We show that an algal transcription factor is involved in photoperiod response, a function conserved in angiosperms despite the wide diversification of these proteins.
Abstract
The response to daylength is a crucial process that evolved very early in plant evolution, entitling the early green eukaryote to predict seasonal variability and attune its physiological responses to the environment. The photoperiod responses evolved into the complex signaling pathways that govern the angiosperm floral transition today. The Chlamydomonas reinhardtii DNA-Binding with One Finger (CrDOF) gene controls transcription in a photoperiod-dependent manner, and its misexpression influences algal growth and viability. In short days, CrDOF enhances CrCO expression, a homolog of plant CONSTANS (CO), by direct binding to its promoter, while it reduces the expression of cell division genes in long days independently of CrCO. In Arabidopsis (Arabidopsis thaliana), transgenic plants overexpressing CrDOF show floral delay and reduced expression of the photoperiodic genes CO and FLOWERING LOCUS T. The conservation of the DOF-CO module during plant evolution could be an important clue to understanding diversification by the inheritance of conserved gene toolkits in key developmental programs.
Due to their sessile lifestyle, vascular plants have evolved intricate genetic regulatory pathways to control essential developmental processes in response to changes in the environment. One of the most distinctive processes is the floral transition (Amasino, 2010), which integrates information from endogenous and external signals to guarantee reproductive success (Valverde, 2011). In Arabidopsis (Arabidopsis thaliana), the CONSTANS (CO) gene plays a central role in the control of photoperiodic flowering by triggering the expression of FLOWERING LOCUS T (FT) in the leaf vascular tissue (An et al., 2004). FT codes for a major component of the florigenic signal that moves through the phloem to induce flower differentiation at the shoot apex (Corbesier et al., 2007; Tamaki et al., 2007). CO and FT are controlled at the transcriptional level by a circadian and photoperiodic signal through the FLAVIN-BINDING, KELCH REPEAT, F-BOX1 (FKF1), GIGANTEA (GI), and CYCLING DOF FACTOR (CDF) genes (Song et al., 2012). In long days (LD), blue light induces the assembly of the FKF1-GI complex that promotes the proteasome-mediated degradation of the CDFs, a four-member family of DNA-binding with One Finger (DOF) transcription factors (TFs) that bind to CO and FT promoters, repressing their expression (Song et al., 2012). The photoperiodic pathway is probably the most conserved of the flowering signaling responses in spermatophytes (Amasino, 2010) and can be traced back to chlorophytes, where CrCO, a CO homologous gene from Chlamydomonas reinhardtii, was identified (Serrano et al., 2009). CrCO is involved in the regulation of starch synthesis and cell growth in the alga (Serrano et al., 2009), two processes that are also regulated by the CONSTANS-LIKE (COL) gene family members in vascular plants (Valverde, 2011; Romero-Campero et al., 2013). In addition, transgenic plants overexpressing CrCO under the control of a constitutive or phloem-specific promoter flowered earlier than wild-type plants and complemented co mutants (Serrano et al., 2009).
CDFs constitute a subgroup of DOF TFs (Imaizumi et al., 2005; Fornara et al., 2009), characterized by the presence of a 52-amino acid DOF domain, a single zinc finger that binds to the DNA consensus sequence AAAG. DOFs can work as transcriptional activators or inhibitors, depending on the protein and the target gene concerned (Noguero et al., 2013). The DOF domain is bifunctional, as it can participate in DNA-binding and protein-protein interactions (Yanagisawa, 1997). DOFs have been described exclusively in plants and are involved in numerous processes, from tissue differentiation to metabolic regulation and seed development. They are widely distributed, with numerous genomic copies, in both gymnosperms and angiosperms and constitute a small gene family in some bryophytes such as the moss Physcomitrella patens. A single-copy CrDOF gene from C. reinhardtii (Moreno-Risueno et al., 2007; Shigyo et al., 2007) represents the ancestor of the DOF TF family. DOFs are absent in heterotrophic eukaryotes, red algae, and heterokonts (Hernando-Amado et al., 2012), so modern DOFs, similar to COLs, constitute a family of TFs exclusive of the green lineage (Serrano et al., 2009).
Here, we show that CrDOF induces CrCO expression in short days (SD) and is a general repressor of algal gene expression in LD. CrDOF expression in Arabidopsis reproduces CDF function by reducing CO and FT transcript levels and delaying flowering. The DOF-CO evolutionarily conserved signaling module constitutes a direct clue to how developmental responses may have evolved from unicellular algae to multicellular plants by the parallel evolution of developmental gene toolkits.
RESULTS
C. reinhardtii CrDOF Defines a Subfamily of DOF TFs Closely Related to CDFs in Arabidopsis
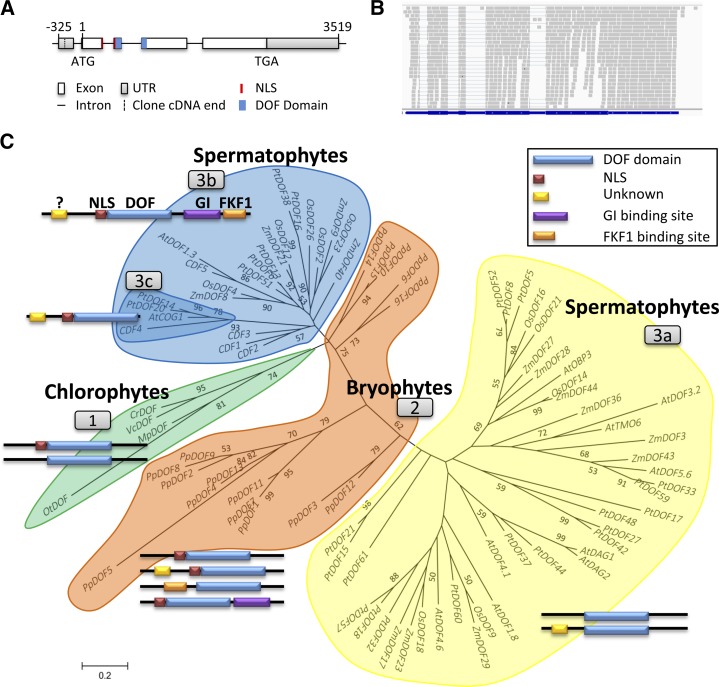
In version 5.5 of the C. reinhardtii genome annotation (http://phytozome.jgi.doe.gov/pz/portal.html), a single-copy gene (Cre12.g521150) at positions 4,424,576 to 4,431,469 on chromosome 12 (CrDOF) showed a significant identity with DOF TFs (Moreno-Risueno et al., 2007). In order to characterize its gene structure, three complementary DNA (cDNA) clones from the Kazusa open reading frame (ORF) collection (http://www.kazusa.or.jp) were sequenced. The CrDOF coding sequence was 1,875 bp long and was identical to the draft genome sequence (Merchant et al., 2007) at the 3′ end but shorter at the 5′ end (Fig. 1A). This was further confirmed by the alignment of the RNA sequencing (RNAseq) data to the predicted gene structure (Fig. 1B).
Figure 1.
Phylogenetic tree and domain evolution of DOF proteins. A, CrDOF gene structure. Squares represent exons, and lines represent introns. The DOF domain is in blue and the nuclear localization signal (NLS) is in red. B, Alignment of sequences obtained from RNAseq (gray) with the CrDOF gene structure (blue). C, Evolutionary relationship of 84 DOF proteins from eight species of the Viridiplantae linage. Three clusters pertaining to evolutionary grade are shown: chlorophytes (group 1), bryophytes (group 2), and spermatophytes (groups 3a–3c). Neighbor joining with the substitution model JTT + G 0.54 was the algorithm employed. The bootstrap number was 500. For abbreviations and protein accession numbers, see Supplemental Table S1. The domain structures of DOF proteins from each cluster are depicted beside each group. DOF domains are shown in blue, NLS in red, an unknown domain in yellow, and GI- and FKF1-binding sites in purple and orange, respectively. Only domains with a significant score in the MEME program are shown.
Using the CrDOF amino acid sequence, a BLAST search in several representative plant species was run and 84 different putative DOF proteins identified (Supplemental Table S1). These sequences were used to construct a phylogenetic tree (Fig. 1C) using the MEGA5 program (Tamura et al., 2011). This phylogenetic tree presents four clusters, including chlorophyte (cluster 1), bryophyte (cluster 2), and spermatophyte (clusters 3a and cluster 3b-3c) sequences. Cluster 3b-3c enclosed the Arabidopsis photoperiodic CDFs (Imaizumi et al., 2005; Fornara et al., 2009), closely aligned with chlorophyte sequences including CrDOF. This protein family also comprised other DOFs from Populus, Zea, and Oryza spp. that may represent putative photoperiodic DOF TFs, such as OsDOF12, which has been shown to be involved in photoperiodic flowering in rice (Oryza sativa; Li et al., 2009). No Physcomitrella spp. protein was included in this cluster; rather, they formed a group on their own, bridging chlorophyte with spermatophyte sequences. Subfamily 3a included DOF proteins from Arabidopsis, poplar (Populus spp.), maize (Zea mays), and rice that have been shown to be involved in diverse processes other than photoperiodic flowering, such as DOF AFFECTING GERMINATION1,2 (AtDAG1,2), involved in germination in Arabidopsis (Papi et al., 2000; Gualberti et al., 2002), or ZmDOF3 (PROLAMIN-BOX BINDING FACTOR1), involved in storage protein synthesis in maize seeds (Vicente-Carbajosa et al., 1997).
CDF proteins have a characteristic domain structure (Fig. 1C, group 3b) with a conserved DOF domain (blue) preceded by an NLS (red). C-terminal domains for GI-binding (purple) and FKF1-binding (orange) sites are specific for CDFs, as they participate in the interactions that control their posttranslational regulation (Kloosterman et al., 2013). A conserved N-terminal domain of unknown function (yellow) is also present. The ancestral algal DOF protein from group 1 (Fig. 1C) contains the conserved DOF domain and the NLS signal but lacks any other clear domain. In P. patens, although GI and FKF1 proteins have not been identified in its genome (Rensing et al., 2008), different DOF proteins (cluster 2) include regions that resemble the GI- and FKF1-binding sites that together with the yellow domains later evolved into the modern plant DOF domains in CDFs (clusters 3b and 3c) and other DOFs (cluster 3a; Kloosterman et al., 2013). Therefore, the original algal DOF TF acquired new domains that defined their regulation and function (Fig. 1C), as suggested by the close association between domain structure and function observed in the phylogenetic tree.
CrDOF Is Differentially Expressed under Diverse Photoperiod Conditions in C. reinhardtii
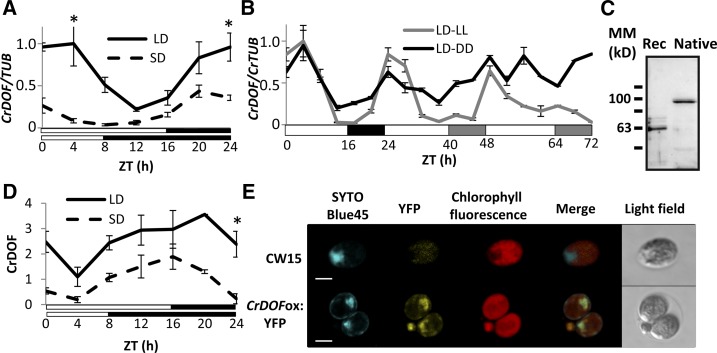
Arabidopsis CDFs are differentially expressed under diverse daylengths in order to correctly time the expression of CO and confer a photoperiodic response to flowering (Imaizumi et al., 2005). CrDOF expression in C. reinhardtii was monitored under LD and SD every 4 h for 24 h starting at Zeitgeber time 0 (ZT0), the moment the lights are switched on. CrDOF mRNA levels were higher in LD than in SD, and the expression profile resembled that of CDFs in Arabidopsis: a peak of expression in the late night and early morning (Fornara et al., 2009; Fig. 2A). When the expression was monitored during a 24-h LD followed by 48 h in continuous light (LL) or continuous dark (DD), the expression of CrDOF continued oscillating (Fig. 2B). This was particularly true for LL, but even a slight oscillation in DD was observed after 48 h, indicating a circadian influence.
Figure 2.
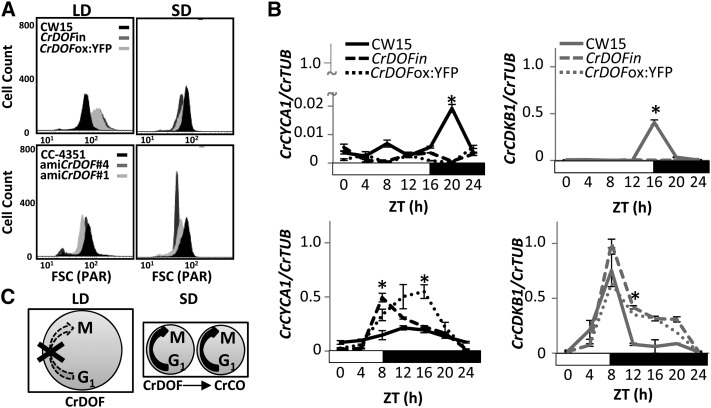
CrDOF expression is influenced by daylength and the circadian clock. A, CrDOF transcript levels in CW15 during a 24-h course in LD (solid line) and SD (dashed line). Data are from three independent experiments ± se. B, Circadian expression of CrDOF in a 24-h LD followed by 48 h in LL (gray) or DD (black). Data are from three independent experiments ± se. C, Immunodetection of CrDOF in a purified recombinant protein fraction (Rec; 1 µg) and algal crude extract (Native; 50 µg). Black lines are MM markers in kD. D, CrDOF protein levels in CW15 in a 24-h course in LD (solid line) and SD (dashed line). Data are from two independent experiments ± se. *, P < 0.05. E, Confocal images of CW15 and CrDOFox:YFP lines. SYTOBlue45 fluorescence around nuclear regions (blue), YFP (yellow), chloroplast (red) are shown. Bars = 5 μm.
To monitor the presence of CrDOF protein, we produced polyclonal antibodies by immunizing rabbits with a recombinant CrDOF purified from Escherichia coli (see “Materials and Methods”). These antibodies were used to detect CrDOF in extracts from the recombinant bacteria and from C. reinhardtii (Fig. 2C). While the recombinant protein showed a molecular mass (MM) of 60 kD, which is close to its predicted size (62 kD), the extract from C. reinhardtii showed a unique band of approximately 100 kD. Because we used SDS-PAGE conditions, the result suggested that the 100-kD band reflected an SDS-resistant posttranslational modification, such as has been shown for other SDS-resistant complexes from plants and animals described previously in the literature (Lüders et al., 2003; Varet et al., 2003; Kubista et al., 2004). Using these antibodies, we followed the presence of CrDOF in 24-h courses in LD and SD (Fig. 2D; Supplemental Fig. S1A) in protein extracts from the same samples as in Figure 2A. CrDOF protein showed a higher accumulation in LD than in SD and a clear circadian oscillation. However, in contrast to the mRNA profile, CrDOF minimal presence was monitored at ZT4 and its maximum was monitored at dusk (ZT16–20), coinciding with the CDF2 stability profile in Arabidopsis (Fornara et al., 2009). Thus, the expression of CrDOF mRNA and its protein stability are displaced in time.
To study CrDOF modifications, protein extracts from C. reinhardtii were incubated with different chemicals and immunodetected with α-CrDOF. When proteins were extracted with 8 m urea (a potent chaotropic agent) or in the presence of 10 mm dithiothreitol (DTT; which breaks Cys disulfide bonds) or a mixture of both (Supplemental Fig. S1B, top), only the 100-kD band was detected. Nevertheless, when proteins were extracted with a buffer containing 6 m guanidine chlorhydrate, a potent ionic chaotrop, the band of 100 kD was replaced by a band of 60 kD (Supplemental Fig. S1B, right). To discard ubiquitin modifications that are involved in Arabidopsis CDF stability (Imaizumi et al., 2005), extracts were incubated with 1 mm of the proteasome inhibitor MG-132. However, the intensity of the signal of the 100-kD band did not increase or show any mobility shift (Supplemental Fig. S1B, bottom right), probably indicating that the proteasome was not involved in CrDOF stability. Therefore, it is possible that CrDOF establishes tight connections with other proteins that control its function. As CDF stability is reduced under blue light in Arabidopsis (Imaizumi et al., 2005), we monitored if CrDOF stability was influenced by light quality. C. reinhardtii cultures were grown under LL, DD, constant blue light, or constant red light, cell extracts were prepared, and immunoblots were run using CrDOF antibodies. No significant difference in CrDOF accumulation was detected in any light condition (Supplemental Fig. S1C), indicating that light quality has no effect on its posttranslational stability.
The presence of CrDOF protein was also monitored using the confocal microscope in an algal line (CrDOFox:YFP; see below) that expressed the yellow fluorescent protein fused to the CrDOF C-terminal end (Fig. 2E). To identify the cell nucleus, we developed a novel method for C. reinhardtii using the nucleic acid-specific dye SYTOBlue45 (Molecular Probes) in order to visualize nuclear material in vivo without interfering with YFP and chloroplast fluorescence (see “Materials and Methods”). SYTOBlue45 dyes in blue a nucleic acid-rich region of CW15, sited in the flagella pole and encircled by the unique cup-shaped structure of the chloroplast, which we identified as the nucleus (Fig. 2E, top). The merged image in the bottom row of Figure 2E shows that the YFP signal coincided with the blue signal from the dye, indicating that the CrDOF:YFP fusion was preferentially localized in the nucleus.
Modification of CrDOF Expression Alters C. reinhardtii Growth
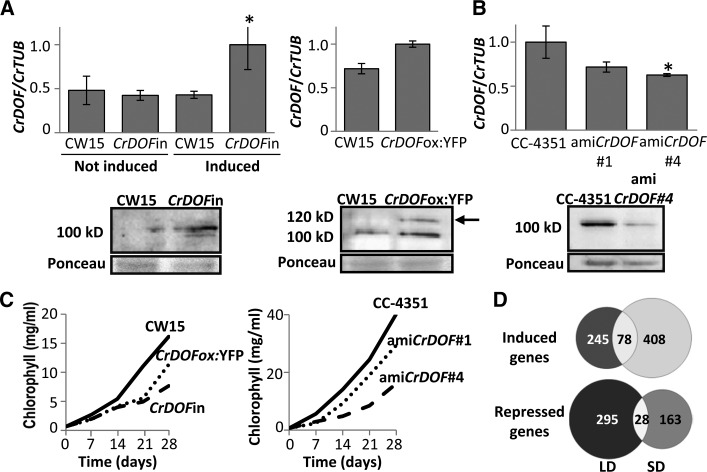
To clarify the role of CrDOF in C. reinhardtii, algal lines with altered levels of CrDOF mRNA were produced using Gateway technology vectors (Invitrogen) containing constitutive or inducible promoters (see “Materials and Methods”). Thus, CrDOF genomic sequence was fused to the pnia2 promoter (CrDOFin) for inducible expression depending on the nitrogen source (Camargo et al., 2007). Figure 3A, top left, shows that CrDOF expression was significantly increased in CW15 transformed with the CrDOFin construct under inductive (+NO3−) conditions, also detecting an increase in protein levels by western blots (Fig. 3A, bottom left). CrDOF expression levels increased when CrDOF was fused to YFP in its C-terminal end and expressed under the control of the chimeric promoter HEAT SHOCK PROTEIN70/RIBULOSE BISPHOSPHATE CARBOXILASE-OXYGENASE SMALL SUBUNIT2 (RbcS2; CrDOFox:YFP) for constitutive expression (Sizova et al., 2001; Fig. 3A, top right). Extracts from these algae presented an extra 120-kD band corresponding to the MM of CrDOF plus the YFP protein (Fig. 3A, bottom right, arrow). CrDOF expression was significantly reduced employing an artificial microRNA (amiCrDOF; see “Materials and Methods”) strategy (Molnar et al., 2009) that was reflected in a decrease in mRNA and protein levels in extracts from the two independent silenced lines (amiCrDOF #1 and #4; Fig. 3B). CrDOF overexpression from the constitutive and the induced promoters had a strong effect on algal growth (Fig. 3C, left). Wild-type and recombinant culture growth in liquid medium (Sueoka NO3−; see “Materials and Methods”) was monitored by measuring chlorophyll content during 4 weeks in LD. Whereas CW15 reached the exponential growth phase in week 2, it was retarded in week 3 in the CrDOFox:YFP line and even further in the CrDOFin line. The amiCrDOF #1 and #4 cultures also grew slower than the CC-4351 wild type (Fig. 3C, right). The growing capacity of the lines was further measured in LD and SD, both on rich (Tris-acetate-phosphate [TAP] NO3−) and normal (Sueoka NO3−) medium agar plates, by droplet growth monitoring (Supplemental Fig. S1D). In all conditions, decreasing or increasing CrDOF transcript levels slowed algal growth, indicating that changes in CrDOF levels affected the growing conditions of the algae.
Figure 3.
Altering CrDOF expression affects algal growth. A, CrDOF transcript levels (graphs at top) and protein (immunoblots at bottom) in CW15 and CrDOFin before and after 2 h of induction and CrDOFox:YFP lines at ZT4 in LD. The arrow marks the 120-kD band corresponding to the CrDOF:YFP fusion. B, Reduction in CrDOF transcript levels (graphs at top) and protein (immunoblots at bottom) in amiCrDOF #1 and #4 compared with CC-4351 at ZT4 in LD. *, P < 0.05. C, Growth curves (chlorophyll content) in CW15 (solid line), CrDOFin (dashed line), and CrDOFox:YFP (dotted line) at left and CC-4351 (solid line), amiCrDOF #1 (dotted line), and amiCrDOF #4 (dashed line) at right in a 4-week course. D, Venn diagrams for induced (top) or repressed (bottom) genes from the RNAseq data comparing CW15 and the intersection between CrDOFox:YFP and CrDOFin lines (Supplemental Fig. S1E) in LD (left) and SD (right).
To assess the systemic effect of CrDOF overexpression in C. reinhardtii, a transcriptomic approach was used. C. reinhardtii cells (CW15) and two overexpression lines (CrDOFin and CrDOFox:YFP) were grown in Sueoka NO3− medium in LD and SD, and RNA was extracted at ZT4 and sequenced using the NGS platform Illumina HiSeq 2000. After all quality parameters were verified, a stepwise protocol to analyze the transcriptome, using the C. reinhardtii genome as reference, was run (see “Materials and Methods”). Several software tools for read mapping (Bowtie and TopHat), transcriptome assembly (Cuffmerge and Cufflinks), and differential gene expression analysis (Cuffdiff and CummeRbund) were used. Differentially expressed genes between CW15 and the two overexpression lines were identified, and Venn diagrams representing these differences were drawn (Supplemental Fig. S1E). The RNAseq data analysis showed that CrDOF overexpression in both lines affected the expression of thousands of genes: 2,399 genes induced and 2,431 repressed in CrDOFin and 3,255 genes induced and 2,632 repressed in CrDOFox:YFP in both LD and SD. When we separately analyzed the intersection between induced and repressed genes of both overexpression lines compared with the wild type in LD and SD and created Venn diagrams (Fig. 3D), a tendency in gene expression, which was daylength dependent, emerged. CrDOF overexpression caused the specific induction of 245 genes in LD and 408 genes in SD, while it caused the specific repression on 295 genes in LD and 163 genes in SD. Therefore, the RNAseq data from the two independent overexpression lines indicated that CrDOF would function preferentially as a repressor of gene expression in LD and as an inducer in SD. These data can be further analyzed on the Web page http://viridiplantae.ibvf.csic.es/crdof_cell_cycle.html.
CrDOF Induces CrCO Expression in C. reinhardtii
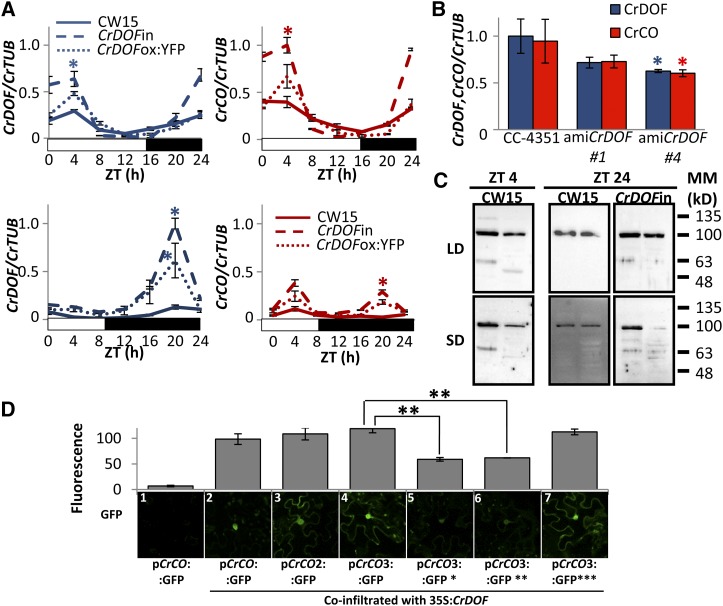
In Arabidopsis and potato (Solanum tuberosum), CDFs reduce CO expression and delay flowering (Imaizumi et al., 2005; Fornara et al., 2009; Kloosterman et al., 2013). To show that CrDOF could have a similar role in algae, CrCO expression was monitored in 24-h experiments in CW15, CrDOFin, and CrDOFox:YFP lines (Fig. 4A). Both in LD and SD, CrDOFin and CrDOFox:YFP showed an increase in CrCO expression during the morning and night. This was also observed in experiments in which CrDOFin lines were transferred from not induced (Supplemental Fig. S2A, left) to induced (Supplemental Fig. S2A, right) conditions for 2 h, resulting in a 3-fold increase in CrCO expression. Constitutive CrDOFox:YFP lines that presented a moderate activation of CrDOF expression also showed a moderate increase in CrCO expression (Supplemental Fig. S2B). On the contrary, in amiCrDOF #1 and #4 (Fig. 4B), reduced levels of CrDOF produced a reduction in CrCO mRNA levels. Thus, both incrementing and reducing CrDOF expression levels had a parallel effect on CrCO, but rather than inhibiting CO expression, as in Arabidopsis, CrDOF behaved as an activator of CrCO expression in C. reinhardtii.
Figure 4.
CrDOF induces CrCO expression. A, A 24-h course of CrDOF (blue) and CrCO (red) expression in LD (top) or SD (bottom) in CW15 (solid lines), CrDOFin (dashed lines), and CrDOFox:YFP (dotted lines). B, Levels of CrDOF (blue columns) and CrCO (red columns) mRNA in CC-4351 and amiCrDOF #1 and #4. Data are from three independent experiments ± se. C, Immunodetection of CrDOF with α-CrDOF in gel filtration fractions 8 (464 kD; left) and 9 (314 kD; right) in protein extracts from cultures in LD (top) or SD (bottom). CW15 at ZT4 (left) and both CW15 and CrDOFin at ZT24 (right) are shown. Black lines are MM markers in kD. D, GFP fluorescence measurements (top) of different pCrCO:GFP constructs in N. benthamiana cells coinfiltrated with 35S:CrDOF. Data are from 100 nuclei in three independent experiments ± se. Results from a significant experiment are shown below each graphic. *, P < 0.05; **, P < 0.01.
As we had observed 60- and 100-kD CrDOF protein bands on C. reinhardtii immunoblots, we wondered if this had a biological significance, so we performed gel filtration experiments in a Superose 12 10/300 GL (GE Healthcare) column attached to an FPLC device (GE Healthcare). Total extracts from CW15 or CrDOFin lines in LD and SD at ZT4 and ZT24 were run in the column, and the elution profile was separated by size and monitored for CrDOF presence (Fig. 4C; Supplemental Table S2). In CW15, a protein complex containing CrDOF eluted only in two fractions corresponding to MM of 464 and 314 kD (Supplemental Fig. S2, C and D). The 100-kD band was observed in all conditions, but the 60-kD protein was only detected at ZT4 in the fraction with the higher MM complex (Fig. 4C). At ZT24, the 60-kD band was absent in CW15 but was present in the CrDOFin extract in both fractions. Because CrCO expression in wild-type cells reaches its highest peak at ZT4 and in CrDOFin is induced at ZT24, we think that this correlates well with the 60-kD band representing the free, active protein that is able to modify CrCO expression. Moreover, the ratio between CrDOF and partners seems to have an important role in the complex size, as the 60-kD band presence always coincided with the higher MM complex.
The capacity to interact with itself was also investigated by bimolecular fluorescence complementation (BiFC; Supplemental Fig. S3A). A translational fusion of CrDOF to the YFP N-terminal part (YFN-CrDOF) was coinfected by Agrobacterium tumefaciens-mediated infiltration in Nicotiana benthamiana leaves together with CrDOF fused to the C-terminal part of the YFP (YFC-CrDOF). The presence of a strong yellow signal in the nucleus of the coinfiltrated N. benthamiana cells showed that CrDOF was able to interact with itself to form dimers or higher MM complexes.
To test if CrDOF could bind to the CrCO promoter, A. tumefaciens-mediated coinfiltration experiments were run. The sequence (846 bp) expanding the intergenic region between CrCO and the upstream gene (Cre06.g278158; Supplemental Fig. S3B) was cloned before the GFP coding sequence in a plant transformation vector (pCrCO:GFP). This construct did not show any expression in N. benthamiana cells using the confocal microscope (Fig. 4D, image 1), indicating that no plant factor could bind to the C. reinhardtii CrCO promoter. When a construct overexpressing CrDOF under the control of a plant constitutive promoter (35S:CrDOF) was coinfiltrated with pCrCO:GFP, a marked increase in GFP fluorescence was observed (Fig. 4D, image 2), demonstrating that CrDOF could bind to the CrCO promoter. In order to quantify the signal, more than 100 nuclei were monitored using the confocal microscope and their fluorescence registered (representing 100% signal). To identify the minimal promoter that still showed CrDOF binding, a 527-bp fragment of pCrCO2 (underlined in Supplemental Fig. S3B) and a 285-bp fragment of pCrCO3 (blue in Supplemental Fig. S3B) were cloned before the GFP coding sequence and coinfiltrated with the 35S:CrDOF construct in N. benthamiana leaves. As observed in Figure 4D (images 3 and 4), GFP fluorescence did not decrease, indicating that the minimal promoter pCrCO3 contained all sequences needed for CrDOF binding. In the 285-bp fragment, three canonical DOF AAAG-binding sites were found (gray in Supplemental Fig. S3B). The three motifs were mutated individually to confer pCrCO3:GFP*, pCrCO3:GFP**, and pCrCO3:GFP*** constructs (Supplemental Fig. S3B). Only pCrCO3:GFP* and pCrCO3:GFP** showed significant decreases in GFP fluorescence when coinfiltrated with 35S:CrDOF, indicating that the motif closer to the CrCO initial ATG codon was not involved in CrDOF binding (Fig. 4D, images 5–7). The two elements with positive binding activity conserved the (A/T)AAAG or (A)CTTT(A) extra nucleotides characteristic of DOF-binding sites (Yanagisawa and Schmidt, 1999) not present in the third site. Therefore, we identified the binding site sequence of the primitive DOF gene and showed that it recognized the same core region as in higher plants. This constitutes additional evidence of the conservation of DOF TF function during plant diversification.
CrDOF Alters Algal Cell Cycle Progression in a Photoperiod-Dependent Manner
CrCO misexpression was shown to affect synchronous division in C. reinhardtii and, thus, the capacity to synchronize cell division and growth in particular environmental and photoperiod conditions (Serrano et al., 2009). As CrDOF overexpression and artificial microRNA constructs showed growth defects and altered CrCO expression, we examined if the cell cycle was also affected in these lines. Cell cycle defects are associated with changes in size due to cell division alterations (Umen and Goodenough, 2001). When we analyzed CrDOFin and CrDOFox:YFP cells in LD by flow cytometry (Fig. 5A, top), they showed an increase in size compared with CW15, whereas in SD they were smaller. On the contrary, amiCrDOF #1 and #4 lines did not show any significant alteration in cell size in LD but showed a marked decrease in SD. Therefore, cell size in C. reinhardtii was severely affected by changes in CrDOF expression and was influenced by photoperiod.
Figure 5.
CrDOF controls the C. reinhardtii cell cycle. A, Flow cytometer cell size from algal cultures in LD (left) and SD (right) in CW15 (black), CrDOFin (dark gray), and CrDOFox:YFP (light gray) at top or CC-4351 (black), amiCrDOF #4 (dark gray), and amiCrDOF #1 (light gray) at bottom. B, Expression of CrCYCA1 (black) and CrCDKB1 (gray) in LD (top) and SD (bottom) in CW15 (solid lines), CrDOFin (dashed lines), and CrDOFox:YFP (dotted lines) in 24-h courses. Data are from three independent experiments ± se. *, P < 0.05. C, Scheme of CrDOF control over cell cycle progression in LD (left) and SD (right). Circle size represents algal size, and number of circles represents growth progression.
Cyclins and cyclin-dependent kinases are key proteins involved in cell cycle progression in eukaryotes, and this function is conserved in green microalgae (Bisova et al., 2005). In C. reinhardtii, it was shown that Cyclin A1 (CrCYCA1) and Cyclin-dependent kinase B1 (CrCDKB1) transcript levels increased when CrCO levels increased in SD in order to promote the highly synchronized mitotic divisions at the end of the day characteristic of synchronous growth (Serrano et al., 2009). In order to assess the role of CrDOF in this process, we measured the capacity to activate the expression of CrCYCA1 and CrCDKB1 in LD and SD in the inducible line. CrDOF induction in two independent lines greatly enhanced the expression of CrCYCA1 in SD (Fig. 5B, bottom left), while in LD it had the opposite effect (Fig. 5B, top left), inhibiting the small peak of expression present in CW15. CrCDKB1 expression showed a similar response, as it was significantly enhanced in SD but was strongly inhibited in LD by CrDOF overexpression (Fig. 5B, right). Therefore, CrDOF overexpression altered cell cycle progression by increasing CrCYCA1 and CrCDKB1 transcript levels in SD and decreasing them in LD. Because CrDOF activates CrCO expression, this effect may be associated with the induction in CrCO mRNA levels observed in CrDOFin during the morning (Fig. 4A), but this could not explain the inhibitory effect in LD. To confirm these results, we performed a clustering and functional enrichment using the RNAseq data from CW15, CrDOFin, and CrDOFox:YFP lines in SD and LD at ZT4. The functional enrichment of genes inhibited by CrDOF overexpression in LD showed that a significant part of the genes were involved in cell cycle progression, including CrCYCA1 and CrCDKB1 (http://viridiplantae.ibvf.csic.es/crdof_cell_cycle.html). Therefore, CrDOF inhibits cell cycle progression in LD independently of CrCO and activates its progression in SD by inducing CrCO expression (Fig. 5C). In this scenario, cells would be larger in LD, as the cell cycle is inhibited during the day (Fig. 5C, left), while in SD cells maintain a small size due to the activation of the cell cycle promoted by CrCO (Fig. 5C, right), as we had shown previously in the flow cytometer experiments.
CrDOF in Arabidopsis Delays Flowering through the Interaction with CDFs
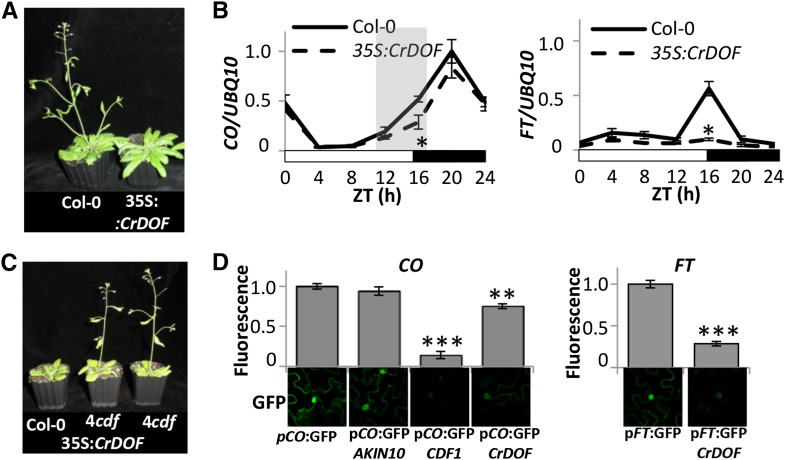
In Arabidopsis, CDFs inhibit CO expression during the morning to avoid FT misexpression outside the evening coincidence window that sets the exact synchronization of flowering by daylength. To test the capacity of CrDOF to alter flowering time in Arabidopsis, we expressed CrDOF under the control of the constitutive promoter 35S (35S:CrDOF). CrDOF heterologous expression in Arabidopsis delayed flowering in LD, 35S:CrDOF plants flowering with 10 leaves more than Columbia-0 (Col-0), and had no effect in SD (Fig. 6A; Table I). We then followed CO and FT expression during a 24-h course in LD (Fig. 6B) and SD (Supplemental Fig. S4A) in Col-0 and 35S:CrDOF plants. While CO expression was reduced during the daytime in LD, particularly during the evening (shadowed in Fig. 6B, left), the expression in SD was not affected (Supplemental Fig. S4A, top). In 35S:CrDOF plants in LD, FT expression was strongly inhibited during the whole photoperiod (Fig. 6B, right), explaining the late-flowering phenotype in LD, but in SD no effect was observed (Supplemental Fig. S4A, bottom).
Figure 6.
CrDOF expression delays flowering in Arabidopsis. A, Late-flowering phenotypes of Arabidopsis 3-week-old plants in LD expressing CrDOF under the control of the 35S promoter (right) compared with Col-0 (left). B, CO and FT 24-h mRNA levels in Col-0 and 35S:CrDOF from 14-d-old plants in LD. Data are from three independent experiments ± se. C, Flowering phenotypes of 4cdf (right), 35S:CrDOF 4cdf (middle), and Col-0 (left) plants grown in soil for 2 weeks in LD. D, Confocal images of 35S:CrDOF-infiltrated N. benthamiana cells together with CO (left) or FT (right) promoter fused to GFP are shown at bottom. CDF1 (positive) and AKIN10 (negative) coinfiltrations were used as controls. GFP fluorescence quantifications are shown at top. Data are from 100 nuclei in three independent experiments ± se. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Table I. Flowering time of Col-0, mutants, and 35S:CrDOF plants in LD and SD.
Data are shown as means ± se of 30 plants from three independent experiments scored for total leaf number at the moment of appearance of the floral bud in LD and SD. Transgenic lines described in this work are T3 plants. **P < 0.01 by Student’s t test.
CDFs work additively to repress CO and FT expression, so that in the quadruple mutant cdf1/cdf2/cdf3/cdf5 (4cdf), CO transcript levels are constitutively high (Fornara et al., 2009). We tested if CrDOF could complement the early-flowering phenotype observed in 4cdf (Fig. 6C; Table I). While the 4cdf mutant flowered earlier than Col-0 in LD and SD, ectopic expression of CrDOF in the 4cdf background had no effect on flowering time. Therefore, CrDOF had no effect on the floral transition in the absence of CDFs. This suggests that CrDOF may interact with the CDFs and enhance their capacity to inhibit CO and FT expression. To test this hypothesis, we performed BiFC experiments in N. benthamiana (Supplemental Fig. S4B). Both CDF1 and CDF2 were able to interact with each other and CDF2 with itself (Supplemental Fig. S4B, images 3 and 4), but they could also interact with CrDOF (Supplemental Fig. S4B, images 1 and 2). When we used CrDOF-specific antibodies in Col-0 nuclear extracts, we could detect several bands around and below 60 kD that could account for Arabidopsis DOF TFs, including CDFs, which present smaller size than other DOFs (Supplemental Fig. S4C, left). In fact, these signals increased in extracts from ZT0 to ZT8, where the maximum amount of CDFs is supposed to accumulate (Fornara et al., 2009). In 35S:CrDOF plants, the immunodetection of the 60-kD band increased significantly both in Col-0 and in the 4cdf mutant background (Supplemental Fig. S4C, right), due to the accumulation of CrDOF, but we could never detect the 100-kD band that we had observed in C. reinhardtii. The exclusive presence of the 60-kD band in the recombinant E. coli cultures (Fig. 2C), the detection of the 60- and 100-kD bands in the gel filtration experiments in C. reinhardtii (Fig. 4C), and the lack of a 100-kD band in Arabidopsis strongly suggest that the higher MM must be a specific characteristic of algae DOFs and that the active form must be the 60-kD protein. Using a 35S:GFP:CrDOF construct, we could also show the nuclear localization of CrDOF in Arabidopsis (Supplemental Fig. S4D).
To further demonstrate the effect of CrDOF over CO and FT expression, we infiltrated N. benthamiana cells with A. tumefaciens carrying constructs expressing GFP under the control of the CO (pCO:GFP) and FT (pFT:GFP) promoters (Fig. 6D). Both constructs showed a clear nuclear GFP signal when infiltrated alone. When a negative control (AKIN10) was cotransformed with pCO:GFP, no reduction in GFP fluorescence was observed. As expected, cotransformation of pCO:GFP with a construct ectopically expressing CDF1 produced a dramatic decrease in GFP fluorescence. Ectopic coexpression of CrDOF had a lower, but still significant, effect in GFP fluorescence. In a similar way to what was shown in the expression experiments, CrDOF also reduced GFP fluorescence when GFP was driven by the FT promoter (Fig. 6D, right), suggesting a role for CrDOF (and CDFs) in FT expression.
DISCUSSION
DOF proteins are present in the genomes of higher plants and unicellular green algae (chlorophytes) and are absent from all other algal divisions and, in fact, from any other prokaryote or non-green-lineage eukaryote (Moreno-Risueno et al., 2007). This way, we could identify CrDOF sequences in several chlorophyte orders, such as Chamydomonadales (C. reinhardtii and Volvox spp.), Trebouxiophyceae (Chlorella and Coccomyxa spp.), Mamiellales (Ostreococcus, Bathyococcus, and Micromonas spp.), and Chlorodendrales (Tetraselmis spp.). CrDOF sequences were absent from other major algal divisions, such as the primary endosymbionts rhodophytes (Cyanidioschyzon spp.) and glaucophytes (Cyanophora spp.), but also from other secondary endosymbionts, such as euglenophytes (Euglena spp.), haptophytes (Emiliania spp.), heteroconts (Phaeodactylum, Thalassiosira, and Navicula spp.), cryptophytes (Guillardia spp.), chloraracniophytes (Bigelowiella spp.), and dynophytes (Symbiodinium spp.). Therefore, they constitute a good model to study the evolution of specific regulatory pathways in plants. DOFs are found as single-copy genes in the genome of chlorophytes, but in P. patens, we have identified 16 genes homologous to CrDOF, while others have identified a family of 19 members (Shigyo et al., 2007). In rice and maize, around 31 and 10 DOF genes have been identified, respectively, while in Arabidopsis and Populus spp., the numbers are even higher: 36 and 61, respectively (Noguero et al., 2013). Therefore, this family has increased from a single-copy gene in algae to a multicopy family with numerous functions, including the control of photoperiodic flowering, in angiosperms. Therefore, the study of the ancestral function of the algal DOF was important to understand how different regulatory pathways are controlled, being the photoperiod pathway of special interest.
In Arabidopsis, potato, and rice, several DOF genes have been classified as CDFs because they are regulated by the clock and at the same time regulate photoperiodic flowering (Imaizumi et al., 2005; Fornara et al., 2009; Li et al., 2009; Kloosterman et al., 2013). In its domain structure, ancient DOF proteins contained a single DOF domain with an NLS in its N-terminal part. This structure is repeatedly found in green microalgae. In the course of evolution, as observed in P. patens DOFs, different domains were incorporated to the basic pattern that added new regulatory elements to the protein and recruited novel regulators to the pathway (Fig. 1C). As a culmination to this evolutionary process, higher plant CDFs possess an established structure that is the combination of several domains present in bryophytes, such as the GI- and FKF1-binding elements, responsible for their complex posttranslational regulation. Therefore, the evolution of DOF factors is a good example of the innovation-amplification-divergence model of evolution by gene duplication described also for the family of COL genes in photosynthetic eukaryotes (Romero-Campero et al., 2013).
DOF TFs have multiple roles in plants, from the control of nitrogen and carbon metabolic balance, as in maize DOF1 (Yanagisawa, 2000), to the control of photoperiodic flowering by Arabidopsis CDFs (Imaizumi et al., 2005; Fornara et al., 2009). At the molecular level, they function as DNA-binding proteins that regulate transcription, either inducing or repressing gene expression (Noguero et al., 2013). Here, we have shown that the C. reinhardtii single-copy DOF gene already had this double function. We provide evidence for gene induction at the individual level, such as CrCO or the repression of CrCYCA1 and CrCDKB1 in CrDOF overexpression lines, but we also provide systemic-level evidence using RNAseq data from wild-type and CrDOF overexpression lines under different photoperiod conditions. We have further shown that CrDOF preferentially induces or represses gene expression depending on photoperiod, such that it is mainly an activator of gene expression in SD and an inhibitor in LD. Indeed, both functions can be recruited to achieve the same goal, such as the regulation of synchronous growth. In C. reinhardtii and other microalgae, cell division is synchronized to the cell cycle (Bišová and Zachleder, 2014), and this is affected by daylength. This way, in LD or LL, with an excess of resources, algae can divide several times and no peak in cell cycle genes is detected. On the contrary, in SD, in stress conditions or nutrient limitation, algae divide once a day and the cell cycle genes peak at dusk (Serrano et al., 2009). Therefore, we can now compare CrDOF overexpression line data with CrCO transgenic line data (Serrano et al., 2009), where CrCO positively regulated CrCYCA1 and CrCDKB1 transcript levels in SD conditions. Thus, we propose that CrDOF induces CrCO expression in SD at the end of the day, promoting the up-regulation of cell cycle factors such as CrCYCA1 and CrCDKB1 (Fig. 7). This peak of expression activates the cell cycle and ensures cell division in that particular time, allowing all algae in the culture to divide synchronously. In LD, on the contrary, CrDOF inhibits the expression of cell cycle genes in specific conditions when no synchronous division is necessary, thus allowing indeterminate cell division throughout the day. In fact, in Arabidopsis, the DOF factor OBF BINDING PROTEIN1 is involved in the control of the cell cycle by inducing the expression of the cyclin CYCD3.3, and its overexpression causes the reduction of G1 phase, accelerating growth (Skirycz et al., 2008), suggesting that this algal DOF function is conserved in plants.
Figure 7.
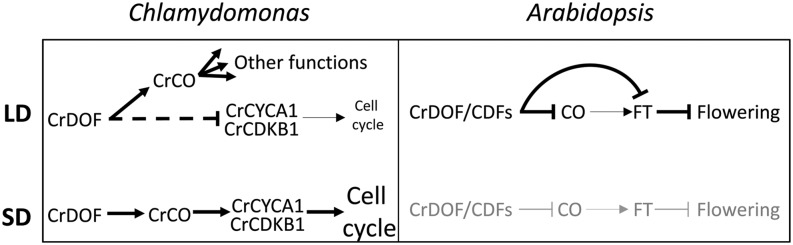
Model for CrDOF function in C. reinhardtii (left) and Arabidopsis (right). In C. reinhardtii, CrDOF in LD is able to induce CrCO expression by binding to its promoter and, moreover, slow down cell cycle progression by inhibiting, directly or indirectly (dashed T line), CrCYCA1 and CrCDKB1 expression. In SD, CrDOF induces cell cycle progression by promoting CrCO expression. In Arabidopsis, the CrDOF/CDF complex delays flowering time by inhibiting CO and FT expression in LD. In SD (gray), CO protein is not present and cannot induce FT expression, so CrDOF ectopic expression does not affect flowering time. Arrows indicate inductive actions, and T lines indicate inhibiting actions.
Our data suggest that a posttranslational regulatory step is necessary to explain CrDOF function, as CrDOF can bind to DNA as a single 60-kD form, but it is mostly detected as a 100-kD band in C. reinhardtii extracts. We propose that the most plausible explanation is that a partner, or more than one, may bind CrDOF in a protein complex. Bound CrDOF would be unable to promote or inhibit gene expression. In the CrDOF-induced line, this regulation is lost, probably due to an excess of CrDOF in the complex, so that free CrDOF could then activate CrCO expression. The experiments in N. benthamiana cells that show that CrDOF can bind directly to the CrCO promoter, the absence of the 100-kD band in plant immunodetection, and the data from the chromatographic experiments in C. reinhardtii, where the 60-kD band appears only when CrDOF induces CrCO expression, suggest that CrDOF is active whenever the 60-kD band is detected. Therefore, we propose that the CrCO mRNA level induction by CrDOF in algae is a direct effect caused by DOF binding to its promoter.
In Arabidopsis, CrDOF overexpression delays flowering by reducing CO and FT levels exclusively in LD and not in SD, reflecting a strict photoperiodic response. In our model, as CrDOF cannot alter flowering time in the 4cdf mutant, it is possible that CrDOF could act in a complex with the CDFs, protecting them from the proteasome degradation triggered by GI-FKF, as CrDOF does not present the binding domains for these proteins (Fig. 7, right). In N. benthamiana experiments, CrDOF also inhibit CO and FT expression, probably through the interaction with the N. benthamiana CDF homologs. This association between CDFs and CrDOF could have interesting applications in plant biotechnology to modify flowering time.
CONCLUSION
It is interesting that both COL and DOF genes seem to have suffered a similar evolutionary process and often cluster together in gene coexpression networks constructed from different organisms representing different steps of plant evolution (Romero-Campero et al., 2013). Both CO and CDF proteins are part of the photoperiod pathway. It is likely that other partners of this route (i.e. FLOWERING BASIC HELIX-LOOP-HELIX [Ito et al., 2012], FKF, and CONSTITUTIVE PHOTOMORPHOGENESIS PROTEIN1) followed a similar coevolution process. If this was the case and, further, could be demonstrated for other routes, it may constitute the basis of a general evolutionarily conserved process in which all members of a regulatory pathway (toolkit) evolve together, restrained and connected by the functional links established early in evolution in the original regulatory network (Romero-Campero et al., 2013; Della Pina et al., 2014; Ichihashi et al., 2014). Our demonstration that the DOF-CO module is conserved from algae to higher plants provides direct evidence that this effect may indeed be taking place. The study of these conserved toolkits throughout the plant evolutionary lineage may provide important hints to understand the processes that allowed the differentiation of higher plants from simple unicellular algae.
MATERIALS AND METHODS
Biological Material and Growth Conditions
Chlamydomonas reinhardtii wild types CW15 (Davies and Plaskitt, 1971) and CC-4351 (from René Matagne, University of Liège, via Michael Schroda, Max Planck Institute of Molecular Plant Physiology) and transgenic lines were grown in flasks with minimal Sueoka medium (Sueoka et al., 1967) or rich TAP medium (Harris, 1989) in LD or SD at 50-µE light intensity with temperatures ranging from 22°C (day) to 18°C (night). For induction of the nia2 promoter, algal cells grown to exponential phase in Sueoka medium supplemented with ammonium were harvested by centrifugation (4 min at 3,200g) and suspended in Sueoka medium supplemented with nitrate.
Arabidopsis (Arabidopsis thaliana) plants were grown in soil for phenotypic analysis or in Murashige and Skoog agar medium supplemented with 1% (w/v) Suc plates for quantitative PCR (QPCR) assays. In both cases, seeds were previously incubated during 4 d at 4°C before sowing. Plants were grown in humidity-controlled (80%) chambers with a 100-µE light intensity and temperatures ranging from 22°C (day) to 18°C (night). Different photoperiods were used: 16 h of light/8 h of dark (LD) and 8 h of light/16 h of dark (SD).
Cloning and Analysis of CrDOF
The complete CrDOF ORF was obtained from the cDNA Collection of the Kazusa DNA Research Centre (http://www.kazusa.or.jp). Sequencing of the cDNA showed 100% identity with the predicted locus Cre12.g521150 (positions 4,425,725 to 4,430,320) from Phytozome version 9.1. The CrDOF deduced protein was 625 amino acids long and contained a DOF-type zinc finger domain (Yanagisawa, 1997; Moreno-Risueno et al., 2007).
Phylogenetic Analysis and Identification of Conserved Motifs
Evolutionary relationships among DOF proteins from the green algae C. reinhardtii, Volvox carteri, Ostreococcus tauri, and Micromonas pusilla, the bryophyte Physcomitrella patens, the monocots rice (Oryza sativa) and maize (Zea mays), and the dicots Arabidopsis and poplar (Populus trichocarpa) were analyzed using the CrDOF full-length amino acid sequence as a query to detect protein homologs in the Phytozome version 9.1 database. All sequences were aligned using the program MUSCLE (Edgar, 2004), and a phylogenetic tree was generated applying the neighbor-joining algorithm (Saitou and Nei, 1987) with the substitution model JTT + G 0.54 (Jones et al., 1992). The number of bootstrap replicates was 500. Phylogenetic analyses were conducted with MEGA5 (Tamura et al., 2011). Accession numbers of the sequences used in the alignment are shown in Supplemental Table S1. A domain analysis of the same 84 DOF proteins in the phylogenetic tree was done using the MEME software (http://meme.nbcr.net/meme/; Bailey et al., 2009). The motif domains were characterized using the Conserved Domain Search Service (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Further identification of the GI- and FKF1-binding domains was done as described (Kloosterman et al., 2013).
Phenotypic Analysis
Growth Curve and Drop Test
Algal cultures were grown to stationary phase in LD conditions and then diluted to 0.5 µg mL−1 chlorophyll with new medium. Culture growth was monitored by chlorophyll measurements once per week for 1 month. For drop tests, the culture in the stationary phase was diluted to 4 to 5 × 106 cells mL−1, and three consecutive 1:10 dilutions were made. Three-microliter drops were grown on agar plates containing Sueoka NO3− or TAP NO3− medium and incubated in LD or SD.
Flow Cytometry
Wild-type and transgenic line samples were taken at ZT4 in LD and SD conditions in Sueoka medium supplemented with NO3− and analyzed on a BD Influx Cell Sorter flow cytometer (Becton Dickinson) equipped with a laser with a 488-nm line at 200 mW and analyzed with BD FACS software.
Analysis of Flowering Time
Flowering time was analyzed in LD and SD conditions in controlled-environment cabinets by scoring the number of rosette (excluding cotyledons) and cauline leaves. Data are from media of at least 30 individuals ± se.
Algal and Plant Transformation
C. reinhardtii
For C. reinhardtii nuclear transformation, a modification of the electroporation protocol from Shimogawara et al. (1998) was used. In brief, about 1 to 5 × 106 cells mL−1 were harvested by centrifugation at 1,500g and suspended in TAP medium supplemented with 60 mm Suc in 1:100th of the original volume. A total of 250 µL of cell suspension and 1 µg of the plasmid were placed into a 0.4-cm gapped cuvette and incubated for 30 min on ice. After an 800-V pulse (15 µF and no shunt resistance) in an electroporator, cells were transferred to 10 mL of TAP medium supplemented with 60 mm Suc and incubated overnight with moderate shaking and light. Cells were harvested by centrifugation, resuspended in 700 µL of TAP medium, and spread into selective TAP agar plates containing hygromycin at 25 µg mL−1. Colonies started to appear after 1 week. For the nitrate-inducible CrDOF expression vector construction, the Gateway-modified version of the pnia2 vector (Serrano et al., 2009) was used. For the constitutive CrDOF vector expression construction, direct gene synthesis from Invitrogen was used. In silico sequencing of the Hsp70A/RbcS2 constitutive promoter, Gateway cassette B, and the 3′-untranslated region of the Rubisco terminator was performed, and restriction enzyme sites present in the Gateway cassette and terminator were changed without modifying the codon use. YFP and hygromycin resistance were cloned into EcoRV/BamHI and KpnI restriction sites, respectively. YFP was adapted to the C. reinhardtii codon preference (Matsuo and Ishiura, 2010).
Arabidopsis
For Arabidopsis Col-0 or 4cdf transformation, the Agrobacterium tumefaciens-mediated floral dip protocol was used. CrDOF cDNA was amplified by PCR (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGGTAGACGGTGGTTCG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCACCTAGCACCCGAGTAAGC-3′ with stop codon and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCCTAGCACCCGAGTAAGCGGC-3′ without stop codon were used for C-terminal fusions) and cloned into the pDONOR 207 plasmid. Finally, CrDOF was cloned in the pEG100 vector to obtain the 35S:CrDOF construct and in the pMDC43 vector to obtain the 35S:GFP:CrDOF construct. Arabidopsis Col-0 plants were transformed with these constructs and plants selected for antibiotic resistance. The 4cdf mutant plants were transformed with the 35S:CrDOF construct, and positive plants were selected by genome PCR tests using 5′-ACTGCAAGACGTGTCAGCG-3′ and 5′-GCCAACTCACTGTTGAACTGC-3′ primers.
RNA Techniques
Expression Analysis by QPCR
Total RNA from C. reinhardtii cells (20 mL of an exponential phase curve) or Arabidopsis seedlings (0.1 g of leaf tissue) was isolated by the Trizol (Invitrogen) procedure following the recommendations of the manufacturer. The final RNA sample was suspended in 30 µL of diethyl pyrocarbonate water, quantified in an ND-1000 spectrophotometer (Nanodrop), and stored at −80°C. One microgram of RNA was used to synthesize cDNA employing the QuantitecReverse Kit (Qiagen), diluted to a final concentration of 10 ng µL−1, and stored at −20°C until QPCR was performed. Specific primers to amplify the 3′ region of each gene analyzed, plus CrTUBULIN and UBIQUITIN10 (Supplemental Table S3) as housekeeping genes, were designed with the Oligo Analyzer program (Integrated DNA Technologies; www.idtdna.com). QPCR was performed with the iQTM5 Multicolor Real-Time PCR Detection System (Bio-Rad) in 10-µL reactions: primer concentration of 0.2 µm, 10 ng of cDNA, and 5-µL SensiFAST SYBR & Fluorescein Kit (Bioline). Each sample was measured in triplicate. The QPCR program consisted of (1) one cycle of 95°C for 2 min; (2) 40 cycles of 95°C for 5 s, 60°C for 10 s, and 72°C for 6 s; and (3) one cycle of 72°C for 6 s. Fluorescence was measured at the end of each extension step, and melting curve analysis was performed between 55°C and 95°C. The initial concentrations of candidate and reference genes were calculated by means of LingRegPCR software version 11.0 (Ruijter et al., 2009). Normalized data were calculated by dividing the average of three replicates of each sample of the candidate and reference genes.
Transcriptomic Analysis by RNAseq
The experimental design consisted of two replicates for each genotype (CrDOFin, CrDOFox:GFP, and wild-type CW15) and growth condition (LD and SD). RNA was extracted from each sample using Trizol. Library preparation was carried out following the manufacturer’s recommendations. Sequencing of RNA libraries was performed with the Illumina HiSeq 2000 sequencer, yielding approximately 40 million 50-bp-long reads for each sample. The software package FastQC (Andrews, 2014) was used for quality control. All sequencing samples were of high quality, and no preprocessing of the reads was required to remove low-quality reads or read fragments. The C. reinhardtii reference genome version 5.3 (Merchant et al., 2007) and annotation were downloaded from the Phytozome database (Goodstein et al., 2012). Mapping of reads to the reference genome, transcript assembly, and differential expression were performed with the software tools Bowtie, TopHat, and Cufflinks (Trapnell et al., 2012) using default parameters. The R package from Bioconductor CummeRbund (Goff et al., 2012) was used for subsequent analysis and graphical representation of the results. Differentially expressed genes were selected as those exhibiting an expression fold change greater than 2 when compared with the wild-type CW15 and P < 0.05. Venn diagrams comparing the different sets of differentially expressed genes were generated with Venn Diagram Plotter (http://omics.pnl.gov/software/venn-diagram-plotter). The RNAseq raw data generated in this study are publicly available from the European Nucleotide Archive database identified with accession number PRJEB6682 (http://www.ebi.ac.uk/ena/data/view/PRJEB6682).
Protein and Immunological Techniques
Purification of Recombinant CrDOF
The CrDOF cDNA fragment was amplified by PCR using specific primers designed with NdeI/BamHI restriction enzyme adaptors (5′-gggcatATGGTAGACGGTGGTTCGCGTG-3′ and 5′-cccggatccTCACCTAGCACCCGAGTAAGC-3′). The lower case portion of the sequences are three bases plus the restriction sequence (total or partial) introduced for PCR amplification and cloning. CrDOF was cloned into pET19b vector, which incorporates an N-terminal His tail (His-CrDOF), and transformed into Escherichia coli BL21 strain. Ten milliliters of an overnight culture of BL21 transformed with His-CrDOF was added to 1 L of Luria-Bertani medium containing 100 µg mL−1 ampicillin and grown at 30°C until the A600 reached 0.7. Recombinant gene expression was induced by the addition of 1 mm isopropylthio-β-galactoside and incubation during 4 h at 30°C. After centrifugation, the pellet was resuspended in lysis buffer (50 mm Tris-HCl, pH 8, 50 mm KCl, 1 mm EDTA, pH 8, 1 mm DTT, 10 mm MgCl2, and 10% [v/v] glycerol) to 3 mL g−1 cells and broken by sonic disruption. After centrifugation, the pellet was solubilized with buffer IB (50 mm Tris-HCl, pH 8, 10 mm EDTA, pH 8, 0.5% [v/v] Triton X-100, and 0.1 mm phenylmethylsulfonyl fluoride [PMSF]), washed twice in the same buffer, and once with TALON K buffer (0.05 m phosphate buffer, pH 8, and 0.3 m KCl). Between each wash, the pellet was sonicated and centrifuged. Finally, the pellet was suspended in TALON buffer including 6 m guanidine chlorhydrate and incubated for 2 h. After this period, the protein extract was disrupted by sonication and centrifuged, and the supernatant was used to purify CrDOF employing BD TALON resin following the recommendations of the manufacturer in denaturing conditions (the presence of 6 m guanidine chlorhydrate). After elution, proteins were precipitated with 100% (v/v) ethanol for 30 min at 4°C. The pellet was washed twice (0.3 m guanidine chlorhydrate in 95% [v/v] ethanol and then 90% [v/v] ethanol). Finally, the pellet was suspended in SDS-PAGE buffer and separated by electrophoresis on 10% (w/v) polyacrylamide gels. One milligram of purified protein was used to produce antibodies.
Protein Extraction and Immunodetection
Cells from 20 mL of exponentially growing C. reinhardtii cultures were centrifuged and suspended in 3 mL g−1 extraction buffer (50 mm Tris-HCl, pH 7.5, 1 mm PMSF, 1 mm DTT, quimostatin, antipain, and leupeptin at 5 µg mL−1, 10 mm β-mercaptoethanol, and 1:1,000 Sigma plant protease inhibitor cocktail). Algae were broken by two cycles of slow freezing to −80°C followed by thawing to room temperature. Cell extracts were centrifuged at 15,000g for 30 min at 4°C, and supernatant was collected. Protein content was estimated by the method of Bradford (1976) using ovalbumin as a standard. Samples were run on SDS-PAGE 10% (w/v) polyacrylamide gels and transferred to nitrocellulose membranes using the Trans-BlotTurboTransfer System (Bio-Rad) according to the manufacturer’s instructions. Western-blot analysis was performed using specific antibodies raised against CrDOF recombinant protein. To disaggregate the CrDOF complex, total protein was processed by different treatments: 8 m urea, 0.1 m DTT, 8 m urea with 0.1 m DTT, and 6 m guanidine chlorhydrate.
Chromatography
Total protein from CW15 and CrDOFin cultures grown in Sueoka medium supplemented with NO3− at LD and SD was extracted as above. One milligram of protein was separated on a Superose 12 10/300 GL column (GE Healthcare). One-milliliter fractions were collected, precipitated with 10% (w/v) TCA, and run on SDS-PAGE gels. Western-blot analysis was performed using anti-CrDOF antibodies. Protein complex MM was calculated using the Gel Filtration Markers Kit (Sigma MWGF1000; Supplemental Fig. S2D).
Nuclear and Protein Extraction
Nuclear isolation of Col-0 and 35S:CrDOF (ZT0 and ZT8) was done as described (Lazaro et al., 2012). Protein extraction was carried out in native conditions using a high-salt buffer (1.6 m KCl, 50 mm HEPES, pH 8, 3 mm MgCl2, 5 mm DTT, 1% [v/v] glycerol, and 1 mm PMSF) and run on SDS-PAGE gels. Western-blot analysis was performed using anti-CrDOF antibodies.
Transitory Expression in Nicotiana benthamiana
BiFC Experiments
To verify CrDOF protein interactions in vivo, CrDOF, CDF1, and CDF2 complete ORFs were cloned in pYFN43 and pYFC43 to produce fusions to the YTP N-terminal part (YFN-CrDOF, YFN-CDF1, and YFN-CDF2) as well as to the YFP C-terminal part (YFC-CrDOF, YFC-CDF1, and YFC-CDF2). Specific primers were used for each gene (CrDOF, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGGTAGACGGTGGTTCG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCCTAGCACCCGAGTAAGCGGC-3′; CDF1, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGCTGGAAACTAAAGATCCTGCG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCACATCTGCTCATGGAAATTGAT-3′; and CDF2, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGGCTGATCCGGCGATTAAGCTC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTATGAGCTCTCATGGAAGTTTGC-3′). These constructs were introduced into A. tumefaciens strain GVG3101 pmp90. Four-week-old N. benthamiana plants were inoculated with the following combinations: YFN-CrDOF and YFC-CrDOF, YFN-CrDOF and YFC-CDF1, YFN-CrDOF and YFC-CDF2, and YFN-CDF2 and YFC-CDF2. As negative controls, pairs of YFC-CrDOF, YFC-CDF1, and YFC-CDF2 with YFN-AKIN10 were used. As positive controls, N- and C-terminal parts of AKIN10 were used, following the protocols described previously (Voinnet et al., 2003). Fluorescent interactions were visualized with a Leica TCS SP2/DMRE confocal microscope using an excitation wavelength of 514 nm.
CrDOF-Binding Assays
In order to assess if CrDOF was able to bind to CrCO, CO, and FT promoters, a similar transitory expression experiment in N. benthamiana leaves was used. In these experiments, 864 nucleotides from the CrCO promoter (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCATGGTGCAGGCATGCCGTGGCTCGGCAAG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCTCGCGTATAGAGGTGTGGT-3′), 1 kb from the AtCO promoter (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCATGGTAGTATAGAGTATCATCATAAACCC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAATAACTCAGATGTAGTAAGTTTG-3′), and 1 kb from the AtFT promoter (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCATGGATTATATGTATAGATAGATTACCG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTTTGATCTTGAACAAACAGG-3′) were cloned first in pDONOR207 and then in pMDC110, which generates C-terminal fusions to GFP (pCrCO:GFP, pCO:GFP, and pFT:GFP). N. benthamiana leaves were inoculated with these constructs, while another set of N. benthamiana leaves were coinoculated with these constructs and 35S:CrDOF. GFP was visualized at 488-nm wavelength. Three independent inoculations were made for each studied condition. For each inoculation, three leaf fragments of the same size were analyzed (total of nine leaf fragments in three biological replicates), in which 11 or 12 randomly picked nuclei with positive signal were chosen, and GFP fluorescence was measured using MBF ImageJ. Therefore, signals from more than 100 nuclei were quantified per condition. Two versions of pCrCO, including 527 nucleotides (pCrCO2) and 285 nucleotides (pCrCO3), were constructed. Mutations at the binding sites were generated with the Muta-Direct Site-Directed Mutagenesis Kit from iNtRON Biotechnology following the instructions of the manufacturer.
C. reinhardtii Microscopy
CW15 and transgenic lines (CrDOFox:YFP) were observed using a confocal microscope (Leica TCS SP2/DMRE) for nuclear localization experiments. To visualize C. reinhardtii nuclei, SYTOBlue45 Fluorescent Nucleic Acid Stains (Molecular Probes) were used. Algae were grown in SD conditions in Sueoka medium supplemented with 10 mm NO3− until lag phase (3–4 µg mL−1 chlorophyll). One milliliter was collected by centrifugation (4 min, 5,500g) and suspended in 1 mL of Tris-buffered saline. One microliter of SYTOBlue45 and 1 and 5 µL of 10% (v/v) Triton X-100 for CW15 cells and transgenic lines, respectively, were added. After incubation for 10 min, cells were centrifuged and suspended in 100 µL of the same buffer. Finally, 3 µL of cells was mixed with 10 µL of 1.2% (w/v) low point fusion agarose at 30°C. The wavelengths used were 514 nm for YFP and 458 nm for SYTOBlue45.
Statistical Analysis
The statistical data presented in the figures and table are marked with asterisks and are means ± se of three biological experiments or two biological experiments. The statistical significance between means of the different samples was calculated using a two-tailed Student’s t test. Differences observed were considered statistically significant at P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Protein characterization and phenotypic analysis of CrDOF.
Supplemental Figure S2. CrDOF functional analysis.
Supplemental Figure S3. CrDOF dimerization and DNA-binding activities.
Supplemental Figure S4. Effect of CrDOF expression in Arabidopsis.
Supplemental Table S1. DOF gene family in plants.
Supplemental Table S2. Presence of CrDOF protein complex in different conditions and lines.
Supplemental Table S3. Primers used in QPCR amplifications.
Supplementary Material
Acknowledgments
We thank Fabio Fornara (University of Milan) for the 4cdf mutant and Alicia Orea for help at the Instituto de Bioquímica Vegetal y Fotosíntesis microscopy/cytometry service.
Glossary
- LD
long days
- TF
transcription factor
- SD
short days
- cDNA
complementary DNA
- ORF
open reading frame
- RNAseq
RNA sequencing
- NLS
nuclear localization signal
- LL
continuous light
- DD
continuous dark
- MM
molecular mass
- DTT
dithiothreitol
- TAP
Tris-acetate-phosphate
- BiFC
bimolecular fluorescence complementation
- Col-0
Columbia-0
- QPCR
quantitative PCR
- PMSF
phenylmethylsulfonyl fluoride
Footnotes
This work was supported by Ministerio de Economía y Competitividad (project nos. BIO2011–28847–C02–00 and BIO2014–52425–P and Juan de la Cierva program funding to F.J.R.-C.), by the Junta de Andalucía (grant no. P08–AGR–03582), by the European Regional Development Fund, and by the Consejo Superior de Investigaciones Científicas-Junta de Ampliación de Estudios and the European Union European Social Fund (fellowship to E.L.-R.).
Articles can be viewed without a subscription.
References
- Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Andrews S. (2014) FastQC a quality-control tool for high-throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (January 2015)
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisova K, Krylov DM, Umen JG (2005) Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiol 137: 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bišová K, Zachleder V (2014) Cell-cycle regulation in green algae dividing by multiple fission. J Exp Bot 65: 2585–2602 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Camargo A, Llamas A, Schnell RA, Higuera JJ, González-Ballester D, Lefebvre PA, Fernández E, Galván A (2007) Nitrate signaling by the regulatory gene NIT2 in Chlamydomonas. Plant Cell 19: 3491–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Davies DR, Plaskitt A (1971) Genetical and structural analyses of cell-wall formation in Chlamydomonas reinhardtii. Genet Res 17: 33–43 [Google Scholar]
- Della Pina S, Souer E, Koes R (2014) Arguments in the evo-devo debate: say it with flowers! J Exp Bot 65: 2231–2242 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KCS, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Goff L, Trapnell C, Kelley D (2013) cummeRbund: analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. R package version 2.10.0. http://www.bioconductor.org/ (January 2015) [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberti G, Papi M, Bellucci L, Ricci I, Bouchez D, Camilleri C, Costantino P, Vittorioso P (2002) Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14: 1253–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Source Book. Academic Press, San Diego [Google Scholar]
- Hernando-Amado S, González-Calle V, Carbonero P, Barrero-Sicilia C (2012) The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biol 12: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y, Aguilar-Martínez JA, Farhi M, Chitwood DH, Kumar R, Millon LV, Peng J, Maloof JN, Sinha NR (2014) Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc Natl Acad Sci USA 111: E2616–E2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T (2012) FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci USA 109: 3582–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Kloosterman B, Abelenda JA, Gomez MdelM, Oortwijn M, de Boer JM, Kowitwanich K, Horvath BM, van Eck HJ, Smaczniak C, Prat S, et al. (2013) Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495: 246–250 [DOI] [PubMed] [Google Scholar]
- Kubista H, Edelbauer H, Boehm S (2004) Evidence for structural and functional diversity among SDS-resistant SNARE complexes in neuroendocrine cells. J Cell Sci 117: 955–966 [DOI] [PubMed] [Google Scholar]
- Lazaro A, Valverde F, Piñeiro M, Jarillo JA (2012) The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Yang C, Li X, Gan Q, Zhao X, Zhu L (2009) Functional characterization of rice OsDof12. Planta 229: 1159–1169 [DOI] [PubMed] [Google Scholar]
- Lüders J, Pyrowolakis G, Jentsch S (2003) The ubiquitin-like protein HUB1 forms SDS-resistant complexes with cellular proteins in the absence of ATP. EMBO Rep 4: 1169–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Ishiura M (2010) New insights into the circadian clock in Chlamydomonas. Int Rev Cell Mol Biol 280: 281–314 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D (2009) Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J 58: 165–174 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Martínez M, Vicente-Carbajosa J, Carbonero P (2007) The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol Genet Genomics 277: 379–390 [DOI] [PubMed] [Google Scholar]
- Noguero M, Atif RM, Ochatt S, Thompson RD (2013) The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci 209: 32–45 [DOI] [PubMed] [Google Scholar]
- Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, Vittorioso P (2000) Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev 14: 28–33 [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Romero-Campero FJ, Lucas-Reina E, Said FE, Romero JM, Valverde F (2013) A contribution to the study of plant development evolution based on gene co-expression networks. Front Plant Sci 4: 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AF (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Serrano G, Herrera-Palau R, Romero JM, Serrano A, Coupland G, Valverde F (2009) Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr Biol 19: 359–368 [DOI] [PubMed] [Google Scholar]
- Shigyo M, Tabei N, Yoneyama T, Yanagisawa S (2007) Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant Cell Physiol 48: 179–185 [DOI] [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, Usuda H (1998) High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P (2001) A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229 [DOI] [PubMed] [Google Scholar]
- Skirycz A, Radziejwoski A, Busch W, Hannah MA, Czeszejko J, Kwaśniewski M, Zanor MI, Lohmann JU, De Veylder L, Witt I, et al. (2008) The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J 56: 779–792 [DOI] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N, Chiang KS, Kates RJ (1967) Deoxyribonucleic acid replication in meiosis of Chlamydomonas reinhardtii. I. Isotopic transfer experiments with a strain producing eight zoospores. J Mol Biol 25: 47–66 [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Goodenough UW (2001) Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev 15: 1652–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F. (2011) CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J Exp Bot 62: 2453–2463 [DOI] [PubMed] [Google Scholar]
- Varet A, Hause B, Hause G, Scheel D, Lee J (2003) The Arabidopsis NHL3 gene encodes a plasma membrane protein and its overexpression correlates with increased resistance to Pseudomonas syringae pv. tomato DC3000. Plant Physiol 132: 2023–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ (1997) A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci USA 94: 7685–7690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. (1997) Dof DNA-binding domains of plant transcription factors contribute to multiple protein-protein interactions. Eur J Biochem 250: 403–410 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. (2000) Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J 21: 281–288 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Schmidt RJ (1999) Diversity and similarity among recognition sequences of Dof transcription factors. Plant J 17: 209–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.