Inactivation of ubiquitin-ligase3 suppresses dwarfism and irregular xylem in an Arabidopsis mutant, generating a line with low acetylated biomass and high glucose release after cellulolysis.
Abstract
eskimo1-5 (esk1-5) is a dwarf Arabidopsis (Arabidopsis thaliana) mutant that has a constitutive drought syndrome and collapsed xylem vessels, along with low acetylation levels in xylan and mannan. ESK1 has xylan O-acetyltransferase activity in vitro. We used a suppressor strategy on esk1-5 to screen for variants with wild-type growth and low acetylation levels, a favorable combination for ethanol production. We found a recessive mutation in the KAKTUS (KAK) gene that suppressed dwarfism and the collapsed xylem character, the cause of decreased hydraulic conductivity in the esk1-5 mutant. Backcrosses between esk1-5 and two independent knockout kak mutants confirmed suppression of the esk1-5 effect. kak single mutants showed larger stem diameters than the wild type. The KAK promoter fused with a reporter gene showed activity in the vascular cambium, phloem, and primary xylem in the stem and hypocotyl. However, suppression of the collapsed xylem phenotype in esk1 kak double mutants was not associated with the recovery of cell wall O-acetylation or any major cell wall modifications. Therefore, our results indicate that, in addition to its described activity as a repressor of endoreduplication, KAK may play a role in vascular development. Furthermore, orthologous esk1 kak double mutants may hold promise for ethanol production in crop plants.
Today, the fields of agriculture and forestry must address challenging issues, particularly in the context of fluctuating environmental conditions, such as ensuring that food and feed production remain efficient, meeting societal demands to reduce inputs, including water, and creating new products, such as biofuel. The demand for biofuel, a renewable alternative to fossil fuel, further increases the need to develop biomass amenable to alcohol fermentation. Second-generation biofuels are based on the fermentation of sugars extracted from lignocellulosic biomass, which is produced from the residues of food crops or from nonfood crops; therefore, its production does not compete with food crops (Sims et al., 2010). This lignocellulosic biomass is made up of secondary cell walls, mostly found in vascular tissues.
Vascular tissues consist of a network of conduits, spanning an entire plant and connecting biosynthetically active leaves to the soil via the root and the shoot. There are two main vascular tissues, the xylem and the phloem, which arise from a lateral meristem called the procambium during primary growth. When dicot plants undergo secondary growth and cell walls are thickening, a secondary meristem, called the cambium or vascular cambium, emerges, giving rise to secondary vascular tissues (Esau, 1965; Buvat, 1989). The xylem is responsible for the upward transport of water and nutrients from the soil to the whole plant. The phloem, positioned parallel to the xylem, supplies sink organs (roots, etc.) with leaf photoassimilates. In angiosperms, such as Arabidopsis (Arabidopsis thaliana), the xylem is composed of two main cellular types, tracheary elements, involved in the transport of water, and fibers, playing a major role in plant support (Turner and Sieburth, 2003). The function of the xylem depends on the plant’s capacity to form thick secondary cell walls that confer mechanical strength to resist gravity and withstand negative pressure, allowing sap to travel upward through vessels. Xylem tissue is a major carbon sink that incorporates sugars into biopolymers (Ragni et al., 2011). Xylem secondary cell walls are mainly composed of cellulose embedded in a matrix of lignin and hemicelluloses. Xylan is a major hemicellulose in monocot and dicot secondary cell walls (Faik, 2010). Xylan has a linear backbone of β-(1,4)-linked d-Xyl residues that can be mono- or di-O-acetylated at positions O-2 and O-3 of the xylosyl residues (Ebringerova and Heinze, 2000).
O-Acetylation of polysaccharides reportedly has a negative effect on the utilization of lignocellulose, such as in the production of paper and bioethanol (Biely, 1985; Grohmann et al., 1989). A major xylan acetyltransferase was recently identified in Arabidopsis: TRICHOME BIREFRINGENCE-LIKE29/ESKIMO1 (TBL29/ESK1; Urbanowicz et al., 2014). esk1 knockout mutants show a 60% reduction in xylan acetylation and a lesser reduction in mannan acetylation (Xiong et al., 2013; Yuan et al., 2013). esk1 has been described previously as a genotype with drought stress symptoms (Bouchabke-Coussa et al., 2008; Lugan et al., 2009); its collapsed xylem vessels (irregular xylem [irx] phenotype) are assumed to be the cause of the drastic hydraulic conductivity drop, and thus the drought stress syndrome, including dwarfism (Lefebvre et al., 2011).
To identify new mutations that restore plant stature but maintain a low xylan O-acetylation level, we explored the possibility of producing this combination by screening an esk1-5 ethyl methanesulfonate (EMS)-mutagenized population for nondwarf phenotypes. The suppressors of esk1 were called beem, for biomass enhancement in esk1-5 mutation background. Here, we describe the identification of one BEEM gene as KAKTUS/UBIQUITIN PROTEIN LIGASE3 (KAK/UPL3), which encodes a protein belonging to the E3-ubiquitin protein ligase family (Downes et al., 2003).
RESULTS
esk1-5 Dwarfism Is Partially Suppressed by kak Mutations
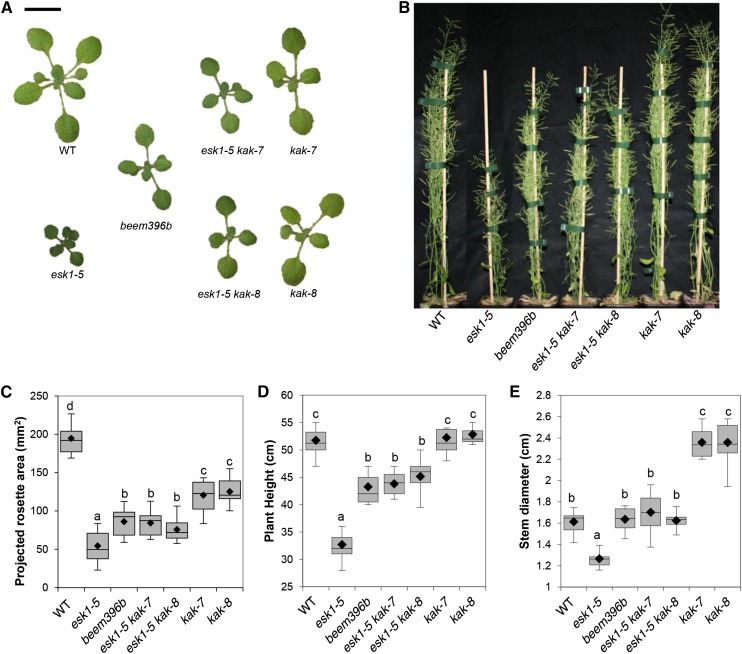
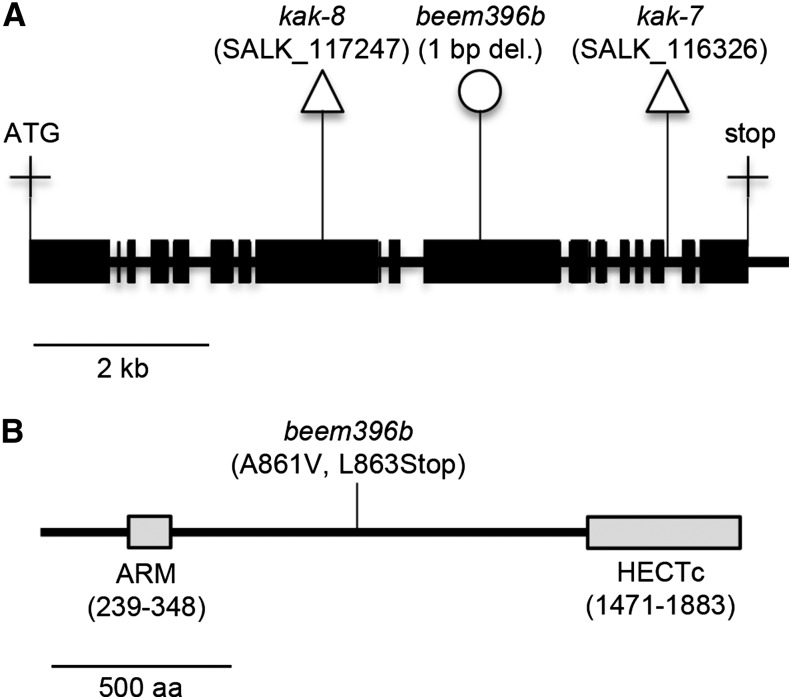
Seeds produced by the esk1-5/tbl29-1 mutant underwent EMS-induced mutagenesis. The resulting M2 population was screened for enhanced rosette size compared with esk1-5. Twelve independent M2 lines with a rosette size intermediate to esk1-5 and the wild type were isolated and named beem. This work will focus on one of these lines, called beem396b (Fig. 1). A mapping-by-sequencing strategy, similar to methods described recently (Abe et al., 2012; Hartwig et al., 2012), was used to identify the causative mutation. The beem396b M2 line was backcrossed twice to its mutant background esk1-5. The resulting F2 population showed a 124:446 segregation (χ2 test for an expected segregation of 1:3; P = 0.0735) for rosette size, suggesting that a single recessive mutation was responsible for the suppressed phenotype. DNA from the 124 plants exhibiting the enlarged rosette phenotype compared with esk1-5 was extracted, pooled in equimolar ratios, and sequenced using Illumina whole-genome sequencing (Supplemental Protocol S1). Predicted variants were ranked by their likelihood of being the causative mutation (Supplemental Protocol S2). The mutation index was mapped along chromosomes and peaked at a unique locus at the bottom of chromosome 4 (base 18045924; The Arabidopsis Information Resource 10 [TAIR10] genome release) with a frequency of 84% (Supplemental Fig. S1; Abe et al., 2012). We attribute the less-than-perfect peak frequency (less than 100%) to the selection of false-positive F2 plants. The mutation with the highest index was a 1-bp deletion causing a frame shift within the KAK/UPL3 (AT4G38600) gene coding sequence. The deletion causes an Ala-to-Val amino acid change at codon 861 of the KAK coding sequence and a premature stop codon six nucleotides after the deletion at codon 863, likely leading to the translation of a truncated KAK protein (Fig. 2; Supplemental Fig. S2).
Figure 1.
Partial suppression of esk1-5 dwarfism by kak mutations. A, Rosettes of 2-week-old plants. Bar = 1 cm. B, Twelve-week-old mature plants. Bar = 5 cm. C, Rosette area of 2-week-old plants. D, Length of the main inflorescence stem of mature 12-week-old plants. E, Diameter of the main inflorescence stem of mature 12-week-old plants. WT, Wild type. In C to E, the box and whisker plots show the distribution of 12, eight, and eight measurements, respectively. Black diamonds represent the means, lines represent medians, top and bottom ends of the boxes represent the first and third quartiles, respectively, and whisker extremities represent maximum and minimum data points. Letters above the boxes indicate groups with significant differences as determined by Tukey’s multiple pairwise comparisons test.
Figure 2.
EMS mutation in beem396b generates a stop codon in the KAK gene. A, KAK gene model. Exons are represented by black boxes, and introns and the noncoding 3′ region are represented by the black line. Triangles indicate T-DNA insertions, and the circle indicates the EMS-induced mutation. B, Position of the EMS-induced mutation within the KAK protein sequence. Gray boxes represent conserved domains: ARM, Armadillo/β-catenin-like repeats motif responsible for the specific interaction of KAK with proteins targeted for degradation; HECTc, catalytic domain responsible for the transfer of polyubiquitin chains from ubiquitin-conjugating enzymes (E2) to proteins targeted for degradation. aa, Amino acids.
KAK has been described as a repressor of endoreduplication in trichomes, with kak mutants exhibiting overbranched trichomes associated with high nuclear DNA content (Perazza et al., 1999; El Refy et al., 2003). In this work, the leaves of kak mutants harbored over 80% of trichomes having four or five branches, while the wild type exhibited mainly three-branched trichomes and a minority of two- and four-branched trichomes (Supplemental Fig. S3). The observation of overbranched trichomes has been correlated with a transfer DNA (T-DNA) insertion in the KAK gene (Perazza et al., 1999; El Refy et al., 2003). Observation of leaf trichomes using scanning electron microscopy showed that beem396b had a typical kak mutant phenotype, whereas the starting esk1-5 mutant had normal trichomes (Supplemental Fig. S3). Among the independent beem lines isolated, only beem308a also presented an overbranched trichome phenotype (Supplemental Fig. S4). Large rosettes and trichome overbranching were observed in F1 plants from a cross between beem396b and beem308a, suggesting that the lines are allelic. Sequencing PCR products (primers are listed in Supplemental Table S1) of the full KAK genomic sequence in beem308a identified a C-to-T transition (position 18043667; TAIR10) within the KAK coding sequence, causing a premature stop codon at codon 1,469 of the KAK protein, likely leading to the translation of a truncated KAK protein (Supplemental Fig. S4). The isolation of the beem308a line in our screen for suppressor lines provided further support to our supposition that KAK inactivation suppresses esk1 dwarfism. Nevertheless, beem308a was not studied further due to the presence of an additional early-flowering mutation that impaired morphometric comparisons between genotypes.
To confirm that KAK inactivation suppresses esk1 dwarfism, we generated and studied esk1-5 kak double mutants with two independent T-DNA insertion lines within the KAK gene (Fig. 2). kak-7 and kak-8 exhibited the typical overbranched trichome phenotype (Supplemental Fig. S3). The rosette surface areas of 2-week-old kak-7 and kak-8 plants were significantly smaller than that of the wild type (Fig. 1), but mature rosette area at bolting (M. Durand-Tardif, unpublished data) and stem height were not significantly different from the wild type (Fig. 1). The stem diameter of kak-7 and kak-8 was significantly larger than in the wild type (Fig. 1). Two double mutants, esk1-5 kak-7 and esk1-5 kak-8, both showed typical kak mutant trichomes (Supplemental Fig. S3) and an enlarged rosette size, as observed in beem396b (Fig. 1). Morphometric parameters, such as rosette area of 2-week-old plantlets and mature rosette size at bolting (M. Durand-Tardif, unpublished data), stem height, and stem diameter (Fig. 1), showed that esk1-5 kak double mutants perform better than esk1-5 but show values similar to those in the beem lines. These data confirm the involvement of kak mutations in the increase in rosette size observed in the esk1-5 mutant. Moreover, kak inactivation did not affect ESK1 transcription, as shown in Supplemental Figure S5; as described elsewhere, the ESK1 transcript is expressed in stems of the wild type and in both kak single mutants (Xin and Browse, 1998). No ESK1 transcript was observed in genotypes carrying the esk1-5 mutation. However, esk1-5 kak double mutants showed significantly smaller rosette areas than the kak single mutants. As expected, the suppression of dwarfism in beem396b is not due to the restoration of ESK1 expression.
The Suppression of Dwarfism Is Not Associated with the Recovery of Cell Wall Acetylation or Major Cell Wall Modifications
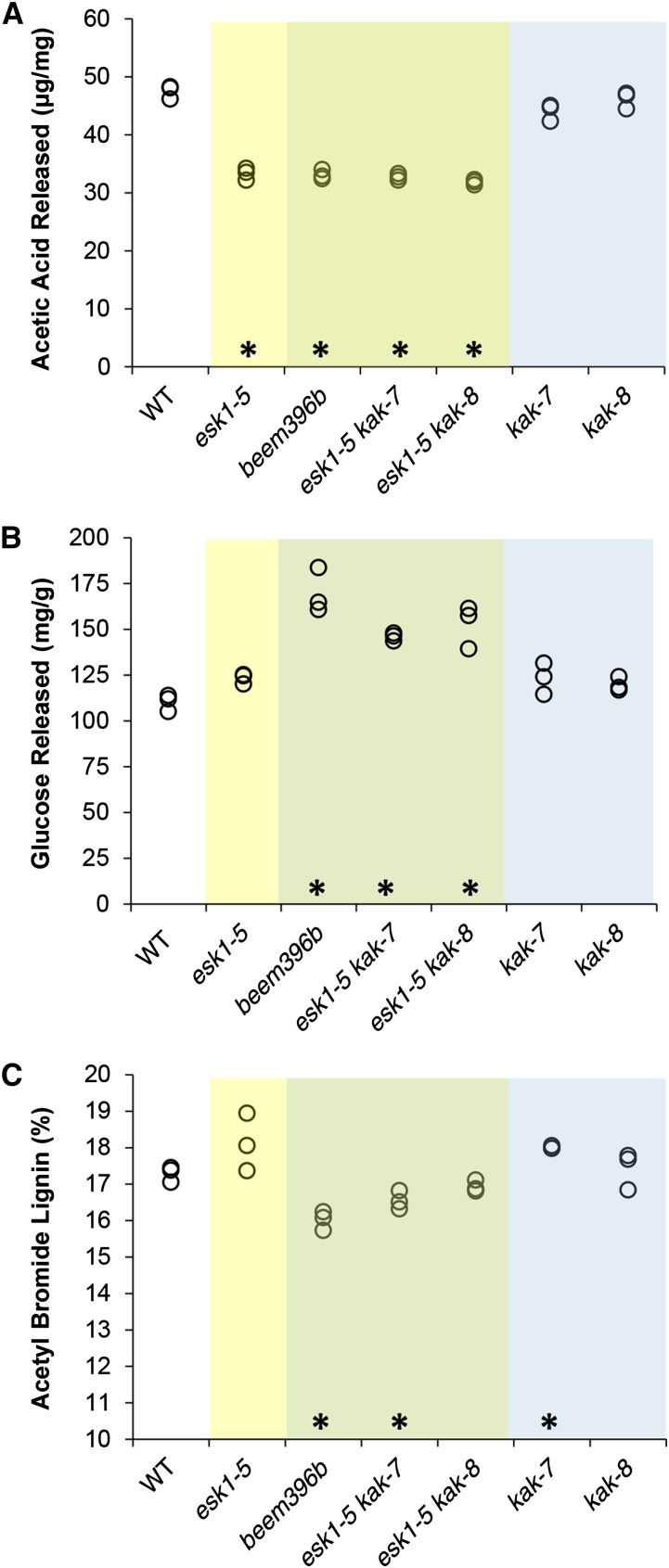
The collapsed xylem phenotype in the esk1-5 mutant is primarily caused by a strong reduction in xylan, and possibly mannan, O-acetylation (Xiong et al., 2013; Yuan et al., 2013). We first compared the alkali-released acetic acid in the cell wall from stems of esk1-5 and beem396b. As described previously, the amount of acetic acid released by esk1-5 was significantly lower than in the wild type (Fig. 3; Xiong et al., 2013; Yuan et al., 2013). The same defect was observed in beem396b and in esk1-5 kak double mutants. In addition, the acetic acid released by kak-7 and kak-8 single mutants was not significantly different from that in the wild type. These results indicate that kak mutations have no effect on the total amount of wall-bound acetate, either in esk1-5 or in a wild-type genetic background.
Figure 3.
esk1 kak double mutants show higher Glc release. The same cell wall samples were analyzed for their acetate content (A), Glc release after cellulase digestion (B), and acetyl bromide lignin (C). The values of three biological replicates are shown (circles). Data from genotypes carrying the esk1 mutation are presented on a yellow background, data from genotypes carrying the esk1 and kak mutations on a green background, and data from genotypes carrying the kak mutation on a blue background. Asterisks over the x axis indicate statistically significant differences compared with the wild type (WT; Dunnett’s test, P < 0.05; n = 3).
To evaluate the saccharification potential of the various mutants, cell wall residues from the two basal internodes on stems of 12-week-old plants were treated with a commercial cellulase preparation (Berthet et al., 2011). We observed that genotypes carrying both esk1-5 and kak mutations released significantly more Glc than the esk1 and kak single mutants or the wild-type line (Fig. 3). In contrast, neither esk1-5 single mutants nor kak single mutants released more Glc than the wild type (Fig. 3). Given that it is well established that lignin has a major detrimental impact on the saccharification of plant cell walls (Chen and Dixon, 2007; Studer et al., 2011; Van Acker et al., 2013), we measured the lignin content of cell wall residues using the acetyl bromide method. The esk1 lignin level was not significantly different from the wild-type level; similarly, the levels of the other single and double mutants were found to be not affected or affected only very weakly (Fig. 3).
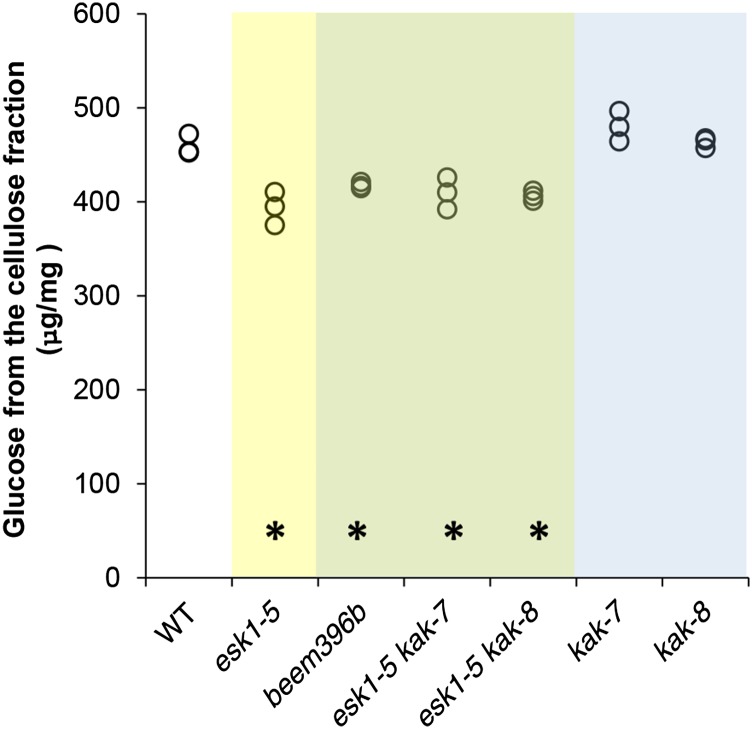
To investigate if kak mutations have other effects on xylem cell wall composition, we then used transmission-mode Fourier transform infrared (FT-IR) microspectroscopy as described previously (Supplemental Fig. S6; Mouille et al., 2003; Lefebvre et al., 2011). The comparison curve of the statistical significance of the difference in absorbance between the wild type and the esk1-5 mutant (Supplemental Fig. S7) showed previously reported differences (Lefebvre et al., 2011). Peaks found at 1,730, 1,369, and 1,230 cm−1 are located at wave numbers related to the reported infrared signature of O-acetyl moieties found in plant cell wall polymers: namely C=O stretching in acetyl groups (1,730 cm−1), deformation of C-H linkages in the methyl group of O-acetyl moieties (1,369 cm−1), and C=O and C-O linkages stretching vibration (1,230 cm−1; Mohebby, 2008). In the right part of the spectrum, between 1,000 and 830 cm−1, modifications are associated with a reduction in cellulose (Mouille et al., 2003). These results, consistent with reported esk1-5 cell wall alterations (Lefebvre et al., 2011; Xiong et al., 2013; Yuan et al., 2013), demonstrate the accuracy of the FT-IR analysis. The comparison curves of FT-IR absorbance spectra of the hypocotyl metaxylem from beem396b (Supplemental Fig. S7) and esk1-5 were very similar, as were the comparison curves from the esk1-5 kak double mutants and esk1-5 (Supplemental Fig. S7). In contrast, the comparison curves of kak-7 and kak-8 were flat, indicating nonsignificant differences with respect to the wild type (Supplemental Fig. S7). Moreover, the gaps between kak-7 and kak-8 curves suggest that the detected differences were due to background noise rather than actual differences in chemical arrangement. We also analyzed the monosaccharides in cell wall residues. As described previously, the Glc mass from the cellulose fraction was significantly lower in esk1-5 knockout genotypes (Fig. 4; Supplemental Table S2), indicating a decrease in crystalline cellulose content (Lefebvre et al., 2011; Xiong et al., 2013; Yuan et al., 2013). As reported by Xiong et al. (2013), we found more Xyl in esk1-5 than in the wild type, but the difference was only slight and had not been observed by Yuan et al. (2013; Supplemental Table S2). We observed higher amounts of Ara, Glc, and Man (except for Man in esk1 kak-7) in the hemicellulose fraction from all mutants compared with the wild type, but we will not discuss these data, nor the weak differences observed in Fuc, Rha, and Gal. Released acetic acid, FT-IR profiles, and the monosaccharide analysis all suggest that kak mutations did not affect xylem cell wall composition in the wild type or in the esk1-5 background. As a result, the suppression of dwarfism in beem and esk1-5 kak double mutants cannot be associated with the recovery of O-acetylation or any other major cell wall modifications in xylem.
Figure 4.
kak does not suppress the cellulose content of esk1-5, as indicated by Glc dosage from the cellulose fraction. The values of three biological replicates are shown (circles). Data from genotypes carrying the esk1 mutation are shown on a yellow background, data from genotypes carrying the esk1 and kak mutations on a green background, and data from genotypes carrying the kak mutation on a blue background. Asterisks over the x axis indicate statistically significant differences compared with the wild type (WT; Dunnett’s test, P < 0.05; n = 3).
The Suppression of Dwarfism Is Associated with the Recovery of Xylem Morphology
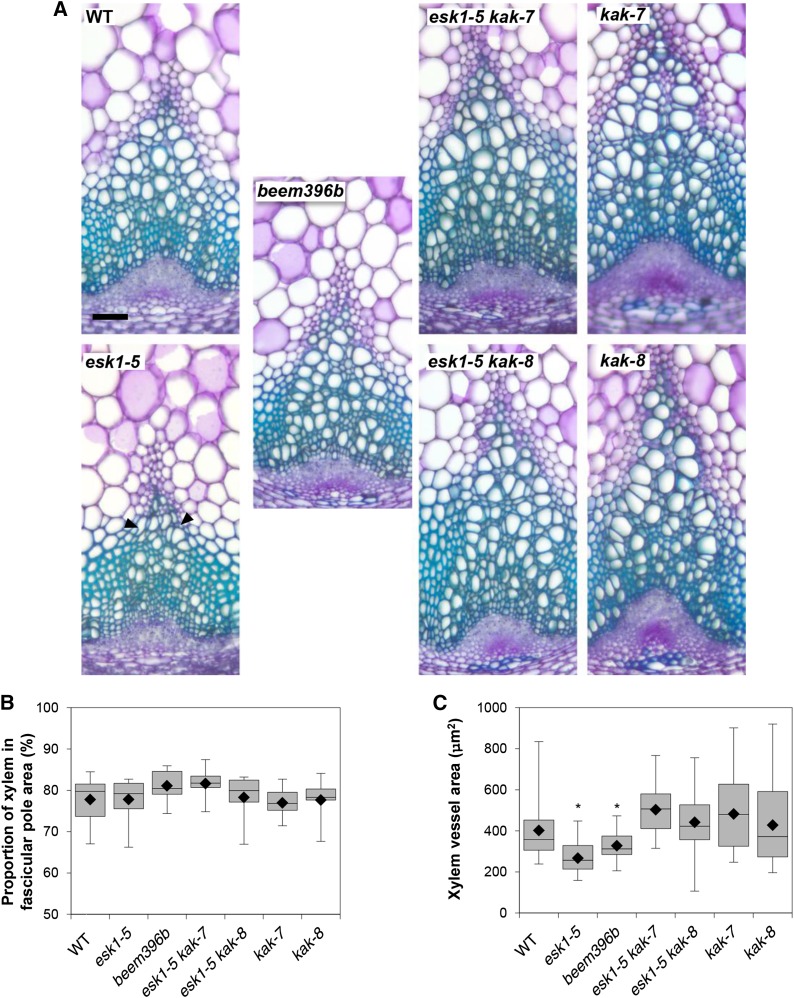
esk1-5 dwarfism is associated with an impairment of water transport due to the deformation of xylem vessels (Lefebvre et al., 2011). Observation with a light microscope of Toluidine Blue Orange-stained cross sections of 10-week-old, fully developed stem bases showed a clear suppression of the irx phenotype in beem396b (Fig. 5). Based on the measurement of 10 to 20 xylem vessel surface areas in 19 to 21 different poles from six different plants (2,299 data points; Supplemental Fig. S8), we observed a bimodal distribution (Supplemental Fig. S9). Consequently, the data were analyzed for two populations: those with large vessels and those with medium-sized vessels. Overall, we observed a pole effect in individual plants and a plant effect in genotypes. Because vessel area was heteroscedastic, we compared vessel area between genotypes with nonparametric tests (permutation-based pairwise Student’s t tests). beem396b presents wild-type-like round-shaped vessels of area intermediate between esk1-5 and the wild type in the large vessel as well as the medium vessel populations (Fig. 5; Supplemental Fig. S9). The double mutants esk1-5 kak-7 and esk1-5 kak-8 also showed suppression of the irx phenotype (Fig. 5), with round-shaped vessels showing an area similar to the wild type, in the large vessel as well as the medium vessel populations (Fig. 5; Supplemental Fig. S9). For kak-7 and kak-8 mutants, the vessel area distribution was large, heteroscedastic, and not significantly different from the wild type, but there was a trend toward higher vessel area, as observed with the wide range of values from the median to the third quartile. These results suggest that rosette enlargement in beem396b and in esk1-5 kak double mutants is associated with the recovery of xylem function. However, the proportion of xylem relative to phloem in the vascular pole area was not significantly different within genotypes (Fig. 5). This recovery of xylem morphology can be viewed as an increase in water transport capacity, which may account for the suppression of dwarfism in the beem line.
Figure 5.
kak mutations have an effect on xylem morphology. A, Cross sections of stem vascular bundles stained with Toluidine Blue. WT, Wild type. Black arrowheads indicate collapsed xylem vessels. Bar = 50 mm. B, Proportion of xylem in the vascular pole area. For each genotype, the areas of xylem and phloem in the fascicular poles from six independent plants were measured as described in Supplemental Figure S8. C, Xylem vessel surface area. Vessel area (from the largest vessel population) measurements were taken for each genotype on 19 to 21 different xylem poles from six independent plants, as described in Supplemental Figure S8. For the box and whisker plots (B and C), black diamonds represent the means, lines in boxes represent medians, top and bottom ends of the boxes represent the first and third quartiles, respectively, and whisker extremities represent maximum and minimum data points. Asterisks indicate the genotypes that are significantly different from the other genotypes (P < 0.05), as attested by pairwise permutational Student’s t tests (999 permutations).
kak Partially Restores Hydraulic Conductivity
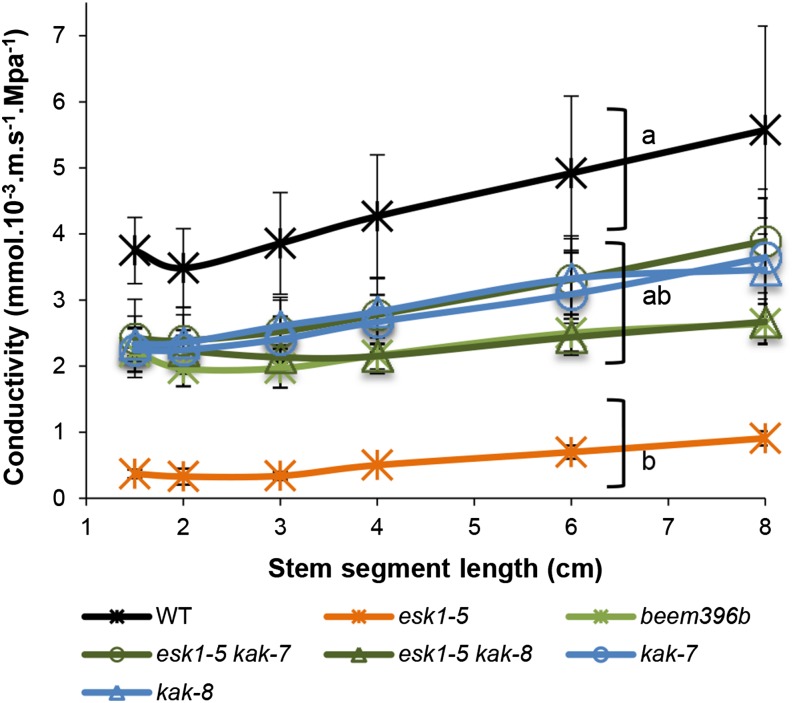
To check if suppression of the irx phenotype was associated with the recovery of sap flow capacity, we measured stem hydraulic conductivity. This trait was measured in segments of basal inflorescence stems sequentially shortened from 8 to 1.5 cm long (Fig. 6). In all samples, conductivity increased with stem segment length, although it is the conductance normalized for length. However, stems have stomata and ramifications, and these features caused leaks in the samples. The volume of these leaks (and thus the relative amount of sap flow) increased with increasing segment length. Conductivity in esk1-5 was significantly lower than in the other genotypes. This difference was found regardless of segment length, ruling out a length effect (P < 0.05 for the esk1-5 effect in a two-way ANOVA, genotype × segment length). esk1 mutants are known to have low hydraulic conductivity (Lefebvre et al., 2011). The wild-type genotype had the highest conductivity, which was not significantly different in kak single and esk1-5 kak double mutants, due to high variance. Nevertheless, for each segment length (except 8 cm), a multiple pairwise comparison test (Tukey’s test, P < 0.05) clustered kak mutants and esk1-5 kak double mutants, with beem396b as intermediate to the esk1-5 single mutant and the wild type.
Figure 6.
kak partially suppress the conductivity default of esk1-5. Hydraulic conductivity was measured on segments of basal inflorescence stems sequentially shortened and plotted as a function of segment length. Each data point represents the mean of two (esk1-5) to six biological replicates. Brackets with different letters indicate genotype groups that are significantly different (P < 0.1), for each segment length except 8 cm, according to Tukey’s multiple pairwise comparisons test. Error bars indicate se. WT, Wild type.
KAK Is Expressed in Vascular Tissues
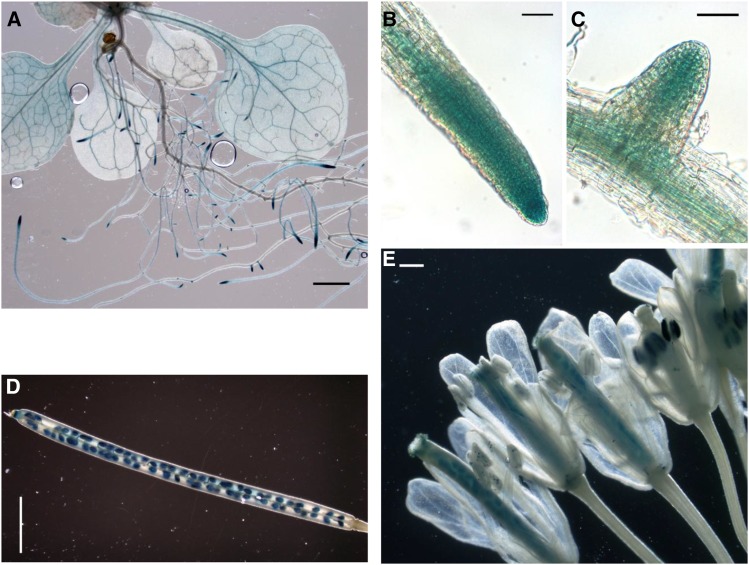
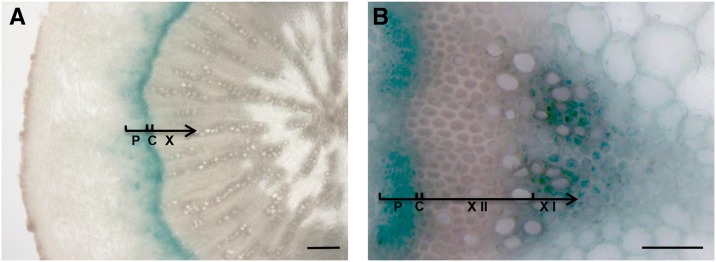
In kak mutants, stem diameter was about 30% larger than in the wild type, consistent with the recovery of stem diameter found in beem396b (Fig. 1). In esk1 kak double mutants, xylem vessels had a round shape, testifying to the absence of collapsed xylem vessels. This suggests a possible involvement of KAK in secondary growth and/or vascular patterning. To investigate this possibility, we used the GUS reporter gene to study the KAK gene expression pattern. A 2-kb sequence upstream from the KAK gene transcription starting site (KAKpro) was fused with the GUS reporter gene. Examination of GUS activity in wild-type plants transformed with the KAKpro:GUS construct showed specific expression in leaf vascular tissues, root tips, and emerging lateral roots of 14-d-old plants (Fig. 7). In fully developed, flowering 8-week-old transgenic plants, GUS activity was detected in reproductive organs (i.e. pollen grains, ovules, and seeds; Fig. 7). Observation of hypocotyl cross sections with a light microscope showed specific GUS activity in the vascular cambium, the meristematic zone that gives rise to secondary phloem and xylem. GUS activity was also observed in the hypocotyl near the cambium on the phloem side and possibly in young secondary xylem cells, although the latter two cell types are difficult to identify because they are small and undifferentiated (Fig. 8; Ye et al., 2002; Turner and Sieburth, 2003). In stem vascular bundles, strong GUS activity was found in the phloem, in the cambium, and in the primary xylem, in cells surrounding the vessels (Fig. 8). The KAK gene expression pattern, as shown via GUS activity, indicated that KAK may play a role in vascular development as well as in male gametophytes, ovules, and developing seeds.
Figure 7.
Expression of KAKpro:GUS in vascular tissues, root tips, pollen, ovaries, and seeds. A to C, Fifteen-day-old seedling. A, Observation with a stereomicroscope of a plantlet showing expression in vascular tissues. Bar = 200 mm. B and C, Observation with a light microscope of a root tip (B) and an emerging lateral root (C) with expression in meristems. Bars = 1 mm. D and E, Eight-week-old flowering plant. D, Maturing siliques showing expression in seeds. Bar = 1 mm. E, Fully developed flower showing expression in ovules and pollen grains. Bar = 100 mm.
Figure 8.
Expression of KAKpro:GUS in the xylem, cambium, and phloem. Hypocotyl (A) and stem (B) cross sections were observed with a light microscope. Phloem (P), cambium (C), xylem (X), secondary xylem (X II), and primary xylem (X I) are indicated. Bars = 20 μm (A) and 5 μm (B).
The observed KAK expression pattern in vascular tissues, therefore, was consistent with the suppressed irx phenotype in esk1 kak double mutants.
DISCUSSION
The Suppression of the Small Rosette Size in esk1-5 Is Indirect
ESK1/TBL29 has O-acetyltransferase activity that operates in xylem tissues (Xiong et al., 2013; Yuan et al., 2013; Urbanowicz et al., 2014). The absence of this function is associated with an irx phenotype; consequently, the esk1 knockout mutants show reduced hydraulic conductivity and dwarfism (Lefebvre et al., 2011). Thus, the suppression of the esk1 dwarf phenotype may be due either to the recovery of xylem function or to an indirect compensatory growth effect.
According to our study, the suppression of the esk1 dwarf phenotype in the beem396b line results from the inactivation of the KAK gene. The effect of kak mutations in beem396b suggests that the effect of the suppression of dwarfism is indirect. kak mutants have previously been described as affecting trichome morphology (Perazza et al., 1999). As expected, beem396b and esk1 kak double mutants both present overbranched trichomes, similar to single insertional kak mutants (Supplemental Fig. S3). beem396b has larger metaxylem vessel areas than esk1-5 but smaller than the wild type, probably due to other, undesirable mutations (Fig. 5). The esk1 kak double mutants have a round shape and larger metaxylem vessel area than esk1, causing the suppression of the irx phenotype (Fig. 5). However, there were no differences in the proportion of the xylem area in fascicular bundles among genotypes (Fig. 5), in accordance with normally coordinated vascular development. Our data are consistent with the previous hypothesis that the collapsed xylem (irx) phenotype causes dwarfism and impairs water uptake from the soil to the aerial organs (Lefebvre et al., 2011). Moreover, the reverse relationship also appears to hold: the suppression of the irx phenotype partially suppresses the dwarf phenotype and restores hydraulic conductivity. The fact that the chemical composition of beem396b is similar to that of genotypes carrying the esk1 mutation, and that the chemical composition of kak mutants is similar to the wild type, confirms this indirect suppression (Figs. 3 and 4; Supplemental Fig. S7). The biological system of the esk1 mutant and its suppressor lines are valuable tools for identifying major genes involved in diverse functions related to water fluxes in planta.
Although kak mutations appear to have no major impact on xylem cell wall composition, the issue of the suppression mechanism of the irx phenotype nevertheless remains. Yuan et al. (2013) attributed the collapsed xylem phenotype of esk1 not only to chemical alterations but also to a reduction in secondary cell wall thickening, which is consistent with a decrease in cellulose content. Thus, a possible explanation for irx suppression by kak mutations involves the recovery or enhancement of secondary cell wall thickening in xylem. However, we did not observe any recovery of cellulose content and only a partial recovery of hydraulic conductivity. Further characterization of esk1-5 kak xylem cell walls with transmission electron microscopy can address the question of secondary wall thickening. Hydraulic conductivity not only depends on vessel diameter and number but on resistance to water flow in the vessel end walls. This latter component of hydraulic conductivity is a function of vessel length and intervessel pits that form thin-walled areas, allowing sap flow between vessels. Thus, a comprehensive study requires a correlation analysis of structural, chemical, and functional parameters. Moreover, vulnerability to cavitation is another avenue for investigation, because this trait relies on vessel structure and is involved in drought tolerance (Tixier et al., 2013).
KAK May Be Involved in Vascular Development
We described new, clear-cut effects of kak inactivation on vascular morphology in an esk1-5 genetic background. kak single mutants show amplified stem diameter (Fig. 1), and the xylem vessel population is made up of larger tracheary elements (Fig. 5). KAK is expressed in trichomes, cotyledons, and roots in 4-d-old plantlets (Patra et al., 2013). We also observed that KAKpro drives gene expression in the vascular cambium, phloem, and primary xylem (Fig. 8). Its expression in cambium, which is the secondary meristem that gives rise to both secondary xylem and the phloem, leads to the hypothesis that KAK may play a role in the balance between xylem and phloem formation. KAK is described as an endoreduplication repressor (Downes et al., 2003; El Refy et al., 2003): it may control cell enlargement in both xylem and phloem, acting with different partners. For example, it interacts with GLABROUS3 (GL3) and ENHANCER OF GLABROUS3 (EGL3) in leaf epidermal cells to repress trichome formation by targeting its partners for the proteasome (Patra et al., 2013). GL3 is also involved in root hair formation through an activation/repression cascade similar to the one involved in epidermal cell differentiation that leads to trichome formation (Ishida et al., 2008). However, we measured root hair length and density in the wild type, esk1-5, beem396b, esk1-5 kak double mutants, and kak single mutants, and there was no genotype effect. Additional evidence provides support for KAK effects in cells other than trichomes. For example, kak mutants exhibit an enhanced endoreduplication level in etiolated hypocotyls, which completely lack trichomes (El Refy et al., 2003), suggesting that KAK plays a broad role in the plant wherever the control of endoreduplication is required. The intense GUS activity in root tips and lateral root primordia (Fig. 7) suggests that KAK is also expressed in apical meristems. In addition, KAK activity occurs in ovules, pollen, and maturing seeds (Fig. 7): cell division versus endoreduplication is probably tightly regulated in these types of cells.
However, it has not yet been demonstrated if the control of endoreduplication by KAK in trichomes is mediated by the degradation of GL3. The possibility that KAK interacts with other target(s) to control endoreduplication through a yet unknown pathway cannot be ruled out. Addressing this issue requires an analysis of esk1 kak gl3 triple mutants.
KAK recognizes the GL3/EGL3/GL1 complex to be degraded by the 26S proteasome, activating GL2 in trichomes, which in turn represses CELLULOSE SYNTHASE5 (CESA5) and activates XYLOGLUCAN ENDOTRANSGLUCOSYLASE17 (XTH17), two genes annotated as being involved in cell wall biogenesis and cell wall organization, respectively (Tominaga-Wada et al., 2009). Currently, we cannot discount the possibility that kak inactivation interferes with this cascade to suppress esk1 dwarfism. Nevertheless, cesa5 mutants have a seed mucilage phenotype (Sullivan et al., 2011), and vegetative plants are indistinguishable from the wild type (Desprez et al., 2007). XTH17 is expressed mostly in roots (Vissenberg et al., 2005; Becnel et al., 2006). Furthermore, our FT-IR and monosaccharide analyses do not support this hypothesis of KAK involvement in cell wall processes, because the results did not show any major differences in cell wall composition or between esk1, beem396b, esk1 kak-7, and esk1 kak-8.
esk1 beem Double Mutants Are Potentially Valuable Genotypes
beem396b and esk1 kak double mutants did not appear to show any cell wall modifications compared with esk1: their FT-IR profiles were similar, and they released less acetate than the wild type, similar to esk1-5. This similarity is consistent with ESK1 having xylan acetyltransferase enzymatic activity and with KAK having no effect on cell wall composition. Nonetheless, beem396b and esk1 kak double mutants are larger than esk1-5 mutants. esk1 kak and other beem lines may be regarded as potentially valuable genotypes, considering their vegetative size, which is close to that of the wild type (Fig. 1), and their lower acetate release (Fig. 3).
We observed enhanced saccharification of cell wall residues from mature 12-week-old stems in esk1 kak double mutants compared with the wild type and esk1 or kak single mutants (Fig. 3). This enhanced saccharification cannot be clearly attributed to a lower lignin level (Fig. 3). The recalcitrance of lignified cell walls to saccharification has a multifactorial origin, and another compositional trait of Arabidopsis cell walls may have a lower degree of acetylation, thus facilitating polysaccharide enzymatic hydrolysis (for review, see Pawar et al., 2013). The lower acetylation level in polymers has been described as beneficial for polymer degradation: polymers are more accessible to degrading enzymes, and degradation is more effective. Stem acetylation is reduced in the esk1 single and double mutants (Xiong et al., 2013). This reduced acetylation is associated with the improved enzymatic degradability of cell wall polysaccharides for the esk1 kak double mutants but not for the dwarf esk1 single mutant. Xiong et al. (2013) reported lower Glc yields for the esk1 mutant in a saccharification assay performed on destarched alcohol-insoluble residues obtained from 7-week-old stems. However, the saccharification conditions employed in that study led to very low Glc yields (close to 20 mg g−1 destarched alcohol-insoluble residues), in contrast to the 5- to 8-fold higher yield obtained in our study. Variation among samples, due to sampling and processing, is very likely responsible for the apparent differences in saccharification performance in the esk1 single mutant. We cannot conclude from our study that acetylation degree is associated with saccharification yield. However, esk1 is severely affected regarding stature, stem mechanical strength, secondary wall thickening (Yuan et al., 2013), and crystalline cellulose content (Lefebvre et al., 2011). These physical and chemical alterations may impair Glc release. Further investigations need to be done to explore saccharification.
Nevertheless, esk1 kak double mutants retain both the advantages of releasing less acetate but more Glc, on the one hand, and having a large vegetative size, on the other hand.
Here, KAK was identified as a suppressor of dwarfism and the collapsed xylem phenotype observed in esk1. Further investigations are required to determine the mechanism behind this suppression. Endoreduplication is currently being explored for its potential involvement in vascular formation. Other mechanisms that may result in cell wall thickening in xylem vessels are also being considered.
The esk1 mutant has a permanent drought syndrome (Bouchabke-Coussa et al., 2008). Given the restored xylem vessel morphology and the partially restored hydraulic conductivity reported in esk1 kak double mutants, it is worth exploring the drought response in beems, esk1 kak double mutants, as well as kak single mutants.
MATERIALS AND METHODS
Screening for esk1-5 Mutant Dwarfism Suppressors (beems)
EMS mutagenesis of Arabidopsis (Arabidopsis thaliana) seeds from the esk1-5 mutant (SALK_089531) was performed according to a previously described protocol (Lightner and Caspar, 1998). About 4,500 M1 plants of the resulting mutagenized population were pooled into bulks of seven plants and advanced to the M2 generation. For each bulk, 200 M2 plants were screened for rosette size compared with esk1-5 in a greenhouse. Theoretically, three to four plants with a suppressed esk1-5 phenotype were expected in a bulk, if one of the seven lines harbored a single recessive causative mutation (Page and Grossniklaus, 2002). Phenotype stability of the beem lines isolated in the M2 population was checked in their M3 progeny and selected if stable. Selected M2 beem lines were then backcrossed twice to their mutant background esk1-5 to segregate away some noncausative EMS-induced mutations.
Direct Mapping and Identification of the beem396b Causative Mutation
The F2 population resulting from the second backcross of the beem396b M2 line was used for mapping by sequencing. Among 570 F2 seeds sown, 124 plants with the largest rosettes were selected, of which 100 were sequenced. Genomic DNA was extracted from flowers of each F2 plant using the protocol described in Supplemental Protocol S1. Briefly, after cetyl-trimethyl-ammonium bromide-based extraction and subsequent RNase treatment, DNA was further purified with phenol-chloroform and quantified using a fluorimetrics-based method (Qubit Fluorimetric Quantification; Life Technologies; http://www.lifetechnologies.com). Equal amounts of genomic DNA from these plants were pooled and sequenced using the Illumina system (http://www.illumina.com/). The generation of sequencing libraries and sequencing were performed at the High-Throughput Sequencing Department at the IMAGIF facility (https://www.imagif.cnrs.fr/plateforme-36-High-throughput_Sequencing_Platform.html) at the Centre National de la Recherche Scientifique. Here, 2 μg of pooled DNA was used to generate a paired-end sequencing library according to Illumina’s protocol. The sequencing library was sequenced using four lanes of a Genome Analyzer IIx flow cell to an expected depth of 100×. A second nonallelic beem line F2 population was also sequenced independently.
The obtained short sequence reads were aligned to the Arabidopsis genome (version TAIR10) to identify mutations using the Illumina package CASAVA 1.8.2., leading to 2.1 × 105 and 2.2 × 105 reads mapping unambiguously on the genome. A homemade pipeline was developed to process the CASAVA output files from both beem lines and identify the causative mutations using a strategy similar to that described by Hartwig et al. (2012; Supplemental Protocol S2).
Confirmation of the beem396b Mutation and Identification of the beem308a Mutation
The EMS-induced kak mutation in beem396b was confirmed by sequencing overlapping PCR products encompassing the full KAK genomic sequence, including a 2-kb upstream transcription start site and a 2-kb downstream transcription termination site, using the primer couples listed in Supplemental Table S1. The same primers were also used to confirm the beem308a mutation.
Determination of Cell Wall-Bound Acetate Content
Inflorescence stems, minus their leaves, were sampled from just above the rosette on 12-week-old greenhouse-grown plants, and all stems were cut at a height corresponding to the second internode on the main stem. Sampled stems were immediately frozen in liquid nitrogen, lyophilized, and then ground to a fine powder in a grinding machine (Mixer Mill MM-2; Retsch; http://www.retsch.com/). Each sample was extracted three times with MilliQ water and three times with 96% (v/v) ethanol at 80°C, then air dried to obtain cell wall residues. The acetate content of cell wall residues was determined using the Megazyme Acetic Acid Kit (K-ACET version 02/11; http://www.megazyme.com/). The assay was downscaled and adapted to a 96-well format and performed essentially as described previously (Gille et al., 2011), with some slight modifications as follows. A total of 2 mg of cell wall residues was suspended in 100 μL of MilliQ water. The cell wall-bound acetate was released by adding 100 μL of 1 m NaOH and incubating for 1 h at room temperature with shaking at 500 rpm. The samples were neutralized with 1 m HCl and centrifuged for 10 min at 14,000 rpm, and 10 μL of the supernatant containing released acetate was transferred to a UV light-capable, 96-well flat-bottom assay plate and diluted with 94 μL of water. The kit content was used as follows. MilliQ water and solutions 2, 3, and 4 were mixed in a ratio of 96.8:2.5:1:1 (115.6 μL + 30 μL + 1.2 µL + 1.2 μL per sample), and 148 μL of this mixture was added to each sample, mixed, and incubated at room temperature for 2 min. The absorbance was read at 340 nm. Solution 5 was diluted 1:10 in water; 12 μL was added to each sample, mixed, and incubated at room temperature for 4 min. The absorbance was read at 340 nm. The amount of acetate in the samples was calculated based on an acetic acid standard curve and according to the manufacturer’s recommendations.
Evaluation of Cell Wall Lignin Content and Enzymatic Degradability
The lignin content of the same cell wall residues used for acetate determination was measured as acetyl bromide lignin and according to a procedure adapted from Fukushima and Hatfield (2001). In brief, 5 to 6 mg of cell wall residues weighed to the nearest 0.01 mg was mixed with 1 mL of freshly prepared 25% (v/v) acetyl bromide in acetic acid in a 2-mL glass vial with a Teflon-lined screw cap. The mixture was then allowed to digest in a thermomixer operated at 55°C and at 600 rpm for 150 min. After cooling, 100 µL of the reaction supernatant was transferred into a 5-mL disposable glass tube together with 1.5 mL of acetic acid:2 m NaOH (50:9, v/v). Then, 0.4 mL of 0.5 m hydroxylamine chlorohydrate and 1 mL of acetic acid were added. The absorbance of the solution was read at 280 nm against a reagent blank. Acetyl bromide lignin content was calculated from this absorbance using an extinction coefficient of 20 g−1 L cm−1. The measurements were performed from biological triplicates.
The enzymatic degradability of the cell wall residues was measured according to a procedure adapted from Berthet et al. (2011). Briefly, 25 to 30 mg of sample weighed to the nearest 0.1 mg was put in a 5-mL disposable plastic tube together with 4 mL of 0.05 m acetate buffer, pH 4.7, containing 4 mg mL−1 of a commercial cellulase preparation (cellulase Onozuka R-10 from Trichoderma viride; Serva) and 0.2 mg mL−1 NaN3. A reagent blank was prepared without cell wall residues. The reaction tubes (biological triplicates for each genotype plus the reagent blank) were placed on a carousel at 45°C for 60 h and then centrifuged (1,500g, 10 min). The Glc released into the reaction mixture was measured using the Biomerieux Glc RTU kit as follows. In a disposable 1-mL spectrophotometry cuvette, 50 µL of the supernatant was mixed with 1 mL of the Biomerieux reagent, and the colorimetric reaction was allowed to proceed for 30 min. Absorbance was read at 505 nm against a cuvette containing 50 µL of water and 1 mL of reagent. This absorbance was corrected for the absorbance obtained for the reagent blank (i.e. for the Glc originating from the Onozuka cellulase preparation). The Glc amounts released by enzymatic hydrolysis of the extract-free samples were calculated after appropriate calibration with standard Glc solutions (prepared in the 0.2–1 mg mL−1 range).
Neutral monosaccharide composition was measured as indicated (Simons et al., 2014).
Hydraulic Conductivity
For hydraulic analyses, seeds were sown on a commercial soil mixture (Humustar) and grown in a growth chamber at 22°C to 20°C under a light intensity of 100 μmol m–2 s–1 (16-h-light/8-h-dark cycle) and with 60% relative humidity. As soon as inflorescence stems showed ripening siliques (i.e. after 7–8 weeks), they were harvested and immediately used for analysis. Hydraulic conductivity (m4 MPa–1 s–1) was measured on segments of basal inflorescence stems using a Xyl’em apparatus (Bronkhorst). Stem segments 8 cm long were cut under water to avoid embolism. Then, the basal ends were gently sealed in a tubing system using seal tape (polytetrafluoroethylene film) to avoid leaking and crushing the samples with the clamps. A solution of KCl (10 mm) and CaCl2 (1 mm) was used to perfuse stems basipetally under 6 to 9 kPa at room temperature, and the flow rate was scored 3 min after starting the perfusion. To evaluate the effect of segment length on conductivity, the stem segment was shortened at its end apex to 6, 4, 3, 2, and 1.5 cm, and the flow rate was measured at each length.
Analysis of KAK Promoter Expression Pattern
The expression pattern of the KAK gene was studied using the GUS reporter gene. A 2-kb sequence upstream from the KAK gene transcription starting site was amplified from wild-type genomic DNA using attB-flanked primers for subsequent Gateway cloning (primers are listed in Supplemental Table S1). The PCR product was first cloned into a pDONR207 vector (Invitrogen; http://www.lifetechnologies.com) with a BP reaction and transferred using an LR reaction into pBI101-R2R3-GUS, a Gateway-compatible binary vector derived from pBI101 (Harscoët et al., 2010), to generate a transcriptional fusion between the KAK promoter (KAKpro) and the GUS reporter gene. The KAKpro:GUS construct was introduced into wild-type plants by Agrobacterium tumefaciens-mediated transformation as described previously (Bechtold and Pelletier, 1998). Eight independent transformed lines with nonsegregating insertions were selected on kanamycin and used for GUS activity assay. The GUS activity assays were performed as reported previously (Marrocco et al., 2003). The eight independent KAKpro:GUS-transformed lines tested showed similar GUS activity patterns.
Cytology and Microscopy
Trichomes were observed on the fourth true leaves of 14-d-old in vitro-grown plants using an SH-1500 scanning electron microscope (Hirox Europe; http://www.hirox-europe.com/).
For Toluidine Blue Orange staining and FT-IR analysis, hypocotyls were collected from 10-week-old greenhouse-grown plants and cleared in 96% (v/v) ethanol. After rehydration, samples were directly glued to a vibratome-compatible disk and cut while immersed in water. The thickness of the cross sections was 20 μm.
Stem basal internodes and hypocotyls were collected from 8-week-old greenhouse-grown plants transformed with the KAKpro:GUS construct.
FT-IR Microspectroscopy
FT-IR analysis on hypocotyl cross sections was performed as described previously (Lefebvre et al., 2011) with the following modifications. The cross section thickness was 20 μm, and the acquisition window of the microspectrophotometer was set to 40 × 40 μm. For each genotype, a total of 45 spectra were acquired from three independent plants. The zone targeted for the acquisition of spectra was the upper region of metaxylem as shown in Supplemental Figure S7.
Statistical and Image Analyses
Statistical analyses were performed using R (http://www.r-project.org/). Nonparametric tests were performed using the packages nparcomp for Tukey’s or Dunnett’s tests (Konietschke et al., 2012) and RVAideMemoire for tests with permutations (Hervé, 2014). Measurements on images were performed with the ImageJ software (http://rsbweb.nih.gov/ij/).
The genetic background of every plant line studied here was Columbia-0. Two independent T-DNA mutants with insertion sites within the KAK/UPL3 gene (At4g38600) were ordered from the Salk Institute Genomic Analysis Laboratory collection. T-DNA mutants were named kak-7 (SALK_116326) and kak-8 (SALK_117247). kak homozygous T-DNA lines were identified by PCR using gene- and T-DNA-specific primers obtained using the T-DNA Primer Design tool provided by the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/tdnaprimers.2.html). Primer sequences are listed in Supplemental Table S1.
The Arabidopsis Genome Initiative locus identifier of the ESK1/TBL29 gene is At3g55990. The sequence data mentioned here can be found in the GenBank data libraries under project accession number PRJNA253006 and sample accession number SAMN02869710.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Mapping of EMS mutation frequency in a beem396b pool.
Supplemental Figure S2. Effect of the beem396b mutation within the KAK coding sequence.
Supplemental Figure S3. Scanning electron micrographs of leaf trichomes.
Supplemental Figure S4. beem308a has a lesion in the KAK gene.
Supplemental Figure S5. Reverse transcription-PCR detection of an ESK1 transcript.
Supplemental Figure S6. Zone targeted for FT-IR microspectroscopy.
Supplemental Figure S7. FT-IR analysis of metaxylem vessels.
Supplemental Figure S8. Measurements of xylem pole, phloem pole, and xylem vessel area.
Supplemental Figure S9. Metaxylem vessel area distribution and analysis.
Supplemental Table S1. List of PCR primers.
Supplemental Table S2. Cell wall monosaccharides.
Supplemental Protocol S1. Preparation and pooling of genomic DNA for Illumina deep sequencing.
Supplemental Protocol S2. High-output sequencing analysis.
Supplementary Material
Acknowledgments
We thank Salem Chabout and Halima Morin for assistance in FT-IR analysis and microscopy, respectively; the Observatoire du Végétal/Centre de Ressources Arabidopsis thaliana team for assistance in screening for the beem lines; Liudmila Chelysheva for image processing; Florian Deligne for mutant genotyping and image analysis; and Herman Höfte, Samantha Vernettes, and Matthieu Reymond for critical reading of the article.
Glossary
- EMS
ethyl methanesulfonate
- TAIR10
The Arabidopsis Information Resource 10
- T-DNA
transfer DNA
- FT-IR
Fourier transform infrared
Footnotes
This work was supported by Gautier Semences (to M.B.), by the Agence Nationale pour la Recherche (grant no. GPLA07–004 to V.L. and J.T.), and by the Bioénergies, Biomolécules et Matériaux Biosourcés du Carbone Renouvelable program Hemicoat (to E.G.).
Articles can be viewed without a subscription.
References
- Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, et al. (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30: 174–178 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Becnel J, Natarajan M, Kipp A, Braam J (2006) Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol 61: 451–467 [DOI] [PubMed] [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, et al. (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biely P. (1985) Microbial xylanolytic system. Trends Biotechnol 3: 286–289 [Google Scholar]
- Bouchabke-Coussa O, Quashie ML, Seoane-Redondo J, Fortabat MN, Gery C, Yu A, Linderme D, Trouverie J, Granier F, Téoulé E, et al. (2008) ESKIMO1 is a key gene involved in water economy as well as cold acclimation and salt tolerance. BMC Plant Biol 8: 125–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvat R. (1989) Ontogeny, Cell Differentiation, and Structure of Vascular Plants. Springer-Verlag, Berlin [Google Scholar]
- Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes BP, Stupar RM, Gingerich DJ, Vierstra RD (2003) The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J 35: 729–742 [DOI] [PubMed] [Google Scholar]
- Ebringerova A, Heinze T (2000) Xylan and xylan derivatives: biopolymers with valuable properties. I. Naturally occurring xylans structures, isolation procedures and properties. Macromol Rapid Commun 21: 542–556 [Google Scholar]
- El Refy A, Perazza D, Zekraoui L, Valay JG, Bechtold N, Brown S, Hülskamp M, Herzog M, Bonneville JM (2003) The Arabidopsis KAKTUS gene encodes a HECT protein and controls the number of endoreduplication cycles. Mol Genet Genomics 270: 403–414 [DOI] [PubMed] [Google Scholar]
- Esau K. (1965) Anatomy of Seed Plants, Ed 2 John Wiley & Sons, New York [Google Scholar]
- Faik A. (2010) Xylan biosynthesis: news from the grass. Plant Physiol 153: 396–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima RS, Hatfield RD (2001) Extraction and isolation of lignin for utilization as a standard to determine lignin concentration using the acetyl bromide spectrophotometric method. J Agric Food Chem 49: 3133–3139 [DOI] [PubMed] [Google Scholar]
- Gille S, Cheng K, Skinner ME, Liepman AH, Wilkerson CG, Pauly M (2011) Deep sequencing of voodoo lily (Amorphophallus konjac): an approach to identify relevant genes involved in the synthesis of the hemicellulose glucomannan. Planta 234: 515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann K, Mitchell DJ, Himmel ME, Dale BE, Schroeder HA (1989) The role of ester groups in resistance of plant cell wall polysaccharides to enzymatic hydrolysis. Appl Biochem Biotechnol 20/21: 45–61 [Google Scholar]
- Harscoët E, Dubreucq B, Palauqui JC, Lepiniec L (2010) NOF1 encodes an Arabidopsis protein involved in the control of rRNA expression. PLoS ONE 5: e12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig B, James GV, Konrad K, Schneeberger K, Turck F (2012) Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol 160: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé M (2014) RVAideMemoire: diverse basic statistical and graphical functions. R package version 0.9-41. http://CRAN.R-project.org/package=RVAideMemoire (March 14, 2015)
- Ishida T, Kurata T, Okada K, Wada T (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Konietschke F, Libiger O, Hothorn LA (2012) Nonparametric evaluation of quantitative traits in population-based association studies when the genetic model is unknown. PLoS ONE 7: e31242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Fortabat MN, Ducamp A, North HM, Maia-Grondard A, Trouverie J, Boursiac Y, Mouille G, Durand-Tardif M (2011) ESKIMO1 disruption in Arabidopsis alters vascular tissue and impairs water transport. PLoS ONE 6: e16645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner J, Caspar T (1998) Seed mutagenesis of Arabidopsis. Methods Mol Biol 82: 91–103 [PubMed] [Google Scholar]
- Lugan R, Niogret MF, Kervazo L, Larher FR, Kopka J, Bouchereau A (2009) Metabolome and water status phenotyping of Arabidopsis under abiotic stress cues reveals new insight into ESK1 function. Plant Cell Environ 32: 95–108 [DOI] [PubMed] [Google Scholar]
- Marrocco K, Lecureuil A, Nicolas P, Guerche P (2003) The Arabidopsis SKP1-like genes present a spectrum of expression profiles. Plant Mol Biol 52: 715–727 [DOI] [PubMed] [Google Scholar]
- Mohebby B. (2008) Application of ATR infrared spectroscopy in wood acetylation. J Agric Sci Technol 10: 253–259 [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Höfte H (2003) Classification and identification of Arabidopsis cell wall mutants using Fourier-transform infrared (FT-IR) microspectroscopy. Plant J 35: 393–404 [DOI] [PubMed] [Google Scholar]
- Page DR, Grossniklaus U (2002) The art and design of genetic screens: Arabidopsis thaliana. Nat Rev Genet 3: 124–136 [DOI] [PubMed] [Google Scholar]
- Patra B, Pattanaik S, Yuan L (2013) Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant J 74: 435–447 [DOI] [PubMed] [Google Scholar]
- Pawar PM, Koutaniemi S, Tenkanen M, Mellerowicz EJ (2013) Acetylation of woody lignocellulose: significance and regulation. Front Plant Sci 4: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Herzog M, Hülskamp M, Brown S, Dorne AM, Bonneville JM (1999) Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152: 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni L, Nieminen K, Pacheco-Villalobos D, Sibout R, Schwechheimer C, Hardtke CS (2011) Mobile gibberellin directly stimulates Arabidopsis hypocotyl xylem expansion. Plant Cell 23: 1322–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Saha R, Amiour N, Kumar A, Guillard L, Clément G, Miquel M, Li Z, Mouille G, Lea PJ, et al. (2014) Assessing the metabolic impact of nitrogen availability using a compartmentalized maize leaf genome-scale model. Plant Physiol 166: 1659–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RE, Mabee W, Saddler JN, Taylor M (2010) An overview of second generation biofuel technologies. Bioresour Technol 101: 1570–1580 [DOI] [PubMed] [Google Scholar]
- Studer MH, DeMartini JD, Davis MF, Sykes RW, Davison B, Keller M, Tuskan GA, Wyman CE (2011) Lignin content in natural Populus variants affects sugar release. Proc Natl Acad Sci USA 108: 6300–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Ralet MC, Berger A, Diatloff E, Bischoff V, Gonneau M, Marion-Poll A, North HM (2011) CESA5 is required for the synthesis of cellulose with a role in structuring the adherent mucilage of Arabidopsis seeds. Plant Physiol 156: 1725–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tixier A, Cochard H, Badel E, Dusotoit-Coucaud A, Jansen S, Herbette S (2013) Arabidopsis thaliana as a model species for xylem hydraulics: does size matter? J Exp Bot 64: 2295–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R, Iwata M, Sugiyama J, Kotake T, Ishida T, Yokoyama R, Nishitani K, Okada K, Wada T (2009) The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. Plant J 60: 564–574 [DOI] [PubMed] [Google Scholar]
- Turner S, Sieburth LE (2003) Vascular patterning. The Arabidopsis Book 2: e0073, doi/10.1199/tab.0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz BR, Peña MJ, Moniz HA, Moremen KW, York WS (2014) Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J 80: 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Oyama M, Osato Y, Yokoyama R, Verbelen JP, Nishitani K (2005) Differential expression of AtXTH17, AtXTH18, AtXTH19 and AtXTH20 genes in Arabidopsis roots: physiological roles in specification in cell wall construction. Plant Cell Physiol 46: 192–200 [DOI] [PubMed] [Google Scholar]
- Xin Z, Browse J (1998) Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc Natl Acad Sci USA 95: 7799–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G, Cheng K, Pauly M (2013) Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29. Mol Plant 6: 1373–1375 [DOI] [PubMed] [Google Scholar]
- Ye ZH, Freshour G, Hahn MG, Burk DH, Zhong R (2002) Vascular development in Arabidopsis. Int Rev Cytol 220: 225–256 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Teng Q, Zhong R, Ye ZH (2013) The Arabidopsis DUF231 domain-containing protein ESK1 mediates 2-O- and 3-O-acetylation of xylosyl residues in xylan. Plant Cell Physiol 54: 1186–1199 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.