An ABA-associated transcription factor regulates lipid mobilization specifically in the embryo to determine proper timing for seed germination.
Abstract
Seed germination is a key developmental transition that initiates the plant life cycle. The timing of germination is determined by the coordinated action of two phytohormones, gibberellin and abscisic acid (ABA). In particular, ABA plays a key role in integrating environmental information and inhibiting the germination process. The utilization of embryonic lipid reserves contributes to seed germination by acting as an energy source, and ABA suppresses lipid degradation to modulate the germination process. Here, we report that the ABA-responsive R2R3-type MYB transcription factor MYB96, which is highly expressed in embryo, regulates seed germination by controlling the expression of ABSCISIC ACID-INSENSITIVE4 (ABI4) in Arabidopsis (Arabidopsis thaliana). In the presence of ABA, germination was accelerated in MYB96-deficient myb96-1 seeds, whereas the process was significantly delayed in MYB96-overexpressing activation-tagging myb96-ox seeds. Consistently, myb96-1 seeds degraded a larger extent of lipid reserves even in the presence of ABA, while reduced lipid mobilization was observed in myb96-ox seeds. MYB96 directly regulates ABI4, which acts as a repressor of lipid breakdown, to define its spatial and temporal expression. Genetic analysis further demonstrated that ABI4 is epistatic to MYB96 in the control of seed germination. Taken together, the MYB96-ABI4 module regulates lipid mobilization specifically in the embryo to ensure proper seed germination under suboptimal conditions.
Seeds are products of plant sexual reproduction and facilitate the spread of offspring. Since plants are sessile organisms with a limited ability to seek favorable growth conditions, seed dispersal is an evolutionarily adaptive trait to disseminate their offspring to optimal locations (Bewley, 1997; Nonogaki, 2014). Thus, seeds enable plants to endure environmental disadvantages and confine growth until they encounter favorable environmental conditions (Bewley, 1997).
Seed embryogenesis is initiated by double fertilization (Gehring et al., 2004; Berger et al., 2008). While the zygote divides asymmetrically to form the diploid embryo, the triploid endosperm constantly proliferates inside the maternal ovule (Natesh and Rau, 1984; West and Harada, 1993). At the end of the embryonic growth phase, cell cycle activities in the embryo are arrested (Raz et al., 2001), and storage molecules, such as proteins, carbohydrates, and lipids, accumulate particularly in the cotyledons (Goldberg et al., 1989; Raz et al., 2001). This phase is referred to as seed maturation. Mature seeds become dehydrated and enter a quiescent state that is tolerant to desiccation (Crouch, 1987; Harada et al., 1988; Kermode, 1990; McCarty and Carson, 1991), establishing primary dormancy.
Seed dormancy can be broken when seeds experience stratification and/or after-ripening (Vleeshouwers et al., 1995; Bewley, 1997; Nonogaki, 2014). Germination is a key developmental transition to determine when to initiate the plant life cycle. Since it is an irreversible transition, seeds have to precisely monitor external environmental factors, such as light, temperature, water and nutrient availability, and high salinity (Huang et al., 2003; Qu et al., 2008; Joosen et al., 2013; Lim et al., 2013). These environmental signals are integrated into internal developmental programs in seeds, which are operated primarily by antagonistic actions of two phytohormones: abscisic acid (ABA) and GA (Holdsworth et al., 1999; Finch-Savage and Leubner-Metzger, 2006; Seo et al., 2006). GA promotes germination, whereas ABA suppresses completion of the developmental transition (Garciarrubio et al., 1997; Debeaujon and Koornneef, 2000; Finkelstein et al., 2002; Peng and Harberd, 2002; Ogawa et al., 2003; Lau and Deng, 2010).
Transcriptional regulation is a fundamental regulation scheme for the seed germination process (Thomas, 1993; Nakabayashi et al., 2005). The GA function in promoting seed germination is accomplished partly through the action of central GA signaling repressors, DELLA-domain proteins, including GIBBERELLIN INSENSITIVE (GAI), REPRESSOR OF GIBBERELLIN1-3 (RGA), RGA-LIKE1 (RGL1), RGL2, and RGL3, all of which act as transcriptional regulators (Hussain and Peng, 2003; Tyler et al., 2004; Cao et al., 2006; Park et al., 2013). They interact with transcription factors to allow DELLAs to regulate gene expression by targeting specific gene promoters (de Lucas et al., 2008; Feng et al., 2008; Arnaud et al., 2010; Gallego-Bartolomé et al., 2010; Hong et al., 2012). Indeed, DELLAs bind to promoters of GIBBERELLIN INSENSITIVE DWARF1, WRKY27, and SCARECROW-LIKE3 to control seed germination (Zentella et al., 2007; Gallego-Bartolomé et al., 2011; Zhang et al., 2011; Stamm et al., 2012). The DELLA proteins are rapidly degraded in response to GA and are thus considered as repressors of seed germination in the absence of GA (Dill et al., 2001; Tyler et al., 2004).
ABA regulation of seed germination also requires ABA-responsive transcription factors. ABSCISIC ACID INSENSITIVE3 (ABI3), ABI4, and ABI5, which encode transcription factors with the B3 domain, APETALA2 domain, and basic Leu zipper domain, respectively, are expressed highly in seeds and are involved in regulating sensitivity to ABA (Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000). Indeed, genetic mutants at these loci exhibit reduced ABA sensitivity as well as accelerated seed germination in the presence of exogenous ABA (Finkelstein and Somerville, 1990). They act in concert with each other and form combinatorial networks to determine the proper time to germinate (Söderman et al., 2000; Nakamura et al., 2001).
In addition to their overlapping roles in ABA-dependent seed germination, ABI3, ABI4, and ABI5 have their own unique functions. ABI4 is particularly interesting in that it plays a unique role in embryonic lipid catabolism during the germination process (Penfield et al., 2006). Seeds utilize lipid reserves to fuel postgerminative seedling growth (Penfield et al., 2005; Quettier and Eastmond, 2009), but the lipid catabolic process is inhibited in the presence of ABA in order to delay seed germination, thereby minimizing environmental damage to early seedlings. Notably, ABA specifically represses embryonic lipid breakdown (Penfield et al., 2006). The differential responses of embryo and endosperm to ABA are accomplished by the confined expression of ABI4 in the embryo (Penfield et al., 2006). However, the mechanisms by which ABI4 is expressed specifically in the embryo and by which specific regulator(s) participate in defining its spatial and temporal expression remain unknown.
We previously showed that the MYB96 transcription factor mediates a variety of plant responses to ABA, including drought resistance, stomatal conductance, lateral root development, anthocyanin accumulation, and cuticular wax biosynthesis (Seo et al., 2009, 2011; Seo and Park, 2010, 2011). The expression of MYB96 is considerably induced by drought stress and ABA (Seo et al., 2009), and it activates the expression of many ABA-responsive genes through direct binding to gene promoters to optimize plant growth and fitness under environmentally unfavorable conditions (Seo et al., 2009, 2011; Seo and Park, 2010).
Here, we report that MYB96 also plays a key role in the ABA control of seed germination in Arabidopsis (Arabidopsis thaliana). The MYB96 gene was significantly expressed in the embryo and conferred embryonic ABA sensitivity in seed germination as well as triacylglycerol (TAG) breakdown. In the presence of ABA, MYB96-overexpressing myb96-ox seeds exhibited delayed germination with reduced lipid breakdown, whereas myb96-1 seeds showed accelerated seed germination and lipid breakdown. The MYB96 regulation of seed germination was largely dependent on ABI4, a key repressor of embryonic lipid mobilization during the germination process. Together, our findings provide evidence that MYB96 is an unequivocal transcriptional regulator of ABI4 and defines the spatial and temporal specificity of its expression to fine-tune seed germination under unfavorable environmental conditions.
RESULTS
MYB96 Is Expressed in Embryos
MYB96 mediates ABA signal transduction in a variety of physiological processes, such as drought response, lateral root development, and cuticular wax accumulation (Seo et al., 2009, 2011). Since seed germination is also a key developmental transition governed by ABA (Bewley, 1997; Nakashima and Yamaguchi-Shinozaki, 2013), it was highly plausible that MYB96 may also regulate seed germination in an ABA-dependent manner.
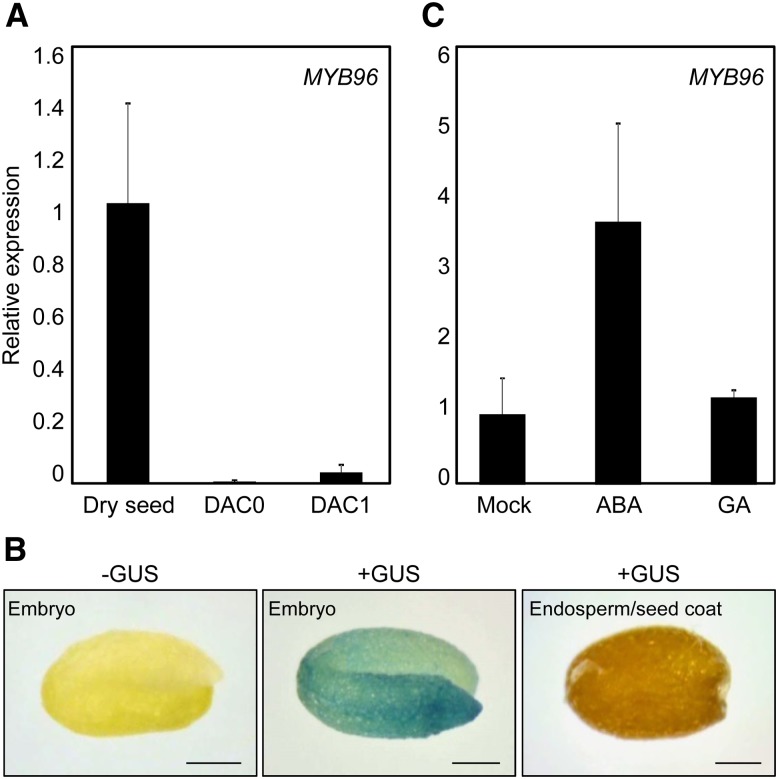
To explore the role of MYB96 in seed germination, we first determined its expression patterns in seeds. Quantitative reverse transcription (qRT)-PCR analysis revealed that MYB96 transcripts accumulated at high levels in dry seeds (Fig. 1A), and its transcript accumulation was drastically reduced after cold stratification and imbibition. The transcript levels of MYB96 were reduced by 98% after stratification compared with those in dry seeds (Fig. 1A) and recovered at seedling stages (Supplemental Fig. S1), implying an inhibitory role of MYB96 in seed germination.
Figure 1.
Expression of MYB96 in seeds. A, MYB96 expression in dry seeds and during seed germination. Wild-type seeds were kept in darkness at 4oC for 72 h (stratification) and transferred to long-day conditions at 22oC for germination. Transcript accumulation was analyzed by qRT-PCR. The EUKARYOTIC TRANSLATION INITIATION FACTOR4A1 (eIF4a) gene (At3g13920) was used as an internal control. Biological triplicates were averaged. Error bars indicate se. DAC, Days after cold stratification. B, Histochemical staining analysis. The pMYB96:GUS transgenic seeds, in which a promoter sequence covering an approximately 2-kb region upstream of the MYB96 transcription start site was transcriptionally fused to a GUS-coding sequence, were subject to GUS staining. Bars = 0.7 μm. C, Induction of MYB96 by ABA in seeds. Stratified wild-type seeds were transferred to MS medium supplemented with 5 μm ABA or GA and incubated for 1 d under long-day conditions. Transcript accumulation was analyzed by qRT-PCR. Biological triplicates were averaged. Error bars indicate se.
Spatial expression patterns of MYB96 were further examined using pMYB96:GUS transgenic seeds, in which a promoter sequence covering an approximately 2-kb region upstream of the MYB96 transcription start site was transcriptionally fused to a GUS-coding sequence. In agreement with the qRT-PCR results, GUS activities were strongly detected in dry seeds (Fig. 1B) and reduced after stratification and imbibition (Supplemental Fig. S2). Moreover, dissection of seeds showed that high levels of GUS expression were observed only in the embryo (Fig. 1B). GUS activity was undetectable in the endosperm and seed coat (Fig. 1B).
MYB96 Is Induced by ABA in Seeds But Not by GA
MYB96 is transcriptionally induced by ABA in seedlings (Seo et al., 2009). Considering that ABA content decreases in seeds following stratification and imbibition (Grappin et al., 2000; Shu et al., 2013), the decreased expression of MYB96 during seed germination seemed likely to correlate with endogenous ABA levels in seeds. Therefore, we asked whether ABA induction of MYB96 is also relevant in seeds.
To examine the effect of ABA on MYB96 expression in germinating seeds, stratified wild-type seeds were plated on Murashige and Skoog (MS) medium supplemented with or without ABA. qRT-PCR analysis showed that transcript levels of MYB96 were significantly increased 3.5-fold by exogenous ABA treatment in wild-type seeds (Fig. 1C; Seo et al., 2009). Histochemical analysis also revealed that the ABA induction of MYB96 was observed mainly in the embryo (Supplemental Fig. S3).
Seed germination is tightly regulated by antagonistic interactions between ABA and GA (Yano et al., 2009; Yaish et al., 2010). Thus, we next asked whether the promotive action of GA in seed germination is also associated with MYB96 expression. Contrary to ABA, MYB96 was unaffected by exogenous GA treatment (Fig. 1C). Furthermore, MYB96 expression was also unchanged in quadruple-DELLA (gai-t6 rga-t2 rgl1-1 rgl2-1) and GA-deficient ga1-3 mutants (Supplemental Fig. S4). These observations suggest that MYB96 plays a primary role in ABA signal transduction in seeds.
MYB96 Regulates ABA-Dependent Seed Germination
Our observation that seed-expressed MYB96 was regulated by ABA suggested that it might have a physiological role in ABA-dependent seed germination. Therefore, we measured the germination rate of seeds of the wild type, the MYB96-overexpressing activation-tagging line myb96-ox (Seo et al., 2009), and the transfer-DNA insertional MYB96-deficient mutant myb96-1 (Seo et al., 2009) at various concentrations of ABA.
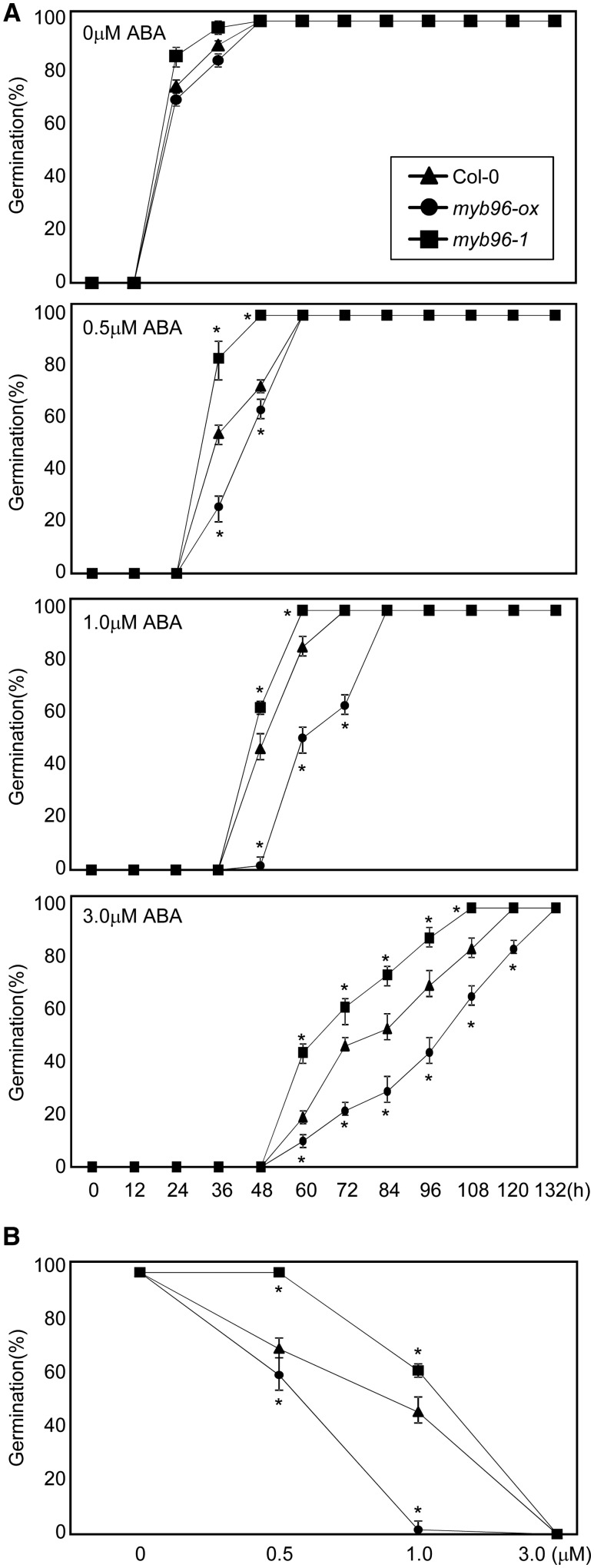
The germination rate of myb96-ox and myb96-1 seeds was similar to that of wild-type seeds in the absence of ABA (Fig. 2A). However, in the presence of ABA, the germination rate was substantially reduced in myb96-ox seeds but elevated in myb96-1 seeds compared with wild-type seeds (Fig. 2A). The differential ABA responses of myb96-ox and myb96-1 seeds were more evident at higher concentrations of ABA (Fig. 2A).
Figure 2.
ABA sensitivity of myb96-ox and myb96-1 in seed germination. A, Germination rate of myb96-ox and myb96-1. Seed germination percentages of the indicated genotypes grown on different concentrations of ABA were quantified after the end of stratification. Radicle emergence was used as a morphological marker for germination. At least 40 seeds per genotype were measured in each replicate. Biological triplicates were averaged. Error bars indicate se. Statistically significant differences between wild-type Columbia-0 (Col-0) and mutants are indicated by asterisks (Student’s t test, *P < 0.05). B, ABA dose-response assay. Wild-type, myb96-ox, and myb96-1 seeds were germinated on an increasing concentration of ABA. Germination percentages were scored 2 d after cold stratification. Biological triplicates were averaged. Error bars indicate se. Statistically significant differences between the wild type and mutants are indicated by asterisks (Student’s t test, *P < 0.05).
For ABA dose-response assays, seeds were germinated on MS medium plates supplemented with different concentrations of ABA, and germination percentages were scored 2 d after cold stratification. These assays clearly revealed that the myb96-ox seeds exhibited increased sensitivity to exogenous ABA and the myb96-1 seeds were hyposensitive to ABA relative to wild-type seeds (Fig. 2B).
Cotyledon opening is also sensitive to exogenous ABA treatment in the association with seed germination. The cotyledon opening of myb96-ox and myb96-1 seeds was indistinguishable from that of wild-type seeds in the absence of ABA (Supplemental Fig. S5). However, in agreement with the seed germination rates, the cotyledon opening of myb96-ox was hypersensitive to ABA, whereas the myb96-1 mutants were hyposensitive (Supplemental Figs. S5 and S6). These observations indicate that MYB96 determines ABA sensitivity and regulates ABA-dependent seed germination.
ABI4 Is Up-Regulated in myb96-ox
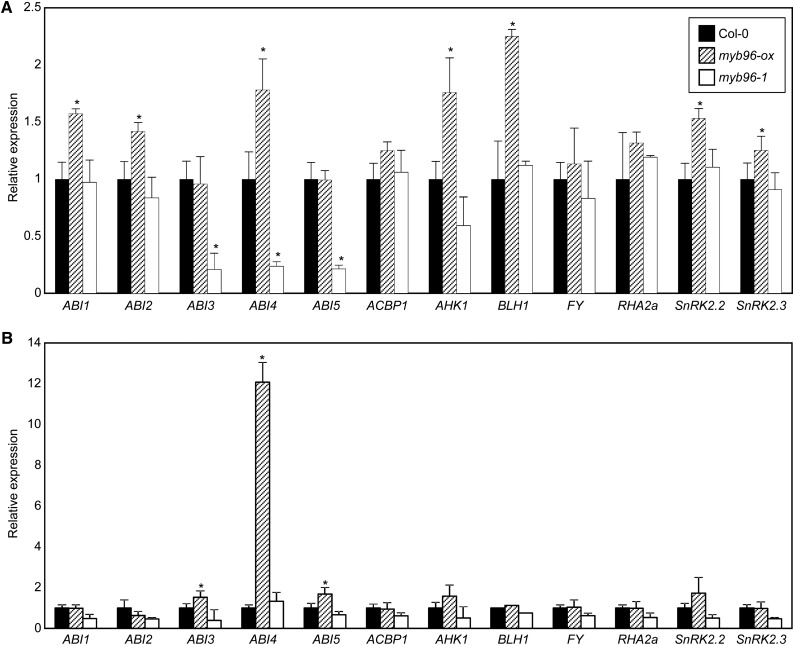
To establish the molecular networks underlying the MYB96 regulation of seed germination, we monitored the expression patterns of genes associated with ABA-dependent seed germination, such as ABI1, ABI2, ABI3, ABI4, ABI5, ACYL-COENZYME A-BINDING PROTEIN1 (ACBP1), ARABIDOPSIS HISTIDINE KINASE1 (AHK1), BEL1-LIKE HOMEODOMAIN1 (BLH1), FLOWERING LOCUS Y (FY), RING-H2 FINGER A2A (RHA2a), SNF1-RELATED PROTEIN KINASE2.2 (SnRK2.2), and SnRK2.3 (Finkelstein and Somerville, 1990; Söderman et al., 2000; Carles et al., 2002; Fujii et al., 2007; Tran et al., 2007; Moes et al., 2008; Li et al., 2011; Jiang et al., 2012; Du et al., 2013; Kim et al., 2013), in the wild type, myb96-ox, and myb96-1, in the absence of ABA. In seeds, many genes were differentially expressed in myb96-ox and myb96-1, but MYB96 regulation of ABI4 was most likely relevant (Fig. 3A). The expression of ABI4 was reduced in myb96-1 seeds, whereas myb96-ox increased ABI4 expression. To further support this observation, we also analyzed the expression of these genes in wild-type, myb96-ox, and myb96-1 seedlings. Most of the genes examined were transcriptionally unchanged in the mutants, but the expression of ABI4 was elevated approximately 12-fold in the myb96-ox mutant (Fig. 3B). The ABI3 and ABI5 genes were also slightly up-regulated in myb96-ox (Fig. 3B). However, their expression was not down-regulated in myb96-1, possibly due to extremely low expression levels of the genes in wild-type seedlings under normal growth conditions.
Figure 3.
Expression of genes involved in ABA-dependent seed germination in myb96-ox and myb96-1 mutants. Germinating seeds (A) and 10-d-old seedlings (B) grown under long-day conditions were harvested for total RNA isolation. Transcript accumulation was analyzed by qRT-PCR. The eIF4a gene was used as an internal control. Biological triplicates were averaged. Error bars indicate se. Statistically significant differences between wild-type Columbia-0 (Col-0) and mutants are indicated by asterisks (Student’s t test, *P < 0.05).
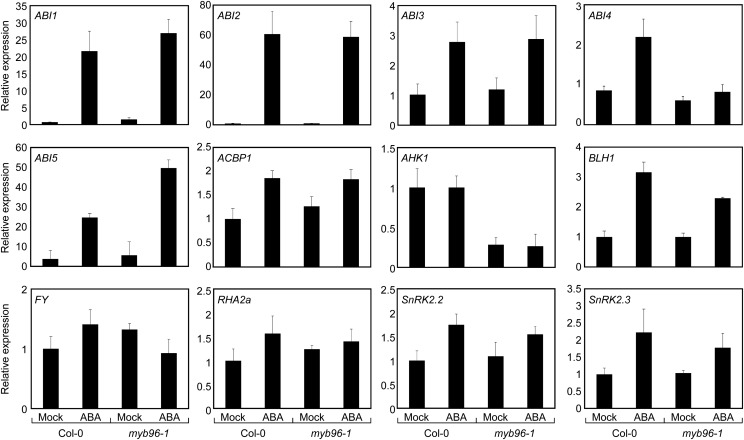
Transcript levels of the same set of genes were analyzed in myb96-1 seeds in the presence of ABA. Most of the genes, except for AHK1, FY, and RHA2a, were induced by ABA. Notably, the ABA induction of ABI4 and BLH1 genes was significantly reduced in the myb96-1 seeds compared with wild-type seeds (Fig. 4), while the ABA induction of the others was not influenced. The expression of two genes was also analyzed in germinating myb96-ox seeds in the presence of ABA. Accordingly, their ABA induction was slightly hypersensitive in myb96-ox (Supplemental Fig. S7). While the expression of BLH1 seems to be dependent upon MYB96, based on the close correlation between ABI4 expression and MYB96 activity (Figs. 3 and 4), we hypothesized that MYB96 is intimately required for the proper expression of ABI4 in the control of ABA-mediated seed germination. Consistent with this, the expression patterns of ABI4 in seeds were similar to those of MYB96. The ABI4 gene was highly expressed in dry seeds, particularly embryo (Söderman et al., 2000; Penfield et al., 2006; Bossi et al., 2009), and its expression was reduced after stratification and imbibition, like the transcript accumulation patterns of MYB96 (Supplemental Fig. S8). Their spatial expression patterns were also similar (Supplemental Fig. S9), further supporting the positive regulation of ABI4 by MYB96.
Figure 4.
Compromised expression of ABI4 in myb96-1 upon exogenous ABA treatment. Germinating seeds were plated on MS medium supplemented with or without 5 μm ABA and incubated for 1 d. Transcript accumulation was analyzed by qRT-PCR. Biological triplicates were averaged. Error bars indicate se. Col-0, wild-type Columbia-0.
MYB96 Binds Directly to the ABI4 Promoter
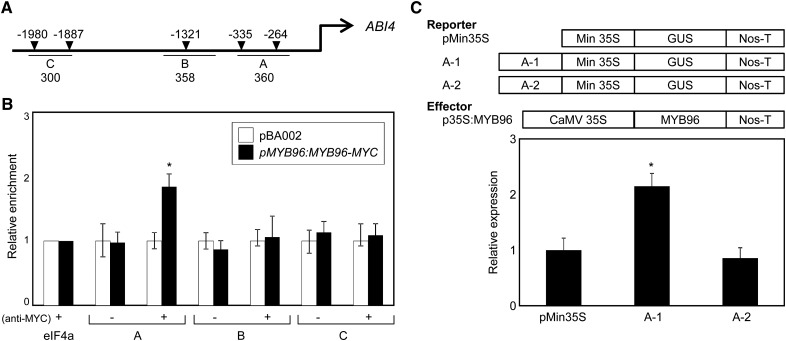
MYB96 is an R2R3-type MYB transcription factor (Seo et al., 2009). Plant R2R3 MYB-DNA-binding domain-containing proteins are known to bind directly to consensus DNA elements that are enriched in A and C residues (Seo et al., 2011; Prouse and Campbell, 2013). Therefore, we asked whether MYB96 binds directly to the consensus motifs on the ABI4 promoter. Sequence analysis revealed that the ABI4 promoter contains five conserved sequence motifs that are analogous to the R2R3-type MYB-binding consensus sequences (Fig. 5A). The presence of MYB-binding cis-elements led us to examine whether MYB96 is targeted to the ABI4 promoter.
Figure 5.
Binding of MYB96 to consensus sequences in the ABI4 promoter. A, R2R3-MYB-binding consensus sequences on the ABI4 promoter. Core binding sequences are marked with arrowheads. Underbars represent amplified genomic regions. The numbers below the underbars indicate amplification product size (bp). B, ChIP assays. Total protein extracts from pMYB96:MYB96-MYC transgenic seeds were immunoprecipitated with an anti-MYC antibody. Fragmented genomic DNA was eluted from the protein-DNA complexes and subjected to quantitative PCR analysis. Biological triplicates were averaged, and statistically significant differences of the measurements were determined using Student’s t test (*P < 0.05). Error bars indicate se. In each experiment, the measurement values in pBA002 were set to 1 after normalization against eIF4a for quantitative PCR analysis. C, Transient expression analysis using Arabidopsis protoplasts. The core promoter sequence elements of ABI4, A-1 (CAAGTAACTTATGA) and A-2 (GCTATAGTTACCCA), were constructed in the reporter plasmid. The recombinant reporter and effector constructs were coexpressed transiently in Arabidopsis protoplasts, and GUS activities were determined fluorimetrically. Luciferase gene expression was used to normalize the GUS activities. The normalized values in control protoplasts were set to 1 and represented as relative activation. Three independent measurements were averaged. Statistically significant differences were determined by Student’s t test (*P < 0.05). Error bars indicate se. CaMV, Cauliflower mosaic virus; Nos-T, nopaline synthase terminator.
To perform chromatin immunoprecipitation (ChIP) assays, we generated pMYB96:MYB96-MYC transgenic plants. Total protein extracts from control pBA002 and pMYB96:MYB96-MYC transgenic seeds were immunoprecipitated with anti-MYC-antibody. DNA bound to epitope-tagged MYB96 proteins was analyzed by quantitative PCR assays. The ChIP analysis showed that the A region of the ABI4 promoter was enriched by MYB96 (Fig. 5B). In contrast, two other genomic fragments containing MYB-binding cis-elements, B and C, were not enriched (Fig. 5B), supporting the specific interaction of MYB96 with the A region of the ABI4 promoter. In addition, control ChIP with resin alone did not enrich the A fragment (Fig. 5B). To confirm the direct binding of MYB96 to the ABI4 promoter, we also generated transgenic plants expressing the R2R3-MYB DNA-binding domain (BD) of MYB96 fused with the MYC-coding sequences (35S:96BD-MYC). ChIP analysis revealed that the MYB DNA-binding domain was sufficient to bind to the ABI4 promoter (Supplemental Fig. S10).
To confirm the MYB96 binding to core sequences of the ABI4 promoter, we performed transient expression analysis using Arabidopsis protoplasts. Two core elements on the ABI4 promoter, A-1 (−264; CAAGTAACTTATGA) and A-2 (−335; GCTATAGTTACCCA), were fused to the 35S minimal promoter. A recombinant reporter plasmid and an effector plasmid, p35S:MYB96, were cotransformed into Arabidopsis protoplasts. Cotransformation with the reporter A-1 increased the GUS activity by 2- to 3-fold, but cotransformation with A-2 did not affect reporter gene expression (Fig. 5C). These results indicate that MYB96 specifically targets proximal sequences upstream of the ABI4 gene to activate its expression.
MYB96 Contributes to Inhibiting TAG Degradation during Germination
Seed germination is elaborately regulated by discrete actions of the embryo and endosperm (Lafon-Placette and Köhler, 2014). An intriguing example is ABA-controlled lipid breakdown and mobilization in the embryo under suboptimal growth conditions (Penfield et al., 2006). Seed TAG degradation is triggered to fuel embryonic growth during germination but is antagonized by ABA specifically in the embryo (Penfield et al., 2006). It is noteworthy that the ABI4 transcription factor is a well-known regulator of this process, which confers differential sensitivity of lipid catabolism to ABA in the embryo and endosperm (Penfield et al., 2006).
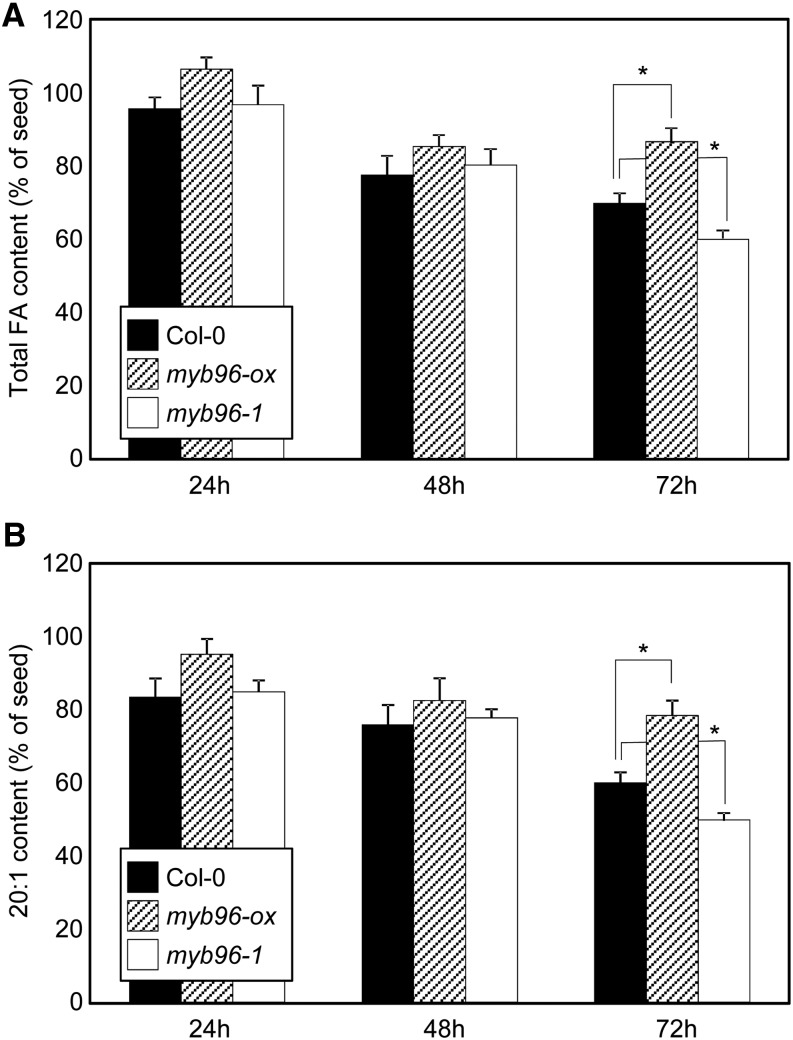
Since ABI4 is a major regulatory target of the MYB96 transcription factor, we hypothesized that MYB96 may also be responsible for the ABA regulation of lipid breakdown during seed germination, similar to the action of ABI4 (Penfield et al., 2006; Wind et al., 2013). To examine this hypothesis, we measured the levels of total fatty acid and eicosenoic acid (20:1) in wild-type, myb96-ox, and myb96-1 seeds for 3 d after cold imbibition in the absence or presence of exogenous ABA. In the absence of ABA, the levels of total fatty acid and eicosenoic acid were largely indistinguishable in myb96-ox and myb96-1 compared with wild-type seeds (Supplemental Fig. S11). However, in the presence of ABA, the levels of total fatty acid and eicosenoic acid were noticeably altered in myb96-ox and myb96-1. Total fatty acid levels were reduced by approximately 30% to 35% 3 d after cold imbibition in wild-type seeds. However, TAG breakdown was largely repressed in the myb96-ox seeds, whereas 43% of total TAG was catabolized in the myb96-1 seeds (Fig. 6A). The levels of eicosenoic acid displayed similar degradation kinetics to total fatty acid (Fig. 6B). These observations demonstrate that MYB96 plays a role in regulating lipid breakdown during seed germination.
Figure 6.
Lipid breakdown in wild-type Columbia-0 (Col-0), myb96-ox, and myb96-1 seeds during seed germination in the presence of ABA. Seeds were germinated on MS plates supplemented with 1 μm ABA and incubated at 23°C under long-day conditions for 3 d. Lipid breakdown was determined by measuring the abundance of total fatty acid (FA; A) and eicosenoic acid (20:1; B). Fatty acid content was expressed as the percentage of the amount present in dry seeds of the indicated genotypes. At least 30 seeds per genotype were measured in each replicate. Biological triplicates were averaged. Error bars indicate se.
ABI4 Is Epistatic to MYB96
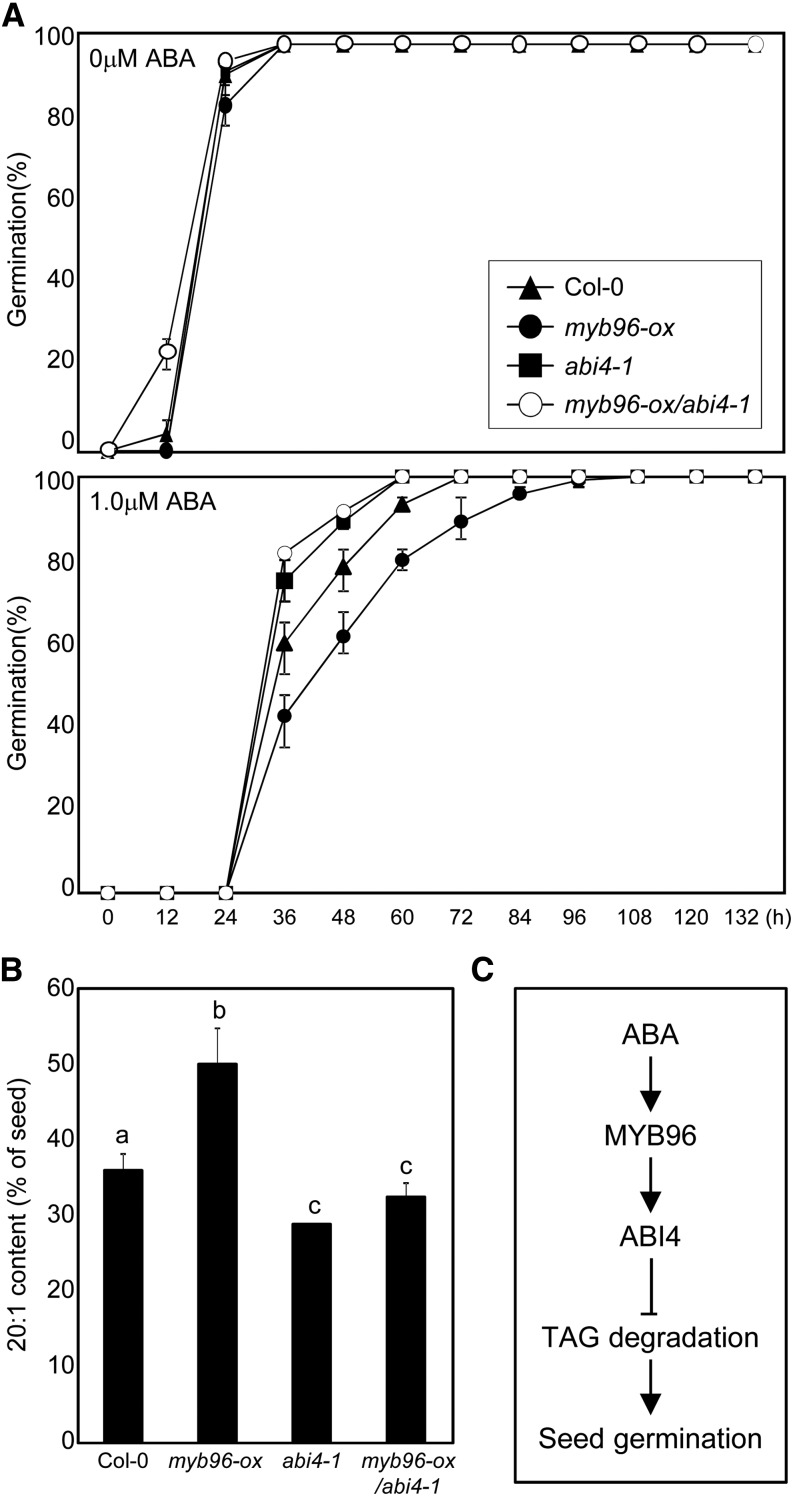
Our results indicated that the MYB96-ABI4 transcriptional cascade is crucial for the ABA regulation of seed germination, probably through lipid mobilization. To confirm the genetic relationship between MYB96 and ABI4, we genetically crossed myb96-ox to the abi4-1 mutant.
We investigated the ABA sensitivity of myb96-ox/abi4-1 seeds during germination. Delayed germination was observed in myb96-ox seeds, whereas the abi4-1 mutation led to accelerated seed germination, in the presence of ABA (Fig. 7A). The germination rate of myb96-ox/abi4-1 seeds was comparable to that of abi4-1 (Fig. 7A), indicating that MYB96 is an upstream regulator of ABI4 and that both genes act in the same pathway.
Figure 7.
Genetic hierarchy of ABI4 and MYB96. A, Germination rate of myb96-ox/abi4-1. The seed germination percentage of each genotype on 1 μm ABA was scored at the indicated time points after cold stratification. At least 50 seeds per genotype were measured in each replicate. Biological triplicates were averaged. Error bars indicate se. B, Lipid breakdown of myb96-ox/abi4-1. Seeds were germinated on MS plates supplemented with 1 μm ABA and incubated at 23°C under long-day conditions for 3 d. Eicosenoic acid (20:1) content was expressed as a percentage of the amount in dry seeds of the indicated genotypes. At least 30 seeds per genotype were measured in each replicate. Biological triplicates were averaged. Error bars indicate se. C, Proposed role of MYB96 during seed germination. The ABA-inducible R2R3-type MYB96 transcription factor regulates ABI4 expression by binding directly to its gene promoter. The MYB96-ABI4 module inhibits lipid mobilization specifically in the embryo, thereby contributing to delaying seed germination under suboptimal conditions. Col-0, Wild-type Columbia-0.
In agreement with the seed germination rate, the eicosenoic acid catabolism of myb96-ox/abi4-1 was also indistinguishable from that of abi4-1 (Fig. 7B). A significant amount of the stored TAG was degraded in both genotypes, even in the presence of ABA. Therefore, we conclude that ABI4 is genetically epistatic to MYB96 with respect to the control of ABA-dependent seed germination as well as TAG catabolism.
Taken together, our observations demonstrate that the MYB96 transcription factor serves as an ABA signaling mediator in modulating seed germination through ABI4-dependent embryonic lipid breakdown (Fig. 7C). MYB96 directly regulates ABI4, a key regulator of embryonic lipid mobilization during seed germination, by binding to its gene promoter. Genetic analyses further bolster this genetic hierarchy in the control of seed germination.
DISCUSSION
MYB96 Regulation of ABI4
ABA is perceived by receptor protein complexes that are composed of PYRABACTIN RESISTANCE (PYR)/PYR1-LIKE (PYL)/REGULATORY COMPONENT OF ABSCISIC ACID RECEPTOR (RCAR), type 2C protein phosphatases (PP2Cs), and SnRK2s (Raghavendra et al., 2010; Weiner et al., 2010). In the presence of ABA, PYR/PYL/RCAR proteins lead to the suppression of PP2C activities, derepressing SnRK2 protein kinases to activate ABA-responsive binding factors/ABA-responsive element-binding proteins (Fujii et al., 2009; Nishimura et al., 2010; Gonzalez-Guzman et al., 2012). Subsequent ABA responses are mediated by a large number of transcription factors, indicating that transcriptional cascades are a fundamental framework in ABA signal transduction (Giraudat et al., 1992; Busk and Pagès, 1998; Finkelstein et al., 1998; Finkelstein and Lynch, 2000; Abe et al., 2003; Pandey et al., 2005; Seo et al., 2009).
In particular, the ABI3, ABI4, and ABI5 transcription factors all play a common role in the ABA regulation of seed germination by constituting combinatorial networks (Söderman et al., 2000; Nakamura et al., 2001). However, despite their functional similarities, the regulatory mechanisms of ABI3, ABI4, and ABI5 differ markedly. The ABI3 protein is proteolytically degraded by a RING E3 ligase, ABI3-INTERACTING PROTEIN2, a negative regulator of ABA signaling (Zhang et al., 2005). Controlled protein degradation also underlies ABI5 activity regulation. The KEEP ON GOING (KEG) E3 ligase executes ABI5 degradation under normal growth conditions (Stone et al., 2006; Liu and Stone, 2010). In the presence of ABA, KEG is rapidly degraded, promoting the accumulation of high levels of ABI5. Additional E3 ligases, such as DWD HYPERSENSITIVE TO ABSCISIC ACID1 (DWA1), DWA2, and SIZ1 (for SAP [scaffold attachment factor, acinus, protein inhibitor of activated signal transducer and activator of transcription] and Miz1 [Msx2-interacting zinc finger]), further fine-tune the protein accumulation of ABI5 (Miura et al., 2009; Lee et al., 2010).
Compared with ABI3 and ABI5, relatively less is known regarding how ABI4 activity is regulated at the molecular level. This study demonstrates that the MYB96 transcription factor regulates ABI4 expression by binding directly to the gene promoter and defines its spatial and temporal expression. Both MYB96 and ABI4 genes are strongly expressed in the embryo and are responsive to exogenous ABA (Penfield et al., 2006). Furthermore, the promotive effect of ABA on ABI4 expression is largely impaired in the myb96-1 mutant, indicating that MYB96 is an unequivocal transcriptional regulator required for the proper expression of ABI4 in an ABA signaling pathway.
The question, then, is how MYB96 activity is regulated by ABA. Given the myriad targets of MYB96, it is likely that MYB96 acts upstream of ABA signaling pathways. It is probable that controlled protein turnover and/or posttranslational modifications mediated by ABA receptor-like complex might underlie the ABA regulation of MYB96 activity, and the ABA induction of MYB96 transcription may be a result of feedback regulation. Further work is required to get a comprehensive view of the signaling network built up from MYB96.
TAG Degradation during Seed Germination
The utilization of seed storage reserves, such as carbohydrates, proteins, and lipids, is crucial to drive germination and postgerminative seedling growth (Penfield et al., 2004, 2006; Quettier and Eastmond, 2009), albeit controversial (Pritchard et al., 2002; Kelly et al., 2011). Arabidopsis seeds mainly store TAG as the major carbon reserve, which accumulate at the later stages of seed maturation (Lu and Hills, 2002; Maeo et al., 2009). Following seed imbibition, fatty acids are degraded through β-oxidation (Footitt et al., 2002) and primarily consumed as a form of soluble carbohydrates that are converted by the glyoxylate cycle and gluconeogenesis (Penfield et al., 2005; Pracharoenwattana et al., 2010). In agreement with this, mutants with defective TAG hydrolysis exhibit delayed germination and perturbed postgerminative seedling growth (Penfield et al., 2005, 2006; Quettier and Eastmond, 2009; Kelly et al., 2011).
Exogenous ABA provokes delayed germination in part by reducing lipid breakdown (Penfield et al., 2006). Genes involved in the conversion of TAG into soluble sugars are down-regulated in response to ABA (Pritchard et al., 2002); thus, the levels of fatty acid-derived sugars in ABA-treated seeds are markedly decreased (Pritchard et al., 2002). Notably, the ABA regulation of lipid breakdown is relevant particularly in embryos (Penfield et al., 2006), indicating the differential sensitivity of embryo and endosperm to ABA. The ABI4 transcription factor is a major regulator in this process. Embryo-expressed ABI4 negatively regulates lipid mobilization and thus seed germination in the presence of ABA (Penfield et al., 2006).
In this study, we discovered the MYB96-ABI4 signaling module that regulates the ABA-dependent seed germination process, possibly through the control of lipid mobilization. MYB96 not only confers ABA sensitivity to embryos but also participates in embryonic lipid mobilization to properly regulate seed germination, similar to the role of ABI4 in seed germination (Penfield et al., 2006). The myb96-1 seeds degrade a significant content of TAG even in the presence of ABA, whereas TAG breakdown is largely impaired in myb96-ox seeds. Given that the MYB96-mediated physiological responses in seeds largely depend on ABI4, MYB96 appears to be an explicit transcriptional regulator of ABI4 to define its expression pattern in seeds and to ensure proper seed germination.
Importance of MYB96-ABI4 Transcriptional Cascades in Seed Germination
The MYB96 transcription factor modulates a variety of plant developmental processes in response to ABA, including shoot development, root architecture remodeling, stomatal closure, cuticular wax accumulation, and hormone biosynthesis (Seo et al., 2009, 2011; Seo and Park, 2010). Based on this study, MYB96 also appears to play a role in ABA-dependent seed germination.
Multiple functions of MYB96 are achieved by regulating myriad transcriptional target genes that underlie a variety of signaling pathways. For example, ABA-induced cuticular wax biosynthesis is mediated by MYB96 regulation of 3-KETOACYL-COENZYME A SYNTHASE genes encoding rate-limiting enzymes of the very-long-chain fatty acid elongation process (Seo et al., 2011). MYB96-GRETCHEN HAGEN3 transcriptional cascades regulate lateral root development under water-deficient conditions by reestablishing auxin homeostasis (Seo et al., 2009). The RESPONSIVE TO DESSICATION22 gene is probably one of the regulatory targets of MYB96 that functions in the control of drought tolerance (Seo et al., 2009). LIPID TRANSFER PROTEIN3 is responsible in part for the MYB96 regulation of freezing and drought tolerance (Guo et al., 2013). In addition, the MYB96-ABI4 transcriptional module is crucial for ABA-dependent seed germination. Genetic analysis reveals that delayed germination of myb96-ox is fully suppressed by the abi4 mutation, indicating that ABI4 is the principal target of MYB96 in the control of seed germination. Collectively, MYB96 is an important ABA signaling mediator that globally links multiple signaling pathways and thus helps plants to trigger efficient physiological and cellular reprogramming for environmental adaptation.
Meanwhile, ABI4 integrates a variety of external and internal signals, such as light, ABA, reactive oxygen species, and soluble sugars (Pesaresi et al., 2007; Shkolnik-Inbar and Bar-Zvi, 2010; Kerchev et al., 2011; Yang et al., 2011b; Foyer et al., 2012), and regulates hormone biosynthesis and sugar and lipid metabolism (Kerchev et al., 2011; Shu et al., 2013; Wind et al., 2013). Thus, ABI4 coordinates developmental programs with environmental signals to optimize plant growth and development processes. MYB96 is particularly relevant in ABA signal transduction to ABI4, and other signaling regulators are likely responsible for the transcriptional regulation of ABI4 with respect to other input signals.
Taken together, the MYB96-ABI4 signaling module is important for the control of ABA-dependent seed germination. MYB96 precisely conveys ABA signaling to ABI4 in embryos during seed germination, and ABI4 in turn integrates multitude environmental and developmental information to determine the proper timing for germination. Multiple transcriptional cascades not only allow for signal amplification but also enable plants to make integrative decisions.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Columbia-0 ecotype was used for all experiments described, unless specified otherwise. Plants were grown under long-day conditions (16-h-light/8-h-dark cycle) with cool-white fluorescent light (100 μmol photons m−2 s−1) at 23°C. The myb96-ox and myb96-1 mutants (GABI_120B05) were reported previously (Seo et al., 2009). The abi4-1 mutant (CS8104) was obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/). The myb96-ox/abi4-1 double mutant was generated by genetic cross of myb96-ox and abi4-1 mutants. Identification of abi4-1 was performed as described previously (Finkelstein et al., 1998).
qRT-PCR Analysis
Total RNA was extracted using TRI agent (Takara Bio) according to the manufacturer’s recommendations. Reverse transcription was performed using Moloney murine leukemia virus reverse transcriptase (Dr. Protein) with oligo(dT)18 to synthesize first-strand complementary DNA (cDNA) from 2 μg of total RNA. Total RNA samples were pretreated with an RNase-free DNase. cDNAs were diluted to 100 μL with Tris-EDTA buffer, and 1 μL of diluted cDNA was used for PCR amplification.
qRT-PCR was performed in 96-well blocks using the Step-One Plus Real-Time PCR System (Applied Biosystems). The PCR primers used are listed in Supplemental Table S1. The value for each set of primers was normalized relative to eIF4a (At3g13920). All qRT-PCRs were performed in triplicate using total RNA samples extracted from three independent biological replicates. The comparative ΔΔCT method, which is a calculation for the relative quantification of target DNA, was employed to evaluate relative quantities of each amplified product in the samples. The threshold cycle was automatically determined for each reaction by the instrument software using default parameters. The specificity of the qRT-PCR was determined by melt-curve analysis of the amplified products using the standard method installed in the system.
Measurement of GUS Activity
Seeds were dissected into embryo and endosperm surrounded with seed coat. For histochemical staining of GUS activity, plant materials were fixed by immersing in 90% (v/v) acetone for 20 min on ice, washed twice with rinsing solution [50 mm sodium phosphate, pH 7, 0.5 mm K3Fe(CN)6, and 0.5 mm K4Fe(CN)6], and subsequently incubated in staining solution containing 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Duchefa) at 37°C for 18 to 24 h, depending on staining intensity (Padmanaban et al., 2007).
Seed Germination Assays
All genotypes were grown at 23°C under long-day conditions, and seeds were collected at the same time. Harvested seeds were dried at room temperature at least 1 month before germination assays. For seed germination assays, 40 to 50 seeds for each line were sterilized and plated on MS medium (one-half-strength MS salts, 0.05% [w/v] MES, pH 5.7, and 0.7% [w/v] agar) supplemented with various concentrations of ABA (0, 0.5, 1, and 3 μm). Plates were stratified in darkness for 3 d at 4°C and transferred to a culture room set at 23°C with a 16-h-light/8-h-dark cycle. Germination was scored at the indicated time points by counting the frequency of radicle emergence from the seed coat and endosperm. For each germination assay, biological triplicates were performed.
To investigate the effects of ABA on cotyledon greening, the percentage of cotyledon greening was scored after the end of stratification. Cotyledon greening was defined as complete expansion of the cotyledon and greening.
ChIP Assays
ChIP assays were performed as described previously (Yang et al., 2011a). pMYB96:MYC-MYB96 transgenic plants, anti-MYC antibodies (Millipore), and salmon sperm DNA/protein A agarose beads (Millipore) were used for ChIP. DNA was purified using phenol/chloroform/isoamyl alcohol and sodium acetate (pH 5.2). The level of precipitated DNA fragments was quantified by quantitative real-time PCR using specific primer sets (Supplemental Table S2). Values were normalized with the level of input DNA. The value in pBA002 control plants was set to 1 after normalization against eIF4a for quantitative PCR analysis.
Transient Gene Expression Assays
For transient expression assays using Arabidopsis protoplasts, reporter and effector plasmids were constructed. The reporter plasmid contains a minimal 35S promoter sequence and the GUS gene. The core elements on the ABI4 promoter were inserted into the reporter plasmid. To construct the p35S:MYB96 effector plasmid, the MYB96 cDNA was inserted into the effector vector containing the cauliflower mosaic virus 35S promoter. Recombinant reporter and effector plasmids were cotransformed into Arabidopsis protoplasts by polyethylene glycol-mediated transformation (Yoo et al., 2007). GUS activities were measured by a fluorometric method. A cauliflower mosaic virus 35S promoter-luciferase construct was also cotransformed as an internal control. The luciferase assay was performed using the Luciferase Assay System kit (Promega).
Fatty Acid Determinations
The total fatty acid content and amount of eicosenoic acid (20:1 n-9) in seeds were measured by gas chromatographic analysis with a known amount of 15:0 fatty acid as an internal standard. Samples were transmethylated at 90°C for 90 min in 0.3 mL of toluene and 1 mL of 5% (v/v) H2SO4 (in methanol). After transmethylation, 1.5 mL of 0.9% (w/v) NaCl solution was added, and the fatty acid methyl esters were transferred to a new tube for three sequential extractions with 1.5 mL of n-hexane. Fatty acid methyl esters were analyzed by gas chromatography using a GC-2010 Plus instrument (Shimadzu) with a 30-m × 0.25-mm (i.d.) HP-FFAP column (Agilent), during which the oven temperature was increased from 170°C to 180°C at 1°C min−1.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ABI1 (At4g26080), ABI2 (At5g57050), ABI3 (At3g24650), ABI4 (At2g40220), ABI5 (At2g36270), MYB96 (At5g62470), ACBP1 (At5g53470), AHK1 (At2g17820), BLH1 (At2g35940), FY (At5g13480), RHA2a (At1g15100), SnRK2.2 (At3g50500), SnRK2.3 (At5g66880), and eIF4a (At3g13920).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Temporal expression of MYB96.
Supplemental Figure S2. Expression of MYB96 in embryo after cold imbibition.
Supplemental Figure S3. ABA induction of MYB96 in embryo.
Supplemental Figure S4. Expression of MYB96 in quadruple-DELLA and GA-deficient mutants.
Supplemental Figure S5. Cotyledon opening of myb96-ox and myb96-1 in the presence of ABA.
Supplemental Figure S6. ABA dose-response assays.
Supplemental Figure S7. Expression of ABI4 and BLH1 in the myb96-ox mutant in the presence of ABA.
Supplemental Figure S8. ABI4 expression in dry seeds and during seed germination.
Supplemental Figure S9. Spatial expression of MYB96 and ABI4.
Supplemental Figure S10. ChIP using 35S:96BD-MYC transgenic plants.
Supplemental Figure S11. Lipid breakdown in wild-type, myb96-ox, and myb96-1 seeds during seed germination in the absence of ABA.
Supplemental Table S1. Primers used in qRT-PCR.
Supplemental Table S2. Primers used in ChIP assays.
Supplementary Material
Glossary
- ABA
abscisic acid
- TAG
triacylglycerol
- qRT
quantitative reverse transcription
- MS
Murashige and Skoog
- ChIP
chromatin immunoprecipitation
- cDNA
complementary DNA
Footnotes
This work was supported by the Basic Science Research (grant no. NRF–2013R1A1A1004831) and Global Research Network (grant no. NRF–2014S1A2A2028392) programs of the National Research Foundation of Korea and by the Cooperative Research Program for Agriculture Science & Technology Development (SSAC, project no. PJ01108101) of the Rural Development Administration (to H.U.K.) as well as by the BK21 PLUS program in the Department of Bioactive Material Sciences (to K.L. and H.G.L.).
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud N, Girin T, Sorefan K, Fuentes S, Wood TA, Lawrenson T, Sablowski R, Østergaard L (2010) Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev 24: 2127–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Hamamura Y, Ingouff M, Higashiyama T (2008) Double fertilization: caught in the act. Trends Plant Sci 13: 437–443 [DOI] [PubMed] [Google Scholar]
- Bewley JD. (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi F, Cordoba E, Dupré P, Mendoza MS, Román CS, León P (2009) The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J 59: 359–374 [DOI] [PubMed] [Google Scholar]
- Busk PK, Pagès M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37: 425–435 [DOI] [PubMed] [Google Scholar]
- Cao D, Cheng H, Wu W, Soo HM, Peng J (2006) Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol 142: 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30: 373–383 [DOI] [PubMed] [Google Scholar]
- Crouch ML. (1987) Regulation of gene expression during seed development in flowering plants. In Browder LW, ed, Developmental Biology: A Comprehensive Synthesis. Plenum Press, New York, pp 367–404 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Koornneef M (2000) Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol 122: 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A, Jung HS, Sun TP (2001) The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZY, Chen MX, Chen QF, Xiao S, Chye ML (2013) Arabidopsis acyl-CoA-binding protein ACBP1 participates in the regulation of seed germination and seedling development. Plant J 74: 294–309 [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Somerville CR (1990) Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol 94: 1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham I, Baker A, Holdsworth M (2002) Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J 21: 2912–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Kerchev PI, Hancock RD (2012) The ABA-INSENSITIVE-4 (ABI4) transcription factor links redox, hormone and sugar signaling pathways. Plant Signal Behav 7: 276–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Alabadí D, Blázquez MA (2011) DELLA-induced early transcriptional changes during etiolated development in Arabidopsis thaliana. PLoS ONE 6: e23918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Marín JA, Prat S, Blázquez MA, Alabadí D (2010) Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol Biol Evol 27: 1247–1256 [DOI] [PubMed] [Google Scholar]
- Garciarrubio A, Legaria JP, Covarrubias AA (1997) Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta 203: 182–187 [DOI] [PubMed] [Google Scholar]
- Gehring M, Choi Y, Fischer RL (2004) Imprinting and seed development. Plant Cell (Suppl) 16: S203–S213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, Barker SJ, Perez-Grau L (1989) Regulation of gene expression during plant embryogenesis. Cell 56: 149–160 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernández MA, Holdsworth MJ, Perez-Amador MA, Kollist H, et al. (2012) Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24: 2483–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grappin P, Bouinot D, Sotta B, Miginiac E, Jullien M (2000) Control of seed dormancy in Nicotiana plumbaginifolia: post-imbibition abscisic acid synthesis imposes dormancy maintenance. Planta 210: 279–285 [DOI] [PubMed] [Google Scholar]
- Guo L, Yang H, Zhang X, Yang S (2013) Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J Exp Bot 64: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada JJ, Baden CS, Comai L (1988) Spatially regulated genes expressed during seed germination and postgerminative development are activated during embryogeny. Mol Gen Genet 212: 466–473 [Google Scholar]
- Holdsworth M, Kurup S, McKibbin R (1999) Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends Plant Sci 4: 275–280 [Google Scholar]
- Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY (2012) Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZY, Zhang XS, Zheng GH, Gutterman Y (2003) Influence of light, temperature, salinity and storage on seed germination of Haloxylon ammodendron. J Arid Environ 55: 453–464 [Google Scholar]
- Hussain A, Peng J (2003) DELLA proteins and GA signalling in Arabidopsis. J Plant Growth Regul 22: 134–140 [Google Scholar]
- Jiang S, Kumar S, Eu YJ, Jami SK, Stasolla C, Hill RD (2012) The Arabidopsis mutant, fy-1, has an ABA-insensitive germination phenotype. J Exp Bot 63: 2693–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosen RV, Arends D, Li Y, Willems LA, Keurentjes JJ, Ligterink W, Jansen RC, Hilhorst HW (2013) Identifying genotype-by-environment interactions in the metabolism of germinating Arabidopsis seeds using generalized genetical genomics. Plant Physiol 162: 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, Quettier AL, Shaw E, Eastmond PJ (2011) Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol 157: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchev PI, Pellny TK, Vivancos PD, Kiddle G, Hedden P, Driscoll S, Vanacker H, Verrier P, Hancock RD, Foyer CH (2011) The transcription factor ABI4 Is required for the ascorbic acid-dependent regulation of growth and regulation of jasmonate-dependent defense signaling pathways in Arabidopsis. Plant Cell 23: 3319–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermode AR. (1990) Regulatory mechanisms involved in the transition from seed development to germination. Crit Rev Plant Sci 2: 155–195 [Google Scholar]
- Kim D, Cho YH, Ryu H, Kim Y, Kim TH, Hwang I (2013) BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J 75: 755–766 [DOI] [PubMed] [Google Scholar]
- Lafon-Placette C, Köhler C (2014) Embryo and endosperm, partners in seed development. Curr Opin Plant Biol 17: 64–69 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoon HJ, Terzaghi W, Martinez C, Dai M, Li J, Byun MO, Deng XW (2010) DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22: 1716–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Jiang H, Bu Q, Zhao Q, Sun J, Xie Q, Li C (2011) The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol 156: 550–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Park J, Lee N, Jeong J, Toh S, Watanabe A, Kim J, Kang H, Kim DH, Kawakami N, et al. (2013) ABA-insensitive3, ABA-insensitive5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25: 4863–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Stone SL (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Hills MJ (2002) Arabidopsis mutants deficient in diacylglycerol acyltransferase display increased sensitivity to abscisic acid, sugars, and osmotic stress during germination and seedling development. Plant Physiol 129: 1352–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476–487 [DOI] [PubMed] [Google Scholar]
- McCarty DR, Carson CB (1991) The molecular genetics of seed maturation in maize. Physiol Plant 81: 267–272 [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes D, Himmelbach A, Korte A, Haberer G, Grill E (2008) Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J 54: 806–819 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K (2013) ABA signaling in stress-response and seed development. Plant Cell Rep 32: 959–970 [DOI] [PubMed] [Google Scholar]
- Natesh S, Rau MA (1984) The embryo. In Johri BM, ed Embryology of Angiosperms. Springer-Verlag, Berlin, pp 377–444 [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H. (2014) Seed dormancy and germination: emerging mechanisms and new hypotheses. Front Plant Sci 5: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanaban S, Chanroj S, Kwak JM, Li X, Ward JM, Sze H (2007) Participation of endomembrane cation/H+ exchanger AtCHX20 in osmoregulation of guard cells. Plant Physiol 144: 82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Nguyen KT, Park E, Jeon JS, Choi G (2013) DELLA proteins and their interacting RING finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell 25: 927–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Graham S, Graham IA (2005) Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem Soc Trans 33: 380–383 [DOI] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Rylott EL, Gilday AD, Graham S, Larson TR, Graham IA (2004) Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16: 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Harberd NP (2002) The role of GA-mediated signalling in the control of seed germination. Curr Opin Plant Biol 5: 376–381 [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Schneider A, Kleine T, Leister D (2007) Interorganellar communication. Curr Opin Plant Biol 10: 600–606 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I, Zhou W, Smith SM (2010) Fatty acid beta-oxidation in germinating Arabidopsis seeds is supported by peroxisomal hydroxypyruvate reductase when malate dehydrogenase is absent. Plant Mol Biol 72: 101–109 [DOI] [PubMed] [Google Scholar]
- Pritchard SL, Charlton WL, Baker A, Graham IA (2002) Germination and storage reserve mobilization are regulated independently in Arabidopsis. Plant J 31: 639–647 [DOI] [PubMed] [Google Scholar]
- Prouse MB, Campbell MM (2013) Interactions between the R2R3-MYB transcription factor, AtMYB61, and target DNA binding sites. PLoS ONE 8: e65132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XX, Huang ZY, Baskin JM, Baskin CC (2008) Effect of temperature, light and salinity on seed germination and radicle growth of the geographically widespread halophyte shrub Halocnemum strobilaceum. Ann Bot (Lond) 101: 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quettier AL, Eastmond PJ (2009) Storage oil hydrolysis during early seedling growth. Plant Physiol Biochem 47: 485–490 [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Raz V, Bergervoet JH, Koornneef M (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128: 243–252 [DOI] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion-Poll A, Sun TP, Koshiba T, et al. (2006) Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 48: 354–366 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23: 1138–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Park CM (2010) MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol 186: 471–483 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park CM (2011) Cuticular wax biosynthesis as a way of inducing drought resistance. Plant Signal Behav 6: 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 151: 275–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Bar-Zvi D (2010) ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 22: 3560–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q (2013) ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet 9: e1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderman EM, Brocard IM, Lynch TJ, Finkelstein RR (2000) Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol 124: 1752–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm P, Ravindran P, Mohanty B, Tan EL, Yu H, Kumar PP (2012) Insights into the molecular mechanism of RGL2-mediated inhibition of seed germination in Arabidopsis thaliana. BMC Plant Biol 12: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas TL. (1993) Gene expression during plant embryogenesis and germination: an overview. Plant Cell 5: 1401–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 104: 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP (2004) Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers LM, Bouwmeester HJ, Karssen CM (1995) Redefining seed dormancy: an attempt to integrate physiology and ecology. J Ecol 83: 1031–1037 [Google Scholar]
- Weiner JJ, Peterson FC, Volkman BF, Cutler SR (2010) Structural and functional insights into core ABA signaling. Curr Opin Plant Biol 13: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Harada JJ (1993) Embryogenesis in higher plants: an overview. Plant Cell 5: 1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wind JJ, Peviani A, Snel B, Hanson J, Smeekens SC (2013) ABI4: versatile activator and repressor. Trends Plant Sci 18: 125–132 [DOI] [PubMed] [Google Scholar]
- Yaish MW, El-Kereamy A, Zhu T, Beatty PH, Good AG, Bi YM, Rothstein SJ (2010) The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet 6: e1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SD, Seo PJ, Yoon HK, Park CM (2011a) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yu X, Song L, An C (2011b) ABI4 activates DGAT1 expression in Arabidopsis seedlings during nitrogen deficiency. Plant Physiol 156: 873–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R, Kanno Y, Jikumaru Y, Nakabayashi K, Kamiya Y, Nambara E (2009) CHOTTO1, a putative double APETALA2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis. Plant Physiol 151: 641–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al. (2007) Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Ogawa M, Fleet CM, Zentella R, Hu J, Heo JO, Lim J, Kamiya Y, Yamaguchi S, Sun TP (2011) Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci USA 108: 2160–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.