, Garab G, Finazzi G, et al. (2014) Chloroplast remodeling during state transitions in Chlamydomonas reinhardtii as revealed by noninvasive techniques in vivo. Proc Natl Acad Sci USA 111: 5042–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath K, Poudyal RS, Eom JS, Park YS, Zulfugarov IS, Mishra SR, Tovuu A, Ryoo N, Yoon HS, Nam HG, et al. (2013) Loss-of-function of OsSTN8 suppresses the photosystem II core protein phosphorylation and interferes with the photosystem II repair mechanism in rice (Oryza sativa). Plant J 76: 675–686 [DOI] [PubMed] [Google Scholar]
- Naumann B, Busch A, Allmer J, Ostendorf E, Zeller M, Kirchhoff H, Hippler M (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7: 3964–3979 [DOI] [PubMed] [Google Scholar]
- Naumann B, Stauber EJ, Busch A, Sommer F, Hippler M (2005) N-terminal processing of Lhca3 is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J Biol Chem 280: 20431–20441 [DOI] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Peltier G, Tolleter D, Billon E, Cournac L (2010) Auxiliary electron transport pathways in chloroplasts of microalgae. Photosynth Res 106: 19–31 [DOI] [PubMed] [Google Scholar]
- Peng L, Shimizu H, Shikanai T (2008) The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J Biol Chem 283: 34873–34879 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Petroutsos D, Busch A, Janssen I, Trompelt K, Bergner SV, Weinl S, Holtkamp M, Karst U, Kudla J, Hippler M (2011) The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell 23: 2950–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroutsos D, Terauchi AM, Busch A, Hirschmann I, Merchant SS, Finazzi G, Hippler M (2009) PGRL1 participates in iron-induced remodeling of the photosynthetic apparatus and in energy metabolism in Chlamydomonas reinhardtii. J Biol Chem 284: 32770–32781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp S, Hambrecht J, Braslavski L, Schroth G, Freichel M, Murakami M, Cavalié A, Flockerzi V (1998) A novel capacitative calcium entry channel expressed in excitable cells. EMBO J 17: 4274–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribil M, Pesaresi P, Hertle A, Barbato R, Leister D (2010) Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol 8: e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Finazzi G, Endler A, Willig A, Baerenfaller K, Grossmann J, Gerrits B, Rutishauser D, Gruissem W, Rochaix JD, et al. (2011) Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc Natl Acad Sci USA 108: 12955–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD. (2007) Role of thylakoid protein kinases in photosynthetic acclimation. FEBS Lett 581: 2768–2775 [DOI] [PubMed] [Google Scholar]
- Rochaix JD. (2014) Regulation and dynamics of the light-harvesting system. Annu Rev Plant Biol 65: 287–309 [DOI] [PubMed] [Google Scholar]
- Rochaix JD, Lemeille S, Shapiguzov A, Samol I, Fucile G, Willig A, Goldschmidt-Clermont M (2012) Protein kinases and phosphatases involved in the acclimation of the photosynthetic apparatus to a changing light environment. Philos Trans R Soc Lond B Biol Sci 367: 3466–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samol I, Shapiguzov A, Ingelsson B, Fucile G, Crèvecoeur M, Vener AV, Rochaix JD, Goldschmidt-Clermont M (2012) Identification of a photosystem II phosphatase involved in light acclimation in Arabidopsis. Plant Cell 24: 2596–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmollinger S, Schulz-Raffelt M, Strenkert D, Veyel D, Vallon O, Schroda M (2013) Dissecting the heat stress response in Chlamydomonas by pharmaceutical and RNAi approaches reveals conserved and novel aspects. Mol Plant 6: 1795–1813 [DOI] [PubMed] [Google Scholar]
- Shapiguzov A, Ingelsson B, Samol I, Andres C, Kessler F, Rochaix JD, Vener AV, Goldschmidt-Clermont M (2010) The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis. Proc Natl Acad Sci USA 107: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. (2014) Central role of cyclic electron transport around photosystem I in the regulation of photosynthesis. Curr Opin Biotechnol 26: 25–30 [DOI] [PubMed] [Google Scholar]
- Soupene E, King N, Feild E, Liu P, Niyogi KK, Huang CH, Kustu S (2002) Rhesus expression in a green alga is regulated by CO2. Proc Natl Acad Sci USA 99: 7769–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht M, Kuhlgert S, Fufezan C, Hippler M (2011) Proteomics to go: Proteomatic enables the user-friendly creation of versatile MS/MS data evaluation workflows. Bioinformatics 27: 1183–1184 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Clowez S, Wollman FA, Vallon O, Rappaport F (2013) Cyclic electron flow is redox-controlled but independent of state transition. Nat Commun 4: 1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Iwai M, Takahashi Y, Minagawa J (2006) Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 103: 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Okamuro A, Minagawa J, Takahashi Y (2014) Biochemical characterization of photosystem I-associated light-harvesting complexes I and II isolated from state 2 cells of Chlamydomonas reinhardtii. Plant Cell Physiol 55: 1437–1449 [DOI] [PubMed] [Google Scholar]
- Tardif M, Atteia A, Specht M, Cogne G, Rolland N, Brugière S, Hippler M, Ferro M, Bruley C, Peltier G, et al. (2012) PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol 29: 3625–3639 [DOI] [PubMed] [Google Scholar]
- Taus T, Köcher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K (2011) Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res 10: 5354–5362 [DOI] [PubMed] [Google Scholar]
- Terashima M, Petroutsos D, Hüdig M, Tolstygina I, Trompelt K, Gäbelein P, Fufezan C, Kudla J, Weinl S, Finazzi G, et al. (2012) Calcium-dependent regulation of cyclic photosynthetic electron transfer by a CAS, ANR1, and PGRL1 complex. Proc Natl Acad Sci USA 109: 17717–17722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima M, Specht M, Hippler M (2011) The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr Genet 57: 151–168 [DOI] [PubMed] [Google Scholar]
- Terashima M, Specht M, Naumann B, Hippler M (2010) Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol Cell Proteomics 9: 1514–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingholm TE, Jensen ON, Robinson PJ, Larsen MR (2008) SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol Cell Proteomics 7: 661–671 [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Gollan PJ, Suorsa M, Kangasjärvi S, Aro EM (2012) STN7 operates in retrograde signaling through controlling redox balance in the electron transfer chain. Front Plant Sci 3: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Grieco M, Kangasjärvi S, Aro EM (2010) Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiol 152: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Nurmi M, Kangasjärvi S, Aro EM (2008) Core protein phosphorylation facilitates the repair of photodamaged photosystem II at high light. Biochim Biophys Acta 1777: 1432–1437 [DOI] [PubMed] [Google Scholar]
- Tokutsu R, Iwai M, Minagawa J (2009) CP29, a monomeric light-harvesting complex II protein, is essential for state transitions in Chlamydomonas reinhardtii. J Biol Chem 284: 7777–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutsu R, Kato N, Bui KH, Ishikawa T, Minagawa J (2012) Revisiting the supramolecular organization of photosystem II in Chlamydomonas reinhardtii. J Biol Chem 287: 31574–31581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutsu R, Minagawa J (2013) Energy-dissipative supercomplex of photosystem II associated with LHCSR3 in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 110: 10016–10021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D, Ghysels B, Alric J, Petroutsos D, Tolstygina I, Krawietz D, Happe T, Auroy P, Adriano JM, Beyly A, et al. (2011) Control of hydrogen photoproduction by the proton gradient generated by cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 23: 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompelt K, Steinbeck J, Terashima M, Hippler M (March 13, 2014) A new approach for the comparative analysis of multiprotein complexes based on 15N metabolic labeling and quantitative mass spectrometry. J Vis Exp http://dx.doi.org/10.3791/51103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkina MV, Blanco-Rivero A, Vainonen JP, Vener AV, Villarejo A (2006) CO2 limitation induces specific redox-dependent protein phosphorylation in Chlamydomonas reinhardtii. Proteomics 6: 2693–2704 [DOI] [PubMed] [Google Scholar]
- Ünlü C, Drop B, Croce R, van Amerongen H (2014) State transitions in Chlamydomonas reinhardtii strongly modulate the functional size of photosystem II but not of photosystem I. Proc Natl Acad Sci USA 111: 3460–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainonen JP, Hansson M, Vener AV (2005) STN8 protein kinase in Arabidopsis thaliana is specific in phosphorylation of photosystem II core proteins. J Biol Chem 280: 33679–33686 [DOI] [PubMed] [Google Scholar]
- van Amerongen H, Croce R (2013) Light harvesting in photosystem II. Photosynth Res 116: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vener AV, van Kan PJ, Rich PR, Ohad I, Andersson B (1997) Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: thylakoid protein kinase deactivation by a single-turnover flash. Proc Natl Acad Sci USA 94: 1585–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. (2001) The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Wang X, Bian Y, Cheng K, Gu LF, Ye M, Zou H, Sun SS, He JX (2013) A large-scale protein phosphorylation analysis reveals novel phosphorylation motifs and phosphoregulatory networks in Arabidopsis. J Proteomics 78: 486–498 [DOI] [PubMed] [Google Scholar]
- Whatley FR, Tagawa K, Arnon DI (1963) Separation of the light and dark reactions in electron transfer during photosynthesis. Proc Natl Acad Sci USA 49: 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. (2014) Mass-spectrometry-based draft of the human proteome. Nature 509: 582–587 [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR, Zielinska DF, Mann M (2011) Comparison of ultrafiltration units for proteomic and N-glycoproteomic analysis by the filter-aided sample preparation method. Anal Biochem 410: 307–309 [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Xue H, Tokutsu R, Bergner SV, Scholz M, Minagawa J, Hippler M (2015) PHOTOSYSTEM II SUBUNIT R is required for efficient binding of LIGHT-HARVESTING COMPLEX STRESS-RELATED PROTEIN3 to photosystem II-light-harvesting supercomplexes in Chlamydomonas reinhardtii. Plant Physiol 167: 1566–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito F, Finazzi G, Delosme R, Nitschke W, Picot D, Wollman FA (1999) The Qo site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J 18: 2961–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.