Abstract
We have applied 57Fe nuclear resonance vibrational spectroscopy (NRVS) for the first time to study the dynamics of Fe centers in Fe-S protein crystals, including oxidized wild type rubredoxin crystals from Pyrococcus furiosus, and the MoFe protein of nitrogenase from Azotobacter vinelandii. Thanks to the NRVS selection rule, selectively probed vibrational modes have been observed in both oriented rubredoxin and MoFe protein crystals. The NRVS work was complemented by extended X-ray absorption fine structure spectroscopy (EXAFS) measurements on oxidized wild type rubredoxin crystals from Pyrococcus furiosus. The EXAFS spectra revealed the Fe-S bond length difference in oxidized Pf Rd protein, which is qualitatively consistent with the X-ray crystal structure.
Keywords: 57Fe, nuclear resonant scattering, nuclear resonant vibrational spectroscopy, NRVS, Mössbauer, synchrotron radiation, EXAFS, normal mode analysis, nitrogenase, rubredoxin
Introduction
Fe-S proteins serve a wide variety of essential tasks in living systems, including electron transfer within and between proteins, catalysis of chemical reactions, sensing the chemical environment, regulation of DNA expression, repair of damaged DNA, and maintenance of molecular structure [1]. The importance of understanding the structure and function of these proteins can hardly be overstated. The synchrotron based methods of EXAFS (Extended X-Ray Absorption Fine Structure) and NRVS (Nuclear Resonance Vibrational Spectroscopy) have both become popular tools for the study of such Fe-S proteins. Although most often used on nonoriented samples, both techniques have an angular dependence that can be employed to extract additional information about a sample. In the case of EXAFS, the modulation χ of the x-ray absorption for atom a due to the presence of backscattering atom b is sensitive to the angle θ between the a-b axis R⃗ab and the electric field polarization E⃗: χ ∝ 3cos2 θ. In contrast, the nuclear absorption probability S(E) (and the NRVS signal) is sensitive to the 57Fe motion e⃗α in a given normal mode α along the beam propagation direction k̂: S(E) ∝ (k̂ • e⃗α)2.
In this paper we report the first single crystal NRVS on two Fe-S proteins – oxidized wild type (WT) Pf Rd and MoFe protein from Av. We demonstrate how the combined application of EXAFS and NRVS to oriented samples can provide additional information about the dynamics of complex Fe-S sites in metalloproteins. The particular samples chosen represent two extremes of complexity. We start by illustrating single crystal spectroscopy of rubredoxin, the simplest known Fe-S protein, with a single approximately tetrahedral FeS4 center (Figure 1). We then illustrate work with nitrogenase, which contains the most complex metal sulfur cluster in biology – the MoFe7S9X FeMo cofactor (Figure 1).
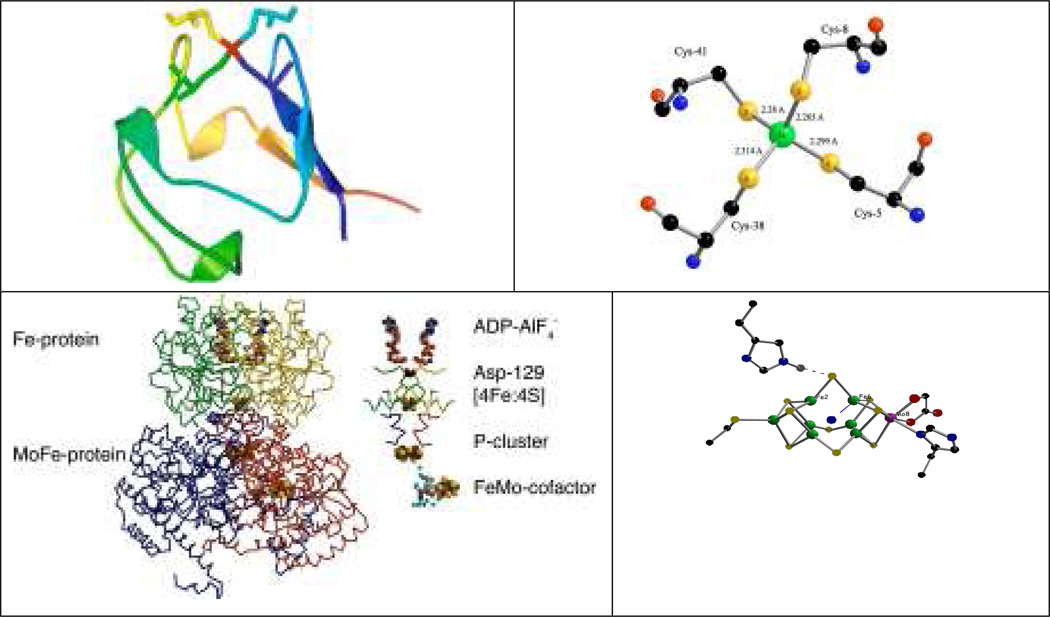
Figure 1.
Top left: a cartoon of oxidized Pf Rd, including sticks for cysteine residues, illustrating exposed location of Fe site (red). Top right: close-up of Fe site showing slight compression of Fe-SCys8 and Fe-SCys41 bonds (PDB Code 1BRF) [2]. Bottom left: N2ase MoFe protein. Bottom right: close-up of FeMo cofactor.
NRVS has rapidly become a popular technique for probing the dynamics of Fe sites in metalloproteins [3–12] as well as Fe containing inorganic complexes [13–24]. NRVS only probes the vibrational modes in which 57Fe nuclei move along the incident beam direction [21,25,26]. This unique selection rule makes NRVS free of any interference from solvent and other protein vibrations (which often limit FT-IR spectroscopy) and independent of particular oxidation states (which often restricts resonance Raman spectroscopy) [5,7]. For bioinorganic applications, NRVS has been used to probe the dynamics of Fe sites in heme proteins [9–12], Fe-S proteins, including rubredoxins [5], [2Fe-2S] and [4Fe-4S] ferredoxins [7], hydrogenase [6], and nitrogenase [3]. Previous experiments with orienting crystal samples have been performed on several heme protein related model compound crystals [20–22] to identify in-plane and out-of-plane vibrations with respect to the porphyrin plane.
The details of NRVS theory have been explained in [21,25,26], the bottom line for the current study is that a NRVS transition for normal mode α contributes a fraction ϕα to the normalized excitation probability S(ν̅) that is directly proportional to the Fe mode composition factor and inversely proportional to ν̅α [21,27]:
| (1) |
In the above equation, ν̅α is the difference between the photon energy and the recoil-free nuclear resonance energy in wave numbers, ν̅R is the recoil energy (~16 cm−1), n̅α =[exp(hcν̅α/kBT) − 1]−1 is the thermal occupation factor for a mode of frequency ν̅α at temperature T [21], and the recoilless fraction f depends on <xFe2>, the mean square fluctuation of the Fe nucleus along the beam direction, via f = exp(−k2<xFe2>). The data are often presented as the 57Fe-centered partial vibrational density of states (PVDOS), DFe (ν̅), using a lineshape function L (ν̅ − ν̅α) [18,21]:
| (2) |
The 57Fe PVDOS for measurements on a perfectly oriented sample takes the form of a series of bands with areas (k̂ • e⃗Fe,α)2 equal to the squared projection of along the beam direction k̂.
Rubredoxins are small (~50 amino acids) electron-transfer proteins that contain a single Fe(S-cys)4 redox center [28]. The high-resolution X-ray crystal structure for oxidized Pyrococcus furiosus rubredoxin (Pf Rd) at 0.95 Å resolution (1BRF) [2] along with several other Rd crystal structures from different bacteria [34,35] reveals a roughly tetrahedral FeS4 site, often described as approaching D2d symmetry via a compression along a S4 axis (Figure 2). In a previous study, we examined Pf Rd still containing the unprocessed N-terminal methionine residue (Pf met-Rd) in frozen solution sample via NRVS. We observed three broad bands of intensity, near 70, 150, and 360 cm−1 [5].
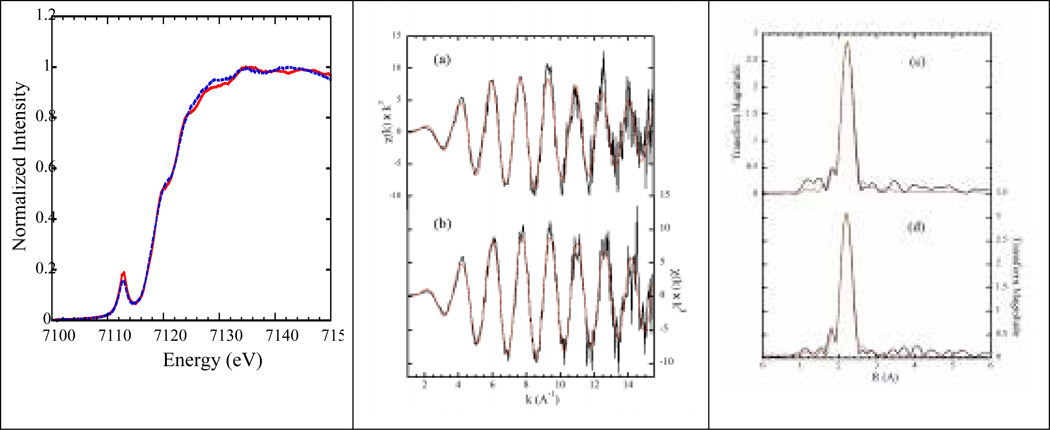
Figure 2.
Single crystal XANES and EXAFS. Left: Fe K-edge of the Pf Rd crystals with E⃗ ‖ a⃗ 2.26 (blue dotted line) and E⃗ ‖ c⃗ 2.28 (red solid line) Middle: k3-weighted EXAFS of the oxidized Pf Rd with E⃗ ‖ a⃗ vs. E⃗ ‖ c⃗. Right: EXAFS Fourier transforms for E⃗ ‖ a⃗ vs. E⃗ ‖ c⃗.
Nitrogenase (N2ase) is an important enzyme that catalyzes the reduction of dinitrogen (N2) to ammonia (NH3) and is ultimately responsible for about half of the food produced globally each year. The Mo version of N2ase from Azotobacter vinelandii consists of two components, the Fe protein (Av2) and the MoFe protein (Av1). Av2 contains one [Fe4S4] cluster, and (coupled with the hydrolysis of MgATP) transfers electrons to Av1. Av1 is an α2β2 tetramer, and it contains two [Fe7MoS9X] FeMo cofactors and two [Fe8S7] P clusters. The FeMo cofactor is the active center of N2ase, and it is the most complicated Fe-S cluster found in biology. In our previous NRVS study [3] of solution has observed a unique spectral feature at ~190 cm−1, which we assigned to FeMo cofactor cluster breathing modes.
Results and Discussion
XANES and EXAFS on oxidized Pf Rd Crystals
We begin by presenting single crystal EXAFS measurements on the same 57Fe-enriched Pf Rd crystals as used for the NRVS work, because the results complement and help interpret the latter experiment. The high-resolution x-ray crystal structure for oxidized Pf Rd at 0.95 Å resolution (1BRF) shows that there are 4 Rd molecules in each crystal unit cell [2]. The Fe-S bonds connected to Cys5 and Cys38 lie close to the bc plane of the crystal with a larger projection onto the c-axis, while the Fe-S bonds involving Cys8 and Cys41 are closer to the ab plane with a larger projection onto the a-axis. Thus, by aligning the x-ray polarization direction along the a-axis (E⃗ ‖ a⃗), the x-ray absorption will mainly probe along the Fe-S bonds for Fe-SCys8 and Fe-SCys41, while with the x-ray polarization along the c-axis (E⃗ ‖ c⃗), the XANES and EXAFS will be more selective for the Fe-S bonds associated with Fe-SCys5 and Fe-SCys38.
The Fe K-edge spectra for Rd crystals with E⃗ ‖ a⃗ and E⃗ ‖ c⃗ are shown in Figure 2. Both spectra have similar features with a strong 1s→3d peak at ~7112.8 eV and a shoulder at ~7119.8 eV, consistent with previous studies on Rd solution samples24,25. The strength of the pre-edge features in both spectra arises from mixing of d and p orbitals in the roughly tetrahedral Fe site geometry. The slight intensity difference of the pre-edge features in E⃗ ‖ a⃗ and E⃗ ‖ c⃗ spectra may come from a slightly different degree of mixing for different orientations.
Figure 2 also presents the EXAFS spectra and Fourier transform for crystals with E⃗ ‖ a⃗ and E⃗ ‖ c⃗ together with representative fits. The fitting parameters are listed in Table 8.2. The Fourier transforms of the EXAFS spectra reveal a single Fe-S peak at ~2.25–2.30 Å from the Fe-SCys interactions. For the E⃗ ‖ a⃗ spectrum, a fit to the k-space EXAFS gave an Fe-S distance of 2.263 Å, while the E⃗ ‖ c⃗ spectrum fit gave a distance of 2.284 Å. The EXAFS results show about the same bond length differential as the x-ray crystal structure, but with slightly shorter average distances. For example, the crystal structure shows that the two shorter (2.273 average) Fe-S bonds (Fe-SCys8, 2.265 Å and Fe-SCys41 2.280 Å) have larger projections onto the a-axis, while the two longer (2.307 average) Fe-S bonds (Fe-SCys5, 2. 299 Å and Fe-SCys38, 2.314 Å) have larger projection onto the c-axis. The slightly longer average crystallographic Fe-S distance (2.29 Å) may in fact be the result of photoreduction in the diffraction measurement.
Single Crystal NRVS on oxidized Pf Rd
NRVS spectra for oxidized Pf Rd single crystals are shown in Figure 3 together with the oxidized Rd in solution [5] from the recombinant rubredoxin containing the unprocessed N-terminal methionine residue (Pf met-Rd) [36]. Overall, the crystal spectra exhibit the same three main regions of intensity, near 70, 140, and 360 cm−1, as the solution NRVS. However, the crystal spectra intensities differ from the solution spectrum, especially near 360 cm−1. This is because the single crystal experiment is selecting particular modes from the overall envelope. In the crystal NRVS spectra, the a-axis and c-axis were aligned along the x-ray beam direction, so that the normal modes with a component of 57Fe motion along the a-axis or c-axis are enhanced.
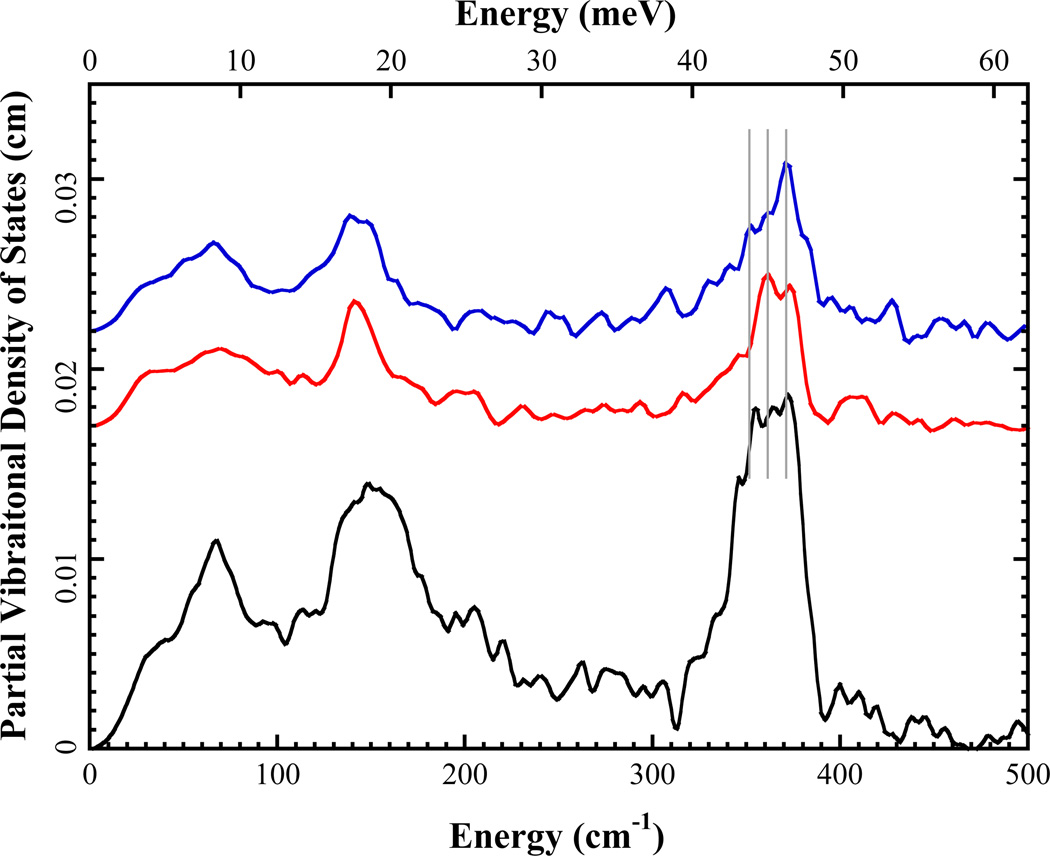
Figure 3.
57Fe PVDOS for top: oxidized Pf Rd with k̂ ‖ c⃗ ( ) or k̂ ‖ a⃗ (
) or k̂ ‖ a⃗ ( ) and bottom: oxidized Pf met-Rd in solution form (6). Alex: normalize all to integrate to 3 for comparison. Overlay aaxis and c-axis data.
) and bottom: oxidized Pf met-Rd in solution form (6). Alex: normalize all to integrate to 3 for comparison. Overlay aaxis and c-axis data.
In previous NRVS studies on oxidized Pf met-Rd, the 360 cm−1 region, which had significant intensity in an envelope between 345 and 375 cm−1, was assigned to asymmetric Fe-S stretching modes. In this region, clear intensity differences can be seen for spectra from different crystal orientations. The k̂ ‖ a⃗ PVDOS shows one strong peak at 371 cm−1 with two lower energy shoulders at ~360 cm−1 and ~350 cm−1, while in the k̂ ‖ c⃗ spectrum, the 360 cm−1 shoulder in becomes the strongest peak in this region, the intensity of 371 (373?) cm−1 peak decreases and 350 cm−1 shoulder disappears. These three features are consistent with the three asymmetric Fe-S stretching modes assigned at 375, 358 and 350 cm−1 in the previous study [5]. The intensity differences of the same peaks in different crystal orientations indicate that the 57Fe motions in these vibrational modes have preferred orientations.
In the bending region (100~200 cm−1), the k̂ ‖ a⃗ and k̂ ‖ c⃗ NRVS spectra show similar features with a strong peak at ~140 cm−1; in the k̂ ‖ a⃗ orientation this peak is slightly split. Both orientations lose intensity in the region between 155 cm−1 and 220 cm−1 compared with the solution Rd spectrum. This indicates that 57Fe motion in vibrational modes of this region have less projection onto the a- and c-axes. However, detailed analysis of this bending region and the region below 100 cm−1 of the crystal spectra is beyond the scope of this work.
For a more quantitative interpretation, we built a simple Fe(SCC)4 model with no symmetry constraints, using the Fe site geometry from the oxidized WT Pf Rd crystal structure. We used force constants from a previous Fe(SCC)4 simulation of solution Pf met-Rd NRVS as a starting point [5]. The three spectra, oxidized solution Pf met-Rd NRVS, k̂ ‖ a⃗ and k̂ ‖ c⃗ WT Pf Rd crystal NRVS, were simultaneously modeled by least squares fitting. (We assume that the oxidized WT Pf Rd NRVS in solution form should be very similar to the oxidized Pf met-Rd NRVS in solution form, at least in the Fe-S stretch region, because of their almost identical Fe site geometry). Since the NRVS intensity below 200 cm−1 involves contributions from cysteine side chain vibrations and motion of the protein backbone in Rd NRVS spectra [5] our Fe(SCC)4 model is too small to capture those features. So, here we focus on the Fe-S stretch region by fixing bending force constants to those for the previous oxidized Pf met-Rd NRVS simulation.
Figure 4 shows the simulations for the crystal spectra. The k̂ ‖ a⃗ simulation shows two Fe-S stretching modes at 357 and 373 cm−1, and the relative intensities are consistent with the experimental data. The 352 cm−1 mode simulated in the solution spectrum has almost disappeared, indicating that the Fe motion in this mode is almost perpendicular to the crystal a axis. In the k̂ ‖ c⃗ simulation, the relative intensities of the modes at 357 and 373 cm−1 are inverted, also consistent with the experimental data. The bottom line is satisfying if not surprising: the highest frequency mode (373 cm−1) has the most intensity in the direction with the shorter (2.263 Å) average Fe-S bond length, while the lower frequency modes are stronger along the longer (2.284 Å) average Fe-S bonds.
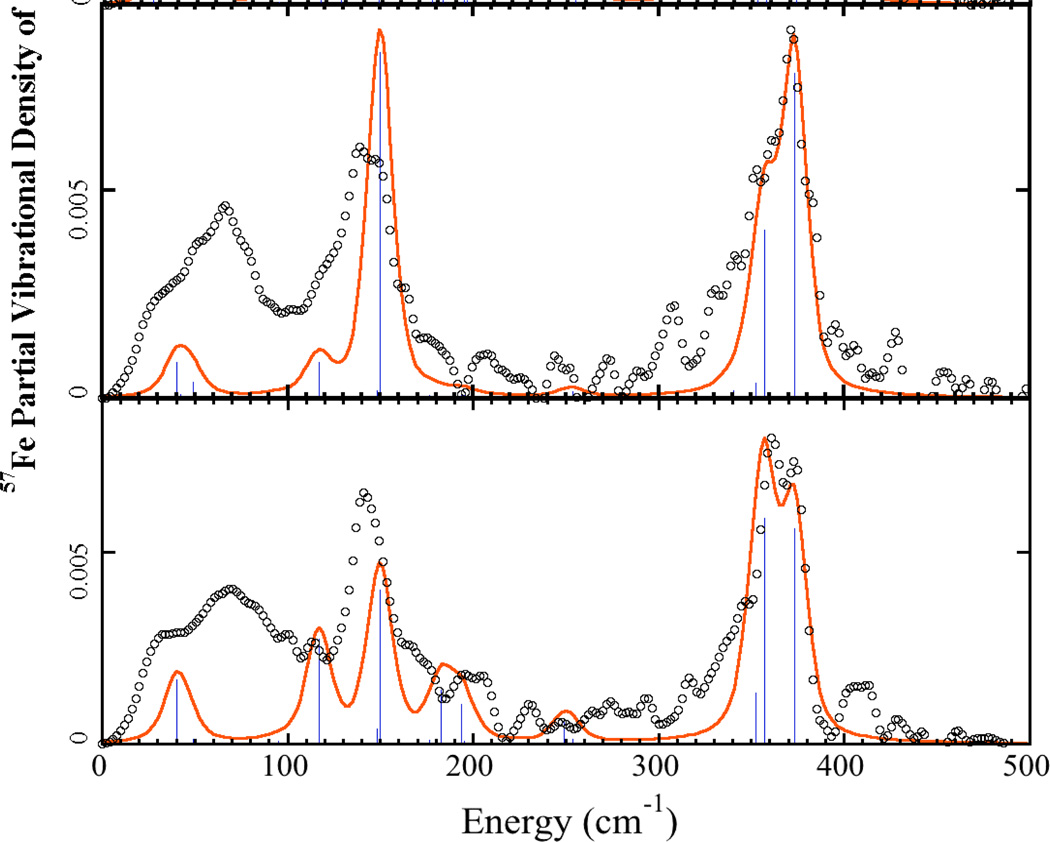
Figure 4.
57Fe PVDOS of Pf Rd crystal data (° ° °) and simulations using Fe(SCC)4 model from 1BRF crystal structure. Sticks represent intensities of individual normal modes. Top: k̂ ‖ a⃗. Bottom: k̂ ‖ c⃗. Scale reduction factors of 1.48 for k̂ ‖ a⃗ and 1.38 for k̂ ‖ c⃗ were used.
Clearly there are additional features in the experimental spectra that are not captured in the bare-bones simulations. Work is being pursued with more complex Fe(S-cysX5)4 models, in which the remaining cysteine atoms are added, as well as the carbonyl C and O of the residue adjacent to the cysteine N, and the amide N of the residue bound to the cysteine carbonyl group
NRVS on Nitrogenase MoFe protein Crystals
Figure 5 shows the NRVS for a MoFe protein crystal with the c-axis aligned along the incident x-ray direction (referred to as k̂ ‖ c⃗ from now on), and the NRVS obtained for the same crystal by turning the sample cell 90° (referred to as k̂ ⊥ c⃗). The sample we used was actually a multi-crystal containing more than one single crystal, the crystal axes of these sub-crystals were roughly aligned with each other. The crystal size was ~0.5–0.7 mm3. Although the statistics and alignment are limited for these preliminary data, we nevertheless observe clear intensity differences for features between 50 and 250 cm−1. (Because of the limited statistics of the current spectra, we do not discuss the region above 250 cm−1.)
Figure 5.
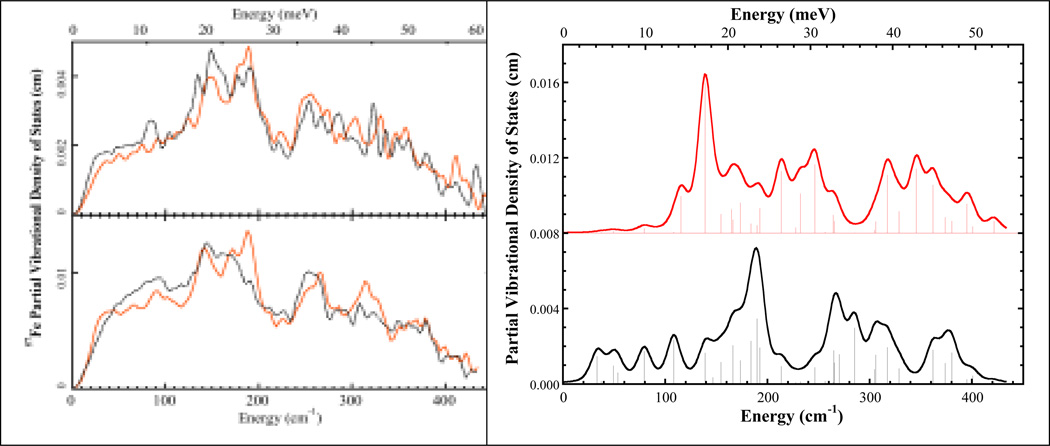
Left. Top: 57Fe PVDOS for MoFe protein crystal with k̂ ‖ c⃗ (---) and k̂ ⊥ c⃗ ( ). Bottom: MoFe protein in solution (
). Bottom: MoFe protein in solution ( ) and P-cluster in ΔnifE MoFe protein (---). Right. Calculated 57Fe PVDOS of FeMo cofactor assuming the C3 axis is parallel to the incident x-ray direction (top) and perpendicular to the incident x-ray direction (bottom). Sticks represent intensities of individual normal modes.
) and P-cluster in ΔnifE MoFe protein (---). Right. Calculated 57Fe PVDOS of FeMo cofactor assuming the C3 axis is parallel to the incident x-ray direction (top) and perpendicular to the incident x-ray direction (bottom). Sticks represent intensities of individual normal modes.
In the k̂ ⊥ c⃗ NRVS, there is an overall maximum at 188 cm−1, a shoulder at 177 cm−1, and a lesser peak at 148 cm−1. In contrast, the k̂ ‖ c⃗ NRVS shows a decrease in the 188 cm−1 peak and increased intensity for the 148 cm−1 peak. The feature at 177 cm−1 also becomes clearly resolved. Another clear difference is at ~84 cm−1, where a distinct peak is seen in the k̂ ‖ c⃗ spectrum, while in the k̂ ⊥ c⃗ spectrum, this feature disappears. Finally, both spectra have a higher energy shoulder observed at 206 cm−1 and another apparent band at ~225 cm−1. Overall, the changes in band intensities prove that the 57Fe nuclear motions in the vibrational modes of this region have preferred directions.
In our previous NRVS study [3], the NRVS spectra of both wild type MoFe protein (Av1) and a ΔnifE mutant protein (ΔnifE:Av1) that only contains P-clusters were reported (Figure 5). The unique feature in the Av1 spectrum compared with the ΔnifE:Av1 spectrum was the peak at ~190 cm−1 with a lower energy shoulder at ~175 cm−1. This feature therefore must belong to a vibrational mode of the FeMo cofactor in MoFe protein.
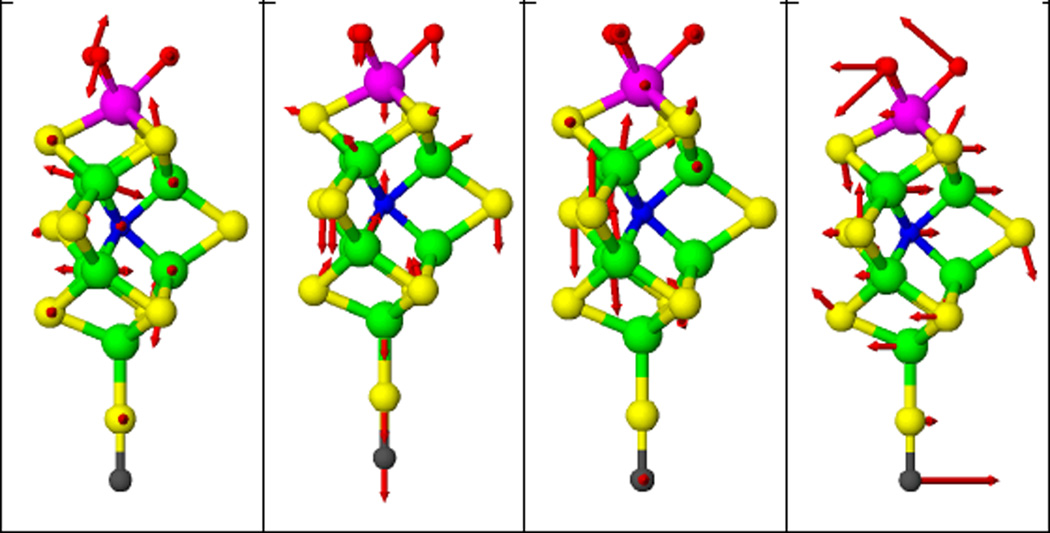
Previously, a C3v symmetry [O3MoFe7NS9(SC)]n− model was used to successfully simulate the FeMo cofactor spectrum [3]. Five normal modes, including 2 A1 modes and 3 E modes, were assigned to the region between 160 and 200 cm−1, and a strong E mode was assigned to the peak at 141 cm−1. Inspection of the five normal modes between 160 and 200 cm−1 shows that the directions of the Fe motions in these modes are closer to the plane perpendicular to the C3 symmetry axis (the line formed by the capping Mo and Fe atoms) than along this axis, while for the E mode at 141 cm−1, the Fe motions are more along the C3 symmetry axis (Figure 6). Figure 5 shows the predicted NRVS spectra of assuming molecular C3 axis parallel to the incident X-ray and perpendicular to the incident X-ray.
Figure 6.
Illustration of molecular motion in FeMo cofactor normal modes, derived from the normal mode calculation using a C3v symmetry [O3MoFe7NS9(SC)]n− FeMo cofactor model (2). Color code: Fe (green), S (yellow), Mo (magenta), O (red), C (black). Left to right: at 188, 177, 141, and 85 cm−1 Figures were made with ATOMS.
In our crystal MoFe protein NRVS experiments, due to the experimental setup limitation, the two crystal orientations we selected actually were k̂ ‖ c⃗ and k̂ ⊥ c⃗, instead of exactly k̂ ‖ b⃗ and k̂ ‖ c⃗. However, since we did our experiment on the same crystal in two different orientations, the spectrum differences we observed should come from the orientation dependence of the vibrational modes. Our experimental data are in line with the predicted spectra shown in Figure 5. In the case of k̂ ‖ c⃗ NRVS, all the C3 axes of FeMo-co clusters are more aligned with the incident x-ray, and a decrease of intensity at ~190 cm−1 and the increase at ~148 cm−1 were observed. This behavior is consistent with the predicted spectrum shown in Figure 5 when C3 axis is aligned with the x-ray direction. Despite their limitations, the current spectra have already shown that single crystal NRVS is a promising technique to selectively probe vibrations from Fe sites even for complicated systems such as N2ase.
Summary
In this work, we have investigated the combined application of single crystal EXAFS and single crystal NRVS on biological Fe-S centers. In the case of rubredoxin, the single crystal EXAFS spectra revealed an Fe-S bond length difference of ~0.02 Å that cannot be resolved in solution data. In the Pf Rd crystal NRVS spectra, a 375 cm−1 mode had more intensity in the k̂ ‖ a⃗ spectrum, corresponding to the shorter bonds seen by EXAFS.
In the MoFe protein crystal NRVS experiment, a multi-crystal MoFe protein was used. The crystal contained more than one single crystal with these sub crystals roughly aligned with each other. In a k̂ ‖ c⃗ vs. k̂ ⊥ c⃗ spectral comparison, significant intensity differences can be seen. Using the simulation result from our previous MoFe protein NRVS studies on the frozen solution form, we can explain these differences based on the different orientations of the Fe motions in the normal modes located at ~150 and ~190 cm−1. Further experiments with additional orientations and using 57Fe enriched FeMo cofactor MoFe protein single crystals would allow for more detailed analyses.
In summary, single crystal NRVS and EXAFS provide a powerful way to selectively probe the Fe centers of metalloproteins. The information obtained from experiments on crystals can help the interpretation of the corresponding frozen solution sample spectra in complicated protein systems. As synchrotron brightness continues to increase, the size of the requisite protein crystals will continue to diminish and the quality of the data will continue to improve.
Experimental Section
Rubredoxin Crystal Sample Preparation
57Fe enriched WT Pf Rd was prepared at the University of Georgia. The experimental (biological) procedures of obtaining and purifying the sample have been described [36]. In this section, we describe the method we used to obtain the large crystals necessary for the NRVS experiment. “X-ray sized” crystals of 57Fe WT Pf Rd were first grown at room temperature using hanging drop method [39] by equilibrating a 4 µl drop against 1ml of deuterated 3.8 M NaK phosphate solution (equimolar of NaD2PO4 and H2DPO4) as precipitating agent. The drop contains 2 µl of 40 mg/ml protein solution and 2 µl of the NaK phosphate solution. The small crystals, which appeared in about 3 days, were first collected and crushed, then used as a concentrated seed stock [39]. The concentrated seed stock was then diluted repeatedly with 3.6 M NaKPO4 solution to reduce the concentration. The concentration was reduced 10-fold in each step: 10µl of the seed stock solution was mixed with 90µl of the 3.6 M NaKPO4 solution and the step was repeated 3 times so that there was a minimal amount of crystal seeds in each drop of the solution. The seed thus serves as a template on which further molecules can assemble, and with time the seed can grow into a large crystal. In order to obtain large, 1mm3-sized crystals, the sitting drop method [39] was used by equilibrating 30 or 40 µl of a mixture of protein and crystallization agent (3:1 and 4:1 ratio) against 1ml of 3.8 NaK phosphate solution. 0.2 µl of the seed stock of the rubredoxin solution was added to seed the sample. If more than one crystal is formed within the sitting drop, a small amount of D2O is used to dissolve most of the crystals, allowing only one crystal per drop to grow. The crystal size used in our experiments is ~0.5–1 mm3, however, using the method described above, crystals can be grown up to ~4 mm3 in the period of 3 months. The crystals were stabilized during growth by increasing the concentration of the reservoir solution from 3.8 M to 4.0 M.
MoFe protein sample preparation
The MoFe protein was concentrated to ~60mg/ml as determined by Bradford assay. MoFe protein crystals were grown under nitrogen atmosphere in a Braun chamber by microcapillary batch diffusion [40]. The precipitating solutions consisted of 30% PEG 4000, 100mM Tris (pH 8.0), and 150–200 mM Na2MoO4. The MoFe crystals were observed to have a dark brown color and grow to the desired size for data collection within 1–2 months. Crystals were harvested on rayon loops and flash cooled in liquid nitrogen.
Crystal Orientation
Oxidized Pf Rd crystals were oriented at Beamline 4.2.2 at the Advanced Light Source (ALS). Crystals were first transferred onto plastic cryo-loops from mother liquid, flash frozen in liquid nitrogen, then placed onto an x-ray diffractometer to find crystal axes. For NRVS samples, the crystal a-axis or c-axis was aligned along incident x-ray direction; for EXAFS samples, the crystal a-axis or c-axis were aligned parallel to the x-ray polarization direction. Aligned crystals were transferred onto custom made Lucite disc cells in liquid nitrogen. The crystals used ranged from 0.5 to 1 mm3, flash freezing crystals of this size did not noticeably crack the crystals, and good quality diffraction pattern can still be obtained. Figure S3 shows the oriented crystal placed in the custom made Lucite NRVS disc cells and the diffraction pattern obtained from this crystal.
Nuclear Resonance Vibrational Spectroscopy
57Fe NRVS data were recorded using published procedures [26] on multiple occasions at Beamline 3-ID at the Advanced Photon Source (APS) [41] and Beamline 9-XU at SPring-8, Japan [42]. Beamline 3-ID provided ~2.5 × 109 photons/sec in 1 meV bandwidth at 14.4125 keV in a 1 mm (vertical) × 3 mm (horizontal) spot, using a water-cooled diamond (1,1,1) double crystal monochromator with 1.1 eV bandpass, followed by separate Si(4,0,0) and Si(10,6,4) channel-cut crystals in a symmetric geometry. The flux at SPring-8 was ~3 × 109 in a 1.1 meV bandwidth, using a LN2-cooled Si(1,1,1) double crystal monochromator followed by asymmetrically cut Ge(4,2,2) and two Si(9,7,5) crystals. During these measurements, crystal samples were maintained at low temperatures using liquid He cryostats. Temperatures were calculated using the ratio of anti-Stokes to Stokes intensity according to: S(−E) = S(E)exp(−E/kT). Spectra were recorded between −20 meV and 80 meV in 0.25 meV step at APS and 0.3meV step at SPring-8. Delayed nuclear fluorescence and Fe K fluorescence were recorded with a single 1 cm2 square avalanche photodiode (APD) at the APS and with an APD array at Spring-8. Each scan took about 40 min, and all scans were added and normalized to the intensity of the incident beam.
X-ray Absorption Spectroscopy
Fe K-edge x-ray absorption data were measured at Stanford Synchrotron Radiation Laboratory beamline 9-3, with a Si 220 double-crystal monochromator and two Rh-coated mirrors: one flat premonochromator mirror for harmonic rejection and vertical collimation and one toroidal postmonochromator mirror for focusing. Fluorescent x-rays were measured using a 30-element Ge fluorescence detector (Canberra Industries), fitted with Soller slits to minimize the relative contribution of scattered radiation. An Oxford Instruments CF1208 liquid He cooled sample cryostat was used to maintain the sample temperature at 9K. The x-ray energy was calibrated using the first inflection point of a standard Fe foil set as 7112 eV. This was measured at the same time as the sample spectrum using two ion chambers positions downstream of the cryostat. To minimize x-ray photoreduction, the sample was moved after each scan so that the beam irradiated a different spot on the sample for each scan – typically two spots were available for each crystal and 3~5 scans were collected requiring reuse of each spot. In addition to that, Al foils were used in front of the incident beam ion chamber to reduce the flux of the incident x-ray beam. The Fe K-edge structure and position were monitored to ensure that no significant photochemistry had occurred.
Normal Mode Calculations
The normal mode calculations were carried out on structural models derived from crystallographic coordinates. A modification of program “VIBRATZ” was used to calculate the normal modes and NRVS spectra [43,44], using Wilson’s GF matrix method and a Urey-Bradley force field. A QR algorithm was used for finding eigenvectors [45]. The NRVS spectra obtained from different orientations were optimized simultaneously.
EXAFS Data Analysis
EXAFS data were analyzed using the EXAFSPAK software suite [46]. As the samples contained no glassing agent, it was first necessary to rigorously screen the data from the individual detector elements to eliminate any diffraction artifacts. Curve fitting used the EXAFSPAK program OPT, with single-scattering phase and amplitude functions calculated using FEFF 7.0 [31].
Supplementary Material
Acknowledgement
This work was funded by NIH GM-65440 (SPC), EB-001962 (SPC), and the DOE Office of Biological and Environmental Research (SPC). Use of the APS is supported by the DOE Office of Basic Energy Sciences. SPring-8 is funded by Japan Synchrotron Radiation Institute (JASRI).
References
- 1.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Ann. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 2.Bau R, Rees DC, Kurtz DM, Scott RA, Huang HS, Adams MWW, Eidsness MK. Crystal structure of rubredoxin from Pyrococcus furiosus at 0.95 Å resolution, and the structures of N-terminal methionine and formylmethionine variants of Pf Rd. Contributions of N-terminal interactions to thermostability. J. Biol. Inorg. Chem. 1998;3:484–493. [Google Scholar]
- 3.Xiao Y, Fisher K, Smith MC, Newton W, Case DA, George SJ, Wang H, Sturhahn W, Alp EE, Zhao J, Yoda Y, Cramer SP. How Nitrogenase Shakes - Initial Information about P-Cluster and FeMo-Cofactor Normal Modes from Nuclear Resonance Vibrational Spectroscopy (NRVS) J. Am. Chem. Soc. 2006;128:7608–7612. doi: 10.1021/ja0603655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann U, Sturhahn W, Linn DE, Jenney FE, Jr, Adams MWW, Rupnik K, Hales BJ, Alp EE, Mayse A, Cramer SP. Observation of Fe-H/D Modes by Nuclear Resonant Vibrational Spectroscopy. J. Am. Chem. Soc. 2003;125:4016–4017. doi: 10.1021/ja028767+. [DOI] [PubMed] [Google Scholar]
- 5.Xiao Y, Wang H, George SJ, Smith MC, Adams MWW, Francis E, Jenney J, Sturhahn W, Alp EE, Zhao J, Yoda Y, Dey A, Solomon EI, Cramer SP. Normal Mode Analysis of Pyrococcus furiosus Rubredoxin via Nuclear Resonant Vibrational Spectroscopy (NRVS) and Resonance Raman Spectroscopy. J. Am. Chem. Soc. 2005;127:14596–14606. doi: 10.1021/ja042960h. [DOI] [PubMed] [Google Scholar]
- 6.Guo Y, Wang H, Xiao Y, Vogt S, Thauer RK, Shima S, Volkers PI, Rauchfuss TB, Pelmenschikov V, Case DA, Alp E, Sturhahn W, Yoda Y, Cramer SP. Characterization of the Fe Site in Iron–Sulfur Cluster-Free Hydrogenase (Hmd) and of a Model Compound via Nuclear Resonant Vibrational Spectroscopy (NRVS) Inorg. Chem. 2008;47:3969–3977. doi: 10.1021/ic701251j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Y, Tan M-L, Ichiye T, Wang H, Guo Y, Smith MC, Meyer J, Sturhahn W, Alp EE, Zhao J, Yoda Y, Cramer SP. Dynamics of Rhodobacter capsulatus [2Fe-2S] Ferredoxin VI and Aquifex aeolicus Ferredoxin 5 via Nuclear Resonance Vibrational Spectroscopy (NRVS) and Resonance Raman Spectroscopy. Biochemistry. 2008;47:6612–6627. doi: 10.1021/bi701433m. [DOI] [PubMed] [Google Scholar]
- 8.George SJ, Igarashi RY, Xiao Y, Hernandez JA, Demuez M, Zhao D, Yoda Y, Ludden PW, Rubio LM, Cramer SP. EXAFS and NRVS Reveal that NifB-co, a FeMo-co Precursor, Comprises a 6Fe Core with an Interstitial Light Atom. J. Am. Chem. Soc. 2008;130:5673–5680. doi: 10.1021/ja0755358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sage JT, Durbin SM, Sturhahn W, Wharton DC, Champion PM, Hession P, Sutter J, Alp EE. Long-Range Reactive Dynamics in Myoglobin. Phys. Rev. Lett. 2001;86:4966–4969. doi: 10.1103/PhysRevLett.86.4966. [DOI] [PubMed] [Google Scholar]
- 10.Zeng WQ, Silvernail NJ, Wharton DC, Georgiev GY, Leu BM, Scheidt WR, Zhao JY, Sturhahn W, Alp EE, Sage JT. Direct probe of iron vibrations elucidates NO activation of heme proteins. 2005;127:11200–11201. doi: 10.1021/ja051052x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng W, Barabanschikov A, Zhang Y, Zhao J, Sturhahn W, Alp EE, Sage JT. Synchrotron-derived vibrational data confirm unprotonated oxo ligand in myoglobin compound II. 2008;130:1816-+. doi: 10.1021/ja077823+. [DOI] [PubMed] [Google Scholar]
- 12.Leu BM, Ching TH, Zhao JY, Sturhahn W, Alp EE, Sage JT. Vibrational Dynamics of Iron in Cytochrome c. 2009;113:2193–2200. doi: 10.1021/jp806574t. [DOI] [PubMed] [Google Scholar]
- 13.Krugh TR, Wittlin FN, Cramer SP. Ethidium Bromide - Dinucleotide Complexes, Evidence for Intercalation and Sequence Preferences in Binding to Double-Stranded Nucleic Acids. Biopolymers. 1975;14:197–210. doi: 10.1002/bip.1975.360140114. [DOI] [PubMed] [Google Scholar]
- 14.Smith MC, Xiao Y, Wang H, George SJ, Coucovanis D, Koutmos M, Sturhahn W, Alp EE, Zhao J, Cramer SP. Normal Mode Analysis of [FeCl4]− and [Fe2S2Cl4]2− via Vibrational Mössbauer, Resonance Raman, and FT-IR Spectroscopy. Inorg. Chem. 2005;44:5562–5570. doi: 10.1021/ic0482584. [DOI] [PubMed] [Google Scholar]
- 15.Xiao Y, Koutmos M, Case DA, Coucouvanis D, Wang H, Cramer SP. Dynamics of an [Fe4S4(SPh)4]2− Cluster Explored via IR, Raman, and Nuclear Resonance Vibrational Spectroscopy (NRVS) – Analysis using 36S Substitution, DFT Calculations, and Empirical Force Fields. Dalton Trans. 2006:2192–2201. doi: 10.1039/b513331a. [DOI] [PubMed] [Google Scholar]
- 16.George SJ, Igarashi RY, Xiao Y, Hernandez JA, Demuez M, Zhao D, Yoda Y, Ludden PW, Rubio LM, Cramer SP. Extended X-ray absorption fine structure and nuclear resonance vibrational Spectroscopy reveal that NifB-co, a FeMo-co precursor, comprises a 6Fe core with an interstitial light atom. 2008;130:5673–5680. doi: 10.1021/ja0755358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell CB, Wong SD, Xiao YM, Klinker EJ, Tenderholt AL, Smith MC, Rohde JU, Que L, Cramer SP, Solomon EI. A Combined NRVS and DFT Study of Fe-IV=O Model Complexes: A Diagnostic Method for the Elucidation of Non-Heme Iron Enzyme Intermediates. 2008;47:9071–9074. doi: 10.1002/anie.200803740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sage JT, Paxson C, Wyllie GRA, Sturhahn W, Durbin SM, Champion PM, Alp EE, Scheidt WR. Nuclear resonance vibrational spectroscopy of a protein active-site mimic. J. Phys.: Condens. Matter. 2001;13:7707–7722. [Google Scholar]
- 19.Rai BK, Durbin SM, Prohofsky EW, Sage JT, Ellison MK, Scheidt WR, Sturhahn W, Alp EE. Iron normal mode dynamics in a porphyrin-imidazole model for deoxyheme proteins. 2002;66 doi: 10.1103/PhysRevE.66.051904. [DOI] [PubMed] [Google Scholar]
- 20.Rai BK, Durbin SM, Prohofsky EW, Sage JT, Ellison MK, Roth A, Scheidt WR, Sturhahn W, Alp EE. Direct determination of the complete set of iron normal modes in a porphyrin-imidazole model for carbonmonoxy-heme proteins: [Fe(TPP)(CO)(1-Melm)] 2003;125:6927–6936. doi: 10.1021/ja028219w. [DOI] [PubMed] [Google Scholar]
- 21.Leu BM, Zgierski MZ, Wyllie GRA, Scheidt WR, Sturhahn W, Alp EE, Durbin SM, Sage JT. Quantitative Vibrational Dynamics of Iron in Nitrosyl Porphyrins. J. Am. Chem. Soc. 2004;126:4211–4227. doi: 10.1021/ja038526h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leu BM, Silvernail NJ, Zgierski MZ, Wyllie GRA, Ellison MK, Scheidt WR, Zhao JY, Sturhahn W, Alp EE, Sage JT. Quantitative vibrational dynamics of iron in carbonyl porphyrins. 2007;92:3764–3783. doi: 10.1529/biophysj.106.093773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulsen H, Schunernann V, Trautwein AX, Winkler H. Mossbauer studies of coordination compounds using synchrotron radiation. 2005;249:255–272. [Google Scholar]
- 24.Winkler H, Chumakov AI, Trautwein AX. Nuclear resonant forward and nuclear inelastic scattering using synchrotron radiation for spin crossover systems. 2004;235:137–152. [Google Scholar]
- 25.Alp E, Sturhahn W, Toellner TS, Zhao J, Hu M, Brown DE. Vibrational Dynamics Studies by Nuclear Resonant Inelastic X-Ray Scattering. Hyp. Interact. 2002;144/145:3–20. [Google Scholar]
- 26.Sturhahn W. Nuclear resonant spectroscopy. J. Phys. Cond. Matt. 2004;16:S497–S530. [Google Scholar]
- 27.Sturhahn W, Toellner TS, Alp EE, Zhang X, Ando M, Yoda Y, Kikuta S, Seto M, Kimball CW, Dabrowski B. Phonon Density of States Measured by Inelastic Nuclear Resonant Scattering. Phys. Rev. Lett. 1995;74:3832–3835. doi: 10.1103/PhysRevLett.74.3832. [DOI] [PubMed] [Google Scholar]
- 28.Meyer J, Moulis J-M. Rubredoxin. In: Messerschmidt A, Huber R, editors. Handbook of Metalloproteins. Vol. 1. New York: Wiley; 2001. pp. 505–517. [Google Scholar]
- 29.Tan M-L, Bizzarri AR, Xiao Y, Cannistraro S, Ichiye T, Manzoni C, Cerullo G, Adams MWW, Francis E, Jenney J, Cramer SP. Observation of Terahertz Vibrations in Pyrococcus furiosus Rubredoxin via Impulsive Coherent Vibrational Spectroscopy and Nuclear Resonance Vibrational Spectroscopy – Interpretation by Molecular Mechanics. J. Inorg. Biochem. 2007;101:375–384. doi: 10.1016/j.jinorgbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 30.Mitra D, Pelmenschikov V, Guo Y, Case DA, Wang H, Jenney FEJ, Adams MW, Cramer SP. Dynamics of the [4Fe-4S] Cluster in Pyrococcus furiosus Ferredoxin via Nuclear Resonance Vibrational Spectroscopy (NRVS) and Resonance Raman Spectroscopy – Quantitative Simulation by Density Functional Theory. 2008 Draft ready for submission. [Google Scholar]
- 31.Rehr JJ, Albers RC. Theoretical approaches to x-ray absorption fine structure. Rev. Mod. Phys. 2000;72:621–654. [Google Scholar]
- 32.Hahn JE, Hodgson KO. Polarized X-ray absorption-spectroscopy. 1983;211:431–444. [Google Scholar]
- 33.Cramer SP, Flank AM, Weininger M, Mortenson LE. Single Crystal EXAFS of Nitrogenase. J. Am. Chem. Soc. 1986;108:1049–1055. [Google Scholar]
- 34.Dauter Z, Wilson KS, Sieker LC, Moulis JM, Meyer J. Zinc- and iron-rubredoxins from Clostridium pasteurianum at atomic resolution: a high-precision model of a ZnS4 coordination unit in a protein. Proc. Natl. Acad. Sci. U. S. A. 1996;93:8836–8840. doi: 10.1073/pnas.93.17.8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonisch H, Schmidt CL. Ultrahigh-resolution study on Pyrococcus abyssi rubredoxin. Å I. 0.69 X-ray structure of mutant W4L/R5S. Acta Crys. D. 2005;61:990–1004. doi: 10.1107/S090744490501293X. [DOI] [PubMed] [Google Scholar]
- 36.Jenney FE, Jr, Adams MWW. Rubredoxin from Pyrococcus furiosus. Methods Enzymol. 2001;334:45–55. doi: 10.1016/s0076-6879(01)34457-9. [DOI] [PubMed] [Google Scholar]
- 37.Saito H, Imai T, Wakita K, Urushiyama A, Yagi T. Resonance Raman Active Vibrations of Rubredoxin. Normal Coordinate Analysis of a 423-Atom Model. Bull. Chem. Soc. Jpn. 1991;64:829–836. [Google Scholar]
- 38.Rotsaert FA, Pikus JD, Fox BG, Markley JL, Sanders-Loehr J. N-isotope effects on the Raman spectra of Fe2S2 ferredoxin and Rieske ferredoxin: evidence for structural rigidity of metal sites. J. Biol. Inorg. Chem. 2003;8:318–326. doi: 10.1007/s00775-002-0417-4. [DOI] [PubMed] [Google Scholar]
- 39.Ducruix A, Giege R. Crystallization of nucleic acid and proteins. Oxford: Oxford University Press; 1992. p. 73. [Google Scholar]
- 40.Georgiadis MM, Komiya H, Chakrabarti P, Woo D, Kornuc JJ, Rees DC. Crystallographic Structure of the Nitrogenase Iron Protein from Azotobacter vinelandli. Science. 1992;257:1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- 41.Alp EE, Mooney TM, Toellner T, Sturhahn W. Nuclear Resonant Scattering Beamline at the Advanced Photon Source. 1994;90:323–334. [Google Scholar]
- 42.Yoda Y, Yabashi M, Izumi K, Zhang XW, Kishimoto S, Kitao S, Seto M, Mitsui T, Harami T, Imai Y, Kikuta S. Nuclear resonant scattering beamline at SPring-8. Nucl. Inst. Meth. A. 2001;467:715–718. [Google Scholar]
- 43.Shape Software. http://www.shapesoftware.com/. [Google Scholar]
- 44.Dowty E. Fully automated microcomputer calculation of vibrational spectra. Phys. Chem. Minerals. 1987;14:67–79. [Google Scholar]
- 45.Engeln-Mueller G, Uhlig F. Numerical Algorithms with C. Springer-Verlag; 1996. [Google Scholar]
- 46.EXAFSPAK: a Suite of Computer Programs for Analysis of X-ray Absorption Spectra. http://ssrl.slac.stanford.edu/exafspak.html. [Google Scholar]
- 47.Kim JK, Rees DC. Crystallographic Structure and Functional Implications of the Nitrogenase Molybdenum-Iron protein from Azotobacter vinelandii. Nature. 1992;360:553–560. doi: 10.1038/360553a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.