Ubiquitin attachment to nonlysine sites on a transcriptional repressor implies a diversity in ubiquitination linkages.
Abstract
Although many ubiquitin-proteasome substrates have been characterized in plants, very little is known about the corresponding ubiquitin attachment(s) underlying regulated proteolysis. Current dogma asserts that ubiquitin is typically covalently attached to a substrate through an isopeptide bond between the ubiquitin carboxy terminus and a substrate lysyl amino group. However, nonlysine (non-Lys) ubiquitin attachment has been observed in other eukaryotes, including the N terminus, cysteine, and serine/threonine modification. Here, we investigate site(s) of ubiquitin attachment on indole-3-acetic acid1 (IAA1), a short-lived Arabidopsis (Arabidopsis thaliana) Auxin/indole-3-acetic acid (Aux/IAA) family member. Most Aux/IAA proteins function as negative regulators of auxin responses and are targeted for degradation after ubiquitination by the ubiquitin ligase SCFTIR1/AFB (for S-Phase Kinase-Associated Protein1, Cullin, F-box [SCF] with Transport Inhibitor Response1 [TIR1]/Auxin Signaling F-box [AFB]) by an interaction directly facilitated by auxin. Surprisingly, using a Histidine-Hemaglutinin (HIS6x-HA3x) epitope-tagged version expressed in vivo, Lys-less IAA1 was ubiquitinated and rapidly degraded in vivo. Lys-substituted versions of IAA1 localized to the nucleus as Yellow Fluorescent Protein fusions and interacted with both TIR1 and IAA7 in yeast (Saccharomyces cerevisiae) two-hybrid experiments, indicating that these proteins were functional. Ubiquitination on both HIS6x-HA3x-IAA1 and Lys-less HIS6x-HA3x-IAA1 proteins was sensitive to sodium hydroxide treatment, indicative of ubiquitin oxyester formation on serine or threonine residues. Additionally, base-resistant forms of ubiquitinated IAA1 were observed for HIS6x-HA3x-IAA1, suggesting additional lysyl-linked ubiquitin on this protein. Characterization of other Aux/IAA proteins showed that they have diverse degradation rates, adding additional complexity to auxin signaling. Altogether, these data indicate that Aux/IAA family members have protein-specific degradation rates and that ubiquitination of Aux/IAAs can occur on multiple types of amino residues to promote rapid auxin-mediated degradation.
The ubiquitin-proteasome system (UPS) is the major pathway for degradation of regulatory proteins in eukaryotic cells. Ubiquitin (UBQ), an approximately 8-kD protein, is covalently ligated to substrates, often with additional UBQs linked to each other to form a polyubiquitin chain. UBQ chains with specific UBQ-UBQ linkages facilitate interaction between substrates and the 26S proteasome, a multifunctional protease. Ligation of UBQ to substrates requires three enzymatic activities in series: E1 UBQ-activating enzyme, E2 UBQ-conjugating enzyme, and an E3 ubiquitin ligase. UBQ attachment begins with the ATP-dependent covalent linkage of UBQ through its carboxy terminus to the E1 active-site Cys. The E1 then transfers thioester-linked activated UBQ to the E2 active-site Cys, forming a UBQ-charged E2. The UBQ-charged E2 interacts with an E3. E3 ligases bind substrate and catalyze transfer of UBQ either indirectly as a scaffold or directly by first accepting UBQ in a covalent thioester linkage from the E2 and then transferring UBQ to the substrate (Smalle and Vierstra, 2004; Vierstra, 2009). One major scaffold-type E3 ligase class is the multisubunit CULLIN1 (CUL1)-based E3 called the SCF. This complex is named for three of its subunits: S-phase kinase-associated protein1 (SKP1), CUL1, and F-box protein, a family of substrate recognition subunits (Vierstra, 2009; Hua and Vierstra, 2011). In Arabidopsis (Arabidopsis thaliana), SKP1 is known as Arabidopsis SKP1 Homolog1.
Identification of UBQ attachment sites contributes to a molecular understanding of the mechanism and regulation of ubiquitination, a highly specific and often rate-limiting step in degradation. E3 ligases typically catalyze ligation of the UBQ carboxy terminus to the ε-NH2 of Lys residues within substrates, forming a peptide-type bond called an isopeptide bond. Experimental determinations of selectivity for specific Lys residues in UPS substrates have produced varying results. For example, some substrates, such as mammalian Nuclear factor of Kappa light polypeptide gene enhancer in B-cells inhibitor, Alpha and Receptor Interacting Protein1 (a protein kinase associated with the tumor necrosis factor receptor), are stabilized when one or two specific lysines are substituted (Scherer et al., 1995; Baldi et al., 1996; Ea et al., 2006). In contrast, detailed studies of Substrate/Subunit inhibitor of cyclin-dependent protein kinase1 (an SCF substrate in budding yeast [Saccharomyces cerevisiae]) and mammalian jun proto-oncogene revealed that no one specific Lys is required for degradation (Treier et al., 1994; Petroski and Deshaies, 2003). Thus, ubiquitin ligation to substrates can be very specific (i.e. at one particular residue) or broader (i.e. within a region termed the ubiquitination zone; Mattiroli and Sixma, 2014).
No consensus sequence for UBQ attachment to substrate Lys residues has been reported for Arabidopsis proteins from a large-scale proteomic study (Kim et al., 2013). Therefore, prediction of Lys ubiquitination sites in this species by bioinformatics is not yet robust. Using prediction programs based on yeast ubiquitination sites as the training data set has yielded limited success when used on plant proteins (Patra et al., 2013; Chen et al., 2014), indicating that, currently, ubiquitination sites for a specific plant protein cannot be predicted with certainty and must be experimentally determined.
Another complexity in substrate ubiquitination is that residues in addition to or instead of Lys can be sites of UBQ attachment. Ubiquitination of the N-terminal amino group was first observed on metazoan myogenic differentiation1 (MyoD; Breitschopf et al., 1998) and subsequently observed on other proteins (for review, see Ciechanover and Ben-Saadon, 2004). Non-Lys ubiquitination was first described on mammalian major histocompatibility complex class I α-chain (Cadwell and Coscoy, 2005) and has been subsequently found on other proteins from metazoans and yeast (Wang et al., 2007, 2012; Ishikura et al., 2010; Shimizu et al., 2010; Domingues and Ryoo, 2012; Kravtsova-Ivantsiv and Ciechanover, 2012; Boban et al., 2015). Such alternative-site ubiquitination involves Ser and/or Thr hydroxyl groups as oxyesters and Cys sulfhydryl groups as thioesters (for review, see Kravtsova-Ivantsiv and Ciechanover, 2012). Given that these studies are with yeast and mammalian proteins, the extent of non-Lys specificity for plant UPS substrates is unknown.
The Auxin/indole-3-acetic acid (Aux/IAA) repressor proteins are critical regulators of auxin-dependent transcriptional regulation. The rapid degradation of Aux/IAA proteins in the presence of auxin is regulated by the SCFTIR1/AFB (for SCF with Transport Inhibitor Response1 [TIR1]/Auxin Signaling F-box [AFB]) family of E3 ligases (Bargmann and Estelle, 2014; Wang and Estelle, 2014). Although conserved sequences within Aux/IAA domain II are required, family members exhibit varied in vivo half-lives, and sequences outside of the domain II are required for rapid degradation in plants (Ramos et al., 2001; Dreher et al., 2006). These results suggest that recognition by the ligase, ubiquitination rates, or sites; interaction with proteins or ligands; and trafficking to and/or interaction with the proteasome are not equivalent among these proteins. Although the canonical model depicts that all domain II-containing Aux/IAA substrates are ubiquitinated by SCFTIR1/AFB, to date, only Arabidopsis IAA3 and IAA12 have been shown to be ubiquitinated by SCFTIR1 in transiently transfected protoplasts (Maraschin Fdos et al., 2009).
Identifying Aux/IAA ubiquitination sites will contribute to an understanding of what regulates ubiquitination and hence, the stability of this family of proteins. Global mass spectrometry-based approaches have not identified the Aux/IAAs or ubiquitinated Aux/IAA peptides (Saracco et al., 2009; Kim et al., 2013), likely because of their low abundance. Determination of Aux/IAA ubiquitination sites requires a concerted effort using a substrate-centric approach. In this study, we showed IAA1 ubiquitination in vivo and characterized modification site(s). Surprisingly, comprehensive substitution of Lys residues in IAA1 with Arg did not result in ubiquitination resistance or dramatic stabilization of the protein. Wild-type IAA1 was ubiquitinated predominantly on lysines but also, on Ser/Thr residues as oxyesters. However, if all IAA1 lysines were substituted, ubiquitination was completely sensitive to alkaline treatment, suggesting modification predominantly as oxyesters. To our knowledge, this study represents the first description of oxyester linkages in plant UPS substrates. We also further expand our understanding of in vivo degradation rates of Aux/IAAs with measured rates for Arabidopsis IAA6, IAA7, and IAA10 exhibiting varied dynamics. Based on these data, we propose that the sites of ubiquitination on the Aux/IAAs can be diverse and that the E3 is promiscuous in site usage.
RESULTS
Arabidopsis Aux/IAA Proteins with Conserved Domain II Sequences Are Rapidly, But Not Equivalently, Degraded in Vivo
The Aux/IAA proteins are represented by a 29-member family in Arabidopsis, and 23 of these proteins have the core domain II-conserved amino acids,GWPPV(I/L), shown to be required for low protein accumulation and/or rapid degradation (Gray et al., 1999, 2001; Worley et al., 2000). Although these sequences are required for interaction with TIR1, other sequences are important for rapid degradation in vivo (Ramos et al., 2001) and high-affinity IAA-TIR interaction in vitro (Calderón Villalobos et al., 2012). To continue to explore the diversity of the Aux/IAA in vivo degradation rates previously observed (Dreher et al., 2006), we experimentally determined the degradation rates of several other Arabidopsis domain II-containing IAA proteins. To quantitatively compare rates between proteins to published studies, IAA6, IAA7, and IAA10 were expressed stably in transgenic Arabidopsis as translational fusions with firefly luciferase (LUC), and their half-lives were determined as previously described (Ramos et al., 2001; Zenser et al., 2001, 2003; Dreher et al., 2006; Gilkerson et al., 2009). At least three independent transgenic lines stably expressing each IAA protein were analyzed.
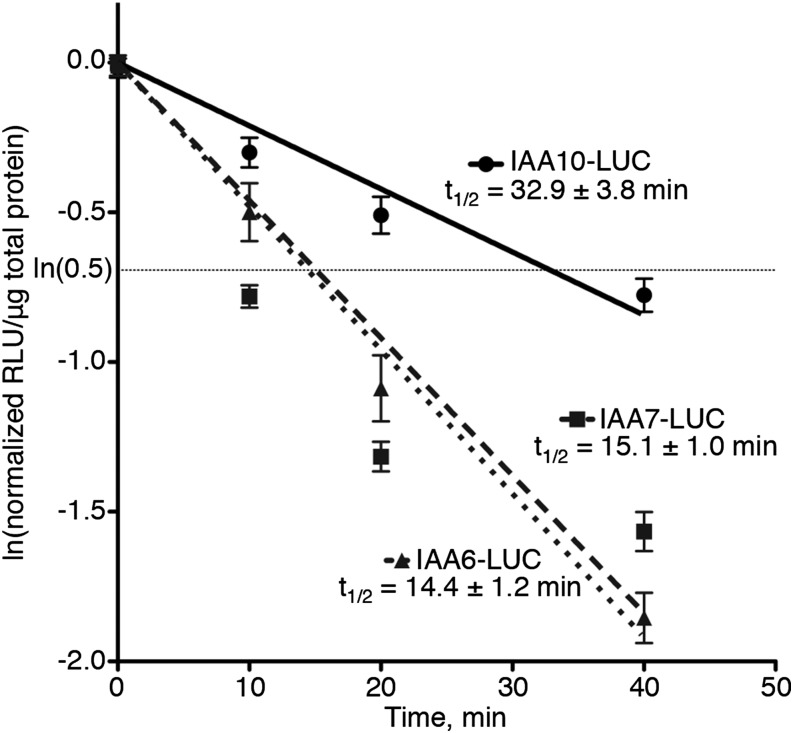
The half-lives for IAA6, IAA7, and IAA10 as LUC fusions in seedlings were determined to be 14.4, 15.1, and 32.9 min, respectively (Fig. 1). IAA6 and IAA7 have very similar half-lives, with overlapping 95% confidence intervals. The half-life of IAA10 is about double that of IAA6 and IAA7, significantly slower. These differences provide additional support for the concept that Aux/IAA proteins containing the consensus core domain II can have very different degradation rates (Dreher et al., 2006) and extend previous results that sequences outside of domain II contribute to the degradation of the Aux/IAAs (Ramos et al., 2001; Dreher et al., 2006).
Figure 1.
Degradation of Arabidopsis IAA proteins as LUC fusions is rapid. Degradation rates were determined using a pooled seedling assay in transgenic Arabidopsis. The degradation line was forced through the origin, and the resulting lines were determined: IAA10-LUC, y = −0.02108x; IAA7-LUC, y = −0.04597x; and IAA6-LUC, y = −0.04801x. Half-lives are given ±95% confidence intervals. Experiments were conducted in biological triplicate, with three technical replicates for each time point and at least three independent lines for each fusion. RLU, Relative light units.
IAA Proteins Contain a Small Number of Strictly Conserved Lys Residues
Although a Lys residue outside of the domain II core motif had already been shown to contribute to Aux/IAA instability in IAA17 (Dreher et al., 2006), its effect on degradation was modest. For this reason, we investigated whether other Lys residues were important for in vivo stability. We focused on Arabidopsis IAA1, because its degradation dynamics are well characterized (Worley et al., 2000; Ramos et al., 2001; Zenser et al., 2001; Dreher et al., 2006; Gilkerson et al., 2009). In addition, in contrast to other family members (Liscum and Reed, 2002), expression of IAA1 domain II mutants in plants results in a mild phenotype (Park et al., 2002; Yang et al., 2004; Ku et al., 2009), indicating that plants expressing long-lived IAA1 proteins could be easily grown and propagated.
Our initial hypothesis was that a few Lys residues, possibly conserved ones, in domain II-containing Aux/IAA proteins would be required for instability by serving as ubiquitination site(s). To identify conserved lysines, we performed a multiple sequence alignment (Supplemental Fig. S1) using 23 domain II-containing Arabidopsis Aux/IAA family members that showed auxin-dependent interaction with the F-box protein TIR1 in yeast (Shimizu-Mitao and Kakimoto, 2014), suggesting that they are ubiquitinated by SCFTIR1 in an auxin-dependent manner in plants. Although IAA1 has a total of 16 Lys residues, only 3 Lys residues are strictly conserved among all 23 IAA proteins (corresponding to K37, K78, and K89 in IAA1; Fig. 2, red; Supplemental Fig. S1). Three other IAA1 Lys residues in domain IV are more than 85% conserved (K134, K155, and K160; Fig. 2, green), whereas two others (K53 and K67; Fig. 2, blue) are 50% to 85% conserved among the sequences. The remaining eight lysines (Fig. 2, purple) were not considered conserved, because they are present in more than 50% of the proteins, although for three, there are Lys residues in other IAAs in close proximity to these IAA1 lysines in the primary sequence (Fig. 2; Supplemental Fig. S1). For reference, the four canonical Aux/IAA domains and two predicted nuclear localization sequences (NLSs; Abel et al., 1995) are also denoted in Figure 2. We generated open reading frames (ORFs) containing IAA1 with subsets of Lys to Arg substitutions (KR) in specific regions of the protein, including one where all of the Lys codons were changed to Arg codons (16KR). The proteins tested in this study are indicated in Table I.
Figure 2.
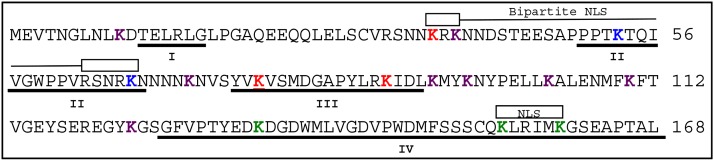
Arabidopsis IAA1 contains conserved and nonconserved lysines. Sequence of IAA1 (At4g14560.1) from The Arabidopsis Information Resource v10 annotated with the four conserved Aux/IAA domains (underlined) and NLSs (sequences below boxes; Abel et al., 1995). The 16 IAA1 Lys residues are in bold colors: red for absolutely conserved, green for highly conserved (more than 85%), blue for moderately conserved (approximately 50%), and purple for nonconserved.
Table I. KR-substituted Aux/IAA Arabidopsis IAA1 nomenclature.
Note that all versions are expressed in Arabidopsis as N-terminal His6x-HA3x epitope-tagged proteins.
| IAA1 Lys-Substituted Protein Namea | Lys Residues Substituted with Arg |
|---|---|
| 2aKR | 53, 67 |
| 2bKR | 78, 89 |
| 3aKR | 89, 93, 96 |
| 3bKR | 134, 155, 160 |
| 6KR | 53, 67, 78, 89, 93, 96 |
| 8KR | 37, 39, 53, 67, 78, 89, 93, 96 |
| 11KR | 37, 39, 53, 67, 78, 89, 93, 96, 134, 155, 160 |
| 16KR | All |
| Wild type | None |
The number indicates the number of Lys (K) to Arg (R) substitutions.
IAA1 Lys to Arg Multiply Substituted Proteins Localize in the Nucleus
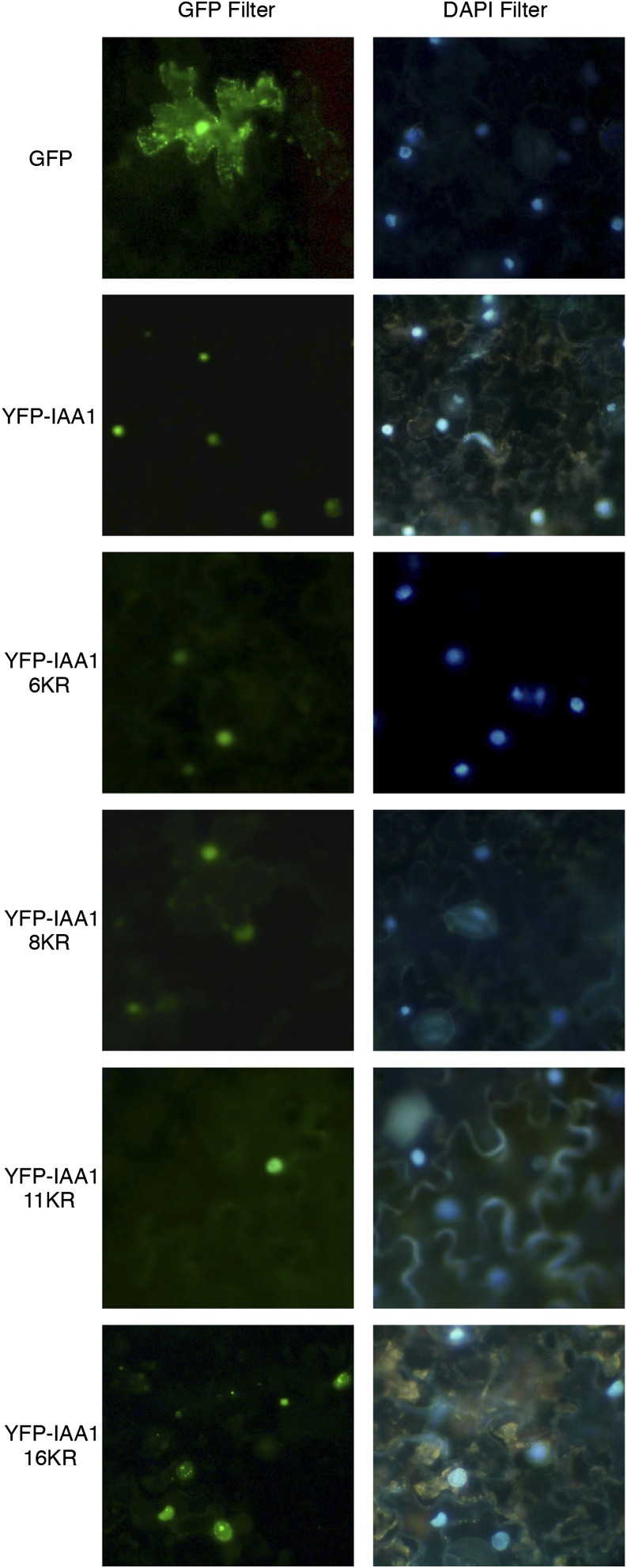
Aux/IAAs are nuclear-localized proteins and contain two NLSs: a bipartite NLS near the N terminus and another NLS in domain IV (Fig. 2). Although the bipartite NLS seems to function as the dominant NLS, together, they have an additive function (Abel and Theologis, 1995). Deletion or mutation of both NLS regions together results in cytosolic localization of Aux/IAA (Abel and Theologis, 1995; Ouellet et al., 2001). Because localization could affect degradation inadvertently because of separation of the E3 from the substrate or other required factors, we evaluated the subcellular localization of IAA1 proteins with Lys substitutions. To test this, we examined localization of Yellow Fluorescent Protein (YFP)-IAA1 KR mutants after transient expression in tobacco (Nicotiana benthamiana) leaves compared with identically fused wild-type IAA1. Native GFP localizes to the cytoplasm and nucleus (Fig. 3). All YFP-IAA1 fusions tested localize to punctate spheres that overlap with 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei, indicating that, as YFP fusions, the IAA1 wild type and the KR-substituted forms all localize to nuclei with no detectable cytoplasmic fluorescence (Fig. 3).
Figure 3.
KR-substituted IAA1 proteins, like wild-type IAA1, localize to the nucleus. Proteins were transiently expressed in tobacco leaves and then visualized in cells of abaxial epidermal peels using a fluorescent microscope with GFP and DAPI filters. Peels were incubated in 2 μg mL−1 DAPI stain to visualize nuclei.
K to R-Substituted IAA1 Proteins Interact with TIR1
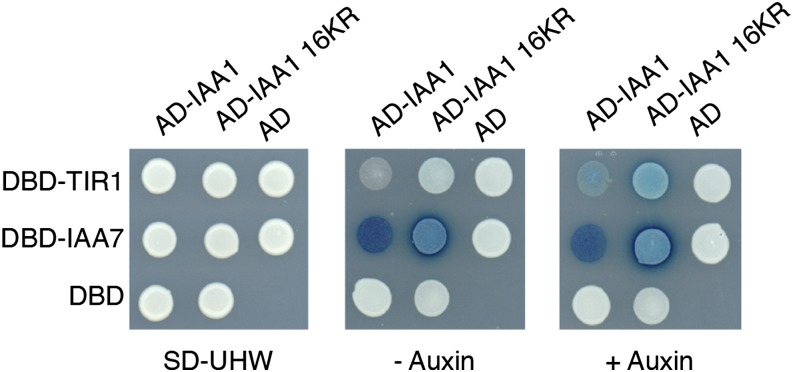
Another unintended consequence of K to R substitutions is stabilization by mechanisms other than loss of ubiquitination sites, such as interfering with ligase binding or aberrant folding. To rule out these possibilities, we tested for interaction with TIR1, an F-box protein shown to directly interact with domain II-containing Aux/IAA proteins (Gray et al., 2001; Kepinski and Leyser, 2004, 2005; Dharmasiri et al., 2005; Tan et al., 2007) and another Aux/IAA protein, representing a known heterodimeric interaction (Kim et al., 1997). Using a yeast two-hybrid assay, wild-type IAA1 and 16KR IAA1 interacted with TIR1 in an auxin-dependent manner as previously reported for other Aux/IAAs (Calderón Villalobos et al., 2012) and interacted with IAA7 in an auxin-independent manner (Fig. 4). This interaction was confirmed on plates by both growth and β-galactosidase activity. All yeast strains express the appropriate recombinant proteins as determined by western-blot analysis (Supplemental Fig. S2). These results indicate that lysines are required for neither binding to the substrate recognition subunit of the known Aux/IAA E3 ligase nor its ability to interact through domains III and IV with other Aux/IAAs and potentially, Auxin Response Factors (ARFs).
Figure 4.
IAA1 16KR binds TIR1 in an auxin-dependent manner and IAA7 in an auxin-independent manner in yeast two-hybrid assays. Yeasts transformed with the following expression plasmids were tested for interaction on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid (X-gal)-containing plates with or without auxin. Blue indicates activation of the β-galactosidase reporter gene. AD, Gal4 activation domain alone; AD-IAA1, full-length IAA1 fused to the Gal4 activation domain; AD-IAA1 16KR, full-length IAA1 16KR fused to the Gal4 activation domain; −auxin, synthetic complete media lacking uracil, His, and Trp supplemented with Gal, raffinose, and X-gal without auxin, as described in “Materials and Methods”; +auxin, synthetic complete media lacking uracil, His, and Trp supplemented with Gal, raffinose, and X-gal without auxin, as described in “Materials and Methods”; DBD, Gal4 DNA binding domain alone; DBD-IAA7, full-length IAA7 fused to the Gal4 DNA binding domain; DBD-TIR1, full-length TIR1 fused to the Gal4 DNA binding domain; SD-UHW, synthetic complete yeast media lacking uracil, His, and Trp to select for both plasmids.
Conserved Lys Residues Are Not Required for Rapid Degradation of IAA1
Because Arg-substituted IAA1 proteins localized normally and interacted with the substrate specificity subunit of the E3 ligase responsible for in vivo degradation of wild-type IAA1, we explored the in vivo effects of Lys loss on IAA1 stability. We expressed substituted Histidine-Hemaglutinin (HIS6x-HA3x)-IAA1 ORFs in multiple independent transgenic Arabidopsis lines driven by the UBQ10 promoter and determined their stability by performing single-time point cycloheximide degradation assays. In these studies, the wild type refers to equivalently epitope-tagged IAA1 with no Lys substitutions expressed in plants as described above for Lys-substituted proteins. Columbia-0 (Col-0) refers to our nontransgenic control line. Although these assays do not allow for direct degradation rate comparisons, we can assess if degradation is dramatically slowed and identify which proteins should be analyzed more thoroughly. The N-terminal HIS6x-HA3x epitope tag does not contain any lysines, and there were no Lys codons introduced in the cloning process (an example of an ORF with translation is in Supplemental Fig. S3).
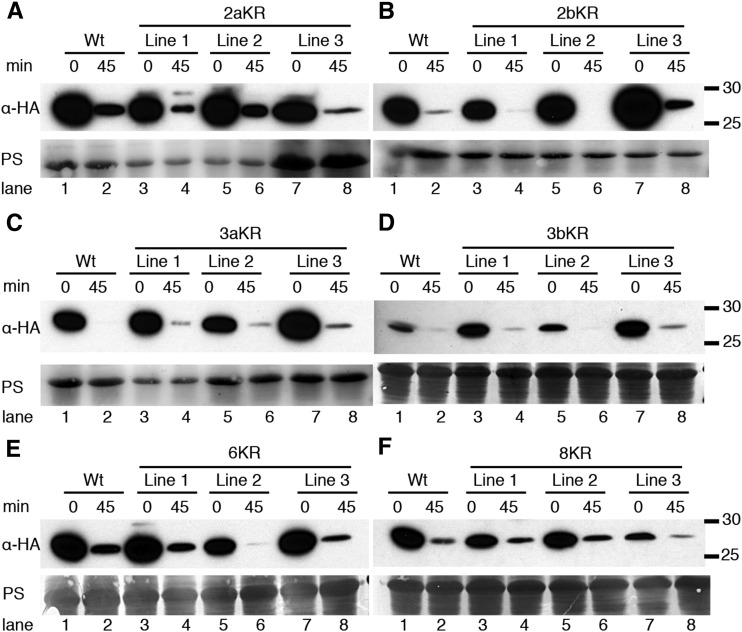
We first tested whether substitution of two lysines positioned close to one another in the primary sequence affected degradation. The stability of 2aKR and 2bKR with Lys substitutions surrounding domain II or in the absolutely conserved lysines in domain III, respectively, was analyzed (Fig. 5; Table I). 2aKR and 2bKR were detectable at the 0 time point, but after 45 min of cycloheximide treatment, their abundance was similarly and dramatically reduced compared with the wild type (Fig. 5, A and B), indicating that degradation of these proteins was rapid. Consequently, we then tested whether a cluster of lysines in domain III (3aKR) or IV (3bKR) was required for degradation; again, these proteins were rapidly degraded (Fig. 5, C and D, respectively). These data suggest that rapid degradation cannot be blocked by substituting two or three Lys residues, even if they are highly conserved (Fig. 2).
Figure 5.
KR-substituted IAA1 proteins are degraded rapidly in vivo. A to D, Three independent lines expressing the indicated HIS6x-HA3x-IAA1 proteins were tested in cycloheximide degradation assays. Plants were treated with 200 μg mL−1 cycloheximide or mock (0) for 45 min, and HIS6x-HA3x-IAA1 was visualized by anti-HA immunoblotting. Ponceau S-stained membranes (PSs) are shown for loading controls. Molecular mass markers (in kilodaltons) are indicated to the right. A, Wild-type (Wt) and 2aKR lines. Lanes 1 to 6 and lanes 7 and 8 have 30 and 150 μg of total protein, respectively. B, Wild-type and 2bKR lines. Lanes 1 to 8 have 30 μg of total protein. C, Wild-type and 3aKR lines. Lanes 1 to 4 and 5 to 8 have 30 and 60 μg of total protein, respectively. D, Wild-type and 3bKR lines. All lanes have 30 μg of total protein loaded. E, Wild-type and 6KR lines. All lanes have 80 μg of total protein. F, Wild-type and 8KR lines. All lanes have 80 μg of total protein.
We generated IAA1 protein with combinations of these Lys substitutions (Table I); for example, the 6KR protein substituted Arg for all domain II- and domain III-localized Lys residues, whereas the 8KR protein additionally substituted two N-terminal lysines, K37 and K39. The stability of these proteins was tested in vivo as above. Treatment with cycloheximide for 45 min reduced the amount of the wild type, 6KR, and 8KR to similar levels (Fig. 5, E and F), suggesting that these versions of IAA1 are degraded at similar rates.
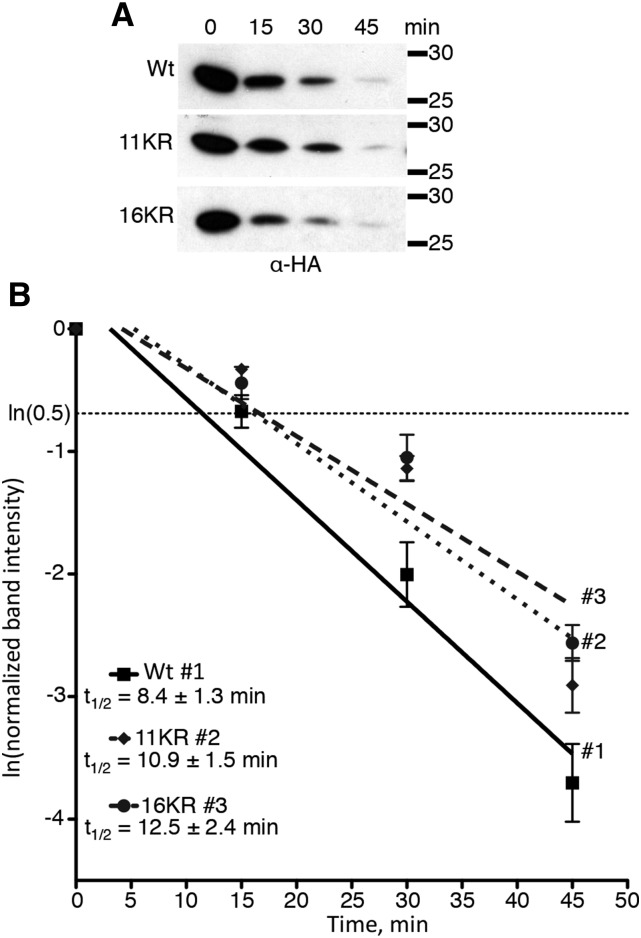
Next, we examined 11KR stability. This protein has all conserved lysines or adjacent lysines substituted with Arg. We used cycloheximide degradation assays to determine the degradation rate of the 11KR protein in direct comparison with the wild type in seedlings (Fig. 6). Relative protein abundance was measured by anti-HA immunoblotting (Fig. 6A) followed by densitometry (Fig. 6B). Data from multiple independent lines and multiple replicas were used to calculate average half-lives of 8.4 ± 1.3 and 10.9 ± 1.5 min for the wild type and 11KR, respectively (Fig. 6B). Half-lives are shown as 95% confidence intervals, and although the 95% confidence intervals for the wild type and 11KR lines overlap, the lines are slightly statistically different by linear regression analysis (P value = 0.01312; α = 0.05). This analysis would suggest that the 11KR protein is degraded slightly more slowly than the wild type but still very rapidly degraded.
Figure 6.
11KR and 16KR IAA1 proteins are degraded rapidly in vivo. A, Western-blot analysis of cycloheximide degradation assays for KR-substituted HIS6x-HA3x-IAA1 proteins. A representative anti-HA blot is shown for each protein; 100 μg of total protein was loaded for each time point. B, The degradation line was determined by regression analysis, and the resulting lines determined are IAA1, y = −0.08273x + 0.2574; 11KR, y = −0.06356x + 0.3358; and 16KR, y = −0.05529x + 0.2309. Half-lives are given ±95% confidence intervals. Numbers indicate corresponding regression lines for given proteins. The lines for 11KR and 16KR are not different from one another (P value = 0.3242). Both the 11KR and 16KR lines differ from wild-type (Wt) IAA1 (P values = 0.01312 and 0.0157, respectively). Experiments were conducted in biological triplicate, with three technical replicates for each time point and at least three independent lines for each.
IAA1 Is Rapidly Degraded, Even When No Lys Residues Are Present
IAA1 11KR protein has five remaining nonconserved lysines distributed throughout the linear sequence that could potentially affect degradation (Fig. 2). To address if these remaining Lys residues affected IAA1 degradation, we generated an ORF with all Lys codons substituted to Arg codons (16KR). To determine whether this protein is functional in vivo, its subcellular localization and interaction with TIR1 were determined. The 16KR protein localizes to the nucleus (Fig. 3) and interacts with TIR1 in an auxin-dependent manner in yeast (Fig. 4). As with the other Lys-substituted proteins, we expressed HIS6x-HA3x-IAA1 16KR in transgenic Arabidopsis lines. Single-time point cycloheximide degradation assays with adult rosette leaves from T1 plants indicated that this protein is short lived in multiple transgenic lines (Supplemental Fig. S4). Using cycloheximide degradation assays as described above, the 16KR half-life for 16KR was 12.5 ± 2.4 min (Fig. 6B). By linear regression analysis, the 16KR half-life is slightly statistically different from that of the wild type (P value = 0.0157; α = 0.05) but not different from that of the 11KR protein (P value = 0.3242; α = 0.05). This shows that, even in the absence of Lys resides, IAA1 protein is very rapidly degraded in vivo and that there is no additive defect in degradation when the remaining five lysines in the 11KR protein are substituted.
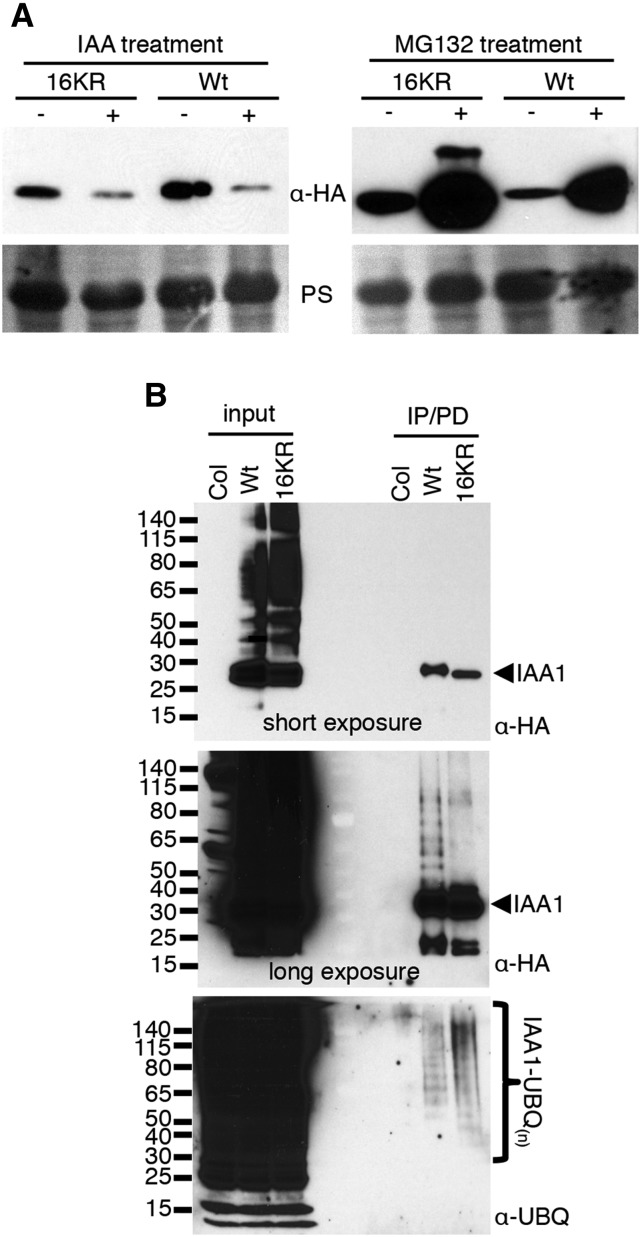
Lys Residues Are Not Required for Auxin-Induced Degradation, Proteolysis by the Proteasome, or in Vivo Ubiquitination of IAA1
Because rapid degradation of 16KR was surprising, we designed experiments to examine the mechanism of degradation. Aux/IAA protein degradation depends on the ubiquitin ligase SCFTIR1/AFB, and exogenous auxin treatment has been shown to reduce steady levels of Aux/IAAs by promoting interaction of IAA proteins with the F-box proteins TIR1/AFB. To examine if SCFTIR1/AFB was involved in degradation of 16KR, we treated the wild type- and 16KR-expressing lines with IAA, the most abundant endogenous auxin, and measured steady-state levels (Fig. 7). IAA treatment of transgenic seedlings reduced the steady-state level of the 16KR similar to that of the wild type (Fig. 7A, left), suggesting involvement of SCFTIR1/AFB. To test if degradation of 16KR was proteasome dependent, we treated seedlings with MG132, a proteasome inhibitor. Both the wild type and 16KR (Fig. 7A, right) strongly accumulated after MG132 treatment. Altogether, this indicated that Lys residues on IAA1 are not required for auxin-stimulated degradation or proteasome-dependent degradation in vivo.
Figure 7.
HIS6x-HA3x-IAA1 16KR protein levels are regulated by auxin and the UPS in vivo. A, 16KR and wild-type (Wt) IAA1-expressing transgenic seedlings were treated with 5 μm IAA or solvent or 50 μm MG132 and corresponding solvent for 6 h. HIS6x-HA3x-IAA1 proteins were visualized by anti-HA immunoblotting, with Ponceau S-stained membranes shown as loading controls; 80 μg of total protein is loaded for each sample. B, In vivo ubiquitination of wild-type and 16KR proteins was examined by DPDs from plant extracts of MG132-treated seedlings. HIS6x-HA3x-IAA1 was immunoprecipitated from approximately 5 mg of total protein using anti-HA beads. Col-0 extract, a nontransgenic line, was used as a negative control. Protein was eluted from anti-HA beads in denaturing urea buffer, recaptured on Co2+ beads, and finally eluted in 1× Laemmli sample buffer; 20% and 80% of the eluted protein were loaded for anti-HA and anti-UBQ, respectively, and 80 μg of total protein was loaded for input for both blots. Unmodified HIS6x-HA3x-IAA1 is indicated with arrows on the right, and molecular mass markers in kilodaltons are labeled on the left. Two exposures for the anti-HA blot are shown to show equal loading of the unmodified forms (short exposure) and visualize the slower migrating species (long exposure).
To determine whether IAA1 lacking all lysines is a substrate for ubiquitination, transgenic seedlings expressing 16KR or the wild type were used to isolate HIS6x-HA3x-IAA1 proteins in a two-step procedure followed by anti-UBQ immunoblotting. The first purification step was an immunoprecipitation (IP) with anti-HA beads. Proteins were then eluted off anti-HA beads in denaturing buffer and recaptured using Co2+ chromatography in the presence of 8 m urea. This approach ensured that only ubiquitin covalently bound to IAA1 would be retained. Both IAA1 versions, the wild type and 16KR, purified higher Mr-ubiquitinated species from these denaturing pull downs (DPDs), indicating that both are ubiquitinated in vivo (Fig. 7B). Equivalent amounts of IAA1 proteins were enriched, which was verified by the anti-HA blot (Fig. 7B, short exposure). Curiously, although high-Mr species are readily visualized with the wild type, the level of similar species in the 16KR sample is much lower (anti-HA blot; Fig. 7B, long exposure). This difference is not understood but could result from interference by UBQ on the anti-HA-horseradish peroxidase (HRP) immunoreactivity with ubiquitinated 16KR protein. However, it is clear from the antiubiquitin blot that both the wild type and 16KR are ubiquitinated in vivo.
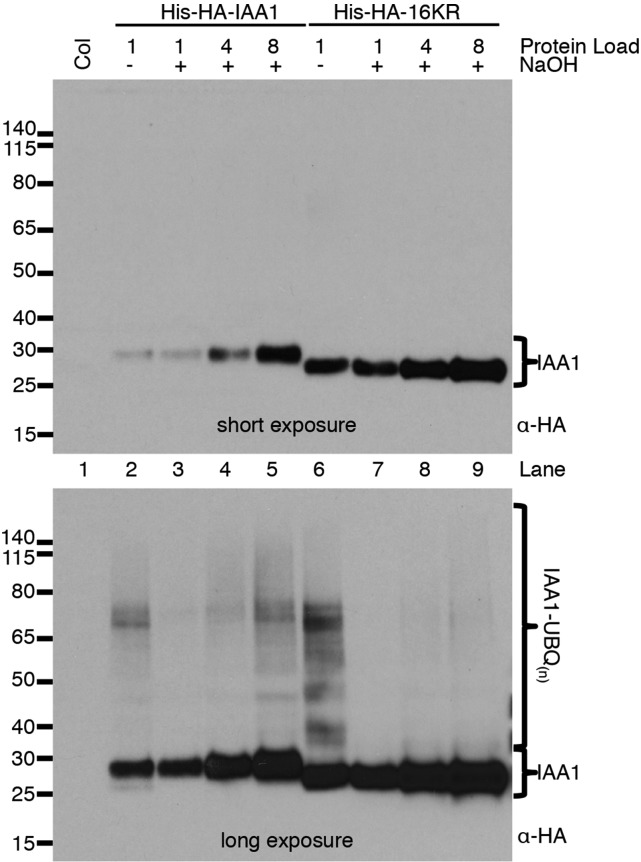
After showing that 16KR was rapidly degraded and ubiquitinated in vivo, we investigated the type of UBQ linkages present. In 16KR, there are no lysyl amino groups available for isopeptide linkages, and the endogenous amino terminus is blocked with the HIS6x-HA3x tag. The IP/pull-down (PD) experiment in Figure 7B performed under strong reducing conditions would destroy any thiol linkages. We were, therefore, left with the strong possibility that 16KR was modified with UBQ by oxyester linkages. Oxyester linkages but not isopeptide linkages are labile in high pH, because hydroxide ions participate in a nucleophilic attack on oxyesters. We incubated protein extracts prepared in a denaturing and thiol-reducing buffer with or without 0.2 m NaOH and assayed for UBQ-modified IAA1 using anti-HA immunoblotting (Fig. 8). Extracts at pH 7.5 retained ubiquitinated forms (Fig. 8, lanes 2 and 6). Treatment with base surprisingly reduced the amount of ubiquitination on wild-type IAA1 (compare lane 2 with lane 3 in Fig. 8) but did not completely abolish it (Fig. 8, lanes 5 and 6). This result suggests that, on wild-type IAA1 protein, both isopeptide and oxyester-linked UBQ contribute to total ubiquitination. Base treatment of the 16KR IAA1 extract completely abolished detectable ubiquitination on that protein (compare lanes 7 and 8 with lane 6 in Fig. 8). We conclude that both oxyester- and isopeptide-linked UBQ are present on wild-type IAA1 and that detectable ubiquitination on 16KR IAA1 is oxyester linked.
Figure 8.
Ubiquitin modifications of HIS6x-HA3x-IAA1 and HIS6x-HA3x-IAA1 16KR are labile at high pH. Protein extracts from Col-0 and HIS6x-HA3x-IAA1 lines were treated with 0.2 m NaOH for 2 h at 37°C. HIS6x-HA3x-IAA1 proteins and UBQ modification were examined by anti-HA immunoblotting. All lanes were loaded with 80 μg of total protein. HIS6x-HA3x-IAA1s were diluted into Col-0 extract to obtain the dilution series. Protein load indicates the concentration of HIS6x-HA3x-IAA1 extracts in the dilution series relative to the respective untreated control sample. Two exposures are shown for the same blot to visual unmodified IAA1s (short exposure) and UBQ-modified species (long exposure). Molecular mass markers (in kilodaltons) are indicated.
DISCUSSION
To be able to describe and predict the developmental and morphological effects of auxin, it is necessary to understand the molecular basis of diversity in the degradation rates of the Aux/IAA proteins that serve as repressors of the transcriptional response to auxin. Aux/IAA protein degradation is regulated by an SCF-type ubiquitin E3 ligase, with TIR1 or a related AFB as the substrate-interacting subunit. Clearly, the nature and abundance of TIR1/AFB are determinants of Aux/IAA degradation (Vernoux et al., 2011; Calderón Villalobos et al., 2012; Havens et al., 2012; Wang and Estelle, 2014; Yu et al., 2015). However, intrinsic properties of Aux/IAA proteins also regulate their degradation (Worley et al., 2000; Gray et al., 2001; Ramos et al., 2001; Dreher et al., 2006).
The Arabidopsis Aux/IAA family of 29 members can be divided into 10 clades based on sequence similarity (Overvoorde et al., 2005). Aux/IAA proteins (IAA20 and IAA29 to IAA34) that lack a complete domain II are contained in three separate clades and do not interact with TIR1 in vitro or in yeast (Calderón Villalobos et al., 2012; Shimizu-Mitao and Kakimoto, 2014). Correspondingly, these Aux/IAAs are long lived (or for IAA31 with a partial domain II, slowly degraded) in vivo (Dreher et al., 2006; Calderón Villalobos et al., 2012; Shimizu-Mitao and Kakimoto, 2014). Because expression of IAA15, the sole member of one clade, is extremely low if expressed at all (Overvoorde et al., 2005), it was not included in this analysis. Together with previously published data, the degradation rate for at least one member of each domain II-containing clade within the Aux/IAA family has been determined using the same experimental methodology, so that rates may be more directly compared (Table II). From these data, we propose that IAAs fall into three broad classes: those very rapidly degraded, with half-lives ranging from 8 to 15 min; those with rapid degradation, with approximately 2- to 3-fold slower half-lives at approximately 20 to 30 min; and finally, a class currently containing one protein with a slow but measurable degradation, with a half-life of approximately 80 min. Although the data are limited, members within the same clade seem to have comparable half-lives, suggesting that shared sequences contribute to their characteristic half-lives. For example, IAA7 and IAA17 in the same clade both fall into the very rapid degradation class, and IAA8 and IAA9, with rapid degradation, are members of a different clade that seem to share degradation properties (Dreher et al., 2006). The degradation rates for additional members need to be measured directly to test this hypothesis.
Table II. Summary of Aux/IAA degradation rates as LUC fusions.
CHX, Cycloheximide; ND, not determined.
| Aux/IAA Member in Clade | Half-Life | Degradation Class | Reference | Comments |
|---|---|---|---|---|
| min | ||||
| IAA1 | 11.8 ± 0.9 | Very rapid | Dreher et al. (2006) | |
| IAA6 | 14.4 ± 1.2 | Very rapid | This work | |
| IAA17, IAA7 | 10.1 ± 0.5; 15.1 ± 1.0 | Very rapid | Dreher et al. (2006); this work | Gray et al. (2001) measured t1/2 for endogenous IAA7 at 10.8 ± 1.1 min |
| IAA8, IAA9 | 19.0 ± 2.3 (IAA9) | Rapid | Dreher et al. (2006) | IAA8 degradation curve not determined, but protein loss with CHX is equivalent to IAA9 in parallel experiment |
| IAA10 | 33 ± 3.8 | Rapid | This work | |
| IAA28 | 79.3 ± 4.4 | Slow | Dreher et al. (2006) | |
| IAA15 | ND | Low expression; not certain to contribute significant protein in cells |
Aux/IAA proteins are degraded by the proteasome, and ubiquitinated species of IAA1 (this work), IAA3, and IAA12 have been detected (Ramos et al., 2001; Maraschin et al., 2009). Typically, ligation of UBQ to most substrates occurs through an isopeptide bond between the carboxy terminus of UBQ and the ε-NH2 of internal Lys residues within the substrate. A few studies have attempted to identify preferred ubiquitination sites within substrates. Such studies are important for determining the promiscuity of the E3 ligase for acceptor sites, especially when one E3 targets numerous substrates, and can also provide information about the number of independent attachment sites that a substrate may have, the kinetics of ubiquitination, and whether accessibility to the lysyl acceptor site(s) could be regulated. For instance, ubiquitin chain attachment to multiple sites simultaneously might be required for very rapid degradation or dissolution of a multiprotein complex where not all proteins are equivalently short lived. Also, blocking all ubiquitination sites could reveal redundant ubiquitin-independent mechanisms of degradation.
In this study, we sought to identify preferred Lys ubiquitination sites in Aux/IAA proteins using IAA1, a member of the very rapidly degraded class, as a model. We initially hypothesized that conserved Lys residues among rapidly degraded Aux/IAAs would serve as ubiquitination sites and that substituting them would strongly stabilize IAA1. We show that Lys substitutions neither affect intracellular localization nor interaction with the ligase. Although evidence suggests that interaction with other Aux/IAAs or ARFs is not important for rapid degradation, because the N-terminal one-half of IAA17 lacking the dimerization domains when fused to LUC has the same degradation as full-length IAA17 (Worley et al., 2000; Ramos et al., 2001; Dreher et al., 2006), 16KR interacts in yeast with another Aux/IAA, IAA7, similarly to wild-type IAA1. These interactions serve to indicate that substitution of lysines in domains III and IV does not abrogate protein-protein interactions or result in misfolding and therefore, should not be a confounding effect in degradation studies. We also showed that degradation of 16KR was affected by MG132, a proteasome inhibitor, and accelerated in the presence of auxin (all traits observed in the wild type), suggesting that both the wild type and 16KR are degraded by the same mechanism.
Remarkably, all Lys-substituted versions of IAA1 tested, including one completely lacking Lys residues, were degraded rapidly in vivo. The 11KR and 16KR proteins had the same degradation rates, slightly slower but comparable with the wild-type protein. This difference likely results from substitution of K37, which we previously showed had an approximately 3-fold effect on degradation in IAA17 (Dreher et al., 2006). We conclude that, aside from this Lys residue, no other Lys residue has a significant effect on Aux/IAA degradation.
Detection of copurifying, higher Mr-ubiquitinated species after denaturing affinity chromatography shows that 16KR is ubiquitinated in vivo. This result suggests that IAA1 can be ubiquitinated on non-Lys sites, with several other types of sites possible: the α-NH2 group, hydroxyl groups of Ser/Thr, and/or thiol groups of Cys residues. The native IAA1 N terminus, however, does not exist in the tagged versions used in our experiments, because the amino acids adjacent to the endogenous α-NH2 group of IAA1 are separated from the N terminus by the HIS6x-HA3x epitope sequence. Thus, if used as a UBQ acceptor site in 16KR, it does not represent a biologically relevant site for endogenous IAA1. In MyoD, the presence of an N-terminal epitope tag, which interestingly, contains a Lys residue, strongly stabilized the protein (Breitschopf et al., 1998), suggesting that adjacent sequences influence N-terminal ubiquitination. If this effect is universal, then the IAA1 N terminus is unlikely to be a site of ubiquitination in 16KR. The reduction of ubiquitination to below detection upon base treatment, to which N-terminal ubiquitination is insensitive, also indicates that the N terminus of 16KR is not ubiquitinated.
Our data support oxyester ubiquitination of IAA1. Although we have not eliminated the possibility of thioesters on Cys residues, this linkage does not contribute to the ubiquitination observed for either the wild type or 16KR because of the strong reducing conditions used. When examined for susceptibility of cleavage by base, a hallmark of oxyester-linked ubiquitination, 16KR ubiquitination was not detectable. One explanation is that, only in the absence of Lys sites, the E3 can just as efficiently ligate ubiquitin to neighboring Ser or Thr residues as oxyesters. However, a large fraction of wild-type IAA1 ubiquitination was susceptible to base cleavage, indicating a mixture of isopeptide and oxyester linkages present on the wild-type protein. This indicates that oxyester linkages on 16KR do not simply substitute for isopeptide linkages in the absence of lysines but rather, make a significant contribution to total ubiquitination, even when lysines are present. Given the nature of these results, we performed IP followed by mass spectrometry experiments with both the wild type and Lys-less IAA1 in an attempt to explicitly map ubiquitination sites. Unfortunately, our concerted efforts did not result in any UBQ-modified peptides for either protein (data not shown), but this result is not surprising given the numerous technical challenges associated with such experiments (Shimizu et al., 2010; Wang et al., 2012; Kim et al., 2013).
CONCLUSION
Given the unexpected findings that Aux/IAA degradation lacks selectivity for specific Lys residue(s) and that ubiquitination occurs as oxyesters in both the presence and the absence of lysines, we propose that ubiquitination and degradation of SCFTIR1/AFB substrates are resilient and robust molecular mechanisms. The experiments presented here add key molecular details of Aux/IAA degradation and the idea that members of the Aux/IAA exhibit complex degradation dynamics. Furthermore, this study expands the knowledge base for mechanisms of plant UPS substrate ubiquitination and extends the paradigm of non-Lys linkages to plants. The significance of these findings warrants additional experimental attention and highlights the need for more ubiquitination sites to be characterized in plant UPS substrates, because their regulated proteolysis is critical for responses to abiotic and biotic stresses, hormones, light, circadian rhythm, and many other processes (for review, see Kelley and Estelle, 2012).
MATERIALS AND METHODS
Molecular Cloning and Plant Material
The AtIAA1 (At4g14560.1) coding sequence (Abel et al., 1995) used in previous degradation studies (Worley et al., 2000; Zenser et al., 2001, 2003; Dreher et al., 2006; Gilkerson et al., 2009) was recombined into pDONR201 after PCR using primers 8-110 and 3-192 by Gateway Technology (Invitrogen). Primer and oligo sequences are indicated in Supplemental Table S1. Site-directed mutagenesis was performed using the QuickChange Site-Directed Mutagenesis Kit (Stratagene) following the manufacturer’s instructions to generate ORFs for double, triple, 6KR, 8KR, and 11KR coding sequences. For expression of HIS6x-HA3x epitope-tagged IAA1 KR mutants without additional Lys residues introduced by epitope tags or vector sequences, an HIS6x-HA3x epitope sequence was cloned into pLIT29 (New England Biolabs) to add the HIS6x-HA3x sequence 5′ of IAA1 ORFs. An HA3x ORF had been previously cloned NcoI/HindIII into pLIT29 by adding the sequence for the HA epitope into the forward primer with an NdeI site separating the HA3x sequence from the ORF (Salmon et al., 2008). The HIS6x sequence was added by annealing oligos 3-250 and 3-254 and ligating them into KpnI/NcoI restriction sites of HA3x ORF pLIT29, making HIS6x-HA3x ORF pLIT29. The IAA1 KR ORFs were PCR amplified from the pDONR201 derivatives described above with primers 3-310 and 3-311 and cloned NdeI/BamHI into similarly digested HIS6x-HA3x ORF pLIT29. The HIS6x-HA3x-IAA1 16KR ORF was chemically synthesized using oligos (GenScript USA Inc.) and cloned KpnI/BamHI into pUC57. The epitope sequences are identical to those in HIS6x-HA3x ORF pLIT29. The IAA1 16KR without the HIS6x-HA3x was cloned into pDONR201. All HIS6x-HA3x-IAA1 KR ORFs were subsequently cloned from the pLIT29-based plasmids using KpnI/BamHI into the plant transformation vector pGreenII 0029 backbone (Hellens et al., 2000), which had been modified in the HindIII/NotI sites with a UBQ10 promoter:(KpnI)ORF(BamHI):nopaline synthase terminator (NOS) expression cassette (Dreher et al., 2006).
The resulting UBQ10:HIS6x-HA3x-IAA1:NOS cassettes in pGreenII 0029 were transformed into Agrobacterium tumefaciens strain AGL1 along with pSOUP (Hellens et al., 2000) for plant transformation. Arabidopsis (Arabidopsis thaliana) Col-0 was transformed using the floral dip method (Clough and Bent, 1998) to generate lines used for experiments. At least three independent lines initially segregating for a single transgene were made homozygous, and T3 and/or T4 generations were analyzed. The ORF was PCR amplified from seedling DNA isolated from the same aliquot of seed analyzed, and the DNA sequence was determined to verify their identities.
For yeast (Saccharomyces cerevisiae) interaction assays, full-length wild-type IAA1 and mutant IAA1 16KR coding sequences were amplified by PCR using Phusion polymerase (New England BioLabs) with primers DK807 and DK808 containing EcoRI (5′) and XhoI (3′) sequences. Digested PCR fragments were ligated into pB42AD (Clontech) using the same restriction enzyme sites to generate pDK193 (pB42AD-IAA1) and pDK194 (pB42AD-IAA1K0). Both clones were sequence verified. pGILDA-TIR1 and pGILDA-IAA7 have been previously described (Yu et al., 2013).
To generate YFP-IAA1 fusions for localization experiments, IAA1 ORFs were recombined from pDONR201 (above) into pEarleyGate104 (Earley et al., 2006) using Gateway Technology (Invitrogen). A. tumefaciens strain AGL1 was transformed with the resulting constructs.
Multiple sequence alignment was performed using Jalview (http://www.jalview.org/).
Immunoblotting for IAA Proteins
Gels containing plant extracts were transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore) for immunoblotting. All membranes were blocked with 5% (w/v) nonfat dry milk in Tris-buffered saline plus Tween 20 (TBST; 50 mm Tris, 200 mm NaCl, and 0.1% [v/v] Tween 20) and 0.165% (v/v) antifoam Y emulsion (Sigma) before incubation with the indicated antibodies. For anti-HA blots, membranes were blocked for 15 min, immunoblotted with 1:1,000 rat anti-HA-peroxidase (3F10 rat monoclonal antibody; Roche) in 5% (w/v) nonfat dry milk in TBST (50 mm Tris, 200 mm NaCl, and 0.1% [v/v] Tween 20) and 0.165% (v/v) antifoam Y emulsion (Sigma) for 2 h, and then washed three times for 5 min. Western blots were developed with Amersham ECL Plus Western Detection System (GE Healthcare) and x-ray film following the manufacturer’s instructions.
Transient Tobacco Expression and Localization Studies
Four-week-old tobacco (Nicotiana benthamiana) leaves were infiltrated with AGL1 cells resuspended to optical density at 600 nm 1.0 in infiltration buffer (10 mm MES, pH 5.6, 10 mm MgCl2, and 150 μm acetosyringone) and incubated for 3 d at room temperature under constant light. YFP-IAA1 localization was observed in cells of abaxial epidermal peels using a fluorescent microscope (Endow GFP BP Filter Set 41017; Carl Zeiss Axioplan Universal Microscope D-7082 Oberkochen; Chroma Technology Corp.). Peels were treated with 2 μg mL−1 DAPI stain to observe nuclei (Zeiss UV-G 365 Filter Set).
Cycloheximide Degradation Assays and Half-Life Calculations
Half-life determinations for all IAA-LUC fusion proteins were performed as described by Dreher et al. (2006). For single-time point degradation assays of HIS6x-HA3x-IAA1, rosette leaves (two to three) from 4- to 5-week-old transgenic plants grown at 20°C were excised and submerged in 1 mL of 200 μg mL−1 cycloheximide in liquid growth media (GM; Gilkerson et al., 2009) for 45 or 60 min as indicated. Mock-treated samples were submerged in GM for the same length of time, which served as the 0 time point. For experiments using lines expressing HIS6x-HA3x-IAA1 3bKR, 6KR, and 8KR, plants were 1-week-old seedlings grown in liquid GM treated the same as described above. For all experiments, tissue was flash frozen in liquid nitrogen and ground in 200 μL of extraction buffer (50 mm Tris, pH 7.2, 150 mm NaCl, 0.5% [v/v] NP-40, 1 mm phenylmethylsulfonyl fluoride, 10 μm MG132, and one tablet per 10 mL of Buffer Complete Mini Protease Inhibitor; Roche). Total protein concentration was determined by Bradford assay, and dilutions were made in extraction buffer and Laemmli Sample Buffer for SDS-PAGE. Samples were separated on 10% (v/v) polyacrylamide gels by SDS-PAGE and transferred to membrane for immunoblotting. Two biological replicates were performed for each plant line, and at least three independent lines per IAA1 protein were analyzed.
For HIS6x-HA3x-IAA1 wild-type, 11 KR, and 16KR half-life experiments, homozygous T4 or T3 seedlings were used. Seeds (approximately 2 mg) were surfaced sterilized and grown in 1 mL of liquid GM in 60- × 15-mm petri dishes for 6 d at 22°C under constant light (approximately 40 μmol s−1 m−2). On the evening of the sixth day, remaining media were removed and replaced with 900 μL of fresh GM. Degradation experiments were always conducted in the morning to control for possible circadian effects on degradation. On the morning of the experiment, 100 μL of 200 μg mL−1 cycloheximide dissolved in water was added to the seedlings and incubated for 0, 15, 30, and 45 min; 0 represents seedlings mock treated with water for 45 min. Seedlings were frozen in liquid nitrogen and processed for SDS-PAGE and immunoblotting as described above. Equal total protein for each time point was loaded onto gels. This experiment was conducted three times for three to four independent transgenic lines for each protein. Each sample was immunoblotted two times as technical replicates. Blots were scanned using a flat-bed scanner without autotoning and saved as 8-bit grayscale tagged image file format images. Spot densitometry was performed using National Institutes of Health ImageJ 1.36 (http://rsb.info.nih.gov/ij/) without background correction to quantify the amount of HIS6x-HA3x-IAA1 at each time point. The amounts of HIS6x-HA3x-IAA1 at 15, 30, and 45 min were normalized to the amount at 0 min, and the ln of that normalized value was calculated to linearize the data. The ln (normalized band intensity) was plotted against time, and the slope of this line was used to calculate the half-life by the following equation: t1/2 = [ln(0.5)]/slope. Data for all blots were combined to draw a single line for the final half-life calculations. The 95% confidence interval was calculated for the slope of the overall line using GraphPad Prism 5 and used to calculate the confidence interval for the half-life. Linear regression analysis was performed using GraphPad Prism 5 to compare slopes of degradation curves.
Yeast Two-Hybrid Assays
pGILDA and pB42AD clones were transformed into yeast strain EGY48[p8op-lacZ] according to the small-scale transformation method in the Yeast Protocols Handbook (Clontech). Yeast colonies harboring both plasmids were selected on synthetic defined medium (SD) SD/−His/−Trp/−Ura plates containing Glc. Protein-protein interactions were assayed by selecting for growth on SD/−His/−Trp/−Ura + X-gal + sodium phosphate buffer, pH 7.0, + Gal + raffinose both with and without 10 μm IAA after 2 d at 30°C. Plates with yeast colonies were imaged using a flat-bed scanner.
Yeast Protein Extraction and Immunoblotting
Total protein was extracted from liquid cultures of inoculated yeast strains grown in 5 mL of SD/−His/−Trp/−Ura + Gal + raffinose overnight at 30°C using Y-PER Yeast Protein Extraction Reagent (Thermo Scientific) supplemented with 1× Complete (Roche) protease inhibitor tablet. Protein concentrations were determined using a BCA Protein Assay (Thermo Scientific). For immunoblotting, 20 µg of total protein per sample was loaded on 12% (v/v) SDS-PAGE gels and transferred to nitrocellulose membranes (GE Healthcare Life Sciences). All membranes were blocked in 5% (w/v) nonfat dry milk in 1× phosphate-buffered saline plus Tween 20 for 1 h at room temperature. For anti-HA blots, membranes were incubated with 1:1,000 anti-HA-HRP-peroxidase high affinity (3F10; Roche) for 1 h and then washed three times for 5 min each. For anti-LexA blots, membranes were incubated with 1:2,000 anti-LexA antibody (Millipore) overnight at 4°C, washed three times for 5 min each followed by incubation with 1:10,000 goat anti-rabbit HRP-conjugated secondary antibody (Sigma) for 1 h and then washed again three times for 5 min each. All membranes were developed with SuperSignal West Dura Chemiluminescent Substrate (Thermo Scientific) and imaged on an ImageQuant LAS 4000 Mini (GE Healthcare Life Sciences).
Auxin, MG132, Immunoprecipitation/Pull-Down, and NaOH Experiments
Approximately 200 seeds were surfaced sterilized, cold stratified for 3 d at 4°C, and plated in 1 mL of liquid GM in 60- × 15-mm petri dishes. Seedlings were grown for 1 week at 22°C under constant white light at approximately 50 µE. Auxin and MG132 treatment of seedlings was processed as described previously (Dreher et al., 2006), except that treatment was for 6 h with 5 µm IAA or 50 µm MG132. For IP, seedlings were treated with 50 µm MG132 for 3 h and then frozen in liquid nitrogen. Seedlings were ground to a powder in liquid nitrogen, mixed, and ground with IP buffer (50 mm Tris-Cl, pH 7.5, 100 mm NaCl, 10% [v/v] glycerol, 1 mm phenylmethylsulfonyl fluoride, 10 µm MG132, 0.5% [v/v] NP-40, and one 10-mL Complete Mini EDTA Free Protease Inhibitor Tablet; Roche) in a 1:1.5 fresh weight (milligrams):buffer (microliters) ratio. Extracts were centrifuged at 15,000g for 10 min at 4°C. The supernatant was removed and spun again at the same speed. Total protein concentration was determined by Bradford assay, and 5 mg of protein from each line was mixed with 25 µL of 1:1 EZview Red Anti-HA Affinity Gel (Sigma) previously equilibrated in IP buffer. Proteins were immunoprecipitated for 2 h at 4°C with constant mixing. Beads were then washed three times for 10 min each in 1 mL of IP buffer. Protein was eluted from the anti-HA beads by incubating the beads in 500 µL of DPD buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10% [v/v] glycerol, 0.5% [v/v] NP-40, 8 m urea, and 0.5 mm Tris(2-carboxyethyl)phosphine hydrochloride [TCEP]) overnight at room temperature followed by heating for 10 min at 100°C; 500 µL of elution was then mixed with 25 µL of 1:1 HisPur Co2+ Resin (Thermo Scientific) equilibrated with DPD buffer and incubated for 1 h at RT with constant mixing. Co2+ resin was then washed three times for 20 min each in DPD buffer. Protein was eluted off by boiling in 50 µL of 1× Laemmli sample buffer supplemented with 1 mm TCEP and 1 m imidazole; 10 and 40 µL of the sample were loaded for anti-HA and anti-UBQ immunoblotting, respectively.
For immunoblotting, proteins were separated by SDS-PAGE on NuPAGE 4% to 12% BisTris polyacrylamide gradient gels (Life Technologies) in MOPS running buffer and then transferred to Whatman Opitran Reinforced Nitrocellulose Blotting Membrane (10439394) using a wet transfer system. For antiblots using anti-HA-peroxidase (3F10; Roche) antibody, membranes were blocked for 15 min in 5% (w/v) nonfat dry milk in 1× TBST and incubated in 1:1,000 antibody for 2 h in the same solution. For anti-UBQ blots (Pratelli et al., 2012), membranes were blocked in 1% (w/v) bovine serum albumin and 1% (w/v) nonfat dry milk in 1× TBST for 30 min before incubation with 1:2,000 antibody in the same solution. UBQ blots were then washed three times for 5 min each in 1× TBST, blocked again for 15 min in 1% (w/v) bovine serum albumin and 1% (w/v) nonfat dry milk in 1× TBST, and incubated for 1 h with 1:5,000 goat anti-chicken peroxidase-conjugated secondary antibody (KPL). All blots were washed three times for 10 min each in 1× TBST before development with SuperSignal West Pico or Dura Extended Duration Chemiluminescent Substrate (Thermo Scientific) on x-ray film. Anti-UBQ blots were autoclaved for 15 min on a liquid cycle before immunoblotting. All steps of the immunoblotting were performed at room temperature.
For assessment of oxyester UBQ linkages to HIS6x-HA3x-IAA1 proteins, seedlings were grown as above for IP experiments. Proteins were extracted under denaturing conditions in IP buffer supplemented with 8 m urea. Extracts were spun at 15,000g for 10 min at room temperature, supernatant was placed in a new tube, and it was spun again. Protein concentration in the final supernatant fraction was determined by Bradford protein assay. The samples were diluted to a uniform concentration of 4 μg μL−1 in 1× Laemmli sample buffer supplemented with 10 mm TCEP. For each HIS6x-HA3x-IAA1 protein, the sample was then split into two tubes: one with NaOH to a final concentration of 0.2 m and one with water to equivalent volumes. The change in pH could be monitored by the change in color of bromphenol blue in the Laemmli sample buffer. These samples were then incubated with mild shaking at 37°C for 2 h. The samples with NaOH were neutralized with concentrated HCl, again by following the color of bromphenol blue. The water controls had an equivalent volume of water added. Proteins from these samples were separated and immunoblotted with anti-HA antibodies as described above. All IP, auxin, and NaOH experiments were performed on at least three independent transgenic lines for each protein, and all blots are representative of at least three biologically independent experiments. Blots for NaOH treatments were repeated on the same extracts at least three times.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of Aux/IAA proteins.
Supplemental Figure S2. Yeast strains used for two-hybrid assays accumulate the expected DNA binding domain and activation domain fusion proteins.
Supplemental Figure S3. Translation of the HIS6x-HA3x-IAA1 16KR ORF.
Supplemental Figure S4. Single time point cycloheximide assays of the 16KR protein.
Supplemental Table S1. Oligonucleotide primers used for cloning.
Supplementary Material
Acknowledgments
We thank the laboratory of Eric Bennett for efforts to detect the sites of IAA1 ubiquitination by mass spectrometry; Britney Moss for work on 16KR in yeast; Amy Zhang for preliminary experiments on Aux/IAA degradation; the laboratory of J.C. and Laurens Pauwels for helpful suggestions; University of California, Davis undergraduates Joey Lasiter, Tania Gonzalez, and Damei Li and technician Meliza Castro for assistance with plant material and experiments; and the University of California, Davis Controlled Environment Facility for growth of plants.
Glossary
- Col-0
Columbia-0
- DAPI
4′,6-diamino-phenylindole
- DPD
denaturing pull down
- GM
growth media
- IP
immunoprecipitation
- NLS
nuclear localization sequence
- ORF
open reading frame
- TBST
Tris-buffered saline plus Tween 20
- TCEP
Tris(2-carboxyethyl)phosphine hydrochloride
- UPS
ubiquitin-proteasome system
- X-gal
5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid
Footnotes
This work was supported by the National Science Foundation (Arabidopsis 2010 grant no. MCB–0929100 to M.E. and J.C.), the National Institutes of Health (grant no. GM43644 to M.E.), the Howard Hughes Foundation (grants to M.E.), and the Gordon and Betty Moore Foundation (grants to M.E.).
Articles can be viewed without a subscription.
References
- Abel S, Nguyen MD, Theologis A (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Abel S, Theologis A (1995) A polymorphic bipartite motif signals nuclear targeting of early auxin-inducible proteins related to PS-IAA4 from pea (Pisum sativum). Plant J 8: 87–96 [DOI] [PubMed] [Google Scholar]
- Baldi L, Brown K, Franzoso G, Siebenlist U (1996) Critical role for lysines 21 and 22 in signal-induced, ubiquitin-mediated proteolysis of I kappa B-alpha. J Biol Chem 271: 376–379 [DOI] [PubMed] [Google Scholar]
- Bargmann BO, Estelle M (2014) Auxin perception: in the IAA of the beholder. Physiol Plant 151: 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boban M, Ljungdahl PO, Foisner R (2015) Atypical ubiquitylation in yeast targets lysine-less Asi2 for proteasomal degradation. J Biol Chem 290: 2489–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitschopf K, Bengal E, Ziv T, Admon A, Ciechanover A (1998) A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J 17: 5964–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Coscoy L (2005) Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309: 127–130 [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhou Y, Zhang Z, Song J (September 10, 2014) Towards more accurate prediction of ubiquitination sites: a comprehensive review of current methods, tools and features. Brief Bioinform 10.1093/bib/bbu031 [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Ben-Saadon R (2004) N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol 14: 103–106 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Domingues C, Ryoo HD (2012) Drosophila BRUCE inhibits apoptosis through non-lysine ubiquitination of the IAP-antagonist REAPER. Cell Death Differ 19: 470–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea C, Deng L, Xia ZP, Pineda G, Chen ZJ (2006) Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell 22: 245–257 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Gilkerson J, Hu J, Brown J, Jones A, Sun TP, Callis J (2009) Isolation and characterization of cul1-7, a recessive allele of CULLIN1 that disrupts SCF function at the C terminus of CUL1 in Arabidopsis thaliana. Genetics 181: 945–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL (2012) A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol 160: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hua Z, Vierstra RD (2011) The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol 62: 299–334 [DOI] [PubMed] [Google Scholar]
- Ishikura S, Weissman AM, Bonifacino JS (2010) Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J Biol Chem 285: 23916–23924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Estelle M (2012) Ubiquitin-mediated control of plant hormone signaling. Plant Physiol 160: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2004) Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci USA 101: 12381–12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kim DY, Scalf M, Smith LM, Vierstra RD (2013) Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravtsova-Ivantsiv Y, Ciechanover A (2012) Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci 125: 539–548 [DOI] [PubMed] [Google Scholar]
- Ku SJ, Park JY, Ha SB, Kim J (2009) Overexpression of IAA1 with domain II mutation impairs cell elongation and cell division in inflorescences and leaves of Arabidopsis. J Plant Physiol 166: 548–553 [DOI] [PubMed] [Google Scholar]
- Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387–400 [PubMed] [Google Scholar]
- Maraschin Fdos S, Memelink J, Offringa R (2009) Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 59: 100–109 [DOI] [PubMed] [Google Scholar]
- Mattiroli F, Sixma TK (2014) Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat Struct Mol Biol 21: 308–316 [DOI] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A (2001) IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell 13: 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, et al. (2005) Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17: 3282–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Kim HJ, Kim J (2002) Mutation in domain II of IAA1 confers diverse auxin-related phenotypes and represses auxin-activated expression of Aux/IAA genes in steroid regulator-inducible system. Plant J 32: 669–683 [DOI] [PubMed] [Google Scholar]
- Patra B, Pattanaik S, Yuan L (2013) Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant J 74: 435–447 [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2003a) Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol Cell 11: 1435–1444 [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2003b) Redundant degrons ensure the rapid destruction of Sic1 at the G1/S transition of the budding yeast cell cycle. Cell Cycle 2: 410–411 [PubMed] [Google Scholar]
- Pratelli R, Guerra DD, Yu S, Wogulis M, Kraft E, Frommer WB, Callis J, Pilot G (2012) The ubiquitin E3 ligase LOSS OF GDU2 is required for GLUTAMINE DUMPER1-induced amino acid secretion in Arabidopsis. Plant Physiol 158: 1628–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon J, Ramos J, Callis J (2008) Degradation of the auxin response factor ARF1. Plant J 54: 118–128 [DOI] [PubMed] [Google Scholar]
- Saracco SA, Hansson M, Scalf M, Walker JM, Smith LM, Vierstra RD (2009) Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J 59: 344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer DC, Brockman JA, Chen Z, Maniatis T, Ballard DW (1995) Signal-induced degradation of I kappa B alpha requires site-specific ubiquitination. Proc Natl Acad Sci USA 92: 11259–11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Okuda-Shimizu Y, Hendershot LM (2010) Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol Cell 40: 917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Mitao Y, Kakimoto T (2014) Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB. Plant Cell Physiol 55: 1450–1459 [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Treier M, Staszewski LM, Bohmann D (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78: 787–798 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wang R, Estelle M (2014) Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr Opin Plant Biol 21: 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Herr RA, Chua WJ, Lybarger L, Wiertz EJ, Hansen TH (2007) Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J Cell Biol 177: 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Herr RA, Hansen TH (2012) Ubiquitination of substrates by esterification. Traffic 13: 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J (2000) Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 21: 553–562 [DOI] [PubMed] [Google Scholar]
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M (2004) The IAA1 protein is encoded by AXR5 and is a substrate of SCF(TIR1). Plant J 40: 772–782 [DOI] [PubMed] [Google Scholar]
- Yu H, Moss BL, Jang SS, Prigge M, Klavins E, Nemhauser JL, Estelle M (2013) Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol 162: 295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Zhang Y, Moss BL, Bargmann B, Wang R, Prigge M, Nemhauser J, Estelle M (2015) Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat Plants 1: 14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser N, Dreher KA, Edwards SR, Callis J (2003) Acceleration of Aux/IAA proteolysis is specific for auxin and independent of AXR1. Plant J 35: 285–294 [DOI] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J (2001) Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci USA 98: 11795–11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.