Nutrient availability and light quality regulate branching by the production and perception of strigolactone.
Abstract
Plants alter their development in response to changes in their environment. This responsiveness has proven to be a successful evolutionary trait. Here, we tested the hypothesis that two key environmental factors, light and nutrition, are integrated within the axillary bud to promote or suppress the growth of the bud into a branch. Using petunia (Petunia hybrida) as a model for vegetative branching, we manipulated both light quality (as crowding and the red-to-far-red light ratio) and phosphate availability, such that the axillary bud at node 7 varied from deeply dormant to rapidly growing. In conjunction with the phenotypic characterization, we also monitored the state of the strigolactone (SL) pathway by quantifying SL-related gene transcripts. Mutants in the SL pathway inhibit but do not abolish the branching response to these environmental signals, and neither signal is dominant over the other, suggesting that the regulation of branching in response to the environment is complex. We have isolated three new putatively SL-related TCP (for Teosinte branched1, Cycloidia, and Proliferating cell factor) genes from petunia, and have identified that these TCP-type transcription factors may have roles in the SL signaling pathway both before and after the reception of the SL signal at the bud. We show that the abundance of the receptor transcript is regulated by light quality, such that axillary buds growing in added far-red light have greatly increased receptor transcript abundance. This suggests a mechanism whereby the impact of any SL signal reaching an axillary bud is modulated by the responsiveness of these cells to the signal.
To survive and reproduce, plants must respond to continual changes in their local environments, often having to integrate multiple simultaneous changes. Each individual plant integrates inputs from the environment with endogenous developmental programs to achieve optimal growth, survival, and reproduction. To achieve this, plants have evolved phenotypic plasticity (Nicotra et al., 2010), the ability to modify growth and development over their entire life span, in some cases many decades or hundreds of years. The control of branch outgrowth is one of the phenotypes modulated to optimize plant growth. Although the growth of a new branch leads to greater persistence, increased light and carbon capture, and may allow greater production of flowers, it is energetically costly. Therefore, the decision to produce a new branch likely involves signaling from multiple pathways.
Branch formation begins with a genetic program that leads to the growth of axillary meristems from the axils of developing leaves (Steeves and Sussex, 1989; Janssen et al., 2014). In many species, these meristems are produced in every leaf axil soon after the boundary layer forms between the developing leaf primordium and the apical meristem (Bell et al., 2012; Gendron et al., 2012; Yang et al., 2012; Naz et al., 2013; Janssen et al., 2014). In a number of species, the growth of these meristems is suppressed based on their position on the plant. In petunia (Petunia hybrida), for example, the axillary meristems in the nodes of the cotyledons and those of the first two leaves usually do not grow into branches, even in environmental conditions greatly favoring vegetative plant growth (Snowden and Napoli, 2003; Snowden et al., 2005). Stopping these four lowest axillary meristems from growing requires the production of the hormone strigolactone (SL) and its reception; in both SL production and signaling mutants, these normally suppressed nodes produce fully elaborated branches (Napoli and Ruehle, 1996; Snowden and Napoli, 2003).
One of the costs of growing a branch is the allocation of mineral nutrients. Two of the major macronutrients that limit growth are nitrogen (N) and phosphorus (P; Marschner, 1995), and the relationship between N, P, and SL is well established (Yoneyama et al., 2007a, 2007b; López-Ráez et al., 2008; Umehara et al., 2010). Even before their identification as SLs, it was shown that a factor involved in Orobanche spp. seed germination was produced in response to low P and N (Nagahashi and Douds, 2000; Yoneyama et al., 2001). Once identified, it was definitively shown that SLs (their production and exudation) were responsive to low N and P (Yoneyama et al., 2007a, 2007b). SLs affect multiple aspects of root development in response to P and sometimes N (Mayzlish-Gati et al., 2010; Kapulnik et al., 2011a, 2011b; Ruyter-Spira et al., 2011; Arite et al., 2012; Rasmussen et al., 2012; Sun et al., 2014). In rice (Oryza sativa), SL may be the primary regulator of root architecture responses to both P and N (Sun et al., 2014). Linking branching to SLs came when it was demonstrated that the dwarf (d) mutants of rice, the ramosus mutants of pea (Pisum sativum), and the more axillary growth (max) mutants of Arabidopsis (Arabidopsis thaliana) were defective in SL biosynthesis or reception (Gomez-Roldan et al., 2008; Umehara et al., 2008). Additional results from Arabidopsis confirmed that, in response to low P, SL increased in root exudates and in xylem sap and branch outgrowth was reduced (Kohlen et al., 2011).
The light required for carbon fixation is also a major limiting factor in the production of new branches. The shade-avoidance syndrome in plants is a collection of responses evolved to maximize the chance of outcompeting other plants in the collection of light: plants typically grow rapidly taller at the same time as limiting secondary growth in the stem; they also suppress branching and decrease leaf angle relative to the stem, all responses evolved to lift leaves above those of neighboring plants (Franklin, 2008; Casal, 2012). The shade-avoidance syndrome is triggered in plants when the red-to-far-red light (R:FR) ratio decreases, for example, when plants are grown in crowded conditions. Plants have a number of red light sensors in the form of the phytochrome proteins, but with respect to biotic competition for light in natural environments, phyB is the dominant player in the signaling pathway (Franklin, 2008; Casal, 2012). Branching is suppressed in low R:FR conditions in a number of species, and in the species studied thus far, a major component of the far-red light-induced suppression of branching are the type II TCP (named for Teosinte branched1 [TB1], Cycloidia, and Proliferating cell factor) transcription factors, including TB1 of maize (Zea mays), FINE CULM1 (FC1)/OsTB1 of rice, SbTB1 of sorghum (Sorghum bicolor), pea BRANCHED1 (PsBRC1), and the BRC1 and BRC2 genes of Arabidopsis (Doebley et al., 1995; Takeda et al., 2003; Aguilar-Martínez et al., 2007; Finlayson, 2007; Kebrom et al., 2010; Minakuchi et al., 2010; Braun et al., 2012). The connection between these genes and phyB signaling is well established; however, whether their mode of action involves SL signaling remains unresolved (Kebrom et al., 2006; Finlayson, 2007; Finlayson et al., 2010; Whipple et al., 2011; Dun et al., 2012; Guan et al., 2012; González-Grandío et al., 2013; Guo et al., 2013; for review, see Janssen et al., 2014).
Recently, we proposed a model bringing together all of the genes that had been suggested to play a role in the SL pathway to show the interactions that had been described (Janssen et al., 2014). Due to the nature of the data, the model was developed incorporating experimental evidence spanning petunia, Arabidopsis, pea, rice, sorghum, and maize. SL is derived from carotenoids via the sequential actions of a carotenoid isomerase, two carotenoid cleavage dioxygenases (CCD7 and CCD8), and a cytochrome P450 (Abe et al., 2014; Janssen et al., 2014; Zhang et al., 2014a). Following production, SLs are exported by an ATP-binding cassette-type transporter, PLEIOTROPIC DRUG RESISTANCE1 (PDR1; Kretzschmar et al., 2012). The increase in SL content and exudation by plants in low-P environments has been linked to increased expression of these biosynthetic genes (Umehara et al., 2010).
Recent work has shown that the SL reception system uses a receptor protein (DECREASED APICAL DOMINANCE2 [DAD2]/DWARF14 [D14]) that hydrolyzes the SL molecule to inactive compounds (Boyer et al., 2012; Hamiaux et al., 2012; Zhao et al., 2013). The SL receptor mutants (dad2 and d14) and the mutants in an F-box protein (max2 and d3) are completely insensitive to SL (Gomez-Roldan et al., 2008; Umehara et al., 2008; Arite et al., 2009; Hamiaux et al., 2012). The F-box Leu-rich repeat protein MAX2 is involved in the signaling pathway by direct interaction with DAD2/D14 at the protein level in response to SL; interestingly, this F-box protein is also involved in the response to another signal (Woo et al., 2001; Stirnberg et al., 2007; Nelson et al., 2011; Hamiaux et al., 2012; Waters et al., 2012). SL-induced interaction between DAD2/D14 and MAX2 results in ubiquitination and proteosome-dependent degradation of D53, an EAR motif-containing protein that, in turn, can interact with the transcriptional repressor TOPLESS (Jiang et al., 2013; Zhou et al., 2013). The receptor protein is also degraded in response to SL in Arabidopsis in a MAX2- and proteosome-dependent manner (Chevalier et al., 2014). This suggests that axillary meristems will only be restricted in their growth under conditions where both SL and receptor protein are constantly generated. This should allow a rapid return to growth upon exposure to a favorable environment.
The connection between light, the TCP genes, and the SL pathway appears to vary between species. In rice, it has been shown that phytochrome signaling affects the SL receptor’s transcript levels by altering the interaction of OsMADS57 and the rice TCP OsTB1 (Guo et al., 2013). This suggests that light quality and TCP proteins are upstream of the SL receptor and that they may control the sensitivity of plants to branch inhibition by SL by altering the availability of the receptor protein. Other reports have suggested a complete disconnect between TCP and SL in maize (Whipple et al., 2011; Guan et al., 2012). By contrast, in Arabidopsis, the BRC1 and BRC2 genes seem to have diversified in their function (Aguilar-Martínez et al., 2007; Finlayson, 2007; González-Grandío et al., 2013). BRC1 appears to act downstream of the SL signal, with transcript levels of BRC1 altered in SL mutants, while BRC2 expression is unaffected in SL production or signaling mutants (Aguilar-Martínez et al., 2007). Both the brc1 and brc2 mutants themselves have increased branching phenotypes, although this is quite mild for brc2. A TCP ortholog from pea, PsBRC1, is responsive to SL (Braun et al., 2012; Dun et al., 2012), with PsBRC1 mRNA levels regulated in a translation-independent manner, suggesting the SL-induced degradation of a transcriptional repressor of PsBRC1.
Here, we report experiments that alter P and N availability and quantify the effect on the growth of petunia plants in both the wild type and SL signaling mutants. In parallel, we altered the light quality experienced by vegetative-phase petunia plants and measured the affect of this environmental variable. Phenotypic measurements in a two-factor array of treatments confirmed that petunia’s response to the environment integrates the combination of inputs to produce a, presumably, optimized architecture. Against the background of these phenotypic changes, we measured the expression of genes known to be in the SL pathway and tested the hypothesis that light quality, perceived at the axillary bud, alters the availability of the receptor complex, thus moderating the effect of nutrient-regulated SL signaling from the roots.
RESULTS
Light Quality and Nutrient Availability Act in Concert to Regulate Axillary Bud Outgrowth
A series of experiments was carried out to determine the effects of altering P, N, crowding, and light quality (as the R:FR ratio) on the growth of petunia. In these experiments, we included a petunia SL biosynthetic mutant (ccd8, also known as dad1-1), an SL receptor mutant (dad2), and the isogenic wild-type plants.
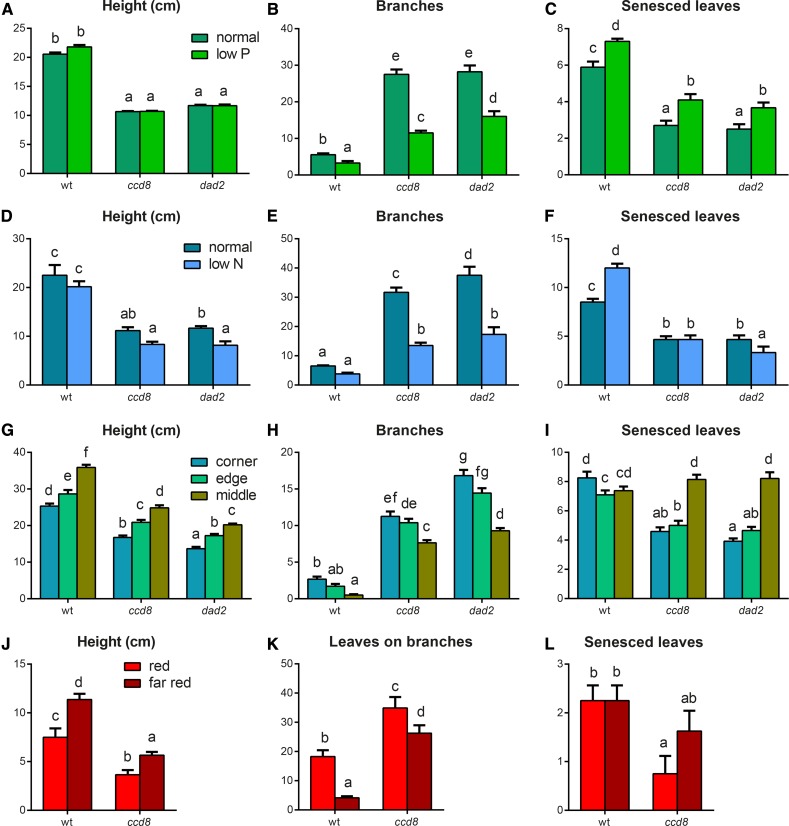
To understand the impacts of nutrients, we examined the effects of nutrient status (altering either P or N availability) and the SL pathway genes on growth responses in hydroponically grown petunia. The three genotypes grew as expected in the control conditions, with the mutant lines being shorter, producing more branches, and having delayed leaf senescence (Fig. 1), as shown previously (Napoli and Ruehle, 1996; Snowden and Napoli, 2003; Snowden et al., 2005). In low-P (Fig. 1, A–C) and low-N (Fig. 1, D–F) conditions, similar trends were observed. We also quantified shoot and root mass and provide more detail on branching into primary and secondary types, and in the N experiment, we measured the affect on branching angle, which is altered in dad mutants (Supplemental Figs. S1 and S2). The wild-type petunia responded to both low P and low N with a reduction in branching and increased leaf senescence but no significant change to plant height (Fig. 1, A–F). The ccd8 and dad2 mutants both showed an increased number of branches under low-N or low-P conditions compared with the wild type; however, both mutant lines were capable of responding to a reduction in nutrients with reduced branching compared with the mutants grown in nutrient-sufficient conditions. Hence, SL signaling is required for normal growth in normal and low-P and low-N conditions, but SL pathway mutants in both biosynthesis and perception are still able to respond to low nutrient availability by suppressing branching (Fig. 1, B and E; Supplemental Figs. S1 and S2).
Figure 1.
Petunia growth responses to individual environmental signals: light and nutrients. All graphs show phenotypic data as indicated. Values are means ± se. A to C, Petunia plants (the wild type [wt; line V26], ccd8, and dad2) were grown in nutrient-controlled hydroponics. After germination, the plants were established in standard-nutrient medium for 4 weeks. The medium was replaced with either standard-nutrient (250 µm P; dark green) or low-nutrient (5 µm P; light green) medium, and growth was continued for 4 weeks (n = 9–10). D to F, Petunia plants (the wild type, ccd8, and dad2) were grown in nutrient-controlled hydroponics. After germination, the plants were established in standard-nutrient medium for 3 weeks. The medium was replaced with either standard-nutrient (750 µm N; dark blue) or low-nutrient (250 µm N; light blue) medium, and growth was continued for 4 weeks (n = 6). G to I, Petunia plants (the wild type, ccd8, and dad2) were grown on soil supplemented with fertilizer for 7 weeks at a peak density of one plant per 25 cm2. The plants were subjected to three levels of crowded conditions for 7 weeks: corner (three neighbors; blue), edge (five neighbors; green), or middle (eight neighbors; mustard; n = 11–20). J to L, Petunia plants (the wild type and ccd8) were germinated and then established in hydroponics in white light for 5 weeks. The lighting was then supplemented with red light (R:FR ratio = 2.3) or far-red light (R:FR ratio = 0.4) for 3 weeks. Bright red bars show red light treatment, and dark red bars show far-red light treatment (n = 8). The statistical significance of differences was determined by ANOVA, and values with no common lowercase identifiers are significantly different from each other (P = 0.05).
To examine the effect of light quality and the SL pathway’s involvement in growth responses, we grew petunia in crowded conditions without nutrient limitation. Any changes observed are the combined actions of the thigmotactic, light quality, and light intensity inputs. The experiment created three classes of crowding, considering the middle, edge, and corner plants of a densely planted stand as separate classes (Supplemental Fig. S3). As the number of neighboring plants increased, all three genotypes had increased plant height and decreased branching (Fig. 1, G and H). The impact on primary versus secondary branching is shown in Supplemental Figure S3. Although the ccd8 and dad2 mutants could still respond to crowding conditions, both the branching and height traits were still significantly different from the wild-type phenotype. Hence, while the SL pathway contributes to the control of height and branching at the different planting densities tested, both biosynthesis and perception mutants remain competent to respond to the proximity of neighboring plants. Interestingly, while the corner and edge (with three and five neighbors, respectively) treatments showed the previously reported (Snowden et al., 2005) delayed leaf senescence in the ccd8 and dad2 mutants (Fig. 1I), when entirely surrounded by competing plants (middle treatment), both of these mutants had a rate of leaf senescence indistinguishable from the wild type. This suggests that some signal in this crowding experiment is able to bypass the mutation in the SL signaling system to induce leaf senescence.
To isolate light quality (as the R:FR ratio) from other signals in our crowding experiment, we altered light quality by specific wavelength addition. Red (660 nm) and far-red (735 nm) light-emitting diode (LED) light sources were used to supplement daylight in a greenhouse, altering the R:FR ratio. The ccd8 mutant displayed the expected decreased height, increased branching, and delayed leaf senescence compared with the wild type, and these differences were apparent in both light conditions (Fig. 1, J–L). As part of the process of developing a more rapid (early-detection) phenotyping technique for branching in petunia, we investigated three alternative measures of branching: branch count data (as used previously in the experiments shown in Fig. 1, B, E, and H), summed branch length (in mm), and leaves-on-branches count data. All three methods were able to distinguish wild-type from ccd8 plants and to detect alterations in branching due to light quality (Supplemental Fig. S4). The leaves-on-branches count gave a robust branching measure and was easily measurable early in development and in plants with limited branch growth; hence, it was used for subsequent experiments. Both wild-type and ccd8 mutant petunia were taller and had fewer branches in the low-R:FR treatment (i.e. far-red light-added treatment; Fig. 1, J and K). However, the magnitude of the branching response in the ccd8 plants was suppressed (4.4-fold in the wild type versus 1.3-fold in ccd8). We measured the diameter of the hypocotyls and stems of the plants in this experiment. In both cases, the ccd8 mutant was insensitive to light quality, whereas wild-type stem growth decreased with the addition of far-red light (Supplemental Fig. S4). Hence, SL signaling is required for normal growth in a range of light quality environments, but some SL-independent systems exist that are able to adjust a range of plant growth characteristics in response to altered R:FR ratios.
Simultaneous Changes in Nutrient and Light Quality Interact to Alter Branching
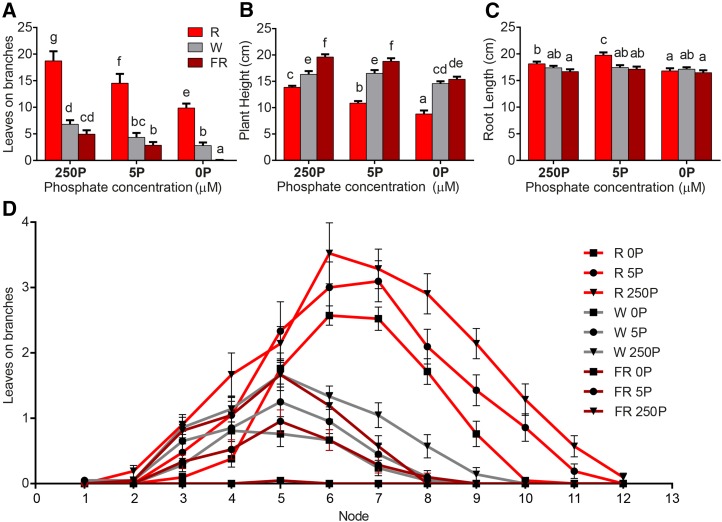
To determine whether one environmental condition might take priority over another, we designed an experiment to alter both nutrient availability and light quality. We regulated both P and the R:FR ratio simultaneously in a three-by-three array of treatments. Both normal amounts of P and increased red light led to higher branch numbers (Fig. 2). These effects are additive, such that the plants grown in red light with sufficient P are the most branched. By contrast, sufficient P and increased far-red light produced the tallest plants (Fig. 2B). Again, the impact of the two environment factors was additive. At 250 and 5 µm P, far-red light increases the height of the plants above that of white light. However, when P is decreased to zero in the medium, this effect is lost, suggesting that P has become limiting. Only very minor changes were observed in root length across the treatments (Fig. 2C). However, a visual inspection of the plants at harvest suggests that root development may have been altered in this experiment in a manner not revealed by a simple measurement of root length (Supplemental Fig. S5).
Figure 2.
Light quality and nutrient status (P) work in concert to control branching in petunia. Petunia plants were germinated and grown on soil in a glasshouse for 20 d and then transferred to hydroponics for 17 d (R:FR ratio = 1; 250 µm P) before being transferred to nine treatment environments for 7 d. The treatments were made up of a three-by-three array of P availability (250, 5, and 0 µm P) by light quality (R:FR ratio = 4–10 for red light [R], 1 for white light [W], and 0.2–0.3 for far-red light [FR]). A to C, The total numbers of leaves on all branches were counted (as a measure of branching), and plant height and root length were measured. The statistical significance of differences was determined by ANOVA, and values with no common lowercase identifiers are significantly different from each other (P = 0.05). D, The numbers of leaves on branches at each node are plotted individually for the nine treatments. Values are means ± se (n = 21).
A detailed characterization of the growth of the axillary buds at each node of the plants was performed (Fig. 2D). The data supported earlier observations that petunia branches in a zone starting from node 3 in growth-promoting conditions (Snowden and Napoli, 2003). Nodes 1 and 2 are kept in a dormant state by developmental or positional cues that are largely insensitive to environmental conditions. Nodes above this are increasingly able to grow out to produce branches, with the most growth observed at nodes 6 and 7 in our experiment, and less branch growth is observed at nodes closer to the apex. Axillary buds at node 7 show a wide range of growth, from none detected in the 0 µm P/far-red light treatment to maximal growth in the 250 µm P/red light treatment. Within each light treatment, the three P treatments alter the growth of the plants such that three distinct patterns of axillary bud growth are seen. Within the P treatments, there is significant overlap in the growth patterns observed in the far-red and white light treatments.
CCD7 Promoter Expression Indicates That Biosynthetic Gene Expression Is Predominantly in Roots and Stems and Is Responsive to Low P in Fine Roots
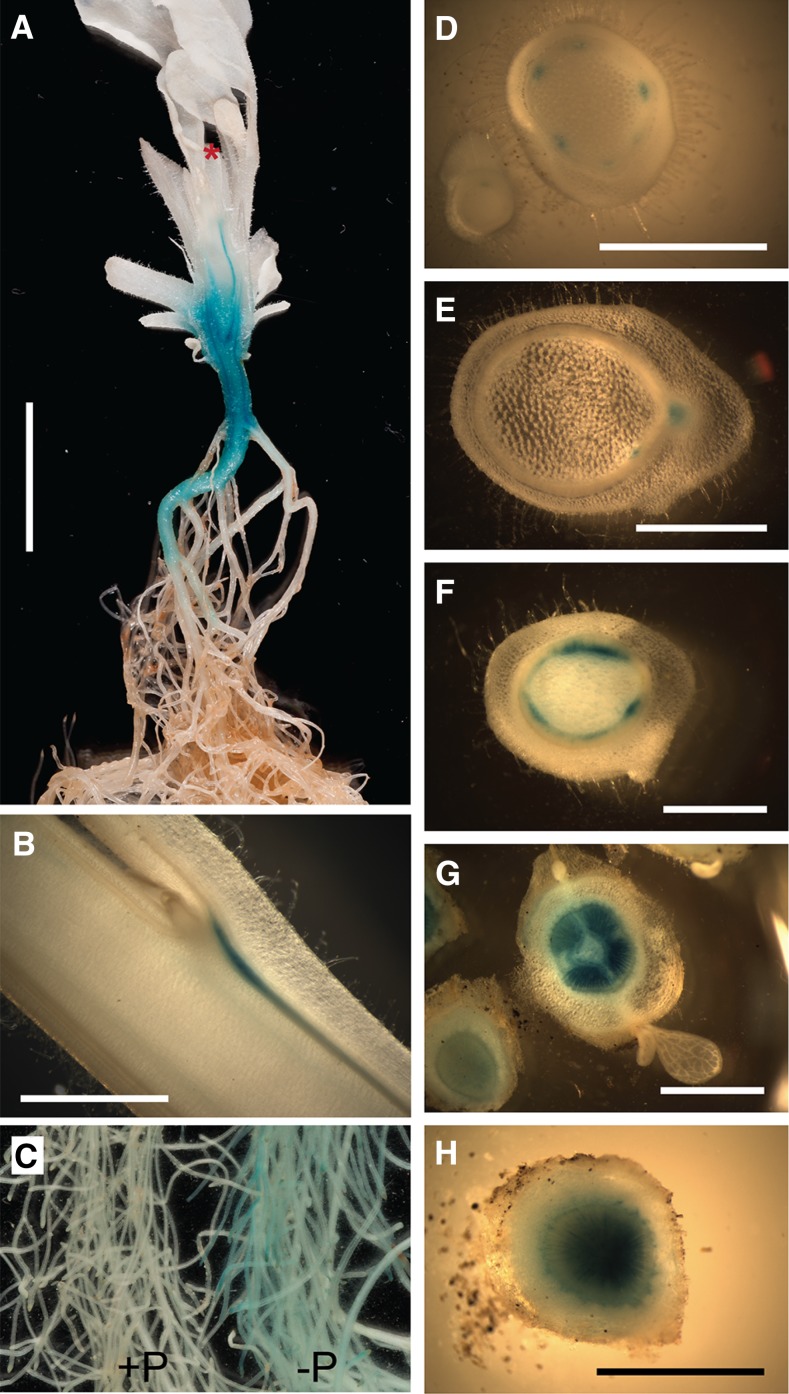
Earlier experiments have shown that the biosynthetic pathway leading to the production of SL is largely localized in the roots and stems of plants (with weak expression in leaves) and that the production of SL is responsive to a number of developmental and environmental factors (López-Ráez et al., 2008; Umehara et al., 2010; Kohlen et al., 2011; Yoneyama et al., 2012; Foo et al., 2013) and the biosynthetic genes respond to nutrient stress (Umehara et al., 2010). These observations indicated which tissues would be most informative for examination of the responses to environmental signals. To define the target tissues further, we examined the CCD7 expression pattern in petunia plants carrying a CCD7 promoter-GUS fusion, showing that this promoter is most strongly active in low stem tissue and in proximal tissues of the main root (Fig. 3). Cross sections of the stem suggest that the expression is widespread near the base but becomes restricted to narrow vascular traces in more acropetal regions of the stem (Fig. 3, A and D–H). Interestingly, there is a tightly localized zone of expression in the vasculature at the junction between the leaf petioles and the stem, immediately adjacent to the axillary bud (Fig. 3B). In conditions of plentiful P, very little GUS staining was observed in fine roots; however, in low-P conditions, staining was more readily apparent (Fig. 3C).
Figure 3.
Expression of the PhCCD7 gene as visualized from a PCCD7-GUS construct in transgenic petunia plants. All plants shown are 6 to 7 weeks old; plants in A and C were grown in hydroponics, and plants shown in B and D to H were soil grown. A, Longitudinal section of the bottom half of the stem of a plant and root system of a plant; most leaves, branches, and the top half of the main shoot have been removed. The red star indicates the position where the stem was cut. B, Longitudinal section through a section of stem showing an axillary bud and subtending leaf. C, Fine roots from two plants that were grown in normal-P (+P; left) or in low-P (−P; right) medium. D to H, Cross sections from a single plant at the upper stem (D), midstem (E; approximately node 12), low stem (F), hypocotyl-low stem junction (G), and main root (H). Bars = 1 cm (A) or 2 mm (B–H).
Three TB1-Like TCP Transcription Factors Were Isolated from Petunia Complementary DNA
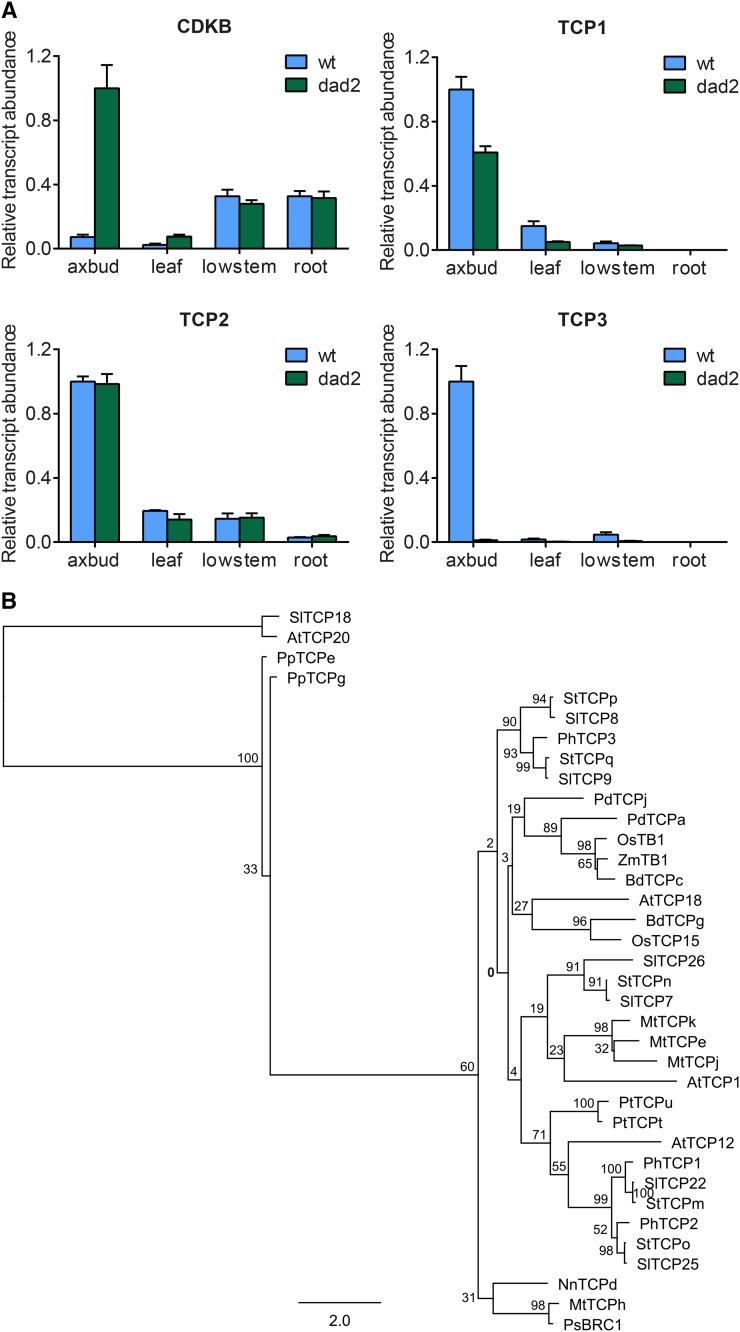
To investigate whether petunia TCP gene family members could be involved in controlling branch growth, particularly under different light conditions, we isolated petunia TCP genes using nested RACE with degenerate primers designed to the conserved TCP domain. Complementary DNA (cDNA) from a mixture of aboveground tissues was used as a template, and three petunia TCP genes (PhTCP1–PhTCP3) with sequence similarity to the TB1-like TCP genes were isolated. The TCP gene family is divided into two distinct clades (classes I and II) with a shared highly conserved TCP domain. A phlyogenetic analysis of the TB1 subclade of class II TCP proteins from rice, Brachypodium distachyon, Arabidopsis, tomato (Solanum lycopersicum), potato (Solanum tuberosum), Medicago truncatula, date palm (Phoenix dactylifera), Lotus spp., Physcomitrella patens, and poplar (Populus tremula) is shown in Figure 4, aligned with the maize TB1, pea BRC1, and three petunia TCP genes (phylogenetic analysis of the entire TCP class II clade is shown in Supplemental Fig. S6; Supplemental Data Set S1). All of the genes known to be involved in branching fall into a single TB1 subclade within the class II genes (Fig. 4B). There is little homology within this subclade between species except for within the three Solanaceae species, where potato and tomato form six pairs of similar genes. For three of the tomato and potato pairs, there is a corresponding petunia TCP, suggesting that there may be further petunia TCP genes to be identified. All the members of the TB1 subclade share a conserved SRXKAR(E/A)RARERT motif in the C-terminal region except for SlTCP25, StTCPo, and PhTCP2; however, the overall feature of this class of genes is the lack of sequence conservation between species apart from the conserved TCP domain.
Figure 4.
Petunia TCP genes involved in the SL signaling pathway. A, Relative transcript abundance in 8-week-old wild-type (wt) and dad2 petunia organs. Values are means ± se (n = 3). Samples were as follows: axbud, axillary bud samples from the four nodes above the highest branch; lowstem, 2 cm of stem above the cotyledons (nodes and internodes); leaf, expanded leaf blade; and root, fine roots. Relative transcript abundance as rescaled against the sample with the greatest expression for each gene is indicated on the graphs. B, Phylogenetic analysis of the TB1 subclade of the class II TCP proteins. Sequences with similarity to known TCP proteins were identified in public databases by iterative BLAST search, and an initial alignment using Geneious was manually edited. Phylogenetic relationships were determined using the PhyML plugin to create a maximum likelihood tree. Numbers are percentage bootstrap values for 100 replicates. Names are given as two letters for the species (At, Arabidopsis; Sl, tomato; St, potato; Ph, petunia; Bd, B. distachyon; Os, rice; Mt, M. truncatula; Ps, pea; Pt, P. tremula; Pd, date palm; Pp, P. patens; Ap, Anemone pulsatilla; Nn, Nelumbo nucifera; Zm, maize) followed by TCP and a number where the gene has been assigned a TCP number in the literature and a letter where such an assignment has not yet been made. AtTCP18 and AtTCP12 are commonly known as BRC1 and BRC2, respectively. SlTCP18 and AtTCP20 are class I TCP proteins and were used as the outgroup.
To examine the involvement of these PhTCP genes in the SL signal transduction pathway, quantitative PCR was used to examine the expression of these genes in wild-type and dad2 mutant petunia. The expression of the PhTCP1 and PhTCP3 genes is decreased in the dad2 mutant, indicating that these genes are likely to be involved in SL signaling (Fig. 4A). The expression of the cell cycle gene CYCLIN DEPENDENT KINASE B (CDKB) is included as a marker for cell division (growth). In the case of PhTCP2, no changes in expression were observed between the wild type and the dad2 mutant. This result limits any function of PhTCP2 in the SL pathway to upstream of DAD2 and further suggests that there is no feedback regulation from the SL pathway regulating this gene. Given the nearly 100-fold change in the expression of PhTCP3 in axillary buds between the wild type and the dad2 mutant, we suggest that PhTCP3 expression is dependent on a functioning SL perception system, and our observations place PhTCP3 downstream of DAD2. These results are consistent with PhTCP3 (and to a lesser extent PhTCP1) being similar in function to AtBRC1 and PhTCP2 being similar to AtBRC2 (Aguilar-Martínez et al., 2007; Finlayson, 2007).
Molecular Responses to P Availability Are Predominantly in Fine Roots
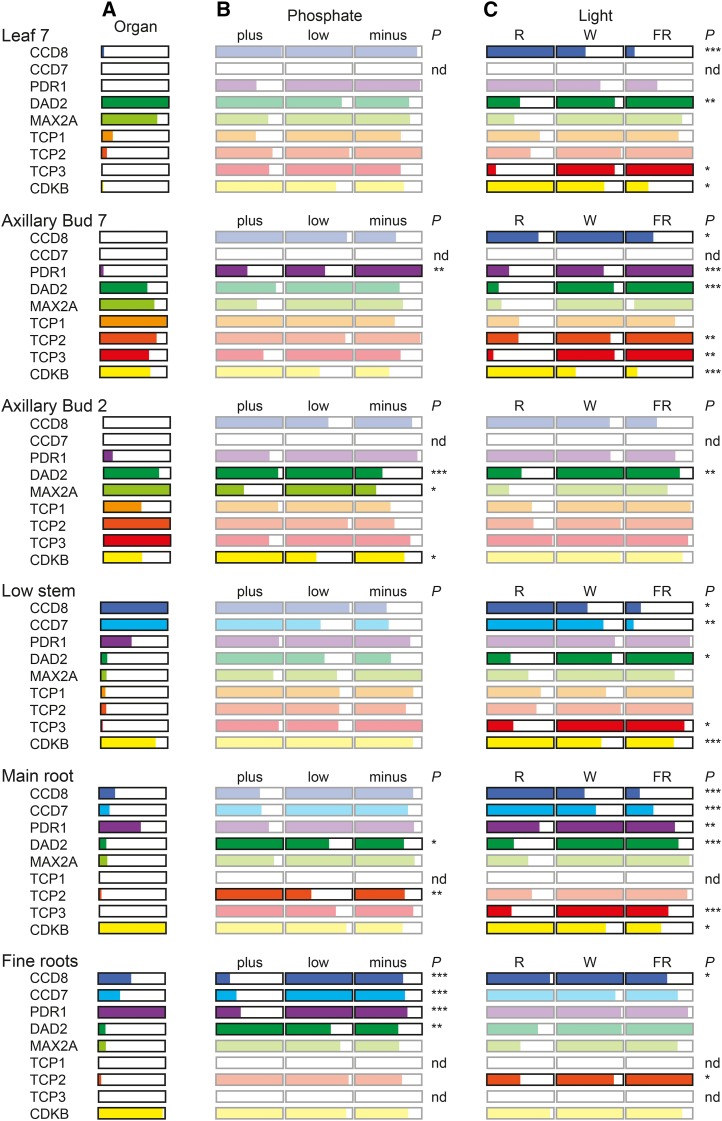
To determine the status of SL signal production and signaling in the plants from the multienvironmental factor experiment, we investigated the expression of the petunia TCP genes as well as other known SL pathway genes. We quantified the relative transcript abundance of these genes in a range of organs, including those shown to express SL biosynthetic (Fig. 3) and perception (Supplemental Data Set S2) genes. In Figure 5, we present the average relative transcript abundances (normalized per gene) grouped by organ (Fig. 5A), nutrient treatment (Fig. 5B), and light treatment (Fig. 5C), with evidence of any changes determined by ANOVA with significance indicated by P < 0.05.
Figure 5.
The transcription of the SL synthesis and reception pathway genes is altered in response to environmental cues. The relative transcript abundance of SL-related genes was quantified in six organs from plants treated in an array of light quality by P availability using quantitative reverse transcription-PCR. The data have been grouped by organ (A), P treatment (B), and light treatment (C), averaged, and within each grouping normalized to 1. Each bar represents average abundance on a linear scale from 0 to 1. The transcript abundance of a given gene (organ average) is directly comparable across all organs within the organ grouping (A) but only within an organ when grouped by P or light (B and C). Transcript abundance is not directly comparable between different genes because of normalization. The statistical significance of any changes seen within the P or light treatments has been tested by ANOVA, and the level of significance is indicated as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Where P > 0.05, the bars have been faded to 20% of their original intensity. For P treatment, plus = 250 µm P, low = 5 µm P, and minus = 0 µm P; for light treatment, R (red light) = R:FR ratio > 4, W (white light) = R:FR ratio = 1, and FR (far-red light) = R:FR ratio < 0.3. Organ average n = 27, P or light n = 9. nd, Transcript not detected.
Previous results suggested that fine roots, the main root, and the low stem would be the major sites of SL synthesis gene expression (Snowden et al., 2005; Drummond et al., 2009). We detected the greatest abundance of CCD7 and CCD8 and also PDR1 transcripts in these organs (Fig. 5A). The transcript of CCD7 was not detectable in the axillary bud or leaf samples. Both CCD8 and PDR1 were detected in these tissues, although at relatively low levels. Prior results also suggested that the genes for the receptor complex (DAD2 and PhMAX2A) would be most abundant in axillary buds and leaves (Drummond et al., 2012; Hamiaux et al., 2012). This can also be observed in the experiment presented here, with the transcripts for these genes also detectable in the remaining organs (at approximately 10% of their maximum). All three of the newly isolated TCP transcription factor transcripts are most abundant in the two axillary bud samples, with either low or nondetectable abundance in the remaining organs. The transcript abundance of CDKB, a marker for cell division, was high in axillary bud 7, low stem, main root, and fine roots. CDKB transcript levels were also moderate in axillary bud 2 and low in leaf 7. Our results suggest that the blade of leaf 7 has largely finished cell division and that axillary bud 2 (usually dormant) may still contain a pool of dividing cells.
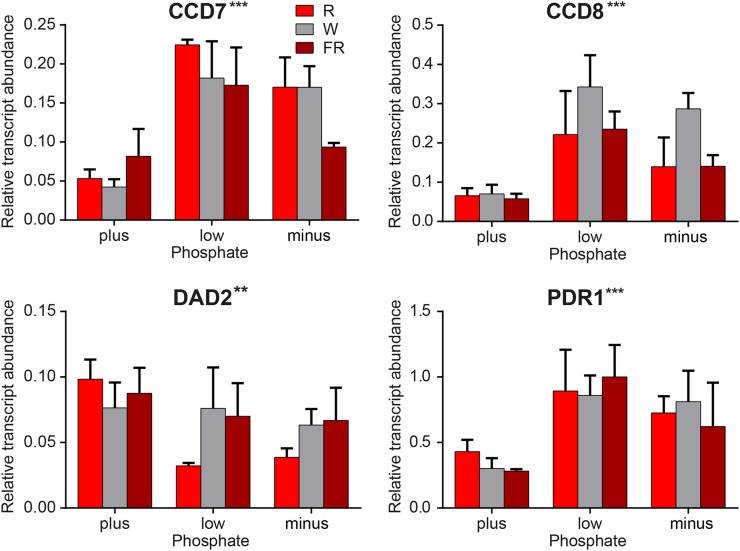
We detected significant increases in the transcript abundance of the SL synthesis and transport genes in fine roots in response to decreased P (Fig. 5B). Conversely, the DAD2 receptor transcript decreased in fine roots, main roots, and axillary bud 2 in response to decreased P. In this analysis, the PhMAX2A transcript abundance was not statistically significantly regulated by P availability or light quality. However, a more detailed analysis of the raw transcript abundance data identified positive correlations (Spearman’s rank correlation coefficient [rs] for different organs of 0.60–0.91; P ≤ 0.001) between these two genes in all the organs tested. The fine root samples showed the most significant changes in gene expression with respect to P; these data are shown in more detail in Figure 6. The CCD7, CCD8, and PDR1 transcript abundances in the low- and minus-P treatments are all greater than those in the plus-P treatment (Fig. 5). With respect to DAD2, the decrease is only significant in the red light treatments (Fig. 5C).
Figure 6.
Details of transcript abundance changes in response to P availability in fine root samples. Relative transcript abundance is shown for selected genes in fine root samples from wild-type petunia treated with varied P availability and light quality. For each gene, the samples are grouped by P treatment, with the colored bars representing light treatment as follows: R:FR ratio > 4 (red; red light [R]), R:FR ratio = 1 (gray; white light [W]), and R:FR ratio < 0.3 (dark red; far-red light [FR]). Values are means with se (n = 3). The values of abundance for each gene are normalized to the greatest abundance of that transcript in any sample. Asterisks next to each gene name indicate the P value in Figure 5.
Molecular Responses to Light Quality Are Widespread But Largely Absent from Axillary Bud 2
In Figure 5C, we also present our quantitative PCR data grouped by light treatment. By contrast to the limited number of changes seen in response to P, the effect of light was seen in a broad set of tissues, particularly axillary bud 7 and the main root. In these two organs, as well as leaf 7 and low stem, the transcript abundance of CDKB decreases with decreasing R:FR ratio. This observation indicates that the light treatments had the expected major impact on the cell cycle in these plants, and this is reflected in the growth phenotypes observed (Fig. 2).
In both the low stem and main root, where the overall relative expression of CCD7 and CCD8 is high, the transcript abundance of these genes decreases as the R:FR ratio decreases. This suggests that the most branched plants might be producing the most SL (assuming that increased expression of CCD7 and CCD8 leads to increased SL production). This positive correlation (rs = 0.45–0.61; P ≤ 0.007) of CCD7 and CCD8 expression with branching is consistent with earlier reports (Foo et al., 2005; Snowden et al., 2005; Johnson et al., 2006; Arite et al., 2007; Simons et al., 2007), where it has been suggested that the relationship was explained by negative feedback from growing branches.
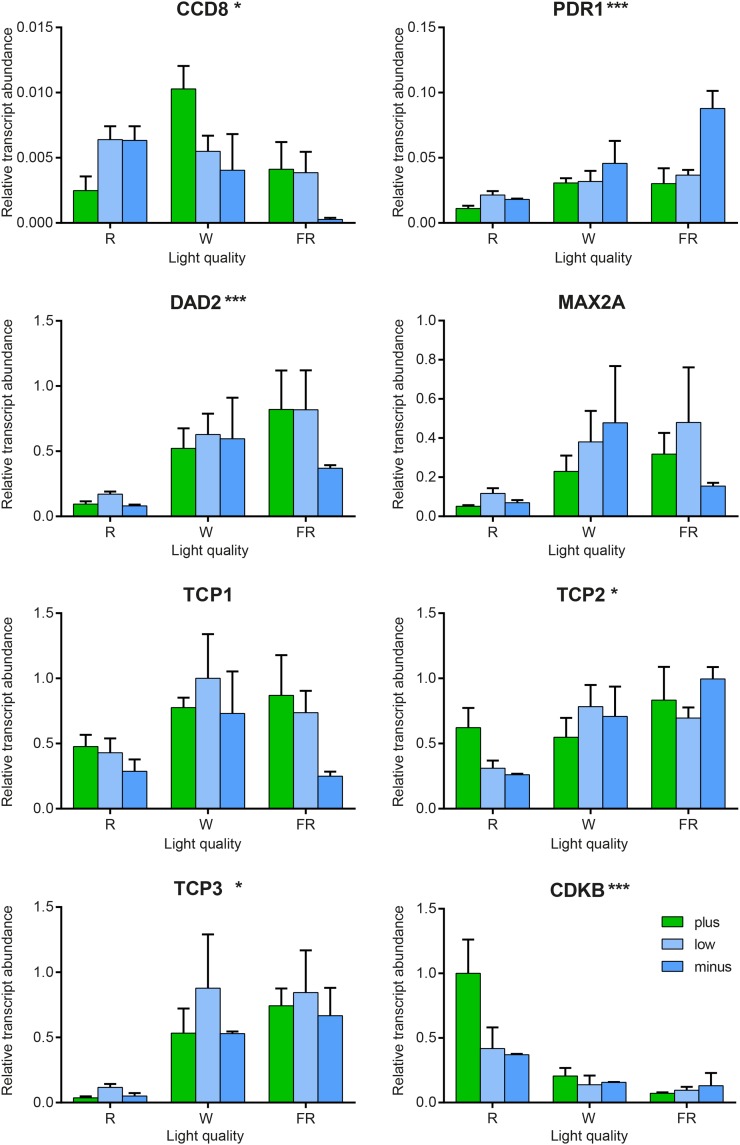
The transcripts for the SL receptor complex and the TCP transcription factors also show significantly altered patterns of abundance in response to the light treatments (Figs. 5C and 7), with increased R:FR ratios correlated with decreased receptor complex gene expression and, hence, decreased sensitivity to SL. By contrast, in axillary bud 2, while the abundances are similar, there is little or no significant regulation by light (Fig. 5C; Supplemental Fig. S7). The relationships between the DAD2 SL receptor transcript and the three TCP transcription factors showed different patterns, with PhTCP3 showing a good correlation with DAD2 in axillary bud 7, PhTCP2 better correlated in axillary bud 2, and PhTCP1 poorly correlated in either sample (Table I).
Figure 7.
Details of transcript abundance changes in response to light quality (R:FR ratio) in axillary bud 7 samples. Relative transcript abundance is shown for the genes tested in the axillary bud 7 samples from wild-type petunia treated with varied P availability and light quality. The CCD7 transcript was not detected in these samples. For each gene, the samples are grouped by light treatment (R, red light; W, white light; and FR, far-red light), with the colored bars representing P availability as follows: green, 250 µm P; blue, 5 µm P; and dark blue, 0 µm P . Values are means with se (n = 3). The values of abundance for each gene are normalized to the greatest abundance of that transcript in any sample. Asterisks next to each gene name indicate the P value in Figure 5.
Table I. rs and corresponding P values for the expression of DAD2 and the three PhTCP genes in buds that are responsive (bud 7) and nonresponsive (bud 2).
Values over 0.7 are shown in bold.
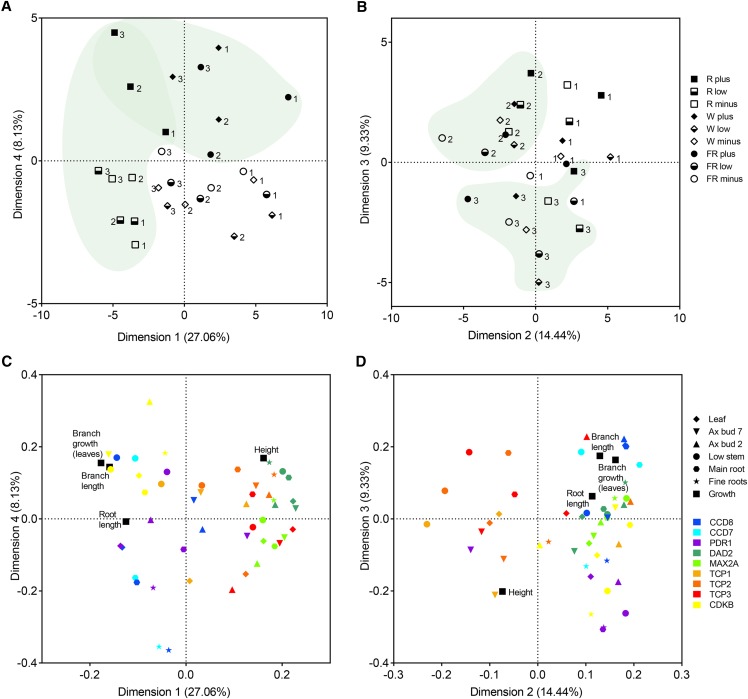
We examined the interrelationships of the molecular data and the phenotypic data using principal component analysis (PCA) to analyze and visualize the data (Fig. 8). By graphing dimensions 1 and 4 of the PCA, we were able to separate red light (squares) from white and far-red light and plus P (black symbols) from low and minus P (Fig. 8A). This suggests that dimension 1, which explains that 27.06% of the data can be attributed to the effect of light quality, and dimension 4, which explains 8.13% of the variation, can be attributed to P availability. Graphing dimensions 2 (14.44%) and 3 (9.33%) allowed some separation of the three replicates (Fig. 8B). Samples were taken over an 8-h period during the middle of an approximately 14-h daylight period, with each replicate completed before the next was started. This PCA suggests that some other minor changes in gene expression in the samples may be occurring that were not directly tested here (e.g. circadian rhythms, temperature, humidity, other light effects).
Figure 8.
PCA of growth and molecular data. Principal component scores (A and B) and latent vectors (C and D) are shown for the first four dimensions from a PCA of growth characteristics (branch growth, root length, and plant height) and gene expression. Gene expression for gene/organ combinations where no expression was detected was excluded from this analysis (i.e. CCD7 expression was not detected in axillary bud or leaf samples, PhTCP1 expression was not detected in root samples, and PhTCP3 expression was not detected in fine roots). Dimensions 1 and 4 (A and C) explain 27.06% and 8.13% of the variance in the data, respectively, and show separation of the environmental treatments, whereas dimensions 2 and 3 (B and D) explain 14.44% and 9.33% of the variance and show some separation of the biological replicates. Light treatments are as follows: R, red light; W, white light; and FR, far-red light.
Using the same PCA axes, we plotted the latent vectors (as terminal points) for the individual factors in the experiment, both molecular and phenotypic (Fig. 8, C and D). This graphing showed clustering between the growth of branches and the expression of CDKB in three of the four dimensions presented, as expected for a gene linked to the cell cycle (Table II). DAD2, PhMAX2A, PhTCP2, and PhTCP3 expression loosely cluster together for dimensions 1 and 4, consistent with the positive correlation results for these genes mentioned above (Fig. 8C; Table I). This cluster of gene expression is separated from the branch growth phenotypic data, which can also be seen with negative correlations, indicating the expected negative control of branching by the SL pathway (Table II). PhTCP3 shows the strongest negative correlation to the branch growth data, indicating that this gene likely has more effect on branch growth than PhTCP1 or PhTCP2. Additionally, clustering of CCD7 with CCD8 indicates that these two genes are likely coordinately expressed in at least root and low stem tissues (as mentioned previously, we did not detect CCD7 expression in axillary bud or leaf samples). Correlations between CCD7 and CCD8 were 0.58 for low stem (P = 0.001), 0.87 for main root (P < 0.001), and 0.88 for fine roots (P < 0.001).
Table II. rs and corresponding P values for expression in axillary bud 7 and overall branch growth.
DISCUSSION
The observation that both light and nutrients affect branch formation is well established. In these experiments, we sought to examine the interplay between environmental cues and the SL pathway in modulating the growth of plants. We found that alterations in nutrient availability and light quality can additively affect the growth of branches and that this control is overlaid on the developmental control of axillary meristem responsiveness to give a range of potential phenotypes for a plant. SL mutants showed increased branching under various nonoptimal growth conditions, indicating a role for the SL hormone in regulating this process.
However, both SL production mutants and reception mutants were still able to modify their phenotype in response to the environmental stimuli, suggesting (unsurprisingly) that the SL pathway is unlikely to be the only regulatory pathway involved in the response to these environmental signals. For example, the delayed leaf senescence phenotype observed in both SL receptor and production mutants is completely absent in the crowded plants, indicating that a leaf senescence signal induced by crowding is independent of the SL pathway. With respect to branching, other signals, including but probably not limited to auxin, also play a role in the plant’s response to light quality and nutrient availability (Casal, 2012; de Jong et al., 2014; Reddy and Finlayson, 2014; Zhang et al., 2014b). A complex interplay between different plant hormone signals is to be expected for a process as energetically costly and developmentally significant as branch growth.
Using conditions identified from preliminary experiments, we designed an experiment that modulated both P levels and R:FR ratios together to produce a range of developmental outcomes, from extreme inhibition of branching to very active outgrowth of axillary meristems to form branches. We were able to establish conditions where the phenotype was significantly altered and determine which nodes were most consistent in their branching response. The length of treatment for the light × P experiment was 7 d. This treatment time is relatively long for a light treatment, as some light responses are known to be fast and transient (Franklin, 2008; Casal, 2012). However, the growth of a branch is more likely to be affected by persistent changes in light conditions (such as shading by neighboring plants) over longer periods of time than by transient changes. Additionally, we aimed to find a time when phenotypic changes to branching were measurable. This also meant that leaf senescence had not occurred within the time frame of this experiment. It should be kept in mind that some additional gene transcription changes could occur in different time frames from those described here. Additional experiments are needed to examine changes over time for the SL pathway genes. An important future goal is to gain an understanding of the dynamics of SL signaling, given that the process of branching at early stages is able to be switched from dormant to actively growing and back (Napoli et al., 1999).
Based on current models of SL signaling, we had some initial predictions, in particular that increased red light might result in a reduction of receptor levels such that the SL pathway was impaired in its response to SL. Our results indicate that light quality does alter receptor complex expression in a manner consistent with the observed phenotype (increasing receptor expression in far-red light and decreasing expression in red light), suggesting that, in petunia, modulation of receptor complex expression is a significant mechanism for branch regulation by light quality, supporting observations reported in other species (Kebrom et al., 2006; Finlayson, 2007; Finlayson et al., 2010; Whipple et al., 2011; Dun et al., 2012; Guan et al., 2012; González-Grandío et al., 2013; Guo et al., 2013).
Neither light quality nor P limitation was entirely dominant over the other signal for branching. This suggests either that both P and R:FR conditions can regulate branching through an SL-independent pathway or that neither environmental condition is able to alter the SL signaling pathway to the point of a completely on or a completely off state. This result suggests that it may be more useful to consider the branching pathway to be regulated by a balance of signals rather than as a simple on/off switch.
One of the aims of these experiments was to see if an analysis of expression could clarify which, if any, of the petunia TCP genes were involved in SL signaling and perhaps place them in the pathway. Results from rice show that the rice ortholog of TB1 (FC1) can bind to and inhibit a transcriptional repressor of rice D14 transcription (Guo et al., 2013). This observation places the rice TCP protein FC1 upstream of the SL receptor in the response to R:FR. It has previously been shown that the steady-state mRNA levels of the sorghum TB1 homolog (SbTb1) are regulated by R:FR ratios in a manner also shown for the Arabidopsis TB1 homologs BRC1 and BRC2 (Kebrom et al., 2006; Aguilar-Martínez et al., 2007). This regulation of BRC1 and BRC2 by R:FR ratios has been shown in Arabidopsis to be dependent on the PhyB gene (Aguilar-Martínez et al., 2007). The above observations strongly suggest that a TCP gene homologous to TB1 functions to connect R:FR signaling from the phytochrome system to the regulation of branching by the SL receptor. However, whereas in monocots there is only one or possibly two TB1 orthologs, in eudicots there are several TCP genes with similarity to TB1. Phylogenetic analysis suggests that three TCP genes in Arabidopsis are closely related to TB1, and in the Solanaceae (e.g. tomato, potato, and petunia), potentially six genes are homologous to TB1. The presence of multiple TB1-like TCP genes, presumably resulting from gene duplication events, allows for the possibility of diversification in function. In our experiments, one TCP gene (PhTCP2) is regulated by R:FR and unaffected by mutations in the SL receptor gene. These observations suggest that PhTCP2 could act to regulate DAD2 levels in response to R:FR ratios in a manner similar to FC1/OsTB1. It is interesting that PhTCP2, as well as homologous tomato and potato proteins, lack the SRXKAR(E/A)RARERT motif in the C-terminal region that is conserved in the rest of the TB1 subclade in both monocots and dicots. Whether this difference plays any role in the function of PhTCP2 in SL signaling is currently unknown.
Results from pea have shown that the expression of a TCP (PsBRC1) in pea is regulated by SL (Braun et al., 2012; Dun et al., 2012). This observation has been interpreted as placing PsBRC1 downstream of the SL receptor in the signal transduction pathway. Our observations show that two of the TCPs in petunia have significantly altered expression in dad2 mutants, and both respond to P and R:FR treatment in a manner consistent with a role in signal transduction downstream of DAD2. While both PhTCP1 and PhTCP3 respond in a similar manner, the changes in PhTCP3 expression have a stronger correlation to branching in our experiments and PhTCP3 expression is affected more in dad2 mutants, suggesting that PhTCP3 may be the primary gene involved in downstream responses to SL. However, PhTCP1 may be sufficiently redundant in function to also fill the role. The difference between monocot and dicot TB1-like TCPs suggests that either one or more duplication events occurred in dicots, with subsequent diversification of TCP function, or that TCP genes were lost from the monocot lineage, with the role(s) of the lost genes taken up by unrelated genes, such as GRASSY TILLERS1 (Whipple et al., 2011).
The observation of multiple TCPs functioning at different points in the SL pathway resolves the apparent paradox in the existing data that suggested that a TCP transcription factor was both upstream and downstream of the DAD2/MAX2 receptor complex. In petunia, PhTCP2 appears to act upstream and PhTCP3 downstream of the DAD2/MAX2 receptor complex, and perhaps this is the case in other species. However, it may be that any differences in TCP gene function play a role in the different architectures between species.
In our experiments, PhMAX2A expression was present in most tissues, and while it did not show statistically significant changes in expression in response to P or R:FR, PhMAX2A expression was highly correlated with DAD2 expression. This suggests that while PhMAX2A expression was variable, it was still responding to environmental signals coordinately with DAD2. Transcriptional responses for DAD2 and MAX2 appear to vary somewhat between species. In petunia, PhMAX2A seems somewhat variable but does appear to respond to environmental and developmental signals (Drummond et al., 2012; this work), whereas DAD2 expression responds significantly (Hamiaux et al., 2012; this work). By contrast, in Arabidopsis and sorghum, MAX2 appears to respond significantly to light signals (Kebrom et al., 2010; Chevalier et al., 2014), and in Arabidopsis, Chevalier et al. (2014) show that D14 mRNA levels do not respond to developmental or light quality signals.
In roots and low stems, the gene expression responses were largely as expected from previous reports and models of SL synthesis (Umehara et al., 2010). Interestingly, the increase in expression of SL synthesis and transport genes in response to P starvation was significant only in the fine roots. We also observed a reduction in the expression of DAD2 and a correlated change in PhMAX2A in response to P starvation in fine roots; a similar reduction in receptor expression has been reported in rice under P and N limitation (Sun et al., 2014), suggesting that an as yet unidentified negative feedback loop regulates the receptor complex. Significant changes in root gene expression were also observed in response to different light conditions, with synthetic genes down-regulated in far-red light where branching is decreased, possibly as a result of some feedback mechanism. Decreased SL biosynthetic gene expression should result in a decrease in SL in far-red light conditions that is presumably balanced by an increase in receptor expression in axillary buds or perhaps by a change in the transport of SL. It remains unclear how SL synthetic genes are regulated; however, the NODULATING SIGNALING PATHWAY1 (NSP1) and NSP2 transcription factors are likely to be involved (Liu et al., 2011).
In these and other experiments (Snowden and Napoli, 2003; Drummond et al., 2012), we observed differential growth of branches within a petunia plant. In this work, the axillary bud at node 2 remains dormant under all conditions, whereas the axillary bud at node 7 is responsive to environmental signals. The basis for this developmental difference is currently unknown. However, the expression of PDR1 and PhTCP3 suggests that they play a role in this difference. PDR1 expression does not change in axillary bud 2, and expression overall is higher in this sample than in axillary bud 7. Within axillary bud 7, we did observe significant decreases in PDR1 expression and, hence, the transport of SL under growth-permitting conditions. Another interesting observation is the high and unchanging level of expression of PhTCP3 at node 2, which is the only tissue where its expression does not correlate with DAD2. Our results suggest that PhTCP3 (like PsBRC1) acts downstream of DAD2/PhMAX2A and that the normal response to SL is to increase the expression of this TCP (Dun et al., 2012) by proteosome-dependent degradation of a repressor of this TCP. With that hypothesis in mind, it is possible that the developmental difference between axillary buds 2 and 7 that prevents growth under any environmental condition is that the putative repressor of PhTCP3 is itself absent in this tissue.
The commitment to the growth of an axillary meristem into a branch is a complex and important point in the development of a plant, especially when environmental conditions are not optimal. The shade-avoidance response results in the suppression of branch growth in petunia, as does the reduction in availability of the key nutrient P. Our understanding of the SL signal production and reception pathway suggested that both nutrient and light quality environmental inputs may be integrated at the point of SL reception, with SL production regulated in response to nutrient availability and the shade-avoidance response regulating the availability of the receptor for SL. By modulating both of these environmental inputs together, we were able to observe a range of developmental outcomes, from extreme inhibition of branching to very active outgrowth of axillary meristems to form branches. Our results show that the SL receptor does act to integrate environmental signals; however, we were not able to completely inhibit the response to either signal, suggesting that it is not an all-or-nothing system where the receptor is completely prevented from responding to input signals.
These results indicate that the genes in the SL pathway are likely to be good targets for the modification of branching in field crops, as the phenotype is robust under diverse conditions while still allowing plants to respond to nonideal growth conditions. The retention of the ability to respond to environmental changes is particularly important for plants that grow for many seasons, where the regulation of branching must be flexible enough to change growth as seasonal and other environmental inputs change.
MATERIALS AND METHODS
Genetic Stocks and Plant Growth Conditions
The wild-type petunia used in these experiments is Petunia hybrida inbred line V26. The dad mutants, derived from this line, were isolated by Napoli and Ruehle (1996). All experiments were conducted in greenhouse conditions under natural light (for some experiments, this was supplemented as described below) and with temperature set at 25°C ± 5°C. All soil-grown plants were grown in commercial potting medium (Yates seed-raising mix) in sixpacks (100-mL size) watered with fertilizer (80 mg L−1 N, 80 mg L−1 P, and 60 mg L−1 potassium) from Wuxal Super 8-8-6 plus micro liquid fertilizer (Aglukon). For crowding experiments, petunia plants were grown in sixpacks placed next to each other to create a six-by-eight grid with two additional two-by-three grids and then grown in the center of a greenhouse unit to avoid any shading from the walls (Supplemental Fig. S3). For hydroponic growth, petunia seeds were germinated on soil or vermiculite, and after 2 weeks, seedlings were transferred to 250 mL of 4- to 8-mm Hydroton clay pebbles that were suspended in nutrient medium. At most, 21 plants were grown in 20 L of constantly aerated (Two AquaOne 1- × 1- × 10-cm airstones) one-half-strength hydroponic salt solution medium (Hamiaux et al., 2012). The P concentration was altered by the variable addition of phosphoric acid. The N concentration was altered by the variable addition of calcium nitrate. The solution volume was maintained at 20 L using medium and replaced only at the time of treatment. The pH of the medium was maintained at 5.7 with the daily addition of hydrochloric acid.
The R:FR ratio was altered by the addition of monochromatic light supplied from LED light sources. All LEDs were purchased from Roithner Lasertechnik. To add red light, two LED660N-66-60 LEDs were used, and to add far-red light, four LED735-66-60 LEDs were used. To achieve appropriate R:FR ratios, for the white, red, and far-red light treatments, the irradiance of the baseline white light was restricted such that photosynthetically active radiation was not greater than 200 µmol photons m−2 s−1. During this experiment, the daylength was approximately 14 h (summer in Auckland). The spectra for the light treatments are shown in Supplemental Figure S8; R:FR ratios were 4 to 10 for the red light treatment, 1 for the white light treatment, and 0.2 to 0.3 for the far-red light treatment. The spectra were measured using an Ocean Optics USB4000 spectroradiometer, and data were captured using the Spectrasuite software following the manufacturer’s instructions (www.oceanoptics.com).
Analysis of the CCD7 Promoter Action in Petunia
Inverse PCR was used to isolate the PhCCD7 promoter using previously described methods (Snowden and Napoli, 1998). A total of 5 kb of sequence upstream of the PhCCD7 ATG was isolated (KR002102), and the entire 5-kb promoter fragment was amplified with PCR using Expand High Fidelity enzyme according to the manufacturer’s instructions and verified by sequencing. The PhCCD7 promoter fragment was cloned into pHEX14, a binary vector containing a promoterless GUS gene (Rae et al., 2014), resulting in a PCCD7-GUS construct. Six transgenic lines were generated with this construct, and the expression pattern for one line is shown (five out of the six lines showed identical GUS expression patterns).
Cloning of the TCP Genes
To identify TCP genes from petunia, nested RACE was performed using degenerate primers designed to the conserved TCP domain. RNA was isolated from 4-week-old wild-type petunia plants (most vegetative organs, although mature leaves were removed so that the material was enriched for stem, shoot apex, and axillary buds). cDNA was generated using a GeneRacer kit from Invitrogen according to the manufacturer’s recommended protocols. The degenerate primer oTB2 (5′-GAYMGICAYWSIAARATITGYACNGC-3′) was first used in combination with the GeneRacer 3′ primer (5′-GCTGTCAACGATACGCTACGTAACG-3′), then a nested PCR used the degenerate primer oTB7 (5′-CARGAYATGYTIGGNTTYGAYAARGC-3′) in combination with the GeneRacer 3′ nested primer (5′-CGCTACGTAACGGCATGACAGTG-3′). Additional sequences for the three TCP genes were isolated through a combination of RACE from a cDNA template and inverse PCR using a genomic DNA template to obtain the full coding sequences of PhTCP1 (KR002103), PhTCP2 (KR002104), and PhTCP3 (KR002105).
Quantitative Reverse Transcription-PCR
The samples from which the data in Figure 4 were produced are axillary buds (from the four nodes above the highest branch), leaf (exposed small leaf near the apex), low stem (the 2 cm of stem above the cotyledons, with axillary buds and leaves removed), and root (fine roots including tips).
The samples from which the data in Figures 5 to 7, Supplemental Figure S7, and Supplemental Data Set S2 were produced are fine roots (including tips), main root (the 2 cm below the root-hypocotyl boundary), low stem (the 2 cm above cotyledons, with axillary buds and leaves removed), the axillary buds from nodes 2 and 7, and leaf blade from leaf 7. Each sample type was represented by three pools of seven plants (except the minus-P samples from axillary buds at node 7, where there were two pools of seven). Each biological replicate for all nine treatment conditions was collected over a 2-h time period before the next biological replicate was started (replicate 1 was collected from 10 am to 12 pm, replicate 2 from 1:15 to 3:10 pm, and replicate 3 from 4:20 to 6 pm). On the day of sample collection, sunrise was at 6:01 am and sunset was at 8:42 pm. The timing of the sample collection matches that of the phenotypic characterization shown in Figure 2: 7 d after the onset of treatments.
Quantitative reverse transcription-PCR was carried out largely as described previously (Drummond et al., 2012). Four reference genes (Actin, Histone, Elongation Factor 1α, and glyceraldehyde-3-phosphate dehydrogenase [GADPH]) were tested, and the two most stable genes as determined by GeNorm were kept for the remaining analysis. The GAPDH primers are described by Kretzschmar et al. (2012). We designed and tested primers to detect the transcripts of PhTCP1 (5′-TGGTTCAAATTCCATATGCAGGT-3′ and 5′-TCCAGTCTATGATGTCTGAAGGA-3′), PhTCP2 (5′-AGCAGCAGTTATTGTAGTACCACT-3′ and 5′-TGCTGGAACTGAAACCTCGAA-3′), and PhTCP3 (5′-TGCAGTCAAGGAGCTGGAAG-3′ and 5′-TATCATTTGTGGCAGATTCGTC-3′). These primers gave sensitive detection of the target transcripts without amplification of any interfering primer dimer. For the experiments shown in Figures 4 to 7, the data come from RNA extracted from three biological replicates of seven pooled individuals, except for Figure 4, where four individuals were pooled.
Phylogenetics, Alignments, and Trees
Sequence data were retrieved from public databases (similarity to existing sequences as determined by BLAST) or generated in this laboratory. All alignments were created using the Geneious Align tool and edited manually. Trees were calculated based on protein sequence alignments using the PhyML plugin for Geneious (Guindon and Gascuel, 2003). Support for the topology was determined by bootstrap with 100 replicates, and percentage support is indicated at each node. All bioinformatic analyses were carried out in Geneious (Biomatters).
Statistical Analysis
Most statistical analyses were performed using the GenStat statistical software package (14th edition). ANOVAs were performed for statistical analyses of phenotypic and expression data. Appropriate transformations were used where necessary to ensure that model assumptions were met. Mean separation tests were performed using Tukey’s lsd test at the 5% level of significance. For any given character in an experiment, values with no common lowercase identifiers are significantly different from each other. For correlations between data, Spearman’s rank correlation test was used. PCA used a correlation matrix with four dimensions. Imputed values for missing data points were calculated in R 3.1.1 using the regularized imputePCA function in the missMDA library (Josse and Husson, 2012).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KR002102, KR002103, KR002104, and KR002105.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Additional phenotypic data for the low phosphate experiment.
Supplemental Figure S2. Additional phenotypic data for the low nitrogen experiment.
Supplemental Figure S3. Additional phenotypic data for the crowding experiment.
Supplemental Figure S4. Additional phenotypic data for the red-to-far-red experiment.
Supplemental Figure S5. Photographs of plants from the phosphate × light experiment.
Supplemental Figure S6. Phylogenetic analysis of TCP proteins.
Supplemental Figure S7. Gene expression in axillary bud 2.
Supplemental Figure S8. Spectra of light for red, white, and far-red treatment.
Supplemental Data Set S1. Nexus file of TCP protein alignment.
Supplemental Data Set S2. QPCR data for the phosphate × light experiment.
Supplementary Material
Acknowledgments
We thank Kevin Reade, Rebecca Turner, Derek White, Sakuntala Karunairetnam, Andrew Gleave, and Jo Putterill for materials, technical assistance, and helpful discussions.
Glossary
- SL
strigolactone
- N
nitrogen
- P
phosphorus
- R:FR
red-to-far-red light
- LED
light-emitting diode
- cDNA
complementary DNA
- rs
Spearman’s rank correlation coefficient
- PCA
principal component analysis
Footnotes
This work was supported by the MeriNet program, the New Zealand Foundation for Research, Science, and Technology, and AgResearch (grant no. C10X0816).
Articles can be viewed without a subscription.
References
- Abe S, Sado A, Tanaka K, Kisugi T, Asami K, Ota S, Kim HI, Yoneyama K, Xie X, Ohnishi T, et al. (2014) Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc Natl Acad Sci USA 111: 18084–18089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Arite T, Kameoka H, Kyozuka J (2012) Strigolactone positively controls crown root elongation in rice. J Plant Growth Regul 31: 165–172 [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50: 1416–1424 [DOI] [PubMed] [Google Scholar]
- Bell EM, Lin WC, Husbands AY, Yu L, Jaganatha V, Jablonska B, Mangeon A, Neff MM, Girke T, Springer PS (2012) Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc Natl Acad Sci USA 109: 21146–21151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer FD, de Saint Germain A, Pillot JP, Pouvreau JB, Chen VX, Ramos S, Stévenin A, Simier P, Delavault P, Beau JM, et al. (2012) Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: molecule design for shoot branching. Plant Physiol 159: 1524–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot JP, Boutet-Mercey S, Dalmais M, Antoniadi I, Li X, Maia-Grondard A, Le Signor C, Bouteiller N, et al. (2012) The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol 158: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. (2012) Shade avoidance. The Arabidopsis Book 10: e0157, doi/10.1199/tab.0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F, Nieminen K, Sánchez-Ferrero JC, Rodríguez ML, Chagoyen M, Hardtke CS, Cubas P (2014) Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26: 1134–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M, George G, Ongaro V, Williamson L, Willetts B, Ljung K, McCulloch H, Leyser O (2014) Auxin and strigolactone signaling are required for modulation of Arabidopsis shoot branching by nitrogen supply. Plant Physiol 166: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Gustus C (1995) teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141: 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RSM, Martínez-Sánchez NM, Janssen BJ, Templeton KR, Simons JL, Quinn BD, Karunairetnam S, Snowden KC (2009) Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiol 151: 1867–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RSM, Sheehan H, Simons JL, Martínez-Sánchez NM, Turner RM, Putterill J, Snowden KC (2011) The expression of petunia strigolactone pathway genes is altered as part of the endogenous developmental program. Front Plant Sci 2: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA (2012) Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson SA. (2007) Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol 48: 667–677 [DOI] [PubMed] [Google Scholar]
- Finlayson SA, Krishnareddy SR, Kebrom TH, Casal JJ (2010) Phytochrome regulation of branching in Arabidopsis. Plant Physiol 152: 1914–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA (2005) The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17: 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Yoneyama K, Hugill CJ, Quittenden LJ, Reid JB (2013) Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol Plant 6: 76–87 [DOI] [PubMed] [Google Scholar]
- Franklin KA. (2008) Shade avoidance. New Phytol 179: 930–944 [DOI] [PubMed] [Google Scholar]
- Gendron JM, Liu JS, Fan M, Bai MY, Wenkel S, Springer PS, Barton MK, Wang ZY (2012) Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc Natl Acad Sci USA 109: 21152–21157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- González-Grandío E, Poza-Carrión C, Sorzano CO, Cubas P (2013) BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25: 834–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JC, Koch KE, Suzuki M, Wu S, Latshaw S, Petruff T, Goulet C, Klee HJ, McCarty DR (2012) Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol 160: 1303–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun 4: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22: 2032–2036 [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Drummond RS, Snowden KC (2014) Regulation of axillary shoot development. Curr Opin Plant Biol 17: 28–35 [DOI] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C (2006) Branching genes are conserved across species: genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol 142: 1014–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse J, Husson F (2012) Handling missing values in exploratory multivariate data analysis methods. Journal de la Societe Francaise de Statistique 153: 79–99 [Google Scholar]
- Kapulnik Y, Delaux PM, Resnick N, Mayzlish-Gati E, Wininger S, Bhattacharya C, Séjalon-Delmas N, Combier JP, Bécard G, Belausov E, et al. (2011a) Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233: 209–216 [DOI] [PubMed] [Google Scholar]
- Kapulnik Y, Resnick N, Mayzlish-Gati E, Kaplan Y, Wininger S, Hershenhorn J, Koltai H (2011b) Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J Exp Bot 62: 2915–2924 [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Brutnell TP, Finlayson SA (2010) Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ 33: 48–58 [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Burson BL, Finlayson SA (2006) Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol 140: 1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska MA, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C (2011) Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol 155: 974–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483: 341–344 [DOI] [PubMed] [Google Scholar]
- Liu W, Kohlen W, Lillo A, Op den Camp R, Ivanov S, Hartog M, Limpens E, Jamil M, Smaczniak C, Kaufmann K, et al. (2011) Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23: 3853–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez JA, Charnikhova T, Gómez-Roldán V, Matusova R, Kohlen W, De Vos R, Verstappen F, Puech-Pages V, Bécard G, Mulder P, et al. (2008) Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol 178: 863–874 [DOI] [PubMed] [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants, Ed 2 Academic Press, London [Google Scholar]
- Mayzlish-Gati E, LekKala SP, Resnick N, Wininger S, Bhattacharya C, Lemcoff JH, Kapulnik Y, Koltai H (2010) Strigolactones are positive regulators of light-harvesting genes in tomato. J Exp Bot 61: 3129–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N, Umehara M, Luo L, Kobayashi K, Hanada A, Ueno K, Asami T, Yamaguchi S, et al. (2010) FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol 51: 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahashi G, Douds DD (2000) Partial separation of root exudate components and their effects upon the growth of germinated spores of AM fungi. Mycol Res 104: 1453–1464 [Google Scholar]
- Napoli CA, Beveridge CA, Snowden KC (1999) Reevaluating concepts of apical dominance and the control of axillary bud outgrowth. Curr Top Dev Biol 44: 127–169 [DOI] [PubMed] [Google Scholar]
- Napoli CA, Ruehle J (1996) New mutations affecting meristem growth and potential in Petunia hybrida Vilm. J Hered 87: 371–377 [Google Scholar]
- Naz AA, Raman S, Martinez CC, Sinha NR, Schmitz G, Theres K (2013) Trifoliate encodes an MYB transcription factor that modulates leaf and shoot architecture in tomato. Proc Natl Acad Sci USA 110: 2401–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 108: 8897–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, Davidson AM, Finnegan EJ, Mathesius U, Poot P, Purugganan MD, Richards CL, Valladares F, et al. (2010) Plant phenotypic plasticity in a changing climate. Trends Plant Sci 15: 684–692 [DOI] [PubMed] [Google Scholar]
- Rae GM, Uversky VN, David K, Wood M (2014) DRM1 and DRM2 expression regulation: potential role of splice variants in response to stress and environmental factors in Arabidopsis. Mol Genet Genomics 289: 317–332 [DOI] [PubMed] [Google Scholar]
- Rasmussen A, Mason MG, De Cuyper C, Brewer PB, Herold S, Agusti J, Geelen D, Greb T, Goormachtig S, Beeckman T, et al. (2012) Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol 158: 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SK, Finlayson SA (2014) Phytochrome B promotes branching in Arabidopsis by suppressing auxin signaling. Plant Physiol 164: 1542–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, et al. (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol 155: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JL, Napoli CA, Janssen BJ, Plummer KM, Snowden KC (2007) Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiol 143: 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Napol CA (1998) Psl: a novel Spm-like transposable element from Petunia hybrida. Plant J 14: 43–54 [DOI] [PubMed] [Google Scholar]
- Snowden K, Napoli C (2003) A quantitative study of lateral branching in petunia. Funct Plant Biol 30: 987–994 [DOI] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ (2005) The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17: 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM (1989) Patterns in Plant Development, Ed 2 Cambridge University Press, Cambridge, UK [Google Scholar]
- Stirnberg P, Furner IJ, Leyser HM (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J 50: 80–94 [DOI] [PubMed] [Google Scholar]
- Sun H, Tao J, Liu S, Huang S, Chen S, Xie X, Yoneyama K, Zhang Y, Xu G (2014) Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J Exp Bot 65: 6735–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J 33: 513–520 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Magome H, Takeda-Kamiya N, Yamaguchi S (2010) Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol 51: 1118–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295 [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP (2011) grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci USA 108: E506–E512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Wang Q, Schmitz G, Müller D, Theres K (2012) The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J 71: 61–70 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Takeuchi Y, Yokota T (2001) Production of clover broomrape seed germination stimulants by red clover root requires nitrate but is inhibited by phosphate and ammonium. Physiol Plant 112: 25–30 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kim HI, Kisugi T, Nomura T, Sekimoto H, Yokota T, Yoneyama K (2012) How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 235: 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kusumoto D, Sekimoto H, Sugimoto Y, Takeuchi Y, Yoneyama K (2007a) Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 227: 125–132 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Yoneyama K, Takeuchi Y, Sekimoto H (2007b) Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta 225: 1031–1038 [DOI] [PubMed] [Google Scholar]
- Zhang Y, van Dijk AD, Scaffidi A, Flematti GR, Hofmann M, Charnikhova T, Verstappen F, Hepworth J, van der Krol S, Leyser O, et al. (2014a) Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat Chem Biol 10: 1028–1033 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ji R, Li H, Zhao T, Liu J, Lin C, Liu B (2014b) CONSTANS-LIKE 7 (COL7) is involved in phytochrome B (phyB)-mediated light-quality regulation of auxin homeostasis. Mol Plant 7: 1429–1440 [DOI] [PubMed] [Google Scholar]
- Zhao LH, Zhou XE, Wu ZS, Yi W, Xu Y, Li S, Xu TH, Liu Y, Chen RZ, Kovach A, et al. (2013) Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res 23: 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.