Two cellulose synthases interact to synthesize Arabidopsis seed coat mucilage cellulose in a unidirectional manner and promote mucilage adherence and expansion.
Abstract
CELLULOSE SYNTHASE5 (CESA5) synthesizes cellulose necessary for seed mucilage adherence to seed coat epidermal cells of Arabidopsis (Arabidopsis thaliana). The involvement of additional CESA proteins in this process and details concerning the manner in which cellulose is deposited in the mucilage pocket are unknown. Here, we show that both CESA3 and CESA10 are highly expressed in this cell type at the time of mucilage synthesis and localize to the plasma membrane adjacent to the mucilage pocket. The isoxaben resistant1-1 and isoxaben resistant1-2 mutants affecting CESA3 show defects consistent with altered mucilage cellulose biosynthesis. CESA3 can interact with CESA5 in vitro, and green fluorescent protein-tagged CESA5, CESA3, and CESA10 proteins move in a linear, unidirectional fashion around the cytoplasmic column of the cell, parallel with the surface of the seed, in a pattern similar to that of cortical microtubules. Consistent with this movement, cytological evidence suggests that the mucilage is coiled around the columella and unwinds during mucilage extrusion to form a linear ray. Mutations in CESA5 and CESA3 affect the speed of mucilage extrusion and mucilage adherence. These findings imply that cellulose fibrils are synthesized in an ordered helical array around the columella, providing a distinct structure to the mucilage that is important for both mucilage extrusion and adherence.
The epidermal cells of Arabidopsis (Arabidopsis thaliana) seed coats produce two distinct secondary cell walls: pectin-rich mucilage and cellulose-rich columellae (Western et al., 2000). When seeds are hydrated, mucilage expands rapidly, rupturing the outer tangential cell wall and forming a mucilage capsule that surrounds the seed. Seed coat mucilage is composed primarily of rhamnogalacturonan I (RG I) and also contains homogalacturonan (HG), hemicelluloses (such as xylans and glucomannans), and cellulose (for review, see Haughn and Western, 2012). Extruded mucilage consists of an outer, nonadherent fraction and an inner, adherent fraction (Western et al., 2000, 2001; Macquet et al., 2007a). The adherent and nonadherent mucilage layers differ in the amount of methylesterified HG (Rautengarten et al., 2008; Saez-Aguayo et al., 2013; Voiniciuc et al., 2013), galactans (Dean et al., 2007; Macquet et al., 2007b), arabinans (Arsovski et al., 2009), mannans (Yu et al., 2014), and cellulose (Harpaz-Saad et al., 2011; Mendu et al., 2011; Sullivan et al., 2011), all of which influence the physical properties of the layers.
Adherent mucilage has a distinct structure, which can be examined using cell wall dyes and antibodies. When treated with cellulose-specific dyes, densely stained rays extend from the top of each columella to the outer edge of the adherent layer, many cell lengths above the seed surface (Mendu et al., 2011; Sullivan et al., 2011). Cytological evidence indicates that cellulose, pectins, and mannans are components of the ray (Haughn and Western, 2012; Griffiths et al., 2014; North et al., 2014; Yu et al., 2014), although the exact manner in which they are assembled is unknown.
Cellulose is abundant in mucilage rays and mediates adherence. Loss-of-function mutations in CELLULOSE SYNTHASE5 (CESA5) result in reduced cellulose levels and increased detachment of mucilage from the seed (Harpaz-Saad et al., 2011; Mendu et al., 2011; Sullivan et al., 2011; Griffiths et al., 2014). How a reduction in cellulose results in a loss of adherence is still unknown, but it likely involves interaction with other mucilage components such as pectin and arabinogalactan proteins (Griffiths et al., 2014). Since cesa5 mutants still have some cellulose in the rays of the adherent mucilage halo (Mendu et al., 2011; Sullivan et al., 2011), additional cellulose synthases must be involved in mucilage cellulose biosynthesis.
The Arabidopsis genome encodes 10 different CESAs (Delmer, 1999; Richmond and Somerville, 2000). Multiple lines of evidence suggest that three different CESAs are required to form one active cellulose synthase complex (CSC; for review, see Somerville, 2006). CSCs are membrane-bound protein complexes that synthesize cellulose microfibrils in the apoplast (for review, see Somerville, 2006; Endler and Persson, 2011; Lei et al., 2012). CESA1, CESA3, and CESA6 are considered the core components of the primary wall CSC (Desprez et al., 2007; Persson et al., 2007). CESA2, CESA5, and CESA9 are partially redundant to CESA6 in primary wall biosynthesis, and genetic evidence suggests that each of these CESA polypeptides can form a functional CSC with CESA3 and CESA1 (Desprez et al., 2007; Persson et al., 2007). CESA10 is expressed in young plants, stems, floral tissue, and the base of rosette leaves (Beeckman et al., 2002; Doblin et al., 2002), but its function in cellulose biosynthesis is unclear. Other cesa mutant lines have been examined for altered mucilage phenotypes (cesa1, radially swollen1 [Burn et al., 2002; Sullivan et al., 2011], cesa2, cesa6, and cesa9 [Mendu et al., 2011]; CESA3, je5 [Sullivan et al., 2011] and cesa10-1 [Sullivan et al., 2011]); to date, only CESA5 has been shown to be required for cellulose biosynthesis during mucilage deposition.
Two mutant alleles of CESA3, isoxaben resistant1-1 (ixr1-1) and ixr1-2, were isolated in a screen for resistance to the herbicide isoxaben (Scheible et al., 2001). Isoxaben inhibits the incorporation of Glc into the emerging cellulose polymer and is considered a potent and specific inhibitor of cellulose biosynthesis (Heim et al., 1990). Homozygous ixr1-1 and ixr1-2 lines show increased resistance to the herbicide, and the mutations causing this resistance were mapped to the genomic locus of CESA3 (Heim et al., 1990; Scheible et al., 2001). The ixr1-1 and ixr1-2 mutations cause amino acid substitutions near the C terminus of the CESA3 protein. ixr1-1 causes a Gly-to-Asn substitution (G998A) located in a transmembrane domain, while ixr1-2 contains a Thr-to-Ile substitution (T942I) in an apoplastic region of the protein between two transmembrane domains (Scheible et al., 2001). Recently, the ixr1-2 allele was shown to affect the velocity of CSCs in the plasma membrane, which consequently modifies cellulose crystallinity in the cell wall (Harris et al., 2012). It is not exactly clear how the ixr1-1 mutation affects cellulose biosynthesis. The effects of either of these mutations on seed coat mucilage have not been investigated.
Since mucilage is composed primarily of pectins with smaller amounts of cellulose, seed coat epidermal cells represent an excellent system to study cellulose biosynthesis and interactions between cellulose and other wall components in muro. In this study, we investigated how cellulose is synthesized and deposited in seed coat epidermal cells. We show that at least three different CESA proteins are highly expressed in the seed coat epidermis during mucilage biosynthesis. These CESAs are oriented and move in a linear fashion around the cytoplasmic column of each cell in an identical pattern to cortical microtubules. In addition, we provide evidence that the adherent mucilage has a helical structure that expands and unwinds during extrusion to form the mucilage ray. We propose that during seed coat epidermal cell development, the biosynthesis of cellulose predetermines the structure of rays in the adherent mucilage layer.
RESULTS
CESA3 and CESA10 Are Highly Expressed during Mucilage Biosynthesis
In order to identify CESAs involved in cellulose biosynthesis during mucilage deposition, we analyzed the expression of all 10 Arabidopsis CESAs during seed development using publicly available Arabidopsis gene expression resources (Supplemental Tables S1–S4; Winter et al., 2007; Bassel et al., 2008; Le et al., 2010; Dean et al., 2011). During seed coat epidermal cell differentiation, mucilage is actively synthesized in large amounts at approximately 5 to 9 DPA (Western et al., 2000). Several genes required for cell wall biosynthesis show increased transcript abundance during this period. The amount of MUCILAGE MODIFIED4 (MUM4) transcript, a gene encoding a Rha synthase required for mucilage synthesis (Usadel et al., 2004; Western et al., 2004, Oka et al., 2007), increases dramatically from 4 to 7 DPA (Western et al., 2004; Dean et al., 2011). Under our growth conditions, seeds at 7 DPA contain an embryo at the bent cotyledon stage and are at the peak of mucilage biosynthesis (Western et al., 2001; Haughn and Chaudhury, 2005). At 10 DPA, the embryo is at the mature green stage when mucilage production has ceased and columella biosynthesis is under way (Western et al., 2000; Haughn and Chaudhury, 2005).
During seed coat development, the expression of CESA3, CESA5, and CESA10 peaks at approximately 7 DPA, similar to MUM4 (Supplemental Tables S1 and S2). CESA2 was more highly expressed at approximately 4 DPA, during the heart embryo stage, whereas CESA1 and CESA6 displayed relatively constant expression levels throughout seed coat development. Whole-seed expression data suggest that CESA3 is the most highly expressed of all CESAs during seed development, based on publicly available microarray data (Supplemental Table S3). We also searched, using the seed compendium data set in the online bioinformatics tool Expression Angler (Toufighi et al., 2005), for CESA genes showing an expression pattern during seed development positively correlated with that of CESA5. The expression patterns of CESA3 (r = 0.733), CESA1 (0.648), and CESA10 (0.633) were positively correlated with CESA5 (Supplemental Table S4). The peak of CESA5, CESA3, and CESA10 expression correlated with mucilage biosynthesis; during this stage, both CESA3 and CESA10 were more highly expressed than CESA5 (Supplemental Table S1).
GFP-Tagged CESA5, CESA3, and CESA10 Are Localized in the Cytoplasmic Column during Mucilage Biosynthesis
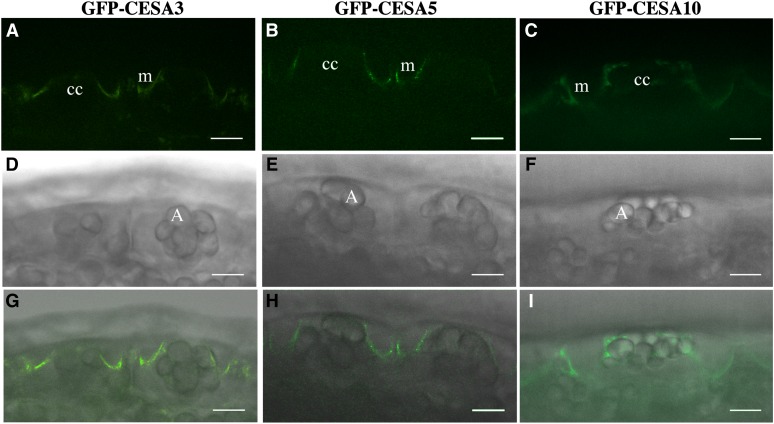
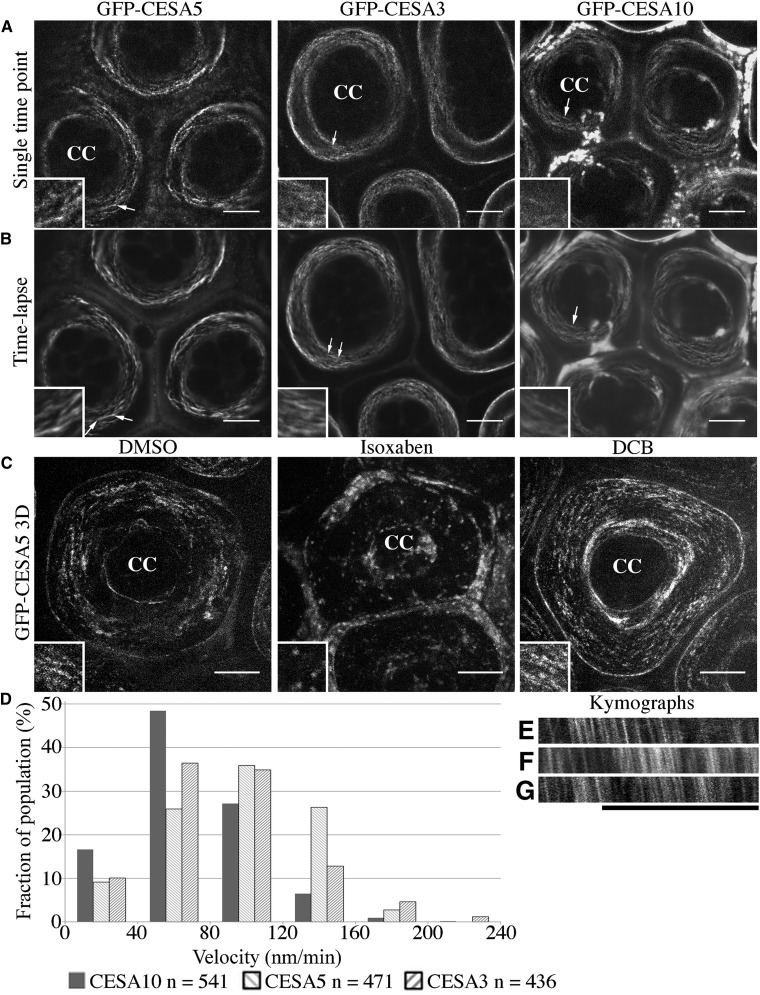
Based on the expression patterns during mucilage biosynthesis (Supplemental Tables S1–S3), we chose to examine the expression and intracellular localization of CESA5, CESA3, and CESA10 in seed coat epidermal cells using CESA-GFP fusion proteins under the control of their native promoters. Previously, GFP-CESA5 was shown to be present in the plasma membrane of seed coat epidermal cells during all stages of seed coat development and to complement the mucilage adherence phenotype of cesa5-1 (Sullivan et al., 2011). GFP-CESA3 can complement the dwarf phenotype of je5, an allele of cesa3 (Desprez et al., 2007), yet je5 does not have any obvious mucilage phenotype (Sullivan et al., 2011). At 7 DPA, GFP-CESA5 was present in seed coat epidermal cells, at the interface between the cytoplasmic column and the mucilage pocket (Fig. 1). At this stage of seed coat development, the subcellular localization of GFP-CESA3 and GFP-CESA10 was nearly identical to that of GFP-CESA5 (Fig. 1). Like GFP-CESA5, GFP-CESA10 and GFP-CESA3 were expressed throughout the period of mucilage and columella biosynthesis (4–10 DPA; Supplemental Fig. S1; Sullivan et al., 2011). GFP-CESA10 colocalized with FM4-64, a plasma membrane marker, in the cytoplasmic column, demonstrating that CESA10 is present in the membrane during mucilage biosynthesis (Supplemental Fig. S2). GFP-CESA10 also partially colocalizes with red fluorescent protein (RFP)-tagged VACUOLAR PROTON ATPASE A1 (VHA-a1), a marker for the trans-Golgi network (Dettmer et al., 2006), suggesting that GFP-CESA10 is secreted and recycled similar to CESAs in other tissues (Supplemental Fig. S2; Supplemental Table S5; Supplemental Video S1; Crowell et al., 2009; Gutierrez et al., 2009). We also examined proCESA6::GFP-CESA6 expression and localization in 7-DPA seed coat epidermal cells, and no fluorescence signal was detected, consistent with the hypothesis that CESA6 is not involved in mucilage biosynthesis in the seed coat (Supplemental Fig. S3; Mendu et al., 2011; Sullivan et al., 2011). These results show that CESA3 and CESA10 proteins, like CESA5, are present in the plasma membrane of the cytoplasmic column of seed coat epidermal cells during mucilage biosynthesis.
Figure 1.
GFP-CESA3 and GFP-CESA10 are expressed in seed coat epidermal cells during mucilage biosynthesis. Seed coat epidermal cells at 7 DPA show GFP-CESA fluorescence adjacent to the mucilage pocket. A to C, GFP channel. D to E, Transmitted light channel. G to I, Overlay of GFP and transmitted light channels. A, Amyloplasts; cc, cytoplasmic column; m, mucilage pocket. Bars = 10 μm.
CESA5 and CESA3 Interact through the Zinc Finger RING-Type Domain
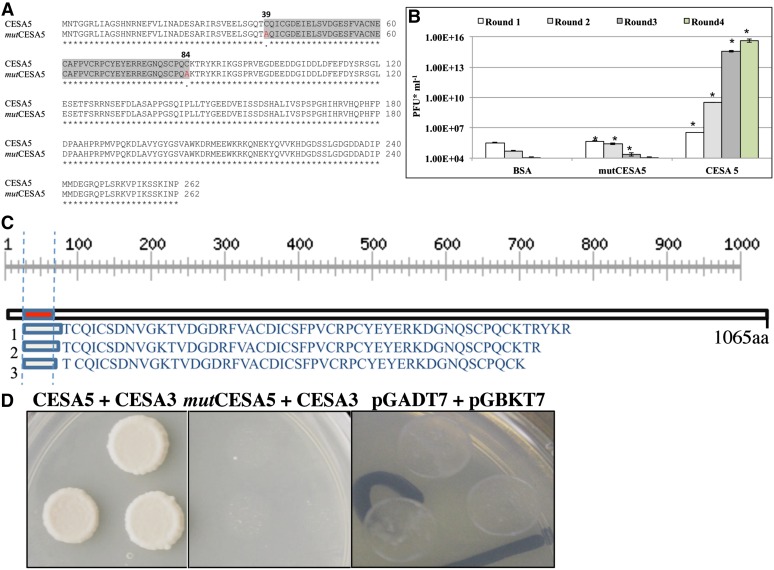
Next, we sought to identify specific protein-protein interactions between CESAs using phage-display technology (Chen et al., 2010; Kushwaha et al., 2012). The N terminus of CESA5, like other CESAs, has a Cys-rich RING-type zinc finger domain that has been implicated in mediating protein-protein interactions (Saurin et al., 1996). Therefore, we cloned the N-terminal 786-bp wild-type CESA5 (N-CESA5) and, as a negative control, a mutCESA5 N-terminal region with mutations in the coding region that result in the substitution of two Cys residues with Ala (C39A and C84A; Fig. 2). These constructs were used as bait in a phage-display assay. We first tested the ability of N-CESA5 and mutCESA5 to bind to zinc using a 4-(2-pyridylazo) resorcinol assay (Supplemental Fig. S4; Louie and Meade, 1998). Zinc binding by N-CESA5 was directly proportional to the protein concentration, while zinc binding of mutCESA5 was not, suggesting that this mutated version of N-CESA5 is incapable of protein-protein interactions. Both constructs were expressed and bound to microtiter plate wells (Supplemental Fig. S4, A and B). The Mr of the protein was as expected for a 262-amino acid protein with a thrombin tag. A seed phage-display library was successively biopanned against the N-CESA5 and mutCESA5 immobilized proteins (Chen et al., 2010). Bovine serum albumin (BSA) and mutCESA5 negative controls failed to bind significant amounts of phage display proteins, whereas N-CESA5 bound to increasing amounts of protein with each successive biopanning round (Fig. 2B; Supplemental Fig. S4C). This indicates that N-CESA5 was selectively binding proteins displayed on phage and that these interactions are specific.
Figure 2.
CESA3 and CESA5 interact in vitro through zinc finger domains. A, CESA5 amino acid sequence alignment of the open reading frame in a 786-bp fragment of the N terminus, with the corresponding sequence from mutCESA5 showing the substitution of Cys (C) at positions 39 and 84 with Ala (A). The highlighted region corresponds to the zinc finger domain, with Ala substitutions in red. B, Phage titer (plaque-forming units [PFU] mL−1) for four rounds of successive biopanning. Asterisks denote significant differences (Student’s t test, P < 0.05) between titers compared with BSA. Error bars indicate se. C, Sequences of three independent CESA3 hits identified through interaction with N-CESA5. The black box represents the N-terminal region of the CESA3 protein, with the zinc finger-like domain of CESA3 shown as a red box. The scale corresponds to the length of the protein in amino acids (aa). D, Yeast (Saccharomyces cerevisiae) two-hybrid assay confirming the interaction between CESA5 and CESA3, but not mutCESA5 and CESA3, with empty vector controls.
Following four successive rounds of biopanning, DNA from randomly selected phage inserts was isolated, amplified, and sequenced. Roughly 60% of identified inserts were found to be in frame. About 40% of the in-frame hits were identified as CESA3 (Fig. 2C). The N-CESA5 peptide binds to a region of approximately 48 to 53 amino acids at the N terminus of CESA3 (Fig. 2C). A yeast two-hybrid assay confirmed the interaction between N-CESA5 and CESA3, while no interaction was detected between CESA3 and mutCESA5 or an empty vector control (Fig. 2D). Therefore, CESA3 and CESA5 are coexpressed in seed coat epidermal cells during mucilage biosynthesis and can interact with one another in vitro to form higher order complexes.
ixr1-1 and ixr1-2 Mutations Alter Polysaccharide Distribution in Mucilage
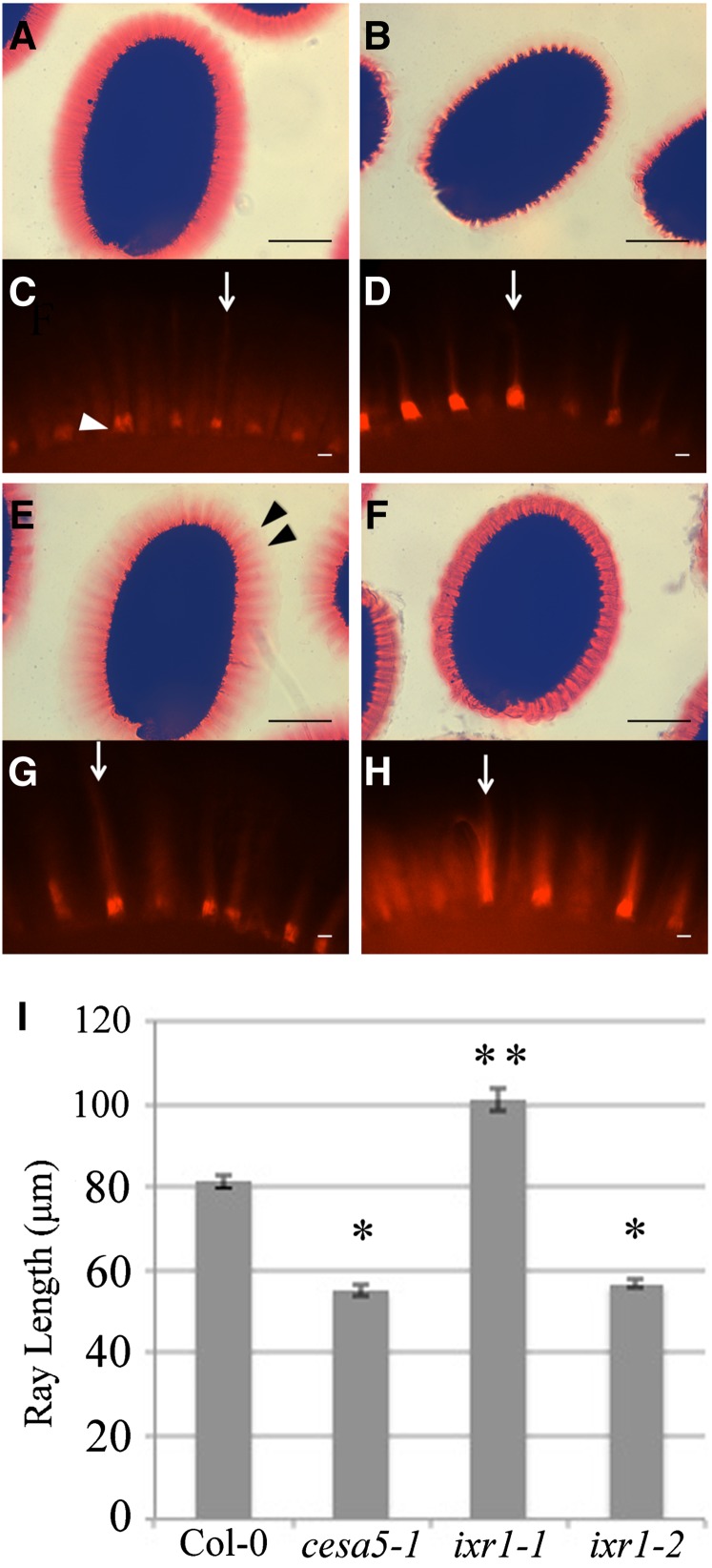
We used a reverse genetics approach to investigate whether mutations in CESA genes result in altered seed coat mucilage. Plants with mutations in CESA1, CESA2, CESA6, CESA9, and CESA10 were previously shown to have normal mucilage (Burn et al., 2002; Mendu et al., 2011; Sullivan et al., 2011). Plants homozygous for mutations in CESA1 (anisotropy1-1; Fujita et al., 2013), CESA3 (ixr1-1 and ixr1-2; Scheible et al., 2001), and CESA10 (cesa10-1 [SALK_150405] and cesa10-2 [SALK_052533]) were obtained (Supplemental Table S6). Seeds were shaken in water, stained with the pectin dye Ruthenium Red (RR; Sterling, 1970), and compared with wild-type and cesa5-1 seeds (Fig. 3; Supplemental Fig. S5). When seeds are hydrated with gentle shaking, mucilage separates into a nonadherent layer and an adherent layer that contains relatively higher amounts of cellulose and remains attached to the parent seed. RR-stained Columbia-0 (Col-0) wild-type seeds showed a large adherent mucilage halo, while cesa5-1 seeds showed an almost complete loss of mucilage RR staining (Fig. 3). The mucilage halo of ixr1-1 seeds was smaller than that of wild-type seeds yet larger than that of cesa5-1 seeds. RR-stained ixr1-1 mucilage had reduced staining in the outer regions of the halo, and the halo margin appeared dentate, with regions of concentrated staining above the columella (Fig. 3E, arrowheads). The ixr1-1 mucilage phenotype segregated as a single recessive nuclear mutation when crossed to Col-0 (3:1 wild type:ixr1-1; χ2 = 1.55 < 3.84; P < 0.05; n = 40) and cosegregated with the ixr1-1 point mutation in the F2 progeny. The ixr1-2 seed mucilage halo was more compact and more intensely stained by RR compared with the wild type (Fig. 3F). Without shaking, no major differences in RR staining were observed between wild-type and mutant seeds (Supplemental Fig. S6, A–D). In addition, both ixr1 mutants have normal epidermal cell morphology and columella size when mature dry seeds were viewed using scanning electron microscopy (SEM; Supplemental Fig. S7). Mutations in the other genes examined, including CESA10, did not show any obvious seed mucilage differences compared with the wild type (Supplemental Fig. S5). We examined the location of the cesa10-2 insertion and tested if this line fails to produce CESA10 transcript. The insertion was located in an intron between the 8th and 9th exons of CESA10 (Supplemental Fig. S5H); however, we were still able to identify transcript in cesa10-2 seeds using primers located 3′ of the insertion (Supplemental Fig. S5E). Additionally, primers spanning the insertion detected a band of unexpected size when amplifying cesa10-2 complementary DNA (Supplemental Fig. S5F). Sequencing of this larger band demonstrates that the insertion is not completely spliced from the CESA10 mRNA and is predicted to cause a premature stop codon in the coding region upstream of the catalytic site that would result in a truncated protein (Supplemental Fig. S5H).
Figure 3.
ixr1 seeds have altered mucilage structure and cellulosic ray density. A, B, E, and F, RR-stained wild-type, cesa5-1, ixr1-1, and ixr1-2 seeds, respectively. Black arrowheads indicate ixr1-1 mucilage rays. Bars = 200 μm. C, D, G, and H, S4B-stained wild-type, cesa5-1, ixr1-1, and ixr1-2 seeds, respectively. White arrowhead indicates columella with primary wall attached. Bars = 10 μm. Arrows indicate rays. I, Ray length of S4B-stained wild-type, cesa5-1, ixr1-1, and ixr1-2 seeds. Error bars indicate se. Single asterisks indicate ray length significantly reduced compared with the wild type, and double asterisks indicate ray length significantly increased compared with the wild type, based on Student’s t test (P < 0.05).
When examining unstained ixr1-1 and ixr1-2 seeds, we were able to clearly observe ray-like structures, which were less visible in the wild type and not visible in the cesa5 mutant (Supplemental Fig. S5, E–H). The altered RR staining of mucilage and increased visibility of rays in ixr1-1 and ixr1-2 seeds suggest altered polysaccharide distribution in mucilage. The distribution of cellulose in the mucilage halo of ixr1 seeds was investigated using Pontamine Fast Scarlet 4B (S4B), a fluorescent stain that binds cellulose (Fig. 3; Anderson et al., 2010; Mendu et al., 2011). Wild-type seeds extruded mucilage and outer tangential primary cell walls, which remain attached to columellae following extrusion, and fluoresced intensely when stained with S4B (Fig. 3C, arrowhead). Two distinct domains were observed in the S4B-stained adherent mucilage halo: intensely staining rays extending from the top of the columellae to the periphery of the mucilage halo (Fig. 3C, arrow), and regions with more diffuse signal between the rays. S4B signals in cesa5-1 mucilage were reduced compared with the wild type, but rays were still observed above the columellae (Fig. 3D, arrow). Interestingly, in ixr1-1 mutants, mucilage had S4B-stained rays that appeared longer and were more intense than in the wild type (Fig. 3G, arrow). Compared with the wild type, ixr1-2 had increased S4B mucilage staining (Fig. 3H, arrow), while cesa5-1 and ixr1-1 had reduced S4B staining.
Ray length in S4B-stained seeds was quantified by measuring the distance from the top of the columellae to the outer edges of the rays. In total, three biological replicates of 20 rays each were measured from multiple seeds for each genotype (Fig. 3I). While cesa5-1 and ixr1-2 mutant seeds had much shorter rays than the wild type, ixr1-1 mutant seeds had much longer rays (Fig. 3I). The RR and S4B staining results are consistent with the idea that ixr1-1 and ixr1-2 mutants have an altered cellulose organization as well as altered structure of the adherent mucilage halo. The two ixr1 missense mutations induce different CESA3 protein changes and appear to have distinct effects on seed mucilage structure.
Next, we isolated a cesa5-1 ixr1-1 double mutant (Supplemental Fig. S8). Only one homozygous ixr1-1 cesa5-1 double mutant was identified in an F2 population of 936 individuals, consistent with CESA3 and CESA5 being closely linked (approximately 1.5-Mb apart) on chromosome III. Based on RR staining, the cesa5-1 ixr1-1 double mutant phenotype appears intermediate between both single mutants (Supplemental Fig. S8, A, B, and D). Mucilage halo widths for the cesa5-1 ixr1-1 double mutant were consistently smaller than those of ixr1-1 and wild-type seeds yet larger than those of cesa5-1 seeds. Since ixr1-1 is not a null mutation and has an effect on ray length opposite to that of cesa5-1, interpretation of this double mutant is difficult. However, based on the intermediate mucilage phenotype of cesa5-1 ixr1-1 seeds, we conclude that both CESA5 and CESA3 are involved in mucilage cellulose synthesis and that they are at most partially redundant. We also isolated four cesa5-1 cesa10-1 double mutants from a segregating F2 population from a cross between both single mutants (n = 72; χ2 = 2.17 < 7.815). No obvious changes in the mucilage phenotype were observed compared with cesa5-1 seeds; therefore, CESA10 is not necessary for mucilage biosynthesis and does not appear to be redundant with CESA5 (Supplemental Fig. S8C).
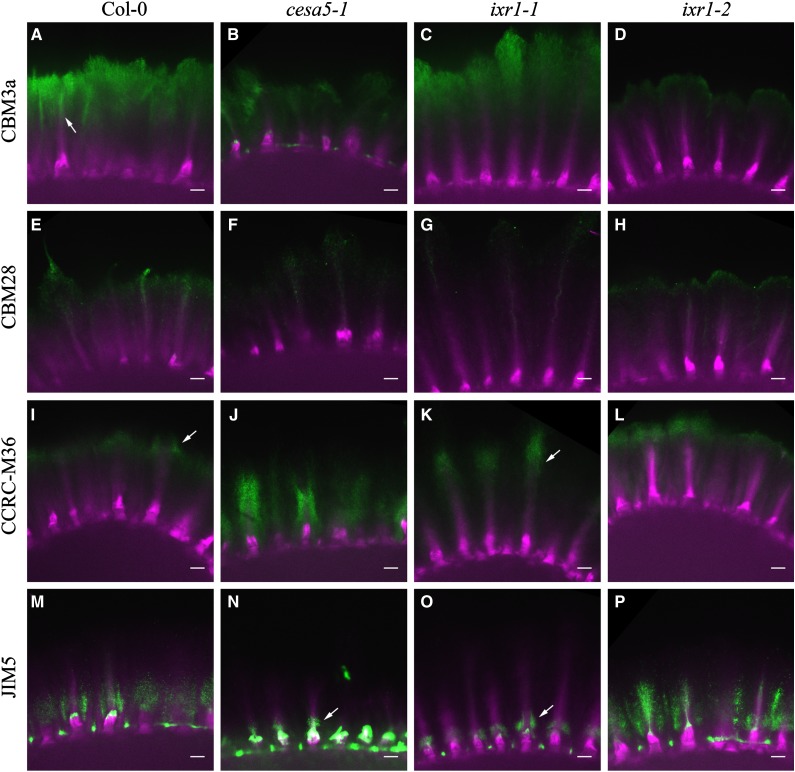
To further characterize the distribution of cellulose in the adherent mucilage layer, seeds were exposed to solutions of proteins carrying one of two carbohydrate-binding modules (CBM3a and CBM28). CBM3a recognizes crystalline cellulose structures, whereas CBM28 recognizes amorphous cellulose structures (Blake et al., 2006; Dagel et al., 2011). Seeds were first hydrated in water for 1 h, exposed to CBM3a or CBM28, and counterstained with S4B to detect the cellulose in the mucilage. Negative controls lacking CBM3a, CBM28, or other primary antibodies showed no detectable fluorescence in the mucilage halo when viewed under similar imaging conditions (Supplemental Fig. S9). Results were similar for multiple seeds from three independent biological replicates. S4B stained the adherent mucilage layer, highlighting the rays above the columella and the diffuse mucilage (Figs. 3 and 4). In contrast, CBM3a most strongly labeled the outer edge of the mucilage halo and the top of the rays (Fig. 4A, arrow; Supplemental Fig. S10). CBM3a signals were reduced in cesa5-1 and ixr1-2 seeds compared with the wild type, while the CBM3a halo size in ixr1-1 seeds was larger than that in the wild type and appeared more concentrated around the rays (Fig. 4, C and D). These results suggest that ixr1-2 has reduced crystalline cellulose in mucilage, while ixr1-1 has an altered distribution of crystalline cellulose in seed mucilage. CBM28 labeling was reduced compared with CBM3a in all genotypes, so laser intensity and detection sensitivity were increased to facilitate imaging. cesa5-1, ixr1-1, and ixr1-2 seeds had reduced CBM28 labeling and altered signal distribution compared with the wild type (Fig. 4; Supplemental Fig. S11), consistent with altered cellulose distribution and amounts in ixr1 mucilage.
Figure 4.
ixr1 seed mucilage has altered cellulose and pectin distribution. Images show CBM or pectin antibody immunolabeling (green) merged with S4B staining (magenta). Seed mucilage was labeled for crystalline cellulose with CBM3a, amorphous cellulose with CBM28, unsubstituted RG I with CCRC-M36, and low-methylesterified HG with JIM5. Bars = 15 μm.
We investigated pectin distribution in ixr1 mucilage with two pectin-specific antibodies (JIM5 and CCRC-M36; Fig. 4; Supplemental Figs. S12 and S13). CCRC-M36 recognizes the unbranched backbone of RG I (Young et al., 2008; Pattathil et al., 2010), whereas JIM5 recognizes partially methylesterified HG (Liners et al., 1989; Knox et al., 1990; Knox, 1997; Willats et al., 2001a; Macquet et al., 2007a). In wild-type seeds, CCRC-M36 labeled the entire adherent mucilage halo, with increased labeling at the periphery of the halo and around the end of the rays (Fig. 4I, arrow; Supplemental Fig. S12). CCRC-M36 signal in cesa5-1 mucilage surrounded the ray (Supplemental Fig. S12, J and N) and was reduced compared with the wild type (Fig. 4J; Supplemental Fig S12). In ixr1-1 mucilage, CCRC-M36 signal was similarly focused at the rays yet farther from the seed surface (Fig. 4K, arrow). The labeling pattern of ixr1-2 seeds was similar to that of wild-type seeds but appeared more intense (Fig. 4L; Supplemental Fig. S12).
JIM5 labeled the inner region of the adherent halo in wild-type seeds and loosely surrounded the rays (Fig. 4M; Supplemental Fig. S13). JIM5 signal in cesa5-1 and ixr1-1 mucilage was reduced compared with the wild type and only observed at the base of the ray, near the columellae (Fig. 4, N and O, arrows). Relative to the wild type, the JIM5 signal appeared to be more abundant in ixr1-2 mucilage and was more closely associated with the rays (Fig. 4P; Supplemental Fig. S13). In summary, the ixr1-1 and ixr1-2 mutations cause distinct changes in the organization of cellulose and pectins in adherent mucilage, suggesting a close association between these cell wall polysaccharides.
ixr1-2 Seeds Have Increased Mucilage Adherence
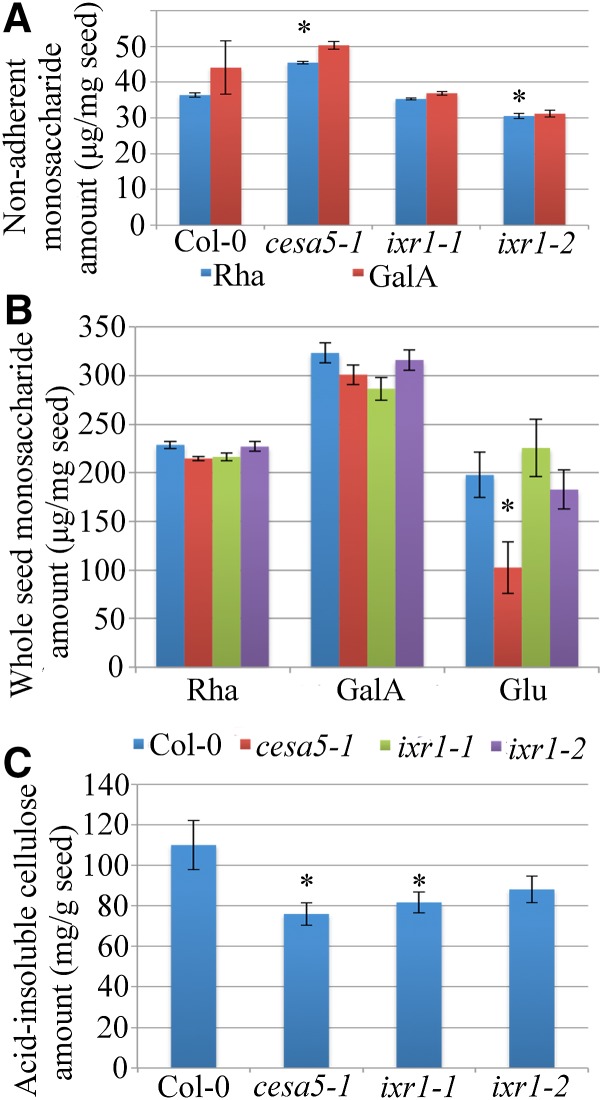
Since the structure of the adherent mucilage halo was changed in ixr1 mutant seeds, soluble mucilage and whole-seed monosaccharide compositions were analyzed to quantify these changes (Fig. 5; Supplemental Table S7). Seeds were shaken in water, and the nonadherent mucilage fraction was isolated, hydrolyzed, and analyzed using high-performance anion-exchange chromatography. Similar to previous reports (Mendu et al., 2011; Sullivan et al., 2011), cesa5-1 mutant seeds had significantly more nonadherent Rha compared with the wild type (Fig. 5A; Student’s t test, P < 0.05), indicating that cesa5-1 mucilage is less adherent than that of the wild type. While ixr1-1 seeds showed no significant change in Rha or GalA amounts, ixr1-2 had a significant reduction in the amount of nonadherent Rha compared with the wild type, suggesting increased mucilage adherence (Fig. 5A; Student’s t test, P < 0.05). GalA amounts in ixr1-2 seeds showed the same trend as Rha, but this reduction was not statistically significant (Supplemental Table S7).
Figure 5.
ixr1 seeds show reduced crystalline cellulose, and ixr1-2 seeds have an increased adherent mucilage layer. A, Rha and GalA amounts in nonadherent mucilage. B, Whole-seed Rha, GalA, and Glu amounts. C, Acid-insoluble cellulose amounts. Error bars indicate se. Asterisks indicate significant differences from the wild type by Student’s t test (P < 0.05).
We next examined whole-seed monosaccharide content in order to determine if the observed defects result from altered mucilage adherence or changes in the amount of mucilage synthesized. Wild-type, cesa5-1, ixr1-1, and ixr1-2 seeds had similar Rha or GalA amounts compared with the wild type, indicating that the biosynthesis of pectin was not affected by mutations in the CESA genes (Fig. 5B). Glc levels were reduced in cesa5-1 seeds but not in ixr1 seeds, suggesting that cellulose biosynthesis is reduced only in cesa5-1 seeds (Fig. 5B). The comparison of whole-seed and mucilage monosaccharide contents suggests that ixr1-2 seeds have increased mucilage adherence.
Crystalline Cellulose Amounts Are Reduced in ixr1 Seeds
We quantified the crystalline cellulose content in whole seeds using the anthrone reagent to determine if ixr1-1 and ixr1-2 affect cellulose biosynthesis (Updegraff, 1969). Crystalline cellulose amounts were significantly lower in cesa5-1 and ixr1-1 seeds compared with wild-type seeds (Fig. 5C; Student’s t test, P < 0.05). The mean amount of crystalline cellulose in ixr1-2 seeds was also lower than in the wild type, but it was not significantly different (P = 0.12; Fig. 5C; Supplemental Table S7). These reductions in crystalline cellulose are consistent with mutations affecting cellulose biosynthesis in mucilage and whole seeds. Unlike cesa5-1, which has reduced crystalline cellulose and less Glc in seeds (Fig. 3, B and C), ixr1-1 and ixr1-2 only show changes in crystalline cellulose levels, suggesting that ixr mutant seeds synthesize cellulose polymers with altered properties.
Cellulose Is Required for Normal Mucilage Extrusion
The structure of RG I in the mucilage pocket has a strong influence on the ability of the mucilage to extrude when exposed to water. Mutations in genes that encode the mucilage-modifying enzymes MUM2 (β-galactosidase) and BETA-XYLOSIDASE1 (BXL1; β-xylosidase/arabinosidase) significantly impair mucilage extrusion (Dean et al., 2007; Macquet et al., 2007b; Arsovski et al., 2009). To investigate if the disruption of cellulose content in mucilage also affects mucilage release, we compared the extrusion rates of cesa5-1 and bxl1-1. When hydrated, all cesa5-1 and wild-type seeds released mucilage very quickly, while bxl1-1 seeds had a delayed mucilage release (Table I). The cesa5-1 bxl1-1 double mutant seeds released mucilage at an even slower rate than the bxl1-1 single mutant (Table I), suggesting that cellulose synthesized by CESA5 facilitates mucilage extrusion. To test this hypothesis, we examined how cesa5-1, ixr1-1, and ixr1-2 seeds extrude mucilage in water-limiting conditions. Compared with the wild type, a lower proportion of cesa5 and ixr1 mutant seeds released mucilage in solutions with increasing concentrations of polyethylene glycol (Table II). Since cesa5-1 enhanced the bxl1 extrusion defect and cesa5-1 and ixr1 mucilage release was delayed compared with wild-type seeds with reduced water availability, cellulose appears to promote mucilage release.
Table I. cesa5-1 enhances the delayed mucilage release phenotype of bxl1-1.
Values are percentages of seeds showing any degree of mucilage release for each genotype when hydrated without shaking for the indicated time or after shaking on a rotator for 120 min. The number of seeds examined for each genotype is shown in parentheses.
| Genotype | Time |
Shaking | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 15 | 30 | 45 | 60 | 90 | 120 | ||
| min | ||||||||
| Col-0 (50) | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 98 |
| cesa5-1 (49) | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| bxl1-1 (47) | 23 | 47.80 | 63 | 60.80 | 72.30 | 76.08 | 80.43 | 87.75 |
| bxl1-1 cesa5-1 (71) | 2.00 | 2.80 | 4.20 | 7.00 | 8.45 | 9.85 | 9.85 | 33.33 |
Table II. Cellulose facilitates mucilage extrusion.
Values indicate the percentage of seeds showing any degree of mucilage release for each genotype. n = 100 for each genotype.
| Genotype | Percentage Polyethylene Glycol |
||||
|---|---|---|---|---|---|
| 0 | 5 | 10 | 15 | 20 | |
| Col-0 | 100 | 90 | 98.40 | 96.88 | 81.36 |
| cesa5-1 | 97.78 | 93.75 | 92.31 | 78.41 | 46.59 |
| ixr1-1 | 95.83 | 92.73 | 92.65 | 80.88 | 60.92 |
| ixr1-2 | 94.55 | 72.22 | 60.00 | 44.68 | 16.84 |
We also examined the ability of ixr1 seeds to extrude mucilage in solutions of calcium chloride and EDTA. Unesterified GalA regions of HG polymers can be cross-linked by calcium bridges (Ca2+), resulting in more cohesive pectin gels (Willats et al., 2001b). Mutant seeds were hydrated in a solution of CaCl2, which is expected to increase cross-linking, or in EDTA, which sequesters divalent cations such as Ca2+ and disrupts cross-linking. EDTA-treated ixr1-1 and ixr1-2 seeds showed mucilage halos similar in size to the wild type, indicating a partial rescue of their mucilage defects (Supplemental Fig. S14). In contrast, CaCl2-treated ixr1-1 and ixr1-2 seeds displayed much smaller mucilage halos than the wild type and frequently failed to release any mucilage (Supplemental Fig. S14), again suggesting that changes in mucilage cellulose can reduce mucilage extrusion.
GFP-CESA5 and GFP-CESA3 Are Localized in Linear Arrays around the Cytoplasmic Column
The rays of extruded mucilage are composed, at least in part, of cellulose and pectins (Figs. 3 and 4). Prior to extrusion, these polymers are tightly packed in the mucilage pockets of the seed coat epidermal cells. While the length of wild-type rays is approximately 80 μm (Fig. 3I), the height of the mucilage pocket is only 15.28 ± 0.74 μm (n = 15). Rays must either expand vertically roughly five to six times the height of the mucilage pocket or be organized in the apoplast around the columella. To understand how cellulose is deposited during mucilage synthesis and ultimately contributes to the formation of rays, we examined the localization and dynamics of GFP-tagged CESA proteins in seed coat epidermal cells (Fig. 6). GFP-CESA5, GFP-CESA3, and GFP-CESA10 are arranged in linear arrays around the cytoplasmic column during mucilage biosynthesis (Fig. 6A, arrows). Time-lapse images of GFP-CESA5, GFP-CESA3, and GFP-CESA10 indicated that all three CESA proteins moved in a circumferential manner around the cytoplasmic column parallel with the outer tangential surface of the cell (Fig. 6, A and B, insets). This pattern was enhanced when imaging GFP-CESA5 seed coat epidermal cells for 10 min at 5 s per image (Supplemental Fig. S15; Supplemental Video S2). GFP-CESA5 was calculated to have a mean velocity of 94.87 ± 29.35 nm min−1 (mean ± sd; n = 471; 22 cells, six seeds), similar to the velocity of unphosphorylated GFP-CESA5 in hypocotyl cells (Bischoff et al., 2011). GFP-CESA3 had a calculated mean velocity of 84.63 ± 38.49 nm min−1 (n = 436; 28 cells from seven seeds), while GFP-CESA10 had a calculated mean velocity of 70.98 ± 35.37 nm min−1 (n = 541; 36 cells from nine seeds). Despite the slight variation in the mean velocity of different CESAs, the three proteins showed a similar overall distribution of velocities (Fig. 6D). Surprisingly, all GFP-CESAs examined moved in a unidirectional, clockwise manner (Fig. 6, E–G).
Figure 6.
CESA5, CESA3, and CESA10 traffic in linear arrays around the cytoplasmic column. A, Single-time point images of GFP-CESAs in seed coat epidermal cells. B, Time-lapse averages (median intensity) of GFP-CESAs in seed coat epidermal cells. Insets show the linear movement of GFP-CESAs around the cytoplasmic column. C, Maximum projection of a z-stack of GFP-CESA5 seeds treated with 0.01% dimethyl sulfoxide (DMSO), 1 μm isoxaben, and 1 μm DCB. CC, Cytoplasmic column. Arrows indicate linear striations and approximate locations of the magnified insets. Insets are approximately 5 μm in height and width. Bars = 10 μm. D, Velocity distribution of GFP-CESAs in seed coat epidermal cells. E to G, Kymographs of GFP-CESAs of images taken every 5 s for 5 min: GFP-CESA5 (E), GFP-CESA3 (F), and GFP-CESA10 (G). Bar = 10 μm.
To test whether this distribution pattern was related to the function of CESA complexes, we treated seeds expressing GFP-CESA5 with two cellulose biosynthesis inhibitors: isoxaben, which induces a rapid internalization of CESAs from the plasma membrane; and 2,6-dichlorobenzonitrile (DCB), which prevents the removal of CESAs from the plasma membrane (for review, see Brabham and Debolt, 2013). Isoxaben treatment abolished the circular distribution of GFP-CESA5 around the cytoplasmic column and increased the amount of intracellular GFP-CESA5 signal, while DCB enhanced the circular pattern and increased the relative signal intensity of GFP-CESA5 in the plasma membrane (Fig. 6C, insets). As a negative control, the seed coat epidermal cell distribution of ECERIFERUM5 (CER5), a membrane-bound ATP-binding cassette transporter involved in cuticular wax biosynthesis (Pighin et al., 2004), was examined. GFP-CER5 had a diffuse distribution along the plasma membrane of seed coat epidermal cells and did not resemble the linear, striated pattern of GFP-CESA3 (Supplemental Fig. S16).
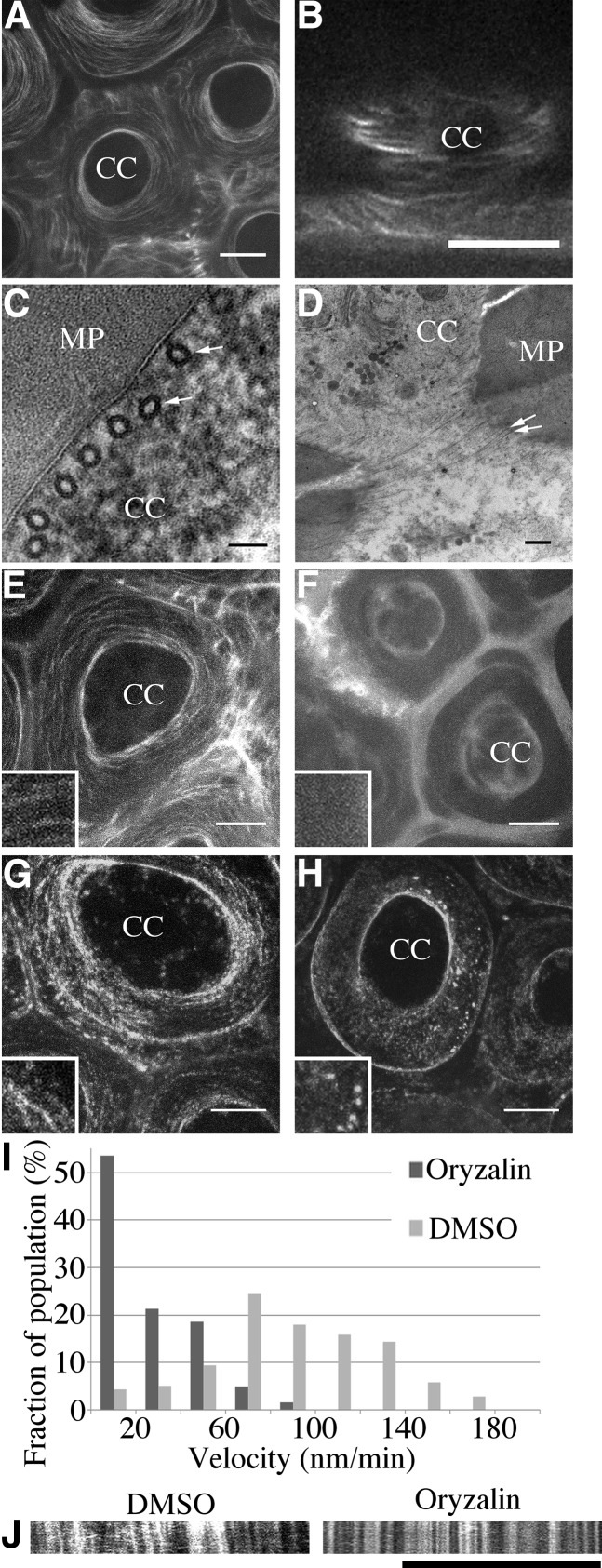
CESA complexes are guided by cortical microtubule arrays (for review, see Bringmann et al., 2012a). In 7-DPA seed coat epidermal cells, microtubules are very prominent, surrounding the cytoplasmic column just beneath the membrane (McFarlane et al., 2008). Using an RFP-TUBULIN6 (TUB6) translational fusion under the control of the UBIQUITIN1 promoter (Ambrose et al., 2011), we further examined this distribution (Fig. 7). RFP-TUB6 appeared in cortical, linear arrays encircling the cytoplasmic column in a pattern that closely resembled the localization of GFP-CESA3, GFP-CESA5, and GFP-CESA10 (compare Fig. 7, A and B, with Fig. 6B). Transmission electron microscopy confirmed this distribution of cortical microtubules, showing a parallel microtubule array perpendicular to the apical-basal axis of the cell (Fig. 7C). Furthermore, in sections just beneath the plasma membrane, we observed microtubules in a cortical, linear array parallel to the tangential primary wall (Fig. 7D, arrows). At 7 DPA, seeds were treated with oryzalin, a microtubule-destabilizing drug, which disrupted RFP-TUB6 localization and reduced the movement of GFP-CESA particles in the plasma membrane (Fig. 7, E–I). The mean velocity of GFP-CESA seeds treated with DMSO was 72.05 ± 37.80 nm min−1 (n = 242; 22 cells from seven seeds), while their mean velocity in oryzalin-treated seeds was 20.74 ± 9.49 nm min−1 (n = 367; 22 cells from six seeds). This suggests that CESA movement during mucilage biosynthesis depends on cortical microtubules. Guided by microtubules, GFP-CESAs appear to circle around the cytoplasmic column, parallel to the outer tangential cell wall.
Figure 7.
Microtubules influence CESA polypeptide movement around the cytoplasmic column of seed coat epidermal cells. A and B, Images of RFP-TUB6 showing microtubule distribution in a circular array around the columella, perpendicular to the surface. A, Maximum projection of a z-stack. Bar = 10 μm. B, Cross section through the cortical region of the cytoplasmic column (CC). Bar = 5 μm. C and D, Transmission electron microscopy images of sections through the cytoplasmic column showing microtubules in the cytoplasm immediately adjacent to the membrane lining the mucilage pocket (MP) and perpendicular to the seed surface. C, Tangential view. Bar = 50 nm. D, Vertical cross section through the cortical region of the cytoplasmic column. Bar = 250 nm. E to H, Maximum projection of z-stack images showing that oryzalin disrupts the arrangement of both microtubules and GFP-CESA3. Bars = 10 μm. E, RFP-TUB6 in seeds treated with 1% DMSO. F, RFP-TUB6 in seeds treated with 1% DMSO and 0.1 mm oryzalin for 2 h. G, GFP-CESA3 in seeds treated with 1% DMSO. H, GFP-CESA3 in seeds treated with 1% DMSO and 0.1 mm oryzalin. Insets are approximately 5 μm in height and width. I, GFP-CESA velocity measurements from DMSO- and oryzalin-treated seeds. J, Kymographs of GFP-CESA3 in DMSO- and oryzalin-treated seeds. Oryzalin significantly decreases the movement of GFP-CESA3. Bar = 10 μm.
Cellulose Is Deposited in a Coiled Pattern That Unwinds during Mucilage Extrusion to Form the Ray
The localization and direction of movement of GFP-CESAs in the plasma membrane suggest that cellulose is deposited in the mucilage pocket in a circular pattern, parallel to the seed surface. However, in extruded mucilage, the cellulose in rays appears straight, extending perpendicularly from the seed surface (Griffiths et al., 2014). These data suggest that the cellulose must be coiled in mature mucilage and unwind during extrusion. We examined the structure of hydrated mucilage using variable pressure scanning electron microscopy (VPSEM; Fig. 8). Mucilage was extruded by exposing seeds to water, and then the organization of partially dehydrated material on the seed surface was examined by VPSEM. The mucilage on the seed surface appeared to be organized in spiral structures around the central columella (Fig. 8A). We next examined S4B-stained mucilage in more detail with confocal microscopy in an attempt to identify similar structures. Although wild-type rays are straight (Griffiths et al., 2014), the longer and thicker rays of ixr1-1 showed a helical distribution of S4B signals (Fig. 8B, arrow), suggesting that the cellulose was coiled prior to extrusion.
Figure 8.
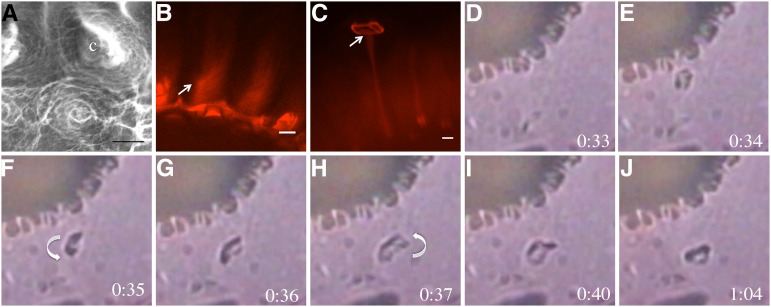
Cellulose and pectin are organized in a spiral around the columella, which uncoils as mucilage expands when hydrated. A, VPSEM of hydrated mucilage. Cell wall material is oriented in spiral structures around the columella (c). B, S4B-stained ixr1-1 mucilage showing the spiraled orientation of cellulose fibrils in rays. The arrow indicates the orientation of the cellulose spiral. C, Maximum projection of a z-stack of S4B-stained fly1-1 seeds. The arrow indicates the point of ray attachment to the disc. Bars = 10 μm. D to J, fly1-1 Seeds hydrated in water, showing fly1 disc rotation during mucilage expansion. Times indicate seconds post hydration. For a clearer image of disc rotation, see Supplemental Video S3. Arrows indicate the direction of rotation.
Since structural elements in mucilage appear to be coiled around the columella but are relatively straight following extrusion in the wild type, we investigated this transition in more detail by observing mucilage during hydration and expansion. Mucilage is normally extruded within approximately 10 s of seed hydration and is difficult to examine. To overcome this limitation, we analyzed the recently characterized flying saucer1 (fly1) mutant, which has seed coat epidermal cells that show delayed outer wall rupture and mucilage release (Voiniciuc et al., 2013). The outer primary walls of fly1 often detach from the columellae as discs that move to the edge of the adherent mucilage halo (Fig. 8; Voiniciuc et al., 2013). Since the fly1 discs remain attached to the top of the rays (Fig. 8C, arrow), they can be used to visualize mucilage expansion and track the formation of rays. We examined the hydration of fly1-1 seeds and observed that 42 out of 45 discs rotated counterclockwise upon mucilage extrusion (Fig. 8, D–J; Supplemental Video S3). The degree of rotation was difficult to quantify, but most discs rotated approximately 360° as mucilage expanded, suggesting that the cellulosic ray they are attached to is initially coiled around the columella and unwinds during extrusion. The rotation of fly1 discs in a predominantly counterclockwise fashion is consistent with a clockwise orientation of cellulose microfibrils, as suggested by the movement of GFP-CESAs.
DISCUSSION
The carbohydrates in the extruded mucilage that tightly adhere to the seed surface form a distinct ray-like structure. Cellulose plays an important role in this structure and is required for adherence. Previous studies have shown that CESA5 has a role in mucilage cellulose biosynthesis, adherence, and ray formation (Harpaz-Saad et al., 2011; Mendu et al., 2011; Sullivan et al., 2011). Here, we show that CESA3, like CESA5, is involved in mucilage cellulose biosynthesis in the seed coat epidermis and is required for proper mucilage structure and adherence. The CESA10 protein, which is also present surrounding the mucilage pocket in seed coat epidermal cells, may function together with CESA5 and CESA3 in mucilage cellulose deposition. However, CESA10 is not essential for the production of cellulose during mucilage biosynthesis. Live-cell imaging of GFP-CESA movement and the rotation of fly1 discs upon hydration strongly suggest that cellulose is deposited by CESA complexes around the cytoplasmic column in a coil-like structure that unwinds during mucilage extrusion to form a cellulosic ray.
CESA3 Is Required for the Synthesis of Cellulose in Seed Mucilage
Several lines of evidence demonstrate that CESA3 is part of a cellulose synthase complex that synthesizes cellulose in the mucilage pocket during mucilage biosynthesis. Based on publicly available microarray data, CESA3 appears to be the most highly expressed CESA gene in the seed coat at 6 to 7 DPA, when mucilage biosynthesis is at its peak (Fig. 1). GFP-CESA3 was localized to the plasma membrane during mucilage deposition (Figs. 1 and 6). CESA3 interacts with CESA5, a protein that was previously shown to be required for mucilage cellulose (Fig. 2; Harpaz-Saad et al., 2011; Mendu et al., 2011; Sullivan et al., 2011). Finally, two mutant alleles of CESA3 showed altered distribution of cellulose in mucilage (Figs. 3 and 4).
CESA10 Is Present, But Not Essential, for Mucilage Biosynthesis
CESA10 is the least well characterized member of the CESA family in Arabidopsis. Clear functions have been attributed to primary cell wall CESAs (CESA1, CESA3, and CESA6; Desprez et al., 2007; Persson et al., 2007), secondary wall CESAs (CESA4, CESA7, and CESA8; Taylor et al., 2003), and the partially redundant CESA6-like group (CESA2, CESA5, and CESA9; Desprez et al., 2007; Persson et al., 2007; Stork et al., 2010; Mendu et al., 2011). No major function has been attributed to CESA10, although CESA10 expression is increased following brassinosteroid treatment (Xie et al., 2011). Here, we show that GFP-CESA10 fusion proteins are expressed in seed coat epidermal cells when cellulose is actively being synthesized in both mucilage and the columella. GFP-CESA10 signal was detected in the plasma membrane of 7-DPA seed coat epidermal cells, similar to that observed for GFP-CESA3 and GFP-CESA5 (Fig. 1; Supplemental Figs. S1 and S2), suggesting that it is actively involved in cellulose synthesis. However, no obvious seed mucilage defects were observed in two independent cesa10 transfer DNA insertion lines (Supplemental Fig. S5), and protein-protein interaction assays only identified an in vitro interaction between the zinc finger-like domains of CESA5 and CESA3, not CESA10. If CESA10 is working in a complex together with CESA5 and CESA3 to synthesize mucilage cellulose, it must interact through a domain other than the zinc finger-like domains and may be redundant with at least one other CESA polypeptide. CESA1 is closely related to CESA10 (Carroll and Specht, 2011) and is expressed in seed coat epidermal cells, making CESA1 a possible candidate for redundancy with CESA10 in this cell type.
Effects of the ixr1 Missense Mutations on Cellulose Biosynthesis
Reduced total Glc and crystalline cellulose contents in cesa5 seeds are consistent with reduced cellulose biosynthesis. The ixr1-1 and ixr1-2 mutations in CESA3 appear to only affect the degree of cellulose crystallinity, as seeds of both mutants result in Glc levels similar to the wild type, but lower amounts of crystalline cellulose were quantified with the Updegraff assay (ixr1-1; Fig. 5) or visualized with CBM3a immunolabeling (ixr1-2; Fig. 4). This is consistent with previous results that showed reduced cellulose crystallinity in ixr1-2 stem tissues compared with the wild type (Harris et al., 2012).
Although both ixr1-1 and ixr1-2 have reduced crystalline cellulose content, they cause distinct mucilage defects. While ixr1-1 seeds have larger mucilage halos than the wild type, ixr1-2 seeds have smaller mucilage halos (Figs. 3 and 5). Additionally, the distribution of pectin epitopes in mucilage is altered when comparing ixr1-1 and ixr1-2 seeds (Fig. 4). CESA proteins contain eight transmembrane helices that form a pore in the plasma membrane through which newly synthesized cellulose passes into the apoplast (for review, see Carpita, 2011). The mutated residue in ixr1-2, located within an extracellular loop between two transmembrane domains, is required for correct formation of the pore and for correct glucan chain alignment to produce a crystalline microfibril (Harris et al., 2012). Since ixr1-1 is located in a transmembrane domain, it is possible that it also affects the formation of the pore and the polymerization and crystallization of cellulose. ixr1-2, and the aegeus mutation in CESA1 located in a transmembrane domain, can affect CESA velocities in the plasma membrane (Harris et al., 2012). Mutations in COBRA-LIKE2 also result in reduced seed cellulose crystallinity and a loss of mucilage adherence (Ben-Tov et al., 2015); however, the mucilage phenotype more closely resembles cesa5-1 than either ixr1 allele, suggesting that the ixr1 alleles could affect other aspects of cellulose biosynthesis aside from crystallinity. Precisely how the ixr1-1 and ixr1-2 mutations modify cellulose deposition and crystallinity to produce unique ray structures remains unclear.
ixr1-1 and ixr1-2 Mutations Impact Mucilage Pectin Organization
Although CESA3 is involved in cellulose biosynthesis in mucilage, changes in the pectin distribution of ixr1-1 and ixr1-2 seed mucilage were also evident (Fig. 4). CCRC-M36 labeling revealed a less adherent and more disorganized pectin localization in ixr1-1 mucilage (Figs. 3 and 4). Changes in the cellulose structure of irx1-1 and ixr1-2 seeds likely modify the distribution and organization of pectins in mucilage. Consistent with this hypothesis, there was a correlation between the distribution of crystalline cellulose (CBM3a; Fig. 4) and RG I in mucilage of both wild-type and mutant seeds. In ixr1-1 seeds, pectins appear to be localized proximal to the rays and are less abundant in the diffuse mucilage region between the rays (Fig. 4). Overall, pectin amounts were unchanged in ixr1-1 and ixr1-2 whole seeds and nonadherent mucilage, suggesting that the impact of these mutations is on the organization and distribution of pectins. Our results are in agreement with previous reports demonstrating a close association between pectins and cellulose (Iwai et al., 2001; Oechslin et al., 2003; Vignon et al., 2004; Zykwinska et al., 2005; Yoneda et al., 2010; Chebli et al., 2012; Wang et al., 2012; Cosgrove, 2014; Griffiths et al., 2014).
Cellulose in Mucilage Is Deposited in a Linear Array around the Cytoplasmic Column
Our results support the hypothesis that cellulose is deposited in the mucilage pockets of seed coat epidermal cells in a unique coiled pattern by CESA complexes. CESA complexes in seed coat epidermal cells move in a linear unidirectional manner, forming circular tracks around the cytoplasmic column, parallel to the outer primary wall (Fig. 7; Supplemental Fig. S12). Cytological analyses of extruded mucilage suggest a coiled or spiral pattern, consistent with the circular trafficking of CESAs and the proposed deposition of mucilage cellulose (Fig. 8).
During cellulose biosynthesis, microtubules guide the orientation of CESA complex movement (Paredez et al., 2006; Chan et al., 2010; Endler and Persson, 2011; Li et al., 2012; Bringmann et al., 2012a, 2012b). In seed coat epidermal cells, microtubules are oriented around the cytoplasmic column in a pattern similar to that observed for the movement of CESA polypeptides. These microtubules are required for directed CESA complex movement and velocity (Figs. 6 and 7), since disrupting them with oryzalin also disrupts the circular, striated pattern of GFP-CESAs and their velocity in seed coat epidermal cells. In addition, previous studies have shown that plants homozygous for mor1-1, a temperature-sensitive allele of the MOR1 gene that encodes a microtubule-organizing protein (Whittington et al., 2001), are also deficient in mucilage extrusion when the mother plant is grown at the nonpermissive temperature (McFarlane et al., 2008), analogous to the effects of the cesa5 and ixr1 mutations on extrusion shown here. These data support the hypothesis that mucilage cellulose is deposited around the cytoplasmic column parallel to the seed surface by CESA complexes guided by microtubules. The fact that CESA movement in seed coat epidermal cells is unidirectional suggests either that cortical microtubules in these cells are organized in a unidirectional manner or that some component or the CSC required for bidirectional movement is absent. Phosphorylation of CESA1 has been shown to influence bidirectional movement (Chen et al., 2010), so it is possible that the phosphorylation status of CESA polypeptides is used to influence the unidirectional movement of CESA complexes observed in seed coat epidermal cells.
Although cellulosic rays in extruded mucilage of wild-type Arabidopsis seeds appear to be straight, they appear as spirals when viewed by VPSEM or in S4B-stained ixr1-1 seeds (Fig. 8). Other species of plants have more obvious helical arrays in their mucilage. For example, a helicoidal array of cellulose material has been reported for quince (Cydonia oblonga) seed mucilage (Abeysekera and Willison, 1990). Spiral and coiled cellulose threads have also been described in mucilage of Artemisia campestris and Salvia spp. (Kreitschitz, 2009). Therefore, deposition of mucilage cellulose in spirals may be a common feature of seed coat epidermal cells.
The Velocity of CESA Polypeptides in Seed Coat Epidermal Cells during Mucilage Biosynthesis Is Distinct
Curiously, during mucilage synthesis in seed coat epidermal cells, GFP-CESAs move much slower than in other tissues. The slower speed is similar to the velocity of dephosphorylated CESA5 in hypocotyls (Bischoff et al., 2011). Thus, it is possible that CESA5 is not phosphorylated in seed coat epidermal cells. The importance of this slower CESA velocity, compared with that in hypocotyls or roots is unclear. The velocity may be necessary to optimize cellulose crystallinity or the degree of cellulose polymerization. Alternatively, velocity might facilitate interactions between the nascent microfibrils and the pectin-rich components of mucilage, thus helping to establish the complex organization of the preray structure prior to mucilage extrusion (Griffiths et al., 2014).
It is interesting that oryzalin treatment nearly abolished both cortical microtubule arrays and CESA movement in seed coat epidermal cells. This differs from previous reports of the effects of oryzalin treatment on CESA velocities, where abolishment of cortical microtubule arrays led to only a 50% reduction in CESA velocities (Li et al., 2012) and in one case to increased velocity (Bischoff et al., 2011). These variations in response might be due to differences in cell type, concentration of oryzalin, or length of treatment.
Function of Cellulose during Mucilage Extrusion
Cellulose strongly influences mucilage adherence in seeds, demonstrating that it plays a role in mucilage retention or resistance to expansion (Harpaz-Saad et al., 2011; Mendu et al., 2011; Sullivan et al., 2011; Griffiths et al., 2014; this study). However, cesa5-1 seeds show a marked reduction in the speed of mucilage extrusion under conditions of reduced water availability (Table II), suggesting that cellulose is necessary for rapid extrusion. One hypothesis consistent with these results is that cellulose, coiled around the columella, can act in a spring-like manner upon hydration. Alternatively, strong interactions between cellulose and pectin may result in a leverage-like effect that directs the force of extrusion upward as the pectin expands. Interactions between pectins and cellulose are consistent with the fact that seeds from a double mutant with loss of function in both CESA5 (cellulose synthesis) and BXL1 (pectin structure) have a mucilage extrusion phenotype much stronger than either single mutant alone (Fig. 5) as well as with a previous study showing a dependence of mucilage adherence on both cellulose and pectins (Griffiths et al., 2014).
A Model for Cellulose Deposition, Structure, and Function in Seed Coat Mucilage
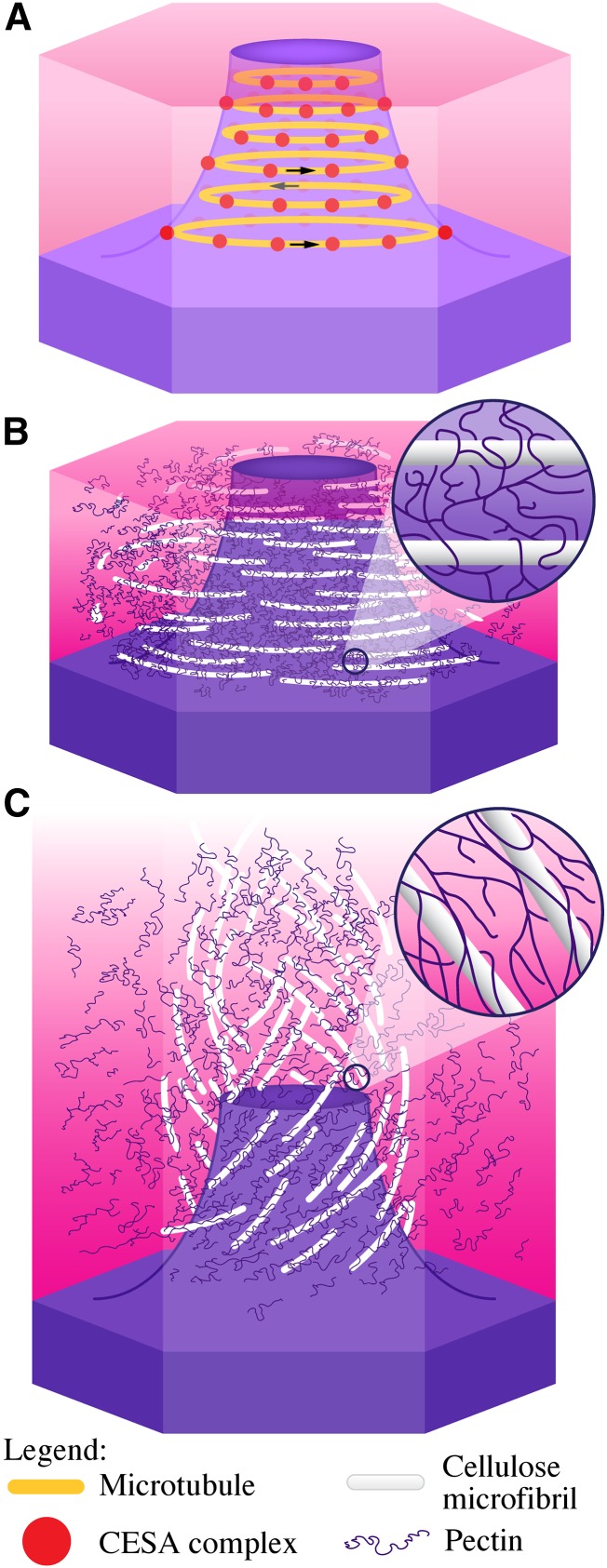
A model illustrating the orientation of cellulose microfibril deposition during mucilage biosynthesis, and the predicted orientation of cellulose microfibrils forming a ray in mature, extruded mucilage, is presented in Figure 9. We propose that microtubule arrays (Fig. 9A) guide the CESA complexes to synthesize cellulose in a coiled array around the cytoplasmic column (Fig. 9B). As yet unspecified interactions between cellulose and pectin in mucilage form a cohesive carbohydrate network adjacent to the cytoplasm and, later in seed coat development, to the columella. Hydration of mucilage forces the rapid expansion of pectins in the mucilage pocket, which ruptures the radial wall above the secondary cell wall thickening (Mendu et al., 2011). This leaves the outer primary wall remnants attached to the columella as the mucilage capsule expands to engulf the seed. As the pectin expands, the cellulose tightly associated with the pectin unwinds to extend upward approximately 80 μm (approximately to the edge of the mucilage halo), forming a ray and establishing the adherent layer (Fig. 9C). Cellulose and pectin not closely associated with the ray may form the diffuse staining region between the rays and/or the pectin in the outer nonadherent layer.
Figure 9.
Model of the orientation of cellulose deposition during mucilage biosynthesis and of mucilage expansion. A, Model of a seed coat epidermal cell at 7 DPA showing CESA complexes (red dots) trafficking around the cytoplasmic column circumferentially, guided by microtubules (orange lines). The direction of CESA complex movement is indicated by the arrows. B, Seed coat epidermal cell at 11 DPA showing cellulose and pectins in mucilage. The inset shows pectins wrapped around cellulose. C, Hydrated mucilage showing the orientation of cellulose microfibrils post hydration. The addition of water forces pectins to expand, pushing and pulling cellulose microfibrils in the process. The inset shows expanded pectins still wrapped around cellulose microfibrils.
This hypothesis agrees with other recent investigations of pectin structure and the relationship between cellulose and pectins (for review, see Cosgrove, 2014). Cellulose-binding assays have demonstrated that pectic RG I side chains can bind to cellulose in vitro (Zykwinska et al., 2005, 2007). NMR analysis of cell walls indicates that pectins can be directly bound to cellulose (Dick-Pérez et al., 2011; Wang et al., 2012). Arabinogalactan proteins can be directly linked to RG I and hemicelluloses, increasing the complexity of the cell wall network (Tan et al., 2013). The effects of cobtorin, a chemical that inhibits the parallel alignment of cellulose microfibrils with microtubules, can be rescued by overexpression of pectin methylesterases and polygalacturonases (Yoneda et al., 2010). The unique deposition of cellulose in mucilage provides an excellent system to further study the interactions between pectins and cellulose and their effects on cell wall rheology. Additional genes that are hypothesized to facilitate cellulose biosynthesis or interactions between pectins and cellulose could be investigated in vivo in the seed coat epidermis using a cell-specific promoter (Esfandiari et al., 2013).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) mutants used in this study were in the Col-0 ecotype, except for bxl1-1, which is in the Wassilewskija background (Arsovski et al., 2009). All transfer DNA insertion and point mutants were obtained from previous studies (Mendu et al., 2011) or the Arabidopsis Biological Resource Center (Alonso et al., 2003; Supplemental Table S6). Seeds were germinated on plates with Arabidopsis medium (Haughn and Somerville, 1986) and 7% (w/v) agar, and seedlings were transferred to soil (Sunshine Mix 4; SunGro) after 7 d. Plants were grown with continuous fluorescent illumination of 80 to 140 μE m−2 s−1 at 20°C to 22°C. Developing seeds were staged as described previously (Western et al., 2001). The ixr1-1 point mutation was screened using ixr1-1 MseI forward primer and ixr1-1 MseI reverse primer derived cleaved-amplified polymorphic sequence markers (Supplemental Table S8).
Microscopy and Image Analysis
Confocal images were acquired on a Perkin-Elmer Ultraview VoX Spinning Disk Confocal system. ProCESA5:GFP-CESA5 (GFP-CESA5) plants were a gift from Helen North and Volker Bischoff (Bischoff et al., 2011; Sullivan et al., 2011). ProCESA3:GFP-CESA3 in je5 (GFP-CESA3; Desprez et al., 2007), ProCESA6:GFP-CESA6 (GFP-CESA6; Desprez et al., 2007), 35S:RFP-VHA-a1 (Dettmer et al., 2006), and ProUBQ1:RFP-TUB6 (RFP-TUB6; Ambrose et al., 2011) plants were kindly provided by the Wasteneys laboratory at the University of British Columbia. Pro:CER5::GFP-CER5 plants was provided by the Kunst laboratory at the University of British Columbia (Pighin et al., 2004). Subcellular localization of GFP was investigated with a 63× oil-immersion objective. For isoxaben and DCB treatments, developing seeds from GFP-CESA5-expressing plants were excised from siliques and treated with 1 μm isoxaben and 1 μm DCB in 0.01% (v/v) DMSO for 1 h at room temperature with orbital shaking. Time-lapse images are projections of multiple time points of six images per min for 5 min. For the determination of the role of microtubules in CESA movement, seeds were dissected from siliques and stabbed to improve infiltration, then treated with 1% (v/v) DMSO or 1% (v/v) DMSO containing 0.1 mm oryzalin (Sigma) for 2 h prior to imaging. Velocity measurements of oryzalin-treated seeds were taken from GFP-CESA3- and GFP-CESA10-expressing seeds. For FM4-64 staining, developing seeds were dissected from siliques and incubated with 10 μg mL−1 FM4-64 for 30 min prior to imaging.
For RR staining, mature dry seeds were hydrated in distilled water, 100 mm CaCl2, or 50 mm EDTA in 50 mm HEPES, pH 6, for 1 to 2 h, rinsed with once with water, and stained with 0.01% (w/v) RR (Sigma-Aldrich) for 60 min or as described while shaking on a rotator. Bright-field micrographs of stained samples were taken with QCapture software and a digital camera (QImaging) equipped on a Zeiss AxioSkop 2 upright light microscope (Carl Zeiss). For cellulose staining, seeds were shaken in water for 1 to 2 h, then stained with 0.01% (w/v) S4B (Sigma-Aldrich Rare Chemical Library; S479896) and 100 mm NaCl for 1 h and rinsed twice before imaging (Anderson et al., 2010; Mendu et al., 2011).
Whole-seed immunolabeling and imaging were conducted according to a previously published method (Griffiths et al., 2014). For VPSEM analysis, hydrated seeds were mounted on carbon stubs and examined with a Hitachi S-4700 scanning electron microscope (Hitachi High-Technologies) at less than 70 atm. SEM images of dry seeds were obtained as described by Voiniciuc et al. (2013). Transmission electron microscopy analysis of microtubules in seed coat epidermal cells was performed as described by Young et al. (2008).
All micrographs were processed with ImageJ (Abramoff et al., 2004) using rolling-ball and kymograph functions. Ray length and mucilage width were also measured using the ImageJ freehand line function and Microsoft Excel to convert pixels to μm. Confocal images containing signals from multiple optical stacks, or time-lapse images, were rendered using the three-dimensional (3D) project brightest point intensity method. Maximum projections of z-stack images were created using the 3D project ImageJ tool from stacks containing 0.5-μm step size. Kymographs were created from time-lapse images at 5-s image intervals using the segmented line tool to outline the direction of GFP-CESA movement, followed by the multiple kymograph plugin for ImageJ (http://imagej.nih.gov/ij/). Time-lapse images were averaged for three frames using the Walking Average program, and then the 3D project tool, using the mean value intensity. The slope of GFP-CESA particle velocity from the kymograph was analyzed using the Read velocities from tsp macro (http://www.embl.de/eamnet/downloads/macros/tsp050706.txt).
Chemical Analysis
Monosaccharide analysis was performed according to published methods (Dean et al., 2007; Mendu et al., 2011; Voiniciuc et al., 2013; Griffiths et al., 2014). Crystalline cellulose was determined based on a previously published method (Updegraff, 1969; Griffiths et al., 2014).
ProCESA10:GFP-CESA10 Construction
To amplify the CESA10 promoter region, primers HindIII_CESA10p-F and XbaI_CESA10p-R were used to amplify a 2,020-bp region upstream from the CESA10 translational start site from wild-type plants using High Fidelity PCR Enzyme Mix (Supplemental Table S8; Thermo Scientific). The PCR product was ligated into pJet1.2 (Thermo Scientific) and sequenced to confirm that no mutations were introduced during PCR. The CESA10 promoter fragment was then excised using HindIII and XbaI and cloned into pGWB6 (Nakagawa et al., 2007) using these restriction enzyme sites to replace the 35S promoter to generate pGWB6-CESA10p. A 5,008-bp DNA fragment containing the genomic region of CESA10 was amplified from wild-type plants with primers CESA10-attB1 and CESA10-attB2 using Phusion polymerase (Supplemental Table S8; Finnzymes). Gateway adapters were added using the adapter protocol (Invitrogen). This 5,008-bp fragment was cloned into pDONR221 using BP Clonase II (Invitrogen) to create pDONR221-CESA10 and was sequenced to confirm that no mutations were introduced during PCR. The fragment was then recombined into the vector pGWB6-CESA10p using LR Clonase II (Invitrogen) to generate pGWB6-CESA10p-GFP-CESA10.
Cloning, Expression, and Purification of Recombinant Protein CESA5
A 786-bp section of the N terminus of CESA5 (nontransmembrane region) was cloned into the pET29b protein expression vector. For this, CESA5 complementary DNA was used as a template. Amplicons were obtained using PCR with Phusion High-Fidelity DNA Polymerase and gene-specific primers (International DNA Technologies) with nonhomologous EcoRI/XhoI restriction endonuclease sites engineered into the forward and reverse primer, respectively (Supplemental Table S8). Amplicons were digested by the appropriate restriction endonuclease, gel purified, and cloned into EcoRI and XhoI sites in precleaved pET29b. N-CESA5 was produced from their respective pET29b plasmids in Rosetta Competent Cell CMD (Millipore) by inoculating 200 mL of Luria-Bertani medium (100 μg mL−1 ampicillin and 34 μg mL−1 chloramphenicol) with 1 mL of starter cultures. Log-phase cells were induced with 1 mm isopropylthio-β-galactoside at room temperature for 8 h, and the cells were collected by centrifugation at 8,000g for 10 min and freeze thawed before lysis. The cells were resuspended in 4 mL of binding buffer (50 mm sodium phosphate and 300 mm NaCl) and sonicated with Tekmar cell disruptor for 15 s. The cell debris was separated from the protein solution by centrifugation (21,000g for 10 min). Equilibrated Talon resin at 0.5 mL was added to the supernatant. The protein was allowed to bind to the resin for 2 h at 4°C and then washed with 15 mL of binding buffer three times. The last wash was performed with binding buffer and 15 mm imidazole in order to get rid of unspecific binding. The protein was eluted in binding buffer + 200 mm imidazole. The eluent was dialyzed against 10 mm Tris-Cl, pH 7.5, buffers overnight. Protein was further analyzed for the appropriate Mr and purity on 12% (w/v) SDS-PAGE gels and stored at −20°C for further use.
Site-Directed Mutagenesis, Cloning, and Purification of Recombinant mutCESA5
The Quick Change Multisite Directed Mutagenesis Kit from Stratagene (200514) was used to substitute the Cys at positions 39 and 84 with Ala (Fig. 2) by using primer CESA5C39 and CESA5C84, respectively (Supplemental Table S8). N-CESA5::pET29b plasmid was used as a template. N-mutCESA5 was further transformed into Rosetta Competent Cell CMD (Millipore). After transformation, N-mutCESA5 was further expressed and purified as N-CESA5. Prior to commencing phage display, we confirmed that proteins could bind to the ELISA plate. A dilution series (10, 1, 0.1, and 0.01 μg mL−1) of recombinant proteins or BSA in 10 mm Tris-HCl, pH 7.5, was introduced into the bottom of microtiter plate wells overnight at 4°C and further processed as described by Kushwaha et al. (2012).
4-(2-Pyridylazo) Resorcinol Assay
Different amounts (40, 80, 100, and 200 μg) of purified recombinant proteins were incubated with 10 μm zinc acetate for 30 min at room temperature in TSD buffer (50 mm Tris-Cl [pH 7.5], 200 mm NaCl, and 1 mm dithiothreitol). Then samples were passed through a Sephadex G-25 (Amersham Biosciences) column to remove unbound divalent cations. For the determination of bound zinc, protein samples were digested with 4 units of Proteinase K (Fisher Scientific) at 60°C overnight. Free zinc ions were quantified colorimetrically according to Hunt et al. (1985). 4-(2-Pyridylazo) resorcinol at 100 μm (Sigma-Aldrich) was added to 100-μL samples, and absorbance was measured at 490 nm.
Biopanning, Titering, Plaque Isolation, PCR, and Sequencing
Biopanning and titration were carried out by the method described previously (Chen et al., 2010; Kushwaha et al., 2012). For each round of biopanning, a 96-well microtiter plate (clear, flat-bottom, standard tissue culture surface; Corning) was used. A 10 μg mL−1 concentration of (1) recombinant N-CESA5, (2) mut-CESA5, or (3) BSA (Sigma-Aldrich), in 100 μL of Tris, pH 7.5, was added to a well for each protein and plate. The plate was covered with plastic wrap, left overnight at 4°C, and further processed as described previously (Chen et al., 2010; Kushwaha et al., 2012). Differences in protein amounts were determined by an ELISA (the average of three replicated wells for each of four protein dilutions), and the alteration in phage titer amounts between N-CESA5, mutCESA5, or BSA (the average of three replications per well and protein for each biopan) were all subjected to Student’s t test at P = 0.05.
At each biopanning round, 18 individual plaques were selected using previously cut, yellow pipet tips from titering plates of sufficient dilution to produce well-separated plaque. The agar/top agarose core containing the plaque was introduced to 100 μL of Tris-HCl, pH 8.5, and vortexed prior to a 90-μL aliquot being retrieved, heated at 65°C for 10 min, and 3 μL being used in PCR with T7 up and down primers (Supplemental Table S8). The 18 PCRs were run on 1% (w/v) agarose gels and visualized using ethidium bromide and transillumination. In the final biopanning round, those amplicons that were not from empty vectors were sequenced. Sequencing was done by Elim Biopharm. The sequences were examined for the linker arms, the orientation of the clone was determined, and the clone was used in WU-BLAST (The Arabidopsis Information Resource) to determine if (1) it was in frame and, if so, (2) whether it encoded part (all) of a coding sequence, and (3) the identity of the encoded protein. In order to identify the protein domains, families, and functional sites, as well as associated patterns and profiles in the in-frame hits, we used the EXPASY Scan Prosite tool (http://www.expasy.org/prosite/). For this, amino acid sequences of in-frame hits in single-letter format were submitted to the Scan Prosite server.
Yeast Two-Hybrid Assay
The yeast two-hybrid test was performed using the Matchmaker Two-Hybrid System 3 (Clontech) according to the manufacturer’s protocols. Yeast (Saccharomyces cerevisiae) strain AH109 was used to determine protein-protein interactions. The GAL4 DNA binding domain vector pGBKT7 and the activation domain vector pGADT7 were used throughout. PCR primers (Supplemental Table S8) were used to amplify binding regions of N-terminal regions of CESA5 and mutCESA5 and the interacting region of CESA3. The PCR products were digested and ligated into the yeast fusion vectors pGADT7 and pGBKT7. Sequence-confirmed plasmids were cotransformed into yeast AH109 and plated on synthetic dextrose/-Trp-Leu medium (Clontech) and grown at 30°C. The grown yeast colonies were replated to synthetic dextrose/-Leu-Trp-His medium to show interaction. Two control empty vectors (pGADT7:pGBKT7 and mutCESA5:CESA3) were used.
Bioinformatics
CESA expression during seed coat development was also examined using the Arabidopsis eFP browser with the developmental map data source and the seed data source (Winter et al., 2007; Bassel et al., 2008) in addition to the Arabidopsis seed coat-specific expression browser (http://bar.utoronto.ca/efp_seedcoat/cgi-bin/efpWeb.cgi; Dean et al., 2011). Coexpression analysis was performed using the Botany Array Resource expression angler (http://bbc.botany.utoronto.ca/ntools/cgi-bin/ntools_expression_angler.cgi; Toufighi et al., 2005).
Arabidopsis Genome Initiative numbers for the genes used in this study can be found in Supplemental Table S6.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GFP-CESA10 and GFP-CESA3 are expressed throughout seed coat development.
Supplemental Figure S2. GFP-CESA10 is localized to the trans-Golgi network and plasma membrane in the cytoplasmic column.
Supplemental Figure S3. GFP-CESA6 is not present in seed coat epidermal cells during mucilage biosynthesis.
Supplemental Figure S4. Phage display demonstrates that N-CESA5 selectively binds to proteins.
Supplemental Figure S5. cesa10 and cesa1 seeds show no major difference in mucilage properties from the wild type.
Supplemental Figure S6. Initial hydration and unstained images of ixr1-1 and ixr1-2 seeds.
Supplemental Figure S7. SEM images of mature ixr1-1 and ixr1-2 seed coat epidermal cells.
Supplemental Figure S8. cesa5-1 ixr1-1 double mutants have an intermediate phenotype.
Supplemental Figure S9. Seed mucilage does not significantly autofluoresce.
Supplemental Figure S10. CBM3a immunolabeling of cesa mutants showing individual light channels and maximum projection of a z-stack.
Supplemental Figure S11. CBM28 immunolabeling of cesa mutants showing individual light channels and maximum projection of a z-stack.
Supplemental Figure S12. CCRC-M36 immunolabeling of cesa mutants showing individual light channels and maximum projection of a z-stack.
Supplemental Figure S13. JIM5 immunolabeling of cesa mutants showing individual light channels and maximum projection of a z-stack.
Supplemental Figure S14. Mutation of cesa influences pectin-mediated mucilage expansion.
Supplemental Figure S15. Images of GFP-CESA5 from a 10-min video (Supplemental Video S2).
Supplemental Figure S16. Maximum projection of a z-stack image of the localization of GFP-CESA3 and GFP-CER5 in seed coat epidermal cells.
Supplemental Table S1. CESA expression in seed coat cells during seed development (Le et al., 2010).
Supplemental Table S2. CESA expression in seed coat cells during development (Dean et al., 2011).
Supplemental Table S3. CESA expression in whole seeds during development (Winter et al., 2007).
Supplemental Table S4. List of the top 30 genes coexpressed with CESA5 in seeds
Supplemental Table S5. Pearson correlations and Manders coefficients for GFP-CESA10 (M1) colocalization with RFP-VHA-a1 (M2) in seed coat epidermal cells.
Supplemental Table S6. Genes, accession numbers, and mutant lines used in this study.
Supplemental Table S7. Monosaccharide composition and whole-seed acid-insoluble cellulose amounts.
Supplemental Table S8. Sequences of primers used in this study.
Supplemental Video S1. GFP-CESA10 is partially localized to the trans-Golgi network.
Supplemental Video S2. GFP-CESA5 traffics in linear arrays around the cytoplasmic column.
Supplemental Video S3. fly1 discs rotate as mucilage expands.
Supplementary Material
Acknowledgments
We thank the University of British Columbia bioimaging facility for technical assistance and equipment use; Miki Fujita (Department of Botany, University of British Columbia) for imaging the GFP-CESA lines; Geoff Wasteneys and Ljerka Kunst (Department of Botany, University of British Columbia), and Volker Bischoff and Helen North (Institute Jean-Pierre Bourgin, Institut National de la Recherche Agronomique) for providing the VHA-1a-RFP, RFP-TUB6, GFP-CER5, and GFP-CESA5 transgenic lines; Dr. Allan Bruce Downie (Department of Horticulture, University of Kentucky) for providing the seed phage-display library; members of the Mansfield laboratory (Faculty of Forestry, University of British Columbia) for aid in performing HPLC; Dr. Erin Gilchrist (Department of Botany, University of British Columbia) for critical reading of the article; and Jozsef Stork and Bruce Downie (Department of Horticulture, University of Kentucky) for technical assistance.
Glossary
- RG I
rhamnogalacturonan I
- HG
homogalacturonan
- CSC
cellulose synthase complex
- BSA
bovine serum albumin
- RR
Ruthenium Red
- Col-0
Columbia-0
- SEM
scanning electron microscopy
- S4B
Pontamine Fast Scarlet 4B
- DCB
2,6-dichlorobenzonitrile
- DMSO
dimethyl sulfoxide
- VPSEM
variable pressure scanning electron microscopy
- 3D
three-dimensional
Footnotes
This work was supported by the National Science and Engineering Research Council of Canada (to J.S.G., P.L., C.V., G.D., S.D.M., and G.W.H.), the Department of Energy (grant no. DOE-FOA 10–0000368 to R.K., M.T., and S.D.), and the National Science Foundation (grant no. IOS–1256029 to R.K., M.T., and S.D.).
Articles can be viewed without a subscription.
References
- Abeysekera RM, Willison JH (1990) Architecture of the fluid cellulosic arrays in the epidermis of the quince seed. Biol Cell 68: 251–257 [Google Scholar]
- Abramoff MD, Magalhaes PJ, Ram SJ (2004) Image Processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ambrose C, Allard JF, Cytrynbaum EN, Wasteneys GO (2011) A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat Commun 2: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CT, Carroll A, Akhmetova L, Somerville C (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Popma TM, Haughn GW, Carpita NC, McCann MC, Western TL (2009) AtBXL1 encodes a bifunctional β-D-xylosidase/α-l-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol 150: 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Fung P, Chow TF, Foong JA, Provart NJ, Cutler SR (2008) Elucidating the germination transcriptional program using small molecules. Plant Physiol 147: 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T, Przemeck GKH, Stamatiou G, Lau R, Terryn N, De Rycke R, Inzé D, Berleth T (2002) Genetic complexity of cellulose synthase A gene function in Arabidopsis embryogenesis. Plant Physiol 130: 1883–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tov D, Abraham Y, Stav S, Thompson K, Loraine A, Elbaum R, de Souza A, Pauly M, Kieber JJ, Harpaz-Saad S (2015) COBRA-LIKE2, a member of the glycosylphosphatidylinositol-anchored COBRA-LIKE family, plays a role in cellulose deposition in Arabidopsis seed coat mucilage secretory cells. Plant Physiol 167: 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff V, Desprez T, Mouille G, Vernhettes S, Gonneau M, Höfte H (2011) Phytochrome regulation of cellulose synthesis in Arabidopsis. Curr Biol 21: 1822–1827 [DOI] [PubMed] [Google Scholar]
- Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, Knox JP (2006) Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem 281: 29321–29329 [DOI] [PubMed] [Google Scholar]
- Brabham C, Debolt S (2013) Chemical genetics to examine cellulose biosynthesis. Front Plant Sci 3: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann M, Landrein B, Schudoma C, Hamant O, Hauser MT, Persson S (2012a) Cracking the elusive alignment hypothesis: the microtubule-cellulose synthase nexus unraveled. Trends Plant Sci 17: 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann M, Li E, Sampathkumar A, Kocabek T, Hauser MT, Persson S (2012b) POM-POM2/cellulose synthase interacting1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24: 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn JE, Hurley UA, Birch RJ, Arioli T, Cork A, Williamson RE (2002) The cellulose-deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J 32: 949–960 [DOI] [PubMed] [Google Scholar]
- Carpita NC. (2011) Update on mechanisms of plant cell wall biosynthesis: how plants make cellulose and other (1→4)-β-d-glycans. Plant Physiol 155: 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll A, Specht CD (2011) Understanding plant cellulose synthases through a comprehensive investigation of the cellulose synthase family sequences. Front Plant Sci 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Crowell E, Eder M, Calder G, Bunnewell S, Findlay K, Vernhettes S, Höfte H, Lloyd C (2010) The rotation of cellulose synthase trajectories is microtubule dependent and influences the texture of epidermal cell walls in Arabidopsis hypocotyls. J Cell Sci 123: 3490–3495 [DOI] [PubMed] [Google Scholar]
- Chebli Y, Kaneda M, Zerzour R, Geitmann A (2012) The cell wall of the Arabidopsis pollen tube: spatial distribution, recycling, and network formation of polysaccharides. Plant Physiol 160: 1940–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Nayak N, Majee SM, Lowenson J, Schäfermeyer KR, Eliopoulos AC, Lloyd TD, Dinkins R, Perry SE, Forsthoefel NR, et al. (2010) Substrates of the Arabidopsis thaliana protein isoaspartyl methyltransferase 1 identified using phage display and biopanning. J Biol Chem 285: 37281–37292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2014) Re-constructing our models of cellulose and primary cell wall assembly. Curr Opin Plant Biol 22: 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Höfte H, Vernhettes S (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagel DJ, Liu YS, Zhong L, Luo Y, Himmel ME, Xu Q, Zeng Y, Ding SY, Smith S (2011) In situ imaging of single carbohydrate-binding modules on cellulose microfibrils. J Phys Chem B 115: 635–641 [DOI] [PubMed] [Google Scholar]
- Dean G, Cao Y, Xiang D, Provart NJ, Ramsay L, Ahad A, White R, Selvaraj G, Datla R, Haughn G (2011) Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol Plant 4: 1074–1091 [DOI] [PubMed] [Google Scholar]
- Dean GH, Zheng H, Tewari J, Huang J, Young DS, Hwang YT, Western TL, Carpita NC, McCann MC, Mansfield SD, et al. (2007) The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19: 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP. (1999) Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50: 245–276 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick-Pérez M, Zhang Y, Hayes J, Salazar A, Zabotina OA, Hong M (2011) Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50: 989–1000 [DOI] [PubMed] [Google Scholar]
- Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP (2002) Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 43: 1407–1420 [DOI] [PubMed] [Google Scholar]
- Endler A, Persson S (2011) Cellulose synthases and synthesis in Arabidopsis. Mol Plant 4: 199–211 [DOI] [PubMed] [Google Scholar]
- Esfandiari E, Jin Z, Abdeen A, Griffiths JS, Western TL, Haughn GW (2013) Identification and analysis of an outer-seed-coat-specific promoter from Arabidopsis thaliana. Plant Mol Biol 81: 93–104 [DOI] [PubMed] [Google Scholar]
- Fujita M, Himmelspach R, Ward J, Whittington A, Hasenbein N, Liu C, Truong TT, Galway ME, Mansfield SD, Hocart CH, et al. (2013) The anisotropy1 D604N mutation in the Arabidopsis cellulose synthase1 catalytic domain reduces cell wall crystallinity and the velocity of cellulose synthase complexes. Plant Physiol 162: 74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths JS, Tsai AYL, Xue H, Voiniciuc C, Šola K, Seifert GJ, Mansfield SD, Haughn GW (2014) SALT-OVERLY SENSITIVE5 mediates Arabidopsis seed coat mucilage adherence and organization through pectins. Plant Physiol 165: 991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Harpaz-Saad S, McFarlane HE, Xu S, Divi UK, Forward B, Western TL, Kieber JJ (2011) Cellulose synthesis via the FEI2 RLK/SOS5 pathway and cellulose synthase 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J 68: 941–953 [DOI] [PubMed] [Google Scholar]
- Harris DM, Corbin K, Wang T, Gutierrez R, Bertolo AL, Petti C, Smilgies DM, Estevez JM, Bonetta D, Urbanowicz BR, et al. (2012) Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc Natl Acad Sci USA 109: 4098–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G, Chaudhury A (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472–477 [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville C (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434 [Google Scholar]
- Haughn GW, Western TL (2012) Arabidopsis seed coat mucilage is a specialized cell wall that can be used as a model for genetic analysis of plant cell wall structure and function. Front Plant Sci 3: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim DR, Skomp JR, Tschabold EE, Larrinua IM (1990) Isoxaben inhibits the synthesis of acid insoluble cell wall materials in Arabidopsis thaliana. Plant Physiol 93: 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt JB, Neece SH, Ginsburg A (1985) The use of 4-(2-pyridylazo) resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal Biochem 146: 150–157 [DOI] [PubMed] [Google Scholar]
- Iwai H, Ishii T, Satoh S (2001) Absence of arabinan in the side chains of the pectic polysaccharides strongly associated with cell walls of Nicotiana plumbaginifolia non-organogenic callus with loosely attached constituent cells. Planta 213: 907–915 [DOI] [PubMed] [Google Scholar]
- Knox JP. (1997) The use of antibodies to study the architecture and developmental regulation of plant cell walls. Int Rev Cytol 171: 79–120 [DOI] [PubMed] [Google Scholar]
- Knox JP, Linstead PJ, King J, Cooper C, Roberts K (1990) Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181: 512–521 [DOI] [PubMed] [Google Scholar]
- Kreitschitz A. (2009) Biological properties of fruit and seed slime envelope: how to live, fly, and not die. In Gorb SN, ed, Functional Surfaces in Biology: Little Structures, Big Effects, Vol 1 Springer, London, pp 11–30 [Google Scholar]
- Kushwaha R, Lloyd TD, Schäfermeyer KR, Kumar S, Downie AB (2012) Identification of Late Embryogenesis Abundant (LEA) protein putative interactors using phage display. Int J Mol Sci 13: 6582–6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Li S, Gu Y (2012) Cellulose synthase complexes: composition and regulation. Front Plant Sci 3: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lei L, Somerville CR, Gu Y (2012) Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc Natl Acad Sci USA 109: 185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liners F, Letesson JJ, Didembourg C, Van Cutsem P (1989) Monoclonal antibodies against pectin: recognition of a conformation induced by calcium. Plant Physiol 91: 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie AY, Meade TJ (1998) A cobalt complex that selectively disrupts the structure and function of zinc fingers. Proc Natl Acad Sci USA 95: 6663–6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Kronenberger J, Marion-Poll A, North HM (2007a) In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol 48: 984–999 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Loudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM (2007b) A naturally occurring mutation in an Arabidopsis accession affects a β-d-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane HE, Young RE, Wasteneys GO, Samuels AL (2008) Cortical microtubules mark the mucilage secretion domain of the plasma membrane in Arabidopsis seed coat cells. Planta 227: 1363–1375 [DOI] [PubMed] [Google Scholar]
- Mendu V, Griffiths JS, Persson S, Stork J, Downie AB, Voiniciuc C, Haughn GW, DeBolt S (2011) Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol 157: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- North HM, Berger A, Saez-Aguayo S, Ralet MC (2014) Understanding polysaccharide production and properties using seed coat mutants: future perspectives for the exploitation of natural variants. Ann Bot 114: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oechslin R, Lutz MV, Amado R (2003) Pectic substances isolated from apple cellulosic residue: structural characterization of a new type of rhamnogalacturonan I. Carbohydr Polym 51: 301–310 [Google Scholar]
- Oka T, Nemoto T, Jigami Y (2007) Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. J Biol Chem 282: 5389–5403 [DOI] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, et al. (2010) A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol 153: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL (2004) Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704 [DOI] [PubMed] [Google Scholar]
- Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T (2008) A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J 54: 466–480 [DOI] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR (2000) The cellulose synthase superfamily. Plant Physiol 124: 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Aguayo S, Ralet MC, Berger A, Botran L, Ropartz D, Marion-Poll A, North HM (2013) PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25: 308–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Borden KLB, Boddy MN, Freemont PS (1996) Does this have a familiar RING? Trends Biochem Sci 21: 208–214 [PubMed] [Google Scholar]
- Scheible WR, Eshed R, Richmond T, Delmer D, Somerville C (2001) Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci USA 98: 10079–10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Sterling C. (1970) Crystal-structure of ruthenium red and stereochemistry of its pectic stain. Am J Bot 57: 172–175 [Google Scholar]
- Stork J, Harris D, Griffiths J, Williams B, Beisson F, Li-Beisson Y, Mendu V, Haughn G, Debolt S (2010) CELLULOSE SYNTHASE9 serves a nonredundant role in secondary cell wall synthesis in Arabidopsis epidermal testa cells. Plant Physiol 153: 580–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Ralet MC, Berger A, Diatloff E, Bischoff V, Gonneau M, Marion-Poll A, North HM (2011) CESA5 is required for the synthesis of cellulose with a role in structuring the adherent mucilage of Arabidopsis seeds. Plant Physiol 156: 1725–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan C, Hao Z, Zhu X, Avci U, Miller JS, et al. (2013) An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25: 270–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Usadel B, Kuschinsky AM, Rosso MG, Eckermann N, Pauly M (2004) RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol 134: 286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignon MR, Heux L, Malainine ME, Mahrouz M (2004) Arabinan-cellulose composite in Opuntia ficus-indica prickly pear spines. Carbohydr Res 339: 123–131 [DOI] [PubMed] [Google Scholar]
- Voiniciuc C, Dean GH, Griffiths JS, Kirchsteiger K, Hwang YT, Gillett A, Dow G, Western TL, Estelle M, Haughn GW (2013) Flying saucer1 is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in Arabidopsis seed mucilage. Plant Cell 25: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zabotina O, Hong M (2012) Pectin-cellulose interactions in the Arabidopsis primary cell wall from two-dimensional magic-angle-spinning solid-state nuclear magnetic resonance. Biochemistry 51: 9846–9856 [DOI] [PubMed] [Google Scholar]
- Western TL, Burn J, Tan WL, Skinner DJ, Martin-McCaffrey L, Moffatt BA, Haughn GW (2001) Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol 127: 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Western TL, Skinner DJ, Haughn GW (2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Young DS, Dean GH, Tan WL, Samuels AL, Haughn GW (2004) MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol 134: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO (2001) MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613 [DOI] [PubMed] [Google Scholar]
- Willats WG, McCartney L, Knox JP (2001a) In-situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta 213: 37–44 [DOI] [PubMed] [Google Scholar]
- Willats WG, McCartney L, Mackie W, Knox JP (2001b) Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 47: 9–27 [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Yang C, Wang X (2011) Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. J Exp Bot 62: 4495–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A, Ito T, Higaki T, Kutsuna N, Saito T, Ishimizu T, Osada H, Hasezawa S, Matsui M, Demura T (2010) Cobtorin target analysis reveals that pectin functions in the deposition of cellulose microfibrils in parallel with cortical microtubules. Plant J 64: 657–667 [DOI] [PubMed] [Google Scholar]
- Young RE, McFarlane HE, Hahn MG, Western TL, Haughn GW, Samuels AL (2008) Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20: 1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Shi D, Li J, Kong Y, Yu Y, Chai G, Hu R, Wang J, Hahn MG, Zhou G (2014) CELLULOSE SYNTHASE-LIKE A2, a glucomannan synthase, is involved in maintaining adherent mucilage structure in Arabidopsis seed. Plant Physiol 164: 1842–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zykwinska A, Thibault JF, Ralet MC (2007) Organization of pectic arabinan and galactan side chains in association with cellulose microfibrils in primary cell walls and related models envisaged. J Exp Bot 58: 1795–1802 [DOI] [PubMed] [Google Scholar]
- Zykwinska AW, Ralet MC, Garnier CD, Thibault JF (2005) Evidence for in vitro binding of pectin side chains to cellulose. Plant Physiol 139: 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.