13C incorporation provides insight into the temporal dynamics of wood formation in hybrid aspen.
Abstract
Wood of trees is formed from carbon assimilated in the photosynthetic tissues. Determining the temporal dynamics of carbon assimilation, subsequent transport into developing wood, and incorporation to cell walls would further our understanding of wood formation in particular and tree growth in general. To investigate these questions, we designed a 13CO2 labeling system to study carbon transport and incorporation to developing wood of hybrid aspen (Populus tremula × tremuloides). Tracking of 13C incorporation to wood over a time course using nuclear magnetic resonance spectroscopy revealed diurnal patterns in wood cell wall biosynthesis. The dark period had a differential effect on 13C incorporation to lignin and cell wall carbohydrates. No 13C was incorporated into aromatic amino acids of cell wall proteins in the dark, suggesting that cell wall protein biosynthesis ceased during the night. The results show previously unrecognized temporal patterns in wood cell wall biosynthesis, suggest diurnal cycle as a possible cue in the regulation of carbon incorporation to wood, and establish a unique 13C labeling method for the analysis of wood formation and secondary growth in trees.
Wood formation in trees depends on photosynthetic CO2 assimilation and subsequent transport of carbon into wood. In several tree species, including aspen (Populus spp.), the developing wood receives most of the carbon as Suc. During wood formation, imported Suc is metabolized to provide precursors for wood cell wall biosynthesis and energy for secondary growth. Structures of the metabolic pathways channeling carbon to cell wall biosynthesis are thought to be relatively well understood (Sharples and Fry, 2007; Alonso et al., 2010). However, the way in which carbon from imported Suc is divided between these metabolic pathways in developing wood is less clear. Temporal dynamics of Suc import, biosynthesis of cell wall polymer precursors, and subsequent carbon incorporation to wood cell walls are also not well understood. In trees, there is evidence suggesting that the rate of carbon incorporation to different cell wall polymers may vary over a diurnal cycle.
The evidence for a diurnal pattern in wood cell wall biosynthesis is mostly anatomical. The structure of the innermost cell wall layer in the secondary cell wall-forming wood fibers of Populus nigra was seen to change in electron microscopy images taken over a 24-h period (Bobák and Nečesaný, 1967). Cellulose microfibrils were visible in samples collected during the day, whereas they were embedded in an amorphous layer during the night (Bobák and Nečesaný, 1967). Bobák and Nečesaný (1967) suggested that this amorphous layer consisted of lignin, because it could be removed using a solution consisting of 0.8 m sodium chlorite in 0.2 m acetic acid. However, it seems unlikely that this solution would specifically remove lignin but may have also removed other cell wall polymers. A similar study of the developing wood tracheids in the conifer Cryptomeria japonica also documented the deposition of an amorphous layer during the night (Yoshida et al., 2000; Hosoo et al., 2002). Immunolabeling of the amorphous layer with xylan- and glucomannan-specific antibodies led Hosoo et al. (2002) to conclude that much of this layer consisted of hemicellulose. In addition to the anatomical evidence, the existence of a diurnal pattern in wood biosynthesis recently received additional support when Solomon et al. (2010) discovered clear diurnal transcript abundance changes in the developing wood of Eucalyptus spp. trees. Transcripts showing diurnal changes in abundance included several genes annotated to be involved in carbohydrate metabolism and cell wall biosynthesis (Solomon et al., 2010). Based on these previous studies, it seems plausible that the diurnal cycle influences wood formation and carbon incorporation to wood. However, it has been difficult to test this hypothesis, because it is technically challenging to measure the diurnal rates of carbon incorporation to different cell wall polymers in developing wood. Analysis of the cell wall polymer content in mature wood does not reveal possible temporal differences in their biosynthesis over a diurnal cycle. In developing wood, the weight proportion of the cell wall synthesized during a single diurnal cycle is small, and consequently, any diurnal differences in the cell wall polymer proportions are not apparent using standard analytical techniques. Therefore, new approaches are needed for the investigation of the temporal dynamics of cell wall biosynthesis in trees.
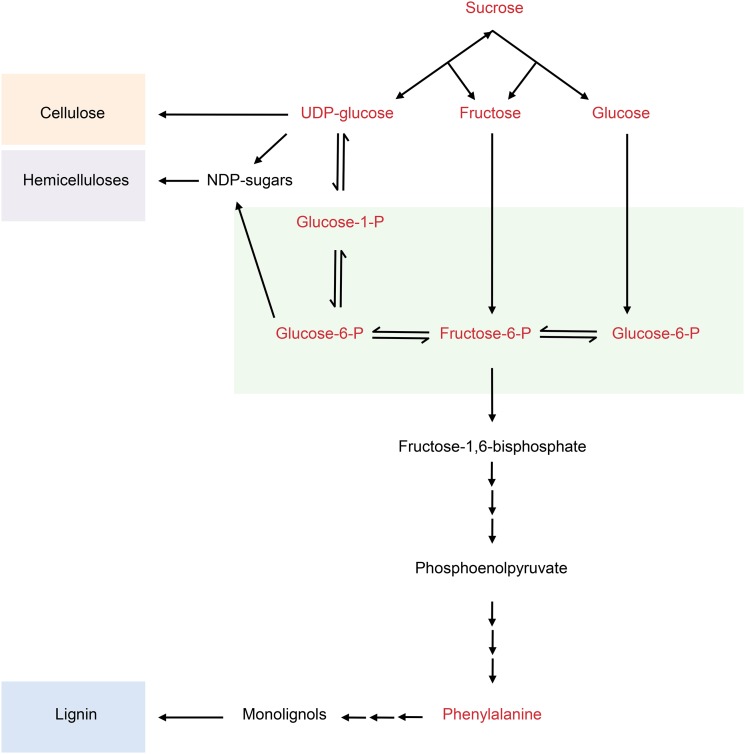
We explored the possibility of monitoring cell wall polymer biosynthesis in the developing wood of 2-month-old greenhouse-grown hybrid aspen (Populus tremula × tremuloides) trees using 13CO2 supply followed by analysis of 13C incorporation into cell walls. The focus was on the main cell wall polymers containing most of the carbon in wood—cellulose, hemicelluloses, and lignin. Cellulose and hemicelluloses are synthesized from the nucleotide sugars derived from the hexose-P pool (Reiter and Vanzin, 2001; Fig. 1). The exception is UDP-Glc, which can also be synthesized directly from Suc by Suc synthase (Kleczkowski et al., 2010; Fig. 1). Carbon for lignin biosynthesis is derived from the amino acid Phe through the monolignol biosynthesis pathway, which produces three hydroxycinnamyl alcohol monomers: p-coumaryl, coniferyl, and sinapyl alcohols (Boerjan et al., 2003; Fig. 1). These monolignols form the p-hydroxyphenyl, guaiacyl (G), and syringyl (S) units, respectively, of the lignin polymer. In Populus spp., wood lignin is predominantly of the G and S types (Mellerowicz et al., 2001).
Figure 1.
A simplified model of the metabolic pathways from Suc to cell wall polymer biosynthesis. 13C incorporation was determined for cellulose, hemicelluloses, lignin, and metabolites (red). Enzymes have been omitted for clarity. NDP-sugars, Nucleotide diphosphate sugars.
The distribution of 13C in metabolism and cell walls was tracked over a diurnal time course using mass spectrometry (MS) and two-dimensional (2D) NMR analysis. We measured the 13C in Suc pools of source leaves, stem phloem, and developing wood. In developing wood, we also monitored the 13C incorporation into the metabolites closely linked to cell wall biosynthesis, namely UDP-Glc, hexose-Ps, and Phe. To investigate the cell wall polymer deposition patterns, the rate of 13C incorporation into different polymers was measured over a diurnal cycle.
RESULTS
Carbon from 13CO2 Is Incorporated into Wood Cell Walls within 24 h
Two-month-old greenhouse-grown hybrid aspen trees were enclosed in a transparent chamber (Supplemental Fig. S1) under controlled growth conditions and supplied with 13CO2 followed by analysis of 13C distribution over a time course. Phloem transport rate in a 4-m-tall gray poplar (Populus tremula × Populus alba) has been shown to be approximately 0.7 m h−1 (Windt et al., 2006), and a similar transport rate may be expected in the hybrid aspens used in this study. In our previous experiments, we could detect 13C in the soluble fraction of developing wood at the end of a 4-h 13CO2 pulse (Mahboubi et al., 2013), but 13C incorporation to wood was not determined. Hence, to establish the timescale from CO2 assimilation to carbon incorporation to stem wood, 13CO2 was applied for 4 h during the light period, and then 13C accumulation in the cell walls was measured at 24, 48, and 72 h using 2D NMR. The results established that the 13C accumulation in the cell walls did not increase markedly from 24 to 72 h after the start of the experiment (Supplemental Fig. S2). Hence, in the following experiments, the 13C flow in metabolism and subsequent incorporation into the developing wood were tracked over a 24-h period.
13C in Suc Peaked First in Leaves and Phloem and Then in Developing Wood
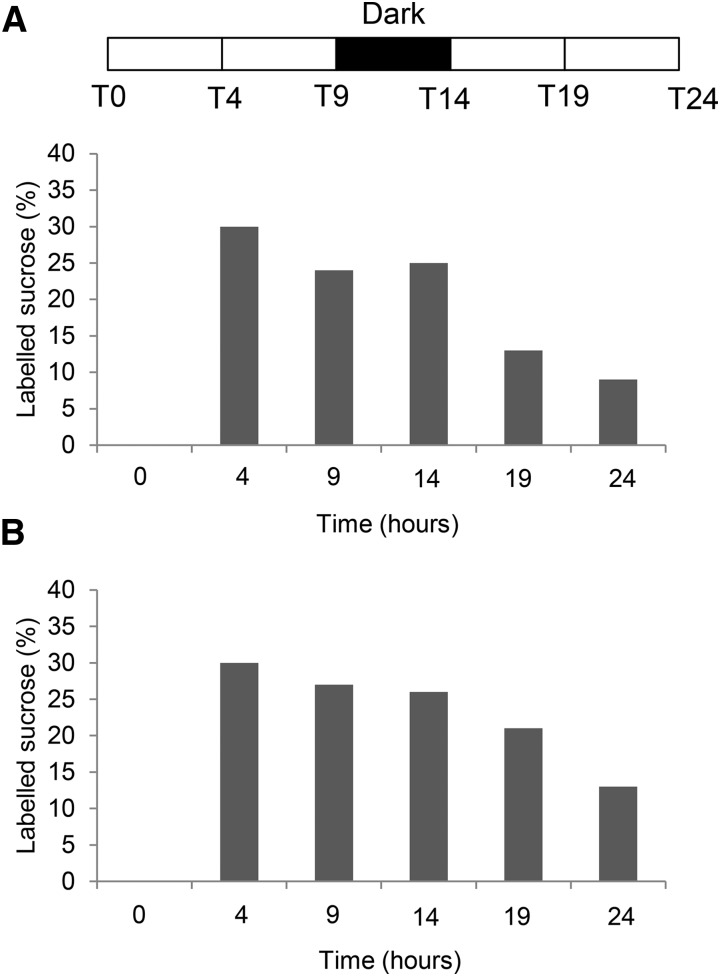
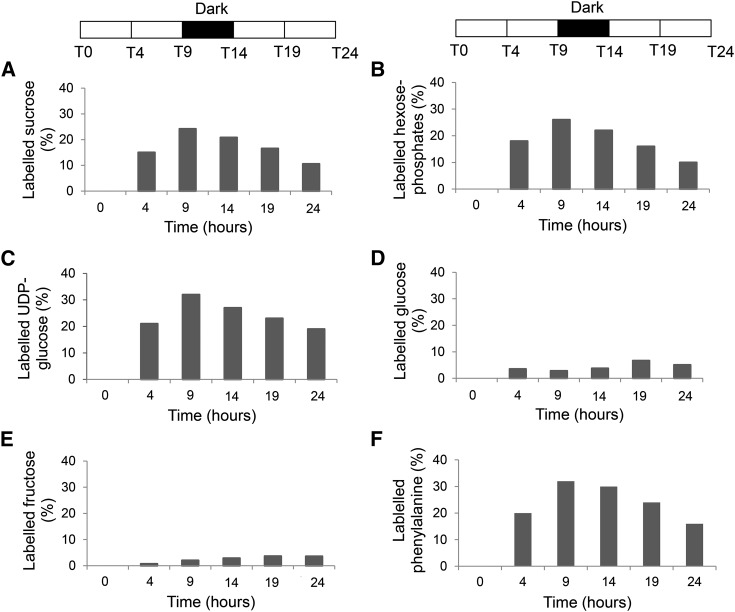
In the first 24-h experiment, 13CO2 was supplied for 4 h, and samples were harvested at 0, 4, 9, 14, 19, and 24 h, with a dark period between 9 and 14 h. Soluble metabolites were extracted from source leaves, stem phloem, and developing wood. The percentage of 13C enrichment in Suc was determined by gas chromatography (GC)-MS. This analysis showed that, in leaves and phloem, 30% of Suc was labeled at the end of the 4-h pulse, and then the 13C fraction decreased to 9% in leaves and 13% in phloem after 24 h (Fig. 2). In the developing wood, Suc labeling reached a maximum of 25% 9 h after the start of the 13CO2 supply and then decreased to 10% after 24 h (Fig. 3A). Suc-derived Glc isotopomers in developing wood were labeled at a similar rate, indicating that the Suc pool was evenly labeled (Supplemental Fig. S3).
Figure 2.
Incorporation of 13C into Suc in source leaves (A) and phloem (B). Two-month-old hybrid aspen trees were supplied with 13CO2 for 4 h and harvested at 4, 9, 14, and 24 h after the start of the experiment. Values are calculated from four biological replicates per time point. The dark period indicated by the black bar was from 9 to 14 h.
Figure 3.
Incorporation of 13C into Suc (A), hexose-Ps (B), UDP-Glc (C), Glc (D), Fru (E), and Phe (F) in developing wood. Two-month-old hybrid aspen trees were supplied with 13CO2 and harvested at 4, 9, 14, and 24 h after the start of the experiment. Values are calculated from four biological replicates per time point. The dark period indicated by the black bars was from 9 to 14 h.
13C Labeling of Central Metabolites in Developing Wood
To investigate the temporal pattern of carbon distribution in the primary metabolism of developing wood, the 13C enrichment in the central metabolites linked to biosynthesis of cell wall polymers was monitored using GC-MS (Suc, Glc, and Fru) and liquid chromatography (LC)-MS (hexose-Ps, UDP-Glc, and phenyl Ala). The position of the analyzed molecules in the metabolic pathways linked to cell wall biosynthesis is shown in Figure 1.
Enrichments of 13C in hexose-Ps and UDP-Glc were similar to each other and Suc, peaking at 9 h after the start of the labeling (Fig. 3, B and C). UDP-Glc reached a maximum enrichment of 32%, decreasing to 19% after 24 h, and hexose-Ps peaked at 26%, decreasing to 10% after 24 h. The similarity of labeling between UDP-Glc, hexose-Ps, and Suc suggested that Suc and cell wall carbohydrate precursor pools in developing wood are closely linked. This was in contrast to Glc and Fru labeling, which differed markedly from the other sugars, reaching modest maximums of 7% and 4%, respectively, of the total after 19 h (Fig. 3, D and E). 13C accumulation in the lignin biosynthesis precursor Phe was similar to Suc and sugar-Ps, reaching a maximum 13C enrichment of 32% after 9 h and then decreasing to 16% at the end of the experiment (Fig. 3F). The isotopomer labeling pattern of Glc, Fru, UDP-Glc, hexose-Ps, and Phe is shown in Supplemental Figure S3.
13C Incorporation into Developing Wood Began after 4 h
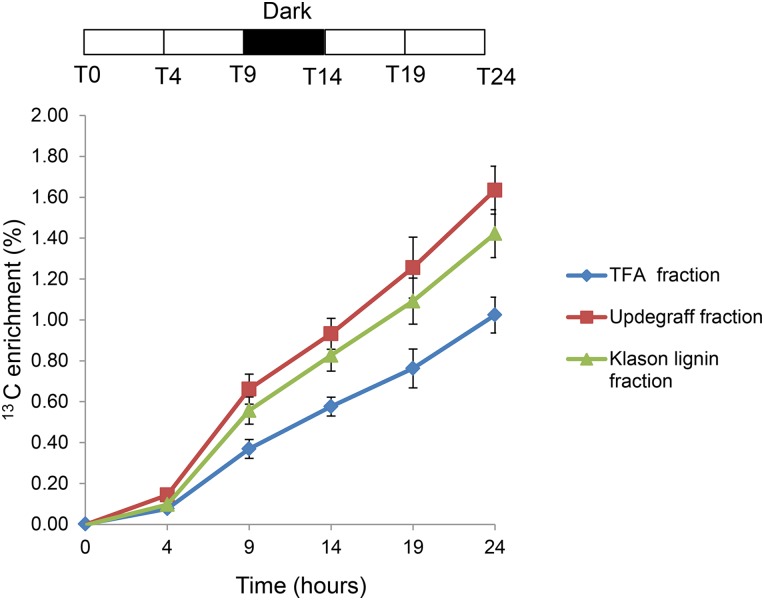
To compare the 13C enrichment patterns of the metabolites with cell wall biosynthesis, the rate of 13C incorporation into the main cell wall polymers in developing wood was measured. Extractive and starch-free cell wall samples from developing wood were prepared and fractionated into a 2 m trifluoroacetic acid (TFA)-soluble fraction containing mostly hemicellulose-derived monosugars, Updegraff cellulose fraction containing crystalline cellulose, and Klason lignin containing 72% (w/w) sulphuric acid-insoluble lignin. The percentage of 13C enrichment in each fraction was measured by elemental analysis-isotope ratio mass spectrometry (EA-IRMS).
According to the EA-IRMS analysis, the 13C enrichment in cell wall polymers was not yet obvious at 4 h but began to increase after this time point (Fig. 4). Updegraff cellulose fraction showed the highest rate of 13C enrichment between 4 and 9 h; then the rate decreased slightly and stayed relatively constant for the remainder of the experiment (Fig. 4). Klason lignin fraction showed a similar pattern of 13C enrichment to Updegraff cellulose, whereas the 2-m TFA hemicellulosic fraction accumulated 13C at a distinctively lower rate compared with the other two fractions (Fig. 4). The highest rate of 13C enrichment in all three fractions occurred between 4 and 9 h, corresponding to the period of 13C Suc increase in developing wood (Fig. 3A).
Figure 4.
Enrichment of 13C in Updegraff cellulose, Klason lignin, and 2 m TFA extracted fraction of developing wood. The 13C enrichment rate of the cell wall fractions is shown as a percentage of the total carbon in each fraction. Error bars represent the sem of four biological replicates.
ANOVA showed significant differences in the 13C enrichment between cell wall fractions as well as between time points (Table I). To investigate the contribution of the different cell wall fractions and time points to the variance in more detail, we used the Tukey’s honestly significant difference test for a pairwise comparison of the data set means. This analysis showed that the Updegraff cellulose and Klason lignin fractions did not differ significantly from each other, whereas the 2-m TFA fraction was significantly different from the other two fractions (Table II). Analysis of the differences between time points showed that the small enrichment increase from 0 to 4 h was not significantly different, whereas the increase from 4 to 9 h was clearly significant at P < 0.001. The differences between 9 to 14 and 14 to 19 h were significant at P < 0.01. The enrichment from 19 to 24 h was again significantly different at P < 0.001 (Table II).
Table I. ANOVA of 13C enrichment between cell wall fractions and time points.
df, Degrees of freedom; ***, P < 0.001.
| Source of Variance | ANOVA |
||||
|---|---|---|---|---|---|
| df | Sum2 | Mean2 | F | P | |
| Cell wall fractions | 2 | 1.13 | 0.56 | 24.14 | 3.23e-08*** |
| Time points | 5 | 16.85 | 3.37 | 143.47 | <2e-16*** |
Table II. Pairwise comparison of means for all cell wall fractions and all time points by Tukey’s honestly significant difference test.
***, P < 0.001; **, P < 0.01.
| Tukey’s Honestly Significant Difference | Mean Difference | Lower Bound | Upper Bound | P |
|---|---|---|---|---|
| Klason lignin versus Updegraff | −0.11 | −0.21 | 0.00 | 0.0517 |
| 2 m TFA versus Updegraff | −0.30 | −0.41 | −0.20 | <0.0001*** |
| 2 m TFA versus Klason lignin | −0.20 | −0.30 | −0.09 | 0.0001*** |
| 0 to 4 h | 0.11 | −0.08 | 0.29 | 0.5358 |
| 4 to 9 h | 0.42 | 0.24 | 0.61 | <0.0001*** |
| 9 to 14 h | −0.25 | −0.43 | −0.06 | 0.0027** |
| 14 to 19 h | 0.26 | 0.07 | 0.44 | 0.0017** |
| 19 to 24 h | 0.32 | 0.14 | 0.51 | <0.0001*** |
The cell wall fractionation and EA-IRMS analysis showed that significant incorporation of 13C to cell walls began 4 h after the start of the 13CO2 supply, which is in accordance with the arrival and metabolism of 13C Suc in developing wood (Fig. 3). Although there was a clear difference in the 13C enrichment rate between the 2-m TFA hemicellulosic fraction and the Updegraff cellulose and Klason lignin fractions, no obvious diurnal effects on cell wall biosynthesis were evident from this analysis. However, as discussed in the introduction, Bobák and Nečesaný (1967) showed that the newly formed cell wall layer in the developing fiber of P. nigra was sensitive to a relatively mild extraction treatment. Hence, we were concerned that the fractionation of the cell wall may have led to the loss of some cell wall constituents and that the most recently synthesized cell wall layer was the most sensitive in this respect. Furthermore, EA-IRMS only provided information about the total 13C enrichment in the isolated cell wall fractions and did not distinguish between different cell wall monomers. Therefore, to obtain a more detailed understanding of cell wall deposition dynamics over the 24-h period, we analyzed the 13C incorporation into developing wood cell walls using 2D NMR.
2D NMR Analysis Revealed Diurnal Patterns of Wood Formation
The 2D NMR analysis was carried out on unfractionated extractive and starch-free developing wood to minimize any modifications or losses caused by sample preparation procedures. NMR peak integrals were assigned to major lignin and polysaccharide monomers based on published literature (Fig. 5; Kim et al., 2008; Kim and Ralph, 2010; Brennan et al., 2012; Mansfield et al., 2012). Similar to EA-IRMS results, the NMR analysis showed that, with the exception of arabinose, the 13C incorporation into the cell walls occurred at the highest rate between 4 and 9 h (Supplemental Fig. S4). However, contrary to EA-IRMS, the 2D NMR analysis showed clear 13C incorporation to cell walls already during the first 4 h of the experiment (Supplemental Fig. S4). This observation supported the concern that some newly formed cell wall polymers may have been lost during the extraction and fractionation procedure of the EA-IRMS samples.
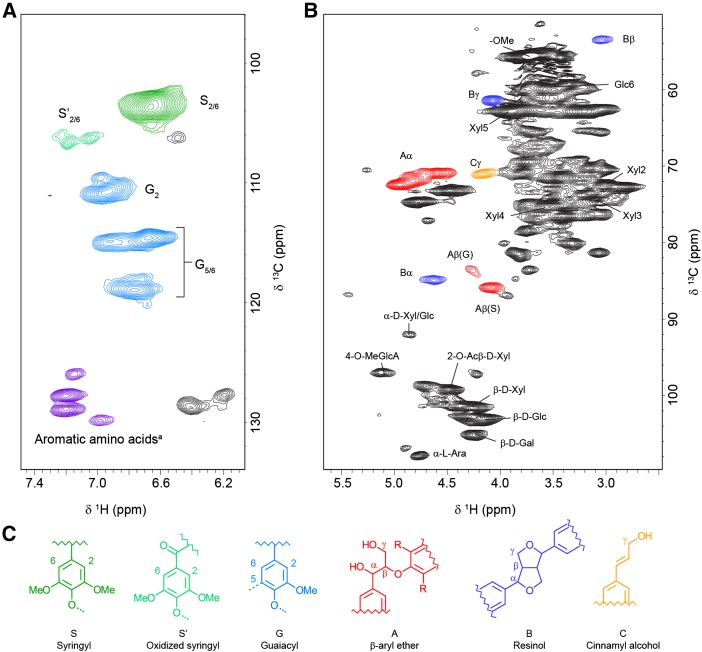
Figure 5.
Peak assignments of the 2D 13C-1H HSQC NMR spectra obtained from the 13C-labeled developing wood of hybrid aspen. A, S and G lignin and aromatic amino acids. B, Polysaccharide units and extra lignin peaks. C, Color-coded chemical structures of the lignin peaks in the spectra. a, Resonances from Phe and Tyr.
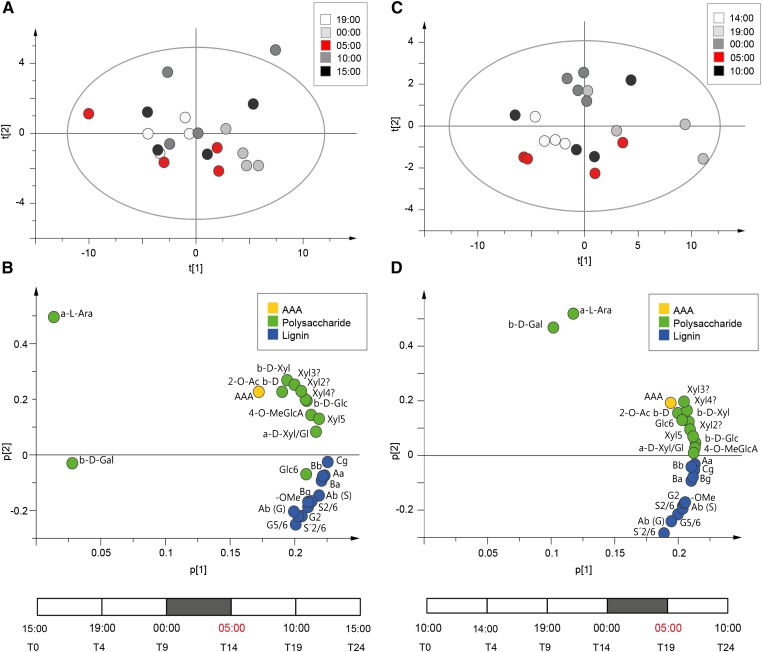
To investigate the correlations between the time points and the NMR peak integrals, we used principle component analysis (PCA). PCA is well suited for analyzing how the time points relate to each other with respect to the NMR peak variables. Before the PCA, values from every time point were subtracted from the previous time point to calculate the percentage of 13C enrichment from one time point to the next. The PCA loading plot clearly showed that the majority of cell wall carbohydrate peaks separated from lignin peaks (Fig. 6B). The contribution of the time points to the separation was not obvious, but especially, Xyl peaks seemed to correlate negatively and S and G lignin peaks seemed to correlate positively with the 14-h time point at the end of the dark period at 5 am (Fig. 6, A and B).
Figure 6.
PCA of the developing wood 13C incorporation patterns analyzed by 2D NMR. PCA score plots show the variation within the time points (A and C), and the loadings show the separation of 2D NMR 13C peak integrals (B and D) for experiments 1 and 2. NMR peaks include cell wall polysaccharides (green), lignin (blue), and aromatic amino acids (yellow). NMR spectra were normalized to a constant sum, and for each peak integral value, the mean value from the previous time point was subtracted. The position of the dark period was different between the two experiments and is shown by the gray bars. Aromatic amino acids (AAAs), G lignin (G5/6 and G2), S lignin (S′2/6 snf S2/6), lignin linkage types (Bα, Aβ [S], Aβ [G], Aα, Cγ, Bγ, and Bβ), lignin methoxy groups (-OMe), α-l-arabinose (α-l-Ara), β-d-Gal, Glc (β-d-Glc, Glc6, and α-d-Xyl/Glc?), Xyl (β-d-Xyl, 2-O-Ac β-d-Xyl, Xyl2, Xyl3, Xyl4, Xyl5, and α-d-Xyl/Glc?), and GlcA (4-0-MeGlcA) are shown. Statistical values for PCA in experiment 1: n = 20, X var = 25, R2X = 0.975, and Q2 = 0.895. Statistical values for PCA in experiment 2: n = 20, X var = 25, R2X = 0.986, and Q2 = 0.943.
We also observed significant label accumulation in two peaks in the aromatic region of the 2D NMR spectra (Fig. 5). The chemical shifts of these peaks are close to those of p-hydroxyphenyl lignin, but in the PCA, these peaks clearly separated from the rest of the lignin peaks, suggesting that they may not be derived from lignin (Fig. 6B). To establish the identity of these peaks, we performed a correlation analysis developed by Öman et al. (2014). This analysis calculates the correlation between a peak of interest in the 2D NMR spectra and all other peaks. High correlation between peaks indicates that they belong to the same molecule. By setting the aromatic region peak at 7.2 or 12.8 ppm as the peak of interest, we discovered that these peaks correlated most highly with a large number of peaks in the aliphatic region of the spectra as well as a number of low-intensity peaks in the regions from 3.6 to 4.5 ppm (1H) and from 40 to 59 ppm (13C; Supplemental Fig. S5). These are spectral regions, where protein-derived signals from the aliphatic amino acid side chains and the amino acid backbone α-carbon are found. This strongly indicated that the peaks in the aromatic region with different 13C labeling rates compared with the lignin peaks actually originated from the aromatic amino acid side chains of Phe and Tyr present in proteins. This interpretation is also in agreement with published 2D NMR spectra values for these amino acids (www.bmrb.wisc.edu). These results suggested that the extractive and starch-free cell wall samples contained some cell wall proteins, which were not removed by the sample preparation process.
Because the end of the night time point at 14 h seemed to contribute to the separation of lignin and carbohydrate peaks in the PCA, we hypothesized that the dark period had an effect on cell wall polymer biosynthesis (Fig. 6, A and B). Alternatively, this effect may have been a coincidence, and instead, the timing of the 13CO2 supply and the arrival of 13C in developing wood contributed to the separation. To distinguish between these two possibilities, we performed a second experiment, in which 13CO2 was supplied 14 h before the start of the dark period. Similar to the first experiment, the highest rate of label incorporation into the cell walls occurred between 4 and 9 h, indicating that the rate of Suc import and metabolism in developing wood was similar between the two experiments (Supplemental Fig. S4B). Notably, the peaks correlating either positively or negatively with the 14-h time point (at 5 am) at the end of dark period in the first experiment showed similar and even clearer correlation with the 19-h time point (at 5 am) in the second experiment (Fig. 6, C and D). In other words, if the dark period did not have an effect, the 19-h time point in the second experiment would be expected to correlate positively with the carbohydrate peaks, similar to experiment 1 (Fig. 6). These results indicated that the diurnal cycle instead of the timing of the 13CO2 supply and 13C import into the developing wood was contributing to the differential 13C incorporation into lignin and cell wall carbohydrates.
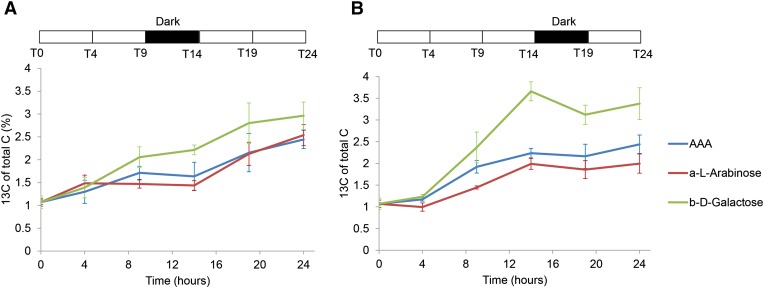
Additional analysis of the 2D NMR spectra also revealed a correlation between the protein-derived amino acid signals and the peak from α-l-arabinose as well as additional carbohydrate peaks that mostly also derive from arabinose (Mansfield et al., 2012; Supplemental Fig. S5). Interestingly, the 13C incorporation to arabinose and aromatic amino acid peaks did not increase during the dark period but continued again in the next light period (Fig. 7). This diurnal pattern was especially clear in experiment 1 (Fig. 7A), but it was also observed in experiment 2 (Fig. 7B). A similar pattern of 13C incorporation was also observed for β-d-Gal in both experiments (Fig. 7).
Figure 7.
13C incorporation pattern into aromatic amino acids (AAAs), α-l-arabinose, and β-d-Gal measured by 2D NMR. A, Experiment 1 with the dark period starting 9 h after the 13CO2 supply. B, Experiment 2 with the dark period starting 14 h after the 13CO2 supply. Error bars represent the sem of four biological replicates. The dark period is indicated by the black bar above the plots.
DISCUSSION
Anatomical differences between day and night in the innermost cell wall layer of developing wood fibers suggested that cell wall biosynthesis is influenced by the diurnal cycle in trees (Bobák and Nečesaný, 1967; Yoshida et al., 2000; Hosoo et al., 2002). This hypothesis was supported by the microarray experiments showing diurnal transcriptional changes in the developing wood of eucalyptus trees (Solomon et al., 2010). However, direct evidence of diurnal effects on the biosynthesis of wood cell walls has been difficult to obtain. To investigate the patterns of wood formation over a diurnal cycle, we designed a 13CO2 feeding chamber for 2-month-old greenhouse-grown hybrid aspen trees (Supplemental Fig. S1). This allowed us to track the assimilated 13C from the source leaves into developing wood cell walls over a diurnal cycle.
To establish the linear phase of 13C incorporation to wood cell walls after 4 h of 13CO2 supply, trees were sampled at 24, 48, and 72 h. 13C enrichment in the developing wood was determined from 2D NMR signature peaks for cellulose, xylan, and lignin. This analysis established that most of the 13C label had reached the cell walls during the first 24 h (Supplemental Fig. S2); therefore, the following experiments focused on identifying the 13C transport, metabolite, and cell wall deposition patterns during a single diurnal cycle.
In source leaves and stem phloem, Suc labeling showed a similar temporal pattern, with a peak at the end of the 4-h 13CO2 supply (Fig. 2). This similarity in Suc labeling between source leaves and stem phloem suggested that Suc pools in the two tissues are closely connected and that the diversion of Suc for export occurs concomitantly with Suc synthesis in the leaves. This observation is in line with the passive phloem loading mechanism in Populus spp., which implies that leaf and phloem Suc pools are directly connected by plasmodesmata. Recent experimental evidence showing that a yeast (Saccharomyces cerevisiae) invertase targeted to the apoplasm of gray poplar had no impact on phloem loading provided strong support for the predominance of the symplasmic phloem loading route in Populus spp. (Zhang et al., 2014). Zhang et al. (2014) also established that 14C Suc could be observed in the source leaf petiole already 45 min after a 15-min 14CO2 pulse. Hence, it is probable that more time points providing higher temporal resolution during the first hours of 13CO2 supply would reveal a difference in the temporal pattern of Suc labeling between source leaves and stem phloem.
13C Suc in the developing wood peaked after 9 h (Fig. 3A), clearly later than in source leaves and stem phloem. There are several transport steps along the radial transport route from phloem to developing wood that may be causing the difference in Suc labeling between developing wood and the adjacent phloem. In Populus spp., Suc export from the phloem and across the cambium is thought to initially follow a symplasmic route along the ray cells (Sauter and Kloth, 1986). Recent experimental evidence has suggested that Suc export from ray cells and import to developing wood fibers require active Suc import from the apoplasm. Reduction of a plasma membrane-localized Sucrose Transporter3 (SUT3) transcript levels using a secondary cell wall promoter-driven RNA interference construct caused a significant reduction in the fiber cell wall thickness (Mahboubi et al., 2013). Mahboubi et al. (2013) also showed that the SUT3RNA interference lines contained more 13C in the soluble fraction of developing wood after 13CO2 supply, implying a blockage in the final step(s) of the Suc import to wood fibers. Previously apoplasmic transport in Populus × canadensis wood was estimated to be approximately 8 to 80 times slower than the symplasmic transport rate (Sauter and Kloth, 1986). Hence, the apoplasmic and active transport steps in the radial phloem to developing wood Suc transport provide one possible explanation for the observed delay in 13C labeling of developing wood Suc compared with the adjacent phloem.
The central role of the plasma membrane-localized SUT3 for carbon import to secondary cell wall-forming wood fibers also implied that the majority of carbon for wood biosynthesis is derived from Suc cleaved inside the developing fibers. After it is inside the cells, Suc can be cleaved by either invertase to Glc and Fru or Suc synthase to UDP-Glc and Fru (Sturm and Tang, 1999). Recently, soluble Suc synthase activity in developing wood of hybrid aspen was shown to play a role in carbon allocation to cell walls, but it was not essential for wood formation per se (Gerber et al., 2014). Transgenic trees with only 4% of wild-type Suc synthase activity in the soluble fraction of developing wood had no obvious growth defects and unchanged proportions of cell wall polymers, suggesting that invertases are the main route of Suc cleavage or that invertases can compensate for the reduced Suc synthase activity in developing wood (Gerber et al., 2014). The rate of 13C incorporation into Glc and Fru over the 24-h experiment was low (Fig. 3, D and F), which could imply that invertases do not occupy a central position in the Suc to cell wall pathway. However, the cytosolic pools of Glc and Fru relevant for cell wall biosynthesis can be expected to quickly convert into hexose-Ps because of the micromolar-range substrate affinities of plant hexo- and fructokinases (Renz and Stitt, 1993; Pego and Smeekens, 2000). The labeling pattern of the developing wood hexose-Ps and UDP-Glc followed the pattern of Suc (Fig. 3, B and C), supporting a model of high-carbon flux rates through the preceding hexose pools (Fig. 1). This model was supported by a recent 13C metabolic flux analysis in Arabidopsis (Arabidopsis thaliana) cell cultures, which estimated that 99% of the carbon for cell wall precursor biosynthesis is derived from invertase cleavage of Suc to Glc and Fru followed by high flux rates into Glc-6-P and Fru-6-P (Chen et al., 2013). However, it should be noted that Chen et al. (2013) studied nonlignifying cells; hence, their carbon flux model may not apply to lignifying tissues, such as developing wood.
Lignin biosynthesis pathway precursor Phe in developing wood showed the highest 13C enrichment at 9 h after the start of the experiment (Fig. 3F). This was similar to the analyzed cell wall carbohydrate precursors, which also peaked at 9 h. In Arabidopsis leaves, the 13C enrichment of Phe after 13CO2 supply was also similar to 13C enrichment in Suc (Szecowka et al., 2013). In photosynthetic tissues, Phe is derived from erythrose-4-P derived directly from the Calvin cycle or the pentose phosphate pathway (Szecowka et al., 2013). In developing wood, carbon for Phe biosynthesis is most likely derived from the imported Suc, which is first metabolized to hexose-Ps and then imported to plastids for aromatic amino acid biosynthesis. The observed similarity of the sugar and Phe labeling patterns is in agreement with this model.
The analysis of central primary metabolism molecules in developing wood established the temporal dynamics of 13C influx to selected precursors of cell wall biosynthesis. To connect the metabolite labeling patterns to cell wall biosynthesis, we analyzed 13C incorporation into extracted cell wall fractions containing Updegraff cellulose, 2 m TFA extracted cell wall monosugars, and Klason lignin using EA-IRMS. With this method, no significant 13C enrichment was detected in the cell walls at 4 h, but after this time point, 13C accumulated over the whole 24-h time course (Fig. 4). Updegraff cellulose fraction accumulated 13C at a similar rate to Klason lignin, whereas both showed significantly higher label accumulation rates compared with 2 m TFA extracted monosugars. The 13C content analysis of cell wall fractions did not find obvious diurnal differences between the fractions. However, the EA-IRMS method did not distinguish between different cell wall constituent monomers and also, raised concerns about the sensitivity of the newly formed cell wall layer to the fractionation procedure involving strong acids.
NMR-based methods have emerged as some of the best analytical tools for obtaining cell wall information without the need of fractionation (Mansfield et al., 2012). Hence, to increase the resolution of the cell wall analysis, 2D NMR was used to investigate the pattern of 13C incorporation to cell wall polymers. In contrast to the EA-IRMS analysis, the 2D NMR data showed clear 13C enrichment in the cell walls already after 4 h of 13CO2 supply, supporting the concern that some recently formed cell wall material was lost because of the fractionation of the cell walls in the EA-IRMS experiment (Supplemental Fig. S4). Analysis of the first time-course experiment suggested that the dark period may have had an effect on the 13C deposition to cell walls, because the time point collected at the end of the dark period (red in Fig. 6A) correlated with the lignin-derived NMR peaks (blue in Fig. 6B). This was confirmed to be the case by supplying 13CO2 5 h earlier in the day during the second time-course experiment, and the time point collected at the end of the dark period (red in Fig. 6C) also showed a correlation with the lignin peaks (blue in Fig. 6D). PCA of both experiments showed a clear separation of lignin peaks from cell wall carbohydrate peaks. The major lignin peaks (S and G peaks) correlated positively with the end of the dark period in both experiments, whereas the cell wall polysaccharides showed negative correlation. The negative correlation with the dark period was most pronounced for the hemicellulosic sugars and less evident for the cellulose-derived Glc signals (Fig. 6). These data suggested that lignin biosynthesis increases relative to hemicellulose biosynthesis during the night. However, the values are relative, meaning that it is not possible to know whether 13C incorporation to lignin increased, if the incorporation to hemicelluloses decreased, or both. Interestingly, Rogers et al. (2005) observed, in Arabidopsis, that the abundance of transcripts encoding for enzymes of monolignol biosynthesis accumulated during the night, lending support to a model where lignin biosynthesis increases during the dark period.
The 2D NMR analysis also revealed that the 13C accumulation into the aromatic amino acids of cell wall proteins ceased during the dark period (Fig. 7). The 13C label was present in the soluble Phe pool (Fig. 3F), whereas the 13C incorporation into aromatic amino acids of the cell wall-localized proteins stopped. This also occurred at the same time as the 13C enrichment of lignin derived from the Phe pool continued (Fig. 4; Supplemental Fig. S4). In other words, 13C-labeled Phe was available but was not incorporated into the cell wall proteins during the dark period. This can mean a general cessation of cell wall protein biosynthesis or export to the cell walls in the dark; alternatively, the cell wall proteins synthesized during the dark may have been more easily released from the wall during the sample preparation. Additional experiments are needed to address these alternatives, but regardless of the explanation, these data provided additional support for the existence of diurnal patterns in wood formation.
Incorporation of 13C into the arabinose and Gal fractions of the cell walls provided another clear temporal pattern example (Fig. 7). Arabinose in cell walls is mainly found in pectin and hemicellulose polymers (Mellerowicz et al., 2001) as well as some cell wall glycoproteins. The correlation of arabinose and protein signals in both experiments (Supplemental Fig. S5) suggested that the arabinose signal may be partly derived from protein-linked polysaccharides, such as the ones found on arabinogalactan proteins (AGPs). AGPs are cell wall glycoproteins with an attached glycan chain rich in arabinose and Gal (Showalter, 2001). In this context, it is interesting to note that, in a proteomics study investigating protein abundance in response to diurnal changes in maize (Zea mays) leaves, the amounts of two Fasciclin-Like Arabinogalactan proteins were shown to decrease at dawn (Riter et al., 2011). Although it is not certain from where cell wall glycans arabinose and Gal signals in our experiments are coming, the data present another example of temporal patterns of cell wall biosynthesis in developing wood.
An emerging concept from our study and the previous anatomical and transcriptome studies is that wood formation in trees is coordinated by the diurnal cycle. It will be interesting to investigate how the diurnal patterns are established in a nonphotosynthetic sink tissue, like developing wood. Solomon et al. (2010) showed that the expression of the central circadian clock genes Circadian Clock Associated1 and GIGANTEA follows a circadian rhythm in constant light in the developing wood of Eucalyptus spp. The presence of a functional circadian clock in the developing wood of trees would provide one obvious candidate mechanism for providing temporal cues. However, other mechanisms may also be operating, such as the observed diurnal changes in the stem diameter corresponding to the photoperiod. It was shown that the stem diameter measured as tangential strain on the inner bark surface increased in C. japonica during the night and decreased during the day (Hosoo et al., 2003). Such volumetric changes are thought to be reflecting changes in the water status of developing cells and could also be involved in establishing diurnal patterns of cell wall biosynthesis.
The aspen 13CO2 labeling system opens the possibility of carbon flux modeling in developing wood and provides a system to study source to sink carbon fluxes in trees. Suc-derived Glc isotopomers were uniformly labeled in developing wood after 13CO2 supply, suggesting that Suc can be considered as a single pool for flux modeling purposes (Supplemental Fig. S3). This should simplify the 13C flux analysis, because no other Suc sources are expected to contribute to the downstream 13C fluxes. However, before metabolic flux analysis in developing wood is feasible, additional method development for the quantification of 13C-labeled wood metabolites is required.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Hybrid aspen (Populus tremula × tremuloides) trees were micropropagated, grown in vitro for 4 weeks, and then transferred to soil. Trees were grown in the greenhouse at a 20°C/15°C light-dark cycle with 50% to 70% humidity and a 18-h-light/6-h-dark photoperiod. Approximately 1 L of 0.2% Rika-S Fertilizer (N:P:K [7:1:5]; Weibulls Horto; http://www.weibullshorto.se) was applied every 7 d.
13CO2 Labeling
Two-month-old hybrid aspen trees were enclosed in a climate-controlled 10-m3 transparent plastic chamber (Supplemental Fig. S1) and labeled with 13CO2 at 25°C and relative humidity of 65% to 75%. For the 72-h experiment, trees were supplied for 4 h with 13CO2, and samples were harvested at 6, 24, 48, and 72 h after the start of the experiment. For the 24-h experiments, trees were supplied for 4 h with 13CO2 and harvested at 4, 9, 14, 19, and 24 h after the start of the experiment. Amount of supplied 13CO2 corresponded to the rate of CO2 consumption in the chamber measured with a WMA-4 CO2 Analyzer (www.ppsystems.com). To determine the rate of CO2 consumption, the CO2 concentration in the chamber was allowed to decrease to 150 ppm before the addition of 13CO2. 13CO2 was then supplied using a 1-L syringe to maintain a CO2 concentration of 350 to 400 ppm during the 4-h labeling period (Supplemental Fig. S1). To ensure efficient mixing of 13CO2 within the chamber, the gas was supplied directly into the air stream from the climate control unit. At each time point, fully expanded source leaves, developing wood, and stem phloem were immediately frozen in liquid nitrogen. Phloem and developing wood samples were prepared by pealing the bark from the bottom one-third of the stem followed by scraping of the exposed phloem and developing wood using a scalpel. Similarly treated unlabeled trees were used as controls in all of the experiments.
GC-MS Analysis
Pestle and mortar homogenized powder from source leaves, phloem, and developing wood (21–25 mg fresh weight) was extracted in 1 mL of extraction mix (CHCl3:MeOH:H2O [20:60:20, v/v], including nine internal standards at 7 ng µL−1 [Pro-13C5, salicylic acid-D6, putrecine-D4, myristic acid-13C3, hexadecanoic acid-13C4, and chlesterol-D7]) in 1.5-mL tubes (reference no. 72.690.007; Sarstedt) on ice followed by derivatization with 30 µL of methoxyamine in pyridine (15 ng µL−1), 30 µL of N-methyl-N-(trimethylsilyl) trifluoroacetamide, 1% trimethylchlorosilane, and 30 µL of heptane with 15 ng µL−1 of methylstearate. GC-MS analysis was performed using an Agilent Technologies 7890A GC System (Agilent Technologies) equipped with a 30-m × 0.250-mm-diameter fused silica capillary column with a bonded 0.25-μm Durabond DB-5MSUI Stationary Phase (part no. 122-5222UI; Agilent J&W GC Columns). All samples were analyzed in both splitless and split 1:5 modes. The injector temperature was 260°C, and the front inlet septum purge flow was 3 mL min−1. The gas flow rate through the column was 1 mL min−1; column temperature was held at 70°C for 2 min, then increased by 20°C per 1 min to 320°C, and held for 12 min. The column effluent was introduced into the ion source of a Pegasus HT GC, High-Throughput TOFMS (LECO Corp.). Data were normalized using internal standards as described by Redestig et al. (2009). In-house software was used to correct for natural abundance of 13C and isotope contribution from trimethylsilyl groups and calculate the percentage of 13C incorporation for each metabolite.
13C Enrichment Calculation from MS Spectra
For each of the derivatized metabolites, the abundance (i.e. relative concentration) was estimated by summarizing the peak areas for a specific fragment isotopic pattern. The degree of 13C incorporation was estimated from a reference fragment isotopic pattern compensating for isotope contributions from the silica (TMS groups) and natural abundances of 13C (approximately 1.1% all carbons; Smith, 1972). Hence, the contribution was expected to be consistently independent, regardless of whether a metabolite was labeled. The reference fragment isotopic pattern was calculated from the unlabeled control samples at baseline, and the relative sd was used for validity assessments of the fragment isotopic pattern. Relative sd above 1% was used to discard the reference fragment isotopic patterns. 13C incorporation was estimated for each ion of a fragment isotopic pattern and then summarized to the percentage of 13C of the total carbon pool. The theory of the 13C calculation is explained in Supplemental Figure S6.
LC-MS Analysis
Pestle and mortar-homogenized developing wood powder (21–25 mg fresh weight) was extracted in 1 mL of extraction mix (80:20 [v/v] ethanol:water, including six internal standards at 7 ng µL−1 [l-Pro-13C5, salicylic acid-D6, utrecine-D4, myristic acid-13C3, hexadecanoic acid-13C4, and chlesterol-D7]) in 1.5-mL tubes (reference no. 72.690.007; Sarstedt) on ice. The LC-MS analysis was carried out on an Agilent 6490 Ultra-HPLC coupled to a triple quadruple MS/MS (Agilent Technologies). Chromatographic separation was performed on a Waters Acquity UPLC BEH Amide 1.7-µm, 2.1- × 50-mm column (Waters Corporation). Mobile phase A contained water and 10 mm ammonium formate (NH4COOH). Mobile phase B contained 90% (v/v) acetonitrile and 10% (v/v) 10 mm ammonium formate in water. Gradient B was 85:85:70:10:10:85:85:85:85, time (in minutes) was 0:0.5:5.5:8:9.5:10:11:14.5:15, and flow was 0.250 L min−1 for the first 10 min and 1.6 L min−1 during equilibration. Sample injection volume was 2 μL. The mass spectrometer was operated in negative electrospray ionization mode with gas temperature of 210°C, gas flow of 11 L min−1, nebulizer at 60 pounds per square inch, sheath gas temperature of 200°C, sheath gas flow of 8 L min−1, capillary voltage of 3,000 V, nozzle voltage of 0 V, iFunnel High-Pressure RF of 90 V, and iFunnel Low-Pressure RF of 60 V. The multiple reaction monitoring was used to analyze sugar-Ps. All multiple reaction monitoring transitions were run in negative mode with dwell time of 50, fragmentor of 380, and cell acceleration voltage of 5. Data were normalized against sample fresh weight and processed using MassHunter Qualitative Analysis and Quantitative Analysis (triple quadruple; Agilent Technologies) and Excel (Microsoft).
Cell Wall Chemical Analysis
Extractive and starch-free developing wood powder was prepared as described by Foster et al. (2010) and used for cellulose, lignin, and cell wall monosaccharide content measurements. Cellulose content was measured using the method by Updegraff (1969) followed by anthrone assay for detection of released Glc (Scott and Melvin, 1953). Lignin content was measured by the Klason lignin method (Fengel and Wegener, 1983). Cell wall monosaccharides were extracted by 2 m TFA hydrolysis of the cell wall material followed by alditol acetate derivatization as described by Fox et al. (1989). The monosaccharide content was determined on a gas chromatograph-mass spectrometer (7890A/5975C; Agilent Technologies; www.agilent.com).
13C to 12C Ratio Analysis of Cell Wall Fractions
The 2-m TFA, Updegraff cellulose, and Klason (H2SO4 insoluble) lignin fractions that were prepared for cell wall chemical analysis were dried and analyzed with an elemental analyzer coupled with an isotope ratio mass spectrometer (www.thermofisher.com) to determine the 13C content. 13C fraction of total carbon was calculated from δ13C based on Vienna Pee Dee Belemnite standard with the reference 13C:12C ratio of 0.0112372.
2D NMR Analysis
Extractive and starch-free cell wall material from developing wood was used for 2D NMR analysis. From 150 to 200 mg of tissue was milled for 5 × 10 min at 500 rpm with 10-min break intervals using a Fritsch Pulverisette 7 Planetary Mill (Fritsch); 50 mg of the resulting wood powder was transferred to an NMR tube, and 600 μL of dimethyl sulfoxide-d6 was added. The suspension was then mixed and sonicated for 4 × 15 min, resulting in a homogenous viscous semitransparent suspension. 2D 1H-13C hetereonuclear single-quantum coherence (HSQC) experiments were performed at 25°C on a Bruker 600 MHz Spectrometer equipped with a 5-mm 1H/13C/31P cryoprobe with z gradients (Bruker Biospin; www.bruker.com). Adiabatic refocusing and inversion pulses were used on the 13C channel (pulse program hsqcetgpsisp2.2). Sweep widths of 8 and 140 ppm were used in the 1H and 13C dimensions, respectively. Twenty-four scans were added for each of 256 increments in F1, and a relaxation delay of 1 s was used, resulting in an experimental time of approximately 2 h per sample. A Gaussian window function was applied in F2, and a 90°-shifted squared sine bell window function was applied in F1 before Fourier transform. Processing and peak volume integration were performed in Topspin (Bruker Biospin; www.bruker.com). The residual dimethyl sulfoxide solvent peak was used as an internal reference (δH, 2.49 ppm; δC, 39.5 ppm). Assignments of peaks were based on previously published studies (Kim et al., 2008; Kim and Ralph, 2010; Brennan et al., 2012; Mansfield et al., 2012). Correlation analysis was performed using a previously described approach (Öman et al., 2014). HSQC spectra were imported to Matlab r2013b (Mathworks Inc.), where they were transformed into row vectors, normalized to a constant sum, and placed in a matrix. Thereafter, a data point (column) in the matrix corresponding to the peak of interest was chosen, and correlations to all other data points were calculated. The resulting row of correlation values was then transformed into a 2D matrix with the same size as the original spectra for visualization of the result. All data points with correlations less than 0.6 were set to 0 to highlight the most important correlations.
Data Analysis
NMR spectra were normalized to a constant sum, and to run the PCA, a unique data set was formed by subtracting the peak value for each replicate in every time point from the mean of the previous time point. The data set was UV scaled and then used for PCA. PCA was done in SIMCA version 13.0 software (Umetrics; http://www.umetrics.com). ANOVA and Tukey’s honestly significant difference tests were also performed in R (http://www.r-project.org) using aov and TukeyHSD functions, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Assembly of the labeling experiment.
Supplemental Figure S2. 13C incorporation to wood over a 72-h period.
Supplemental Figure S3. 13C labeling pattern of analyzed metabolite isotopomers.
Supplemental Figure S4. 2D NMR-derived 13C labeling patterns of cell wall monomers.
Supplemental Figure S5. Correlation of cell wall amino acid and arabinose 2D NMR spectra.
Supplemental Figure S6. Example of the 13C incorporation calculation.
Supplementary Material
Glossary
- 2D
two dimensional
- EA-IRMS
elemental analysis-isotope ratio mass spectrometry
- G
guaiacyl
- GC
gas chromatography
- HSQC
hetereonuclear single-quantum coherence
- LC
liquid chromatography
- MS
mass spectrometry
- PCA
principle component analysis
- S
syringyl
- TFA
trifluoroacetic acid
Footnotes
This work was supported by the Swedish Research Council FORMAS Bioimprove Program, the Umeå Plant Science Centre Berzelii Centre for Forest Biotechnology funded by VINNOVA, Trees and Crops for Future, the Swedish Research Council, and Bio4Energy (Swedish Programme for Renewable Energy).
References
- Alonso AP, Piasecki RJ, Wang Y, LaClair RW, Shachar-Hill Y (2010) Quantifying the labeling and the levels of plant cell wall precursors using ion chromatography tandem mass spectrometry. Plant Physiol 153: 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobák M, Nečesaný V (1967) Changes in the formation of the lignified cell wall within a twenty-four hour period. Biologia Plantarum 9: 195–201 [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Brennan M, McLean JP, Altaner CM, Ralph J, Harris PJ (2012) Cellulose microfibril angles and cell-wall polymers in different wood types of Pinus radiata. Cellulose 19: 1385–1404 [Google Scholar]
- Chen X, Alonso AP, Shachar-Hill Y (2013) Dynamic metabolic flux analysis of plant cell wall synthesis. Metab Eng 18: 78–85 [DOI] [PubMed] [Google Scholar]
- Fengel D, Wegener G (1983) Wood: Chemistry, Ultrastructure, Reactions. Kessel, Remagen, Germany [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010) Comprehensive Compositional Analysis of Plant Cell Walls (Lignocellulosic Biomass) Part II: Carbohydrates. JoVE. http://www.jove.com/index/Details.stp?ID=1837 (May 15, 2015) [DOI] [PMC free article] [PubMed]
- Fox A, Morgan S, Gilbart J (1989) Preparation of alditol acetates by gas chromatography and mass spectrometry. In Bermann J, McGinnis G, eds, Analysis of Carbohydrates. CRC Press, Boca Raton, FL, pp 87–117 [Google Scholar]
- Gerber L, Zhang B, Roach M, Rende U, Gorzsás A, Kumar M, Burgert I, Niittylä T, Sundberg B (2014) Deficient sucrose synthase activity in developing wood does not specifically affect cellulose biosynthesis, but causes an overall decrease in cell wall polymers. New Phytol 203: 1220–1230 [DOI] [PubMed] [Google Scholar]
- Hosoo Y, Yoshida M, Imai T, Okuyama T (2002) Diurnal difference in the amount of immunogold-labeled glucomannans detected with field emission scanning electron microscopy at the innermost surface of developing secondary walls of differentiating conifer tracheids. Planta 215: 1006–1012 [DOI] [PubMed] [Google Scholar]
- Hosoo Y, Yoshida M, Imai T, Okuyama T (2003) Diurnal differences in the innermost surface of the S2 layer in differentiating tracheids of Cryptomeria japonica corresponding to a light-dark cycle. Holzforschung 57: 567–572 [Google Scholar]
- Kim H, Ralph J (2010) Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d(6)/pyridine-d(5). Org Biomol Chem 8: 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ralph J, Akiyama T (2008) Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d 6. BioEnergy Res 1: 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski LA, Kunz S, Wilczynska M (2010) Mechanisms of UDP-glucose synthesis in plants. Crit Rev Plant Sci 29: 191–203 [Google Scholar]
- Mahboubi A, Ratke C, Gorzsás A, Kumar M, Mellerowicz EJ, Niittylä T (2013) Aspen SUCROSE TRANSPORTER3 allocates carbon into wood fibers. Plant Physiol 163: 1729–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SD, Kim H, Lu F, Ralph J (2012) Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc 7: 1579–1589 [DOI] [PubMed] [Google Scholar]
- Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W (2001) Unravelling cell wall formation in the woody dicot stem. Plant Mol Biol 47: 239–274 [PubMed] [Google Scholar]
- Öman T, Tessem MB, Bathen TF, Bertilsson H, Angelsen A, Hedenström M, Andreassen T (2014) Identification of metabolites from 2D (1)H-(13)C HSQC NMR using peak correlation plots. BMC Bioinformatics 15: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pego JV, Smeekens SC (2000) Plant fructokinases: a sweet family get-together. Trends Plant Sci 5: 531–536 [DOI] [PubMed] [Google Scholar]
- Redestig H, Fukushima A, Stenlund H, Moritz T, Arita M, Saito K, Kusano M (2009) Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics data. Anal Chem 81: 7974–7980 [DOI] [PubMed] [Google Scholar]
- Reiter WD, Vanzin GF (2001) Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol Biol 47: 95–113 [PubMed] [Google Scholar]
- Renz A, Stitt M (1993) Substrate-specificity and product inhibition of different forms of fructokinases and hexokinases in developing potato-tubers. Planta 190: 166–175 [Google Scholar]
- Riter LS, Jensen PK, Ballam JM, Urbanczyk-Wochniak E, Clough T, Vitek O, Sutton J, Athanas M, Lopez MF, MacIsaac S (2011) Evaluation of label-free quantitative proteomics in a plant matrix: a case study of the night-to-day transition in corn leaf. Anal Methods 3: 2733–2739 [Google Scholar]
- Rogers LA, Dubos C, Cullis IF, Surman C, Poole M, Willment J, Mansfield SD, Campbell MM (2005) Light, the circadian clock, and sugar perception in the control of lignin biosynthesis. J Exp Bot 56: 1651–1663 [DOI] [PubMed] [Google Scholar]
- Sauter JJ, Kloth S (1986) Plasmodesmatal frequency and radial translocation rates in ray cells of poplar (Populus x canadensis Moench ‘robusta’). Planta 168: 377–380 [DOI] [PubMed] [Google Scholar]
- Scott TA Jr, Melvin EH (1953) Determination of dextran with anthrone. Anal Chem 25: 1656–1661 [Google Scholar]
- Sharples SC, Fry SC (2007) Radioisotope ratios discriminate between competing pathways of cell wall polysaccharide and RNA biosynthesis in living plant cells. Plant J 52: 252–262 [DOI] [PubMed] [Google Scholar]
- Showalter AM. (2001) Arabinogalactan-proteins: structure, expression and function. Cell Mol Life Sci 58: 1399–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BN. (1972) Natural abundance of the stable isotopes of carbon in biological systems. Bioscience 22: 226–231 [Google Scholar]
- Solomon OL, Berger DK, Myburg AA (2010) Diurnal and circadian patterns of gene expression in the developing xylem of Eucalyptus trees. S Afr J Bot 76: 425–439 [Google Scholar]
- Sturm A, Tang GQ (1999) The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci 4: 401–407 [DOI] [PubMed] [Google Scholar]
- Szecowka M, Heise R, Tohge T, Nunes-Nesi A, Vosloh D, Huege J, Feil R, Lunn J, Nikoloski Z, Stitt M, et al. (2013) Metabolic fluxes of an illuminated Arabidopsis rosette. Plant Cell 25: 694–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Windt CW, Vergeldt FJ, de Jager PA, van As H (2006) MRI of long-distance water transport: a comparison of the phloem and xylem flow characteristics and dynamics in poplar, castor bean, tomato and tobacco. Plant Cell Environ 29: 1715–1729 [DOI] [PubMed] [Google Scholar]
- Yoshida M, Hosoo Y, Okuyama T (2000) Periodicity as a factor in the generation of isotropic compressive growth stress between microfibrils in cell wall formation during a twenty-four hour period. Holzforschung 54: 469–473 [Google Scholar]
- Zhang C, Han L, Slewinski TL, Sun J, Zhang J, Wang ZY, Turgeon R (2014) Symplastic phloem loading in poplar. Plant Physiol 166: 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.