Abstract
Background:
Nin one binding protein (NOB1) was identified as a potential oncogene in human glioma and miR-646 plays an important role in human growth and development. However, the underlying molecular mechanisms of NOB1 in tumorigenicity and its correlation with miR-646 in renal cell carcinoma (RCC) have not been investigated.
Methods:
We performed bioinformatic analysis to explore miRNA targeting NOB1. The expression of NOB1 and miR-646 from 100 cases of clear cell RCC (ccRCC) and 30 cases of adjacent non-tumour tissues were detected by quantitative real-time PCR. The expression of miR-646 was correlated with NOB1 expression, tumour features and patient metastasis-free survival. The effect of overexpression of mir-646 on renal cancer cell proliferation was detected by colony formation in soft agar. Using a xenograft tumour model, we observed the in vivo tumorigenesis effect of miR-646 and NOB1.
Results:
miR-646 negatively regulated NOB1 and inhibited the proliferation and migration of renal cancer cells. There was a significant upregulation of NOB1 in ccRCC and it was further increased in metastatic cases, while miR-646 was downregulated in tumour tissues and further decreased in metastatic ccRCC. Additionally, expression of miR-646 was inversely correlated with the expression of NOB1. The downregulation of miR-646 also indicated a higher probability of developing metastasis. Most importantly, miR-646 expression was an independent predictor of ccRCC metastasis by the univariate analysis and binary logistic regression model (both P<0.05). Colony formation in soft agar and xenograft tumour model suggested that miR-646 and NOB1 are required for tumorigenesis in vitro and in vivo. Furthermore, suppression of NOB1 increased the phosphorylation of several proteins in MAPK pathway.
Conclusions:
Downregulated miR-646 in ccRCC was associated with tumour metastasis through MAPK pathway by targeting NOB1. miR-646 and NOB1 may play an important role in the development of ccRCC.
Keywords: renal cell carcinoma, nin one binding protein, miR-646, mitogen-activated protein kinases
Renal cell carcinoma (RCC) is a common urologic malignancy and accounts for ∼3% of adult malignancies and causes ∼90 000 deaths worldwide annually (Jemal et al, 2011). Clear cell RCC (ccRCC) is the largest subtype of RCC and accounts for ∼70% of these tumours. Apart from surgery, it is both radiotherapy and chemotherapy resistant. Biomarkers for early detection and follow-up of the disease are not available at present. Overall, 30% of patients with newly diagnosed disease have evidence of metastases at presentation (Hutson and Figlin, 2007). In the case of metastasis, few patients achieve a durable remission with currently available therapies, with a response rate of ∼15–25% and overall median survival of <12 months (Gupta et al, 2008; Motzer et al, 2013). The early identification of ccRCC metastatic potential may be beneficial for a more precise prediction of clinical outcomes and may ultimately be used to identify subsets of patients that may benefit from specific targeted therapies. MicroRNAs (miRNAs) function as regulatory molecules by inhibiting protein translation, which play important roles in differentiation, development, proliferation, and apoptosis (Bartel, 2004). Downregulation of miRNAs has been suggested to play a critical role in cancer progression (Aguda, 2013; Farazi et al, 2013; Fendler and Jung, 2013).
miR-646 was first identified in a study of miRNAs expressed in human cerebral cortical grey and white matter. miR-646 is known to be mammalian specific, and it only appears in humans and chimpanzees but has not been found in rhesus monkeys (Finnerty et al, 2010). miR-646 belongs to the miR-15/107 gene group; in fact, the normal functions of miR-15/107 gene group members seem to have roles in cell division, angiogenesis, and metabolic pathways. However, dysfunction in miR-107 expression may contribute to neoplasia, neurodegeneration, cardiovascular dysfunction, and other diseases (Gatsiou et al, 2012). However, the dysregulation of miR-646 and its possible involvement in ccRCC has not been reported.
The yeast nin one binding protein (NOB1) is required for the biogenesis and function of the 26S proteasome and plays a role in RNA metabolism. The NOB1 gene encodes a 50 KDa protein consisting of a PIN (PilT amino terminus) domain and a zinc ribbon domain. NOB1 protein is mainly expressed in the spleen, liver and lung. The human ortholog of the NOB1 gene was cloned several years ago (Zhang et al, 2005). To date, the physiological and pathological functions of NOB1 remain unclear, and its relationship with miR-646 has not been reported.
miRNAs were shown to negatively regulate gene expression at the post-transcriptional level by binding to the 3′ untranslated region (3′-UTR) of target mRNAs (Zantarpia et al, 2010). miRNAs have been shown to be involved in tumour progression and metastasis in kidney and other carcinomas (Huang et al, 2009). Thus, we searched for miRNAs targeting NOB1 using miRNA target prediction algorithms such as TargetScan, miRDB and PicTar. miR-646 was found to be a potential target for NOB1. However, experimental validation of miR-646 targeting NOB1 has not yet been documented, and the role of miR-646 in ccRCC needs to be elucidated.
In this study, we show that miR-646 could directly regulate NOB1. Furthermore, we investigated the role in MAPK pathway in ccRCC and found that miR-646 inhibits the activation of the MAPK pathway by suppressing NOB1. In addition, miR-646 overexpression impaired cell viability and invasiveness of cancerous cells. Thus, we elucidated the role of miR-646 as a regulator in NOB1 expression and MAPK pathway activity in human ccRCC, which could be a potential therapeutic target for cancer therapy.
Materials and methods
Ethics statement
Written Informed Consent was obtained from all patients. This study was approved by the Protection of Human Subjects Committee, Tongji University Affiliated Shanghai Tenth People's Hospital.
Tissue samples
A total of 100 cases of ccRCC and 30 cases of adjacent non-tumour kidney tissues were obtained from our hospital (Table 1). All ccRCC cases were confirmed by a senior pathologist, and staged based on the 2011 Union for International Cancer Control TNM classification of malignant tumours. The pathological diagnoses of all enrolled patients were confirmed by two different pathologists, according to the WHO grading system (Gelb, 1997).
Table 1. Characteristics of patients with RCC and associations with clinicopathologic variables.
| Variable | Total (%) | Variable | Total (%) |
|---|---|---|---|
| Gender | Grade | ||
| Male | 79 (79) | 1 | 65 (65) |
| Female | 21 (21) | 2 | 27 (27) |
| Age | 3 | 8 (8) | |
| ⩽45 | 9 (9) | Clinical stage | |
| 45–55 | 61 (61) | I | 39 (39) |
| >55 | 30 (30) | II | 13 (13) |
| BMI | III | 20 (20) | |
| <25 | 54 (54) | IV | 28 (28) |
| ⩾25 | 46 (46) | Metastatic status | |
| Tumour size (cm) | NM | 72 (72) | |
| ⩽4 | 23 (23) | LM | 7 (7) |
| 4–7 | 31 (31) | DM | 21 (21) |
| 7–10 | 28 (28) | T stage | |
| >10 | 18 (18) | T1 | 46 (46) |
| Tumour Necrosis | T2 | 20 (20) | |
| No | 48 (48) | T3 | 29 (29) |
| Yes | 52 (52) | T4 | 5 (5) |
Abbreviations: BMI=body mass index; DM=tumours involving distant metastases; LM=tumours involving lymphatic metastases; NM=tumours involving non-metastasis; RCC=renal cell carcinoma.
Cell culture and cell transfection
HEK-293T cells and human renal cancer cells 786-O, ACHN and Caki-2 (American Type Culture Collection, Manassas, VA, USA) were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 100 U ml−1 penicillin and 100 mg ml−1 of streptomycin. Cells were cultured at 37 °C in a humidified atmosphere of 5% CO2.
HEK-293T, 786-O, ACHN and Caki-2 cells were seeded in 24-well plates and incubated overnight, then transiently transfected with miR-646 precursor, control miR-646 antisense oligonucleotide or siRNA oligos using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Precursor miRNA and control oligos were obtained from Applied Biosystems (Carlsbad, CA, USA). The scrambled shRNA (stem–loop–stem structure) targeting NOB1 sequence was designed and synthesised (NOB1-shRNA: 5′-AAGGTTAAGGTGAGCTCAT-3′). The corresponding sequences of the negative control shRNA (control siRNA) was: 5′-AATTCTCCGAACGTGTCACGT-3′. Approximately, 48 h after transfection, the effects of gene silencing were measured via western blotting and real-time PCR analysis. The full-length NOB1 (NM_014062.2) cDNA containing plasmid was ordered from IBSbio (IBS Solutions Co. Ltd, Shanghai, China). The complete NOB1 cDNA was reverse transcribed by PCR and ligated into the pcDNA3.1 mammalian expression vector (Invitrogen). This pcDNA3.1-NOB1 plasmid consisted of the entire 1239 bp ORF (LNA-anti-miR-NC: 5′-UCUACUCUUUCUAGGAGGUUGUGA-3′ LNA-anti-miR-646–5′-GCCUCAGAGGCAGCUGCUU-3′).
Prediction of miRNAs targeting NOB1
miRNA target predicting algorithms miRDB (http://mirdb.org/miRDB/), TargetScan (http://www.targetscan.org/), and PicTar (http://pictar.mdc-berlin.de/) were used to predict miRNAs targeting NOB1 and the binding regions.
Plasmids constructs and luciferase reporter assay
3′-UTR of NOB1 and a mutation sequence were amplified by PCR using the primers that included a Bgl II restriction site on the 5′ and 3′ strands. The PCR products were inserted into the Bgl II sites of the pGL3-control vector (Promega, Madison, WI, USA) and identified by DNA sequencing. The wild-type plasmid was created containing the 3′-UTR of NOB1 with complementary sequence of miR-646 (pGL3-NOB1 3′-UTR wild), and a mutant plasmid with the mutation sequence without complementary sequence of miR-646 (pGL3-NOB1 3′-UTR mut). Following primers were used: (Forward) NOB1-3′-UTR wild-F, 5′-CAAGCTTAGCGAGTTCCCGCAGGCAAAT-3′ (Reverse) NOB1-3′-UTR wild-R, 5′-CTCTAGACATGATCTCTGGGCACAC-3′ (Forward) NOB1-3′-UTR mut-F, 5′-CAAGCTTAGCGAGTTCCCGCAGGCAAAT-3′ (Reverse) NOB1-3′-UTR mut-R, 5′-CTCTAGACATGATCTCTTTTCACACAGC-3′.
For the luciferase reporter assay, the human malignant renal cell line 786-O was seeded on 24-well plates and co-transfected using Lipofectamine 2000 (Invitrogen) with 100 ng per well of the resulting luciferase UTR-report vector, 2 ng per well of pRLCMV vector (internal control, Promega) and 20 ng per well of miR-646 precursor molecules or control precursor (Applied Biosystems) following the manufacturer's instructions. After 24 h the cells were lysed and the relative luciferase activity was assessed with the Dual-Luciferase Assay Reporter System (Promega).
Total RNA extraction and quantitative real-time PCR
Total RNAs containing miRNAs were extracted from frozen tissue samples or the harvested cells using 1 ml of Trizol reagent (Invitrogen) following manufacturer's instructions. cDNA was synthesised from total RNA (2 mg) using M-MLV Reverse Transcriptase Kit (Promega). cDNA (20 ng) was mixed with SYBR GreenMasterMix (Bio-Rad, Hercules, CA, USA) and amplified in CFX96 real-time detection system (Bio-Rad) according to the manufacturer's protocol (Wang et al, 2011). Each sample was run in triplicate for each gene. Relative expression levels of NOB1 mRNA were calculated by normalising to the level of GAPDH expression by using comparative threshold cycle method, in which fold difference is 2–(Δct of target gene–Δct of reference). Following primers were used: (Forward) NOB1-F, 5′-ATCTGCCCTACAAGCCTAAAC-3′ (Reverse) NOB1-R, 5′-TCCTCCTCCTCC TCCTCAC-3′ (Forward) GAPDH-F, 5′-GAAGGTGAAGGTCGGAGTC-3′ and (Reverse) GAPDH-R, 5′-GAAGATGGTGATGGGATTTC-3′.
Protein extraction and western blotting
Proteins were extracted from human renal cancer tissues or subconfluent culture of cells, and were then characterised by using western blot analysis. Cells were lysed in lysis buffer, resolved in SDS–PAGE gel, and transferred onto an Immobilon-P membrane (Millipore, Billerica, MA, USA). The membrane was blocked with 5% nonfat milk in PBST for 1 h at room temperature. The membrane was probed with primary antibody against NOB1 (1:10 000, Abcam, Cambridge, MA, USA) or GAPDH (1:5000, Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and was incubated overnight at 4°C. After extensive washing, the membrane was incubated with secondary antibody conjugated with horseradish peroxidase (1:10 000; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. Blots were developed using ECL (PE LifeScience, Waltham, MA, USA).
Analysis of cell cycle by flow cytometry
Different cell cycle phases (G1, S or G2/M phase) were characterised by different DNA contents. Fluorescence dyepropidium iodide (PI; Sigma, Santa Clara, CA, USA) was used to analyse the DNA in cell cycle phase at a ratio of 1 : 1. Cells were serum starved for 24 h to synchronise the cells, and media was replaced with complete medium containing growth factor. After 48 h of incubation, cells were harvested with trypsin-EDTA, washed with chilled phosphate-buffered saline (PBS) twice and fixed with 70% ethanol at −20°C for 2 h. The cells were harvested and resuspended in PI/RNase/PBS (100 μg ml−1 PI and 10 μg ml−1 RNase A) for 30 min at 37°C in dark, and filtered through a nylon mesh (400 screen meshes, Metal Enterprise Co. Hebei, China). No less than 10 000 cells were analysed in each sample. The percentage of cells in G0/G1, S and G2/M phases was determined by (fluorescence-activated cell sorting) FACS Calibur flow cytometer (BD Biosciences, San Jose, CA, USA). The experiment was repeated thrice and the results were averaged.
Analysis of cell proliferation in vitro
The in vitro cell proliferation of RCC cell lines was measured using the MTT method (Ai et al, 2012). In brief, cells were seeded into 96-well plates and transfected with miR-646 precursor, control precursor, NOB1-shRNA, control shRNA or nothing for 72 h. In the indicated time periods, 0.1 ml of spent medium was replaced with an equal volume of fresh medium containing MTT 0.5 mg ml−1. Plates were incubated at 37°C for 4 h, and then the medium was replaced with 0.1 ml of DMSO (Sigma, St Louis, MO, USA) and plates were agitated at room temperature for 10 min. The absorbance was measured at 490 nm. BrdU assay was also used for cell proliferation measurement. After 72 h of transfection, RCC cell lines were given a 2-h pulse of BrdU (Sigma) at 4 mg ml−1. Visualisation of new DNA synthesis was done by indirect immunohistochemistry on adherent cultures using primary anti-BrdU antibody followed by a secondary antibody conjugated with horseradish peroxidase. TMB Peroxidase substrate kit (Vector, Burlingame, CA, USA) was used for detection. The absorbance rate at 450 nm was recorded using a 96-well plate reader.
Wound-healing assay and invasive assay
786-O cells was cultured in 25 cm2 culture flask to ∼80–90% confluency, collected by digestion and centrifugation, and then seeded into six-well plates at 2.5 × 105 cells per well. LNA-anti-miR-646 or LNA-anti-miR-NC or pcDNA3.1-NOB1 was transfected into 786-O cells. The floating cells were removed by washing with PBS, and the width of scratch was observed at 0 and 48 h using inverted microscope (50-fold). Each experiment was repeated three times.
Transwell chambers precoated with Matrigel (BD Bioscience) were used to perform the invasion assay. Cells were cultured in serum-free medium in the upper chambers of the Transwell (Corning, Union City, CA, USA) insert (1 × 105 cells per chamber), which are separated from the lower chambers with permeable 8 μm polycarbonate membranes. Medium containing 10% FBS served as the attractant in the lower chambers. After 12 h, the cells were fixed with 75% ethanol and stained with crystal violet. Non-migrating cells on the upper side of the membrane were gently wiped off, and the stained cells on the lower side were observed under a microscope. The number of migrating cells in five fields per chamber were counted and average values were calculated.
Analysis of apoptosis by Annexin V-FITC and PI
The Annexin V-FITC Apoptosis Detection kit (Calbiochem, Darmstadt, Germany) and PI (Sigma) was used to assess the apoptotic effect of miR-646. ACHN cells with different treatments were suspended at a concentration of 1 × 106 cells per millilitre, centrifuged and washed by PBS. The cells were resuspended in 0.5 ml cold Binding Buffer (Santa Cruz Biotechnology, Inc.) with 1.25 μl Annexin V-FITC, and incubated for 15 min at room temperature in the dark and centrifuged for 5 min. Cells were resuspended in 0.5 ml cold Binding Buffer with 10 μl PI, incubated for 4 h and analysed by flow cytometry.
Nude mouse tumorigenicity assay
Nude mouse tumorigenicity assay were performed as previously reported (Hillman et al, 1994). Briefly, specific pathogen-free six-week-old female BALB/C-nu/nu mice were purchased from the Cancer Research Center (Shanghai, China) and maintained under specific pathogen-free conditions in Tongji University. When the female BALB/cnu mice were ∼7–8 weeks old, each mouse was inoculated with 1.5 × 107 ACHN cells transfected with miR-646 or miR-control or NOB1-shRNA in 0.2 ml of PBS subcutaneously in the forelimb with the mice being injected with mock-infected cells as control. Tumour sizes were measured every 5 days in two dimensions using a caliper, and the volume was calculated using the formula: V=width2 × length × 0.5 (mm3). The tumours were excised and weighed after 35 days. All animal experiments were undertaken in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals, with the approval of Animal Care and Use Committee in Tongji University.
Measuring phosphorylation of signalling proteins
The phosphorylation changes of selected proteins involved in signalling pathways were analysed with Proteome Profiler Array kit (ARY003; R&D Systems, Minneapolis, MN, USA). 786-O and ACHN cells were treated with miR-646 precursor, control precursor or NOB1-shRNA. At the designated times, each dish was washed twice with PBS and processed to harvest 1 to 5 million cells. Incubation was done with an array that contained 300 μg of lysate protein. Net integrated pixel density for each spot (an average of duplicate spots after subtraction of average background density) was determined by densitometry and analysed using Quantity One software (ISBE, Sheffield, UK). The data were normalised to net integrated pixel density of kit-supplied internal positive controls.
Immunohistochemical staining and clinical and pathological evaluation
Immunohistochemical staining for NOB1 protein was performed on the validating set of renal cancer patients. In brief, paraffin embedded slides were treated with hydrogen peroxide (H2O2) to block endogenous peroxidase activity, and washed with ddH2O and PBS. Diluted Rabbit polyclonal antibody to NOB1 (Abcam, San Francisco, CA, USA) was added to it and incubated at room temperature for 60 min. The slides were washed with PBS, incubated with biotinylated secondary antibody (Abcam), and treated with Immunopure Metal enhanced DAB substrate (Pierce, Rockford, IL, USA). Staining was categorised into four grades according to immunohistochemical scores. For each slide, 10 randomly selected fields of view under a light microscope were examined for the average intensity of positive cells and the intensity scores were assigned to each sample as follows: none (−), none/weak (+−), weak (+), intermediate (++), and strong (+++).
Statistical analysis
The relative quantification of gene expression detected by real-time PCR was log10 transformed and analysed by Student's t-test or ANOVA. Linear or rank correlation analysis was performed to determine the correlation between the gene expression levels. The univariate analysis and binary logistic regression model were performed to select the risk factors associated with metastasis. Briefly, Fisher's exact test was used to compare the immunolabelling results of NOB1 between high-grade and low-grade ccRCC. All covariables were transformed into binary data, and the tumour metastasis status (metastasis or not) was selected as dependent variable. The Enter method was used in this model. The Kaplan–Meier method and log-rank test were used to estimate and compare the probability of metastasis-free survival. SPSS 15.0 (SPSS Inc, Chicago, IL, USA) was used for the statistical analysis and a significance level of P<0.05 was used to evaluate the difference between groups.
Results
NOB1 is the target of miR-646
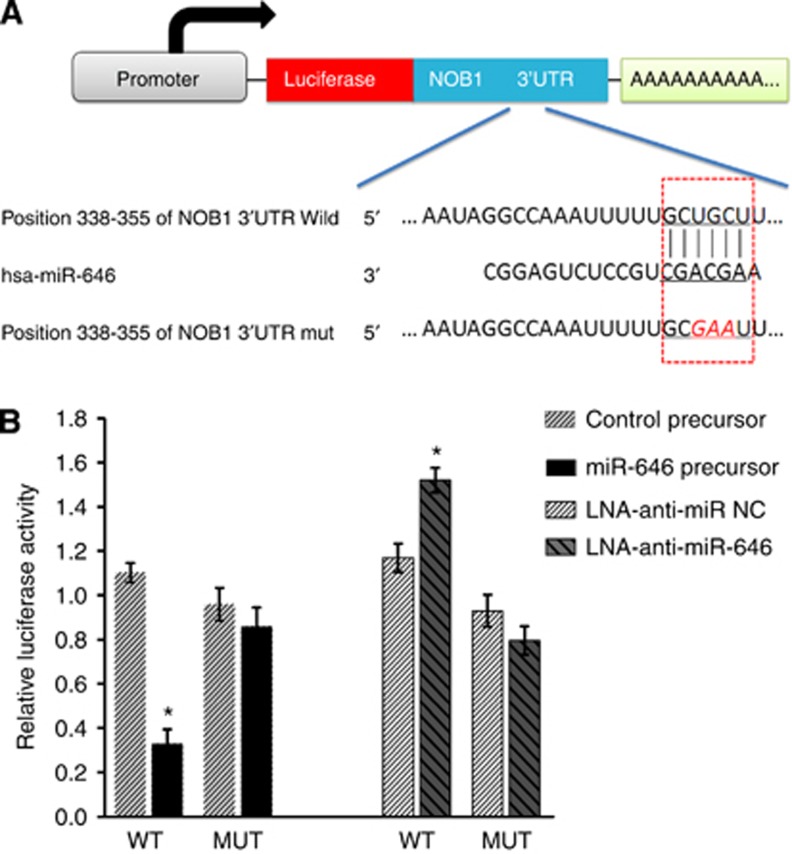
Using TargetScan software (Lewis et al, 2005), NOB1 was verified as a likely target of miR-646 because it contains a putative miR-646 target site in the 3′-UTR (Figure 1A). To confirm this, the luciferase reporter vector was constructed containing oligonucleotides fully complementary to the 3′-UTR of wild-type NOB1, or its relevant mutant. Pre-hsa-miR-646 RNAs or non-functional control miR-NC RNAs or LNA-anti-miR-646 or LNA-anti-miR-NC were co-transfected with the reporter vectors into the human renal cancer cell line 786-O. The miR-646 target sequences and wild-type NOB1 3′-UTR reduced the relative luciferase activity only when miR-646 was present, but not when the corresponding mutant was introduced with miR-646 (Figure 1B). The luciferase reporter data showed that NOB1 is a specific target of miR-646.
Figure 1.
miR-646 targets the 3′-UTR of NOB1 in luciferase reporter assay. (A) Ideograph of NOB1 mRNA. The predicted miR-646-binding site in the NOB1 3′-UTR. The sequence of wild-type (WT) and mutant (MUT) miR-646 target sites in the NOB1 3′-UTR shown in frame. A point mutation (underlined) was made in the seed region to block the binding between miR-646 and mRNA. (B) The luciferase reporter assay was used to confirm the contribution of the four miR-646 target sites. ACHN cells were co-transfected with luciferase reporter plasmids containing either WT or MUT miR-646 target sites and miR-646, control precursors, LNA-anti-miR NC and LNA-anti-miR-646. miR-646 and full-length wild-type NOB1 3′-UTR decreased luciferase activity. All results were derived from independent experiments performed in triplicate (*P<0.01).
MiR-646 inhibits the proliferation of renal cancer cells and directly targets NOB1
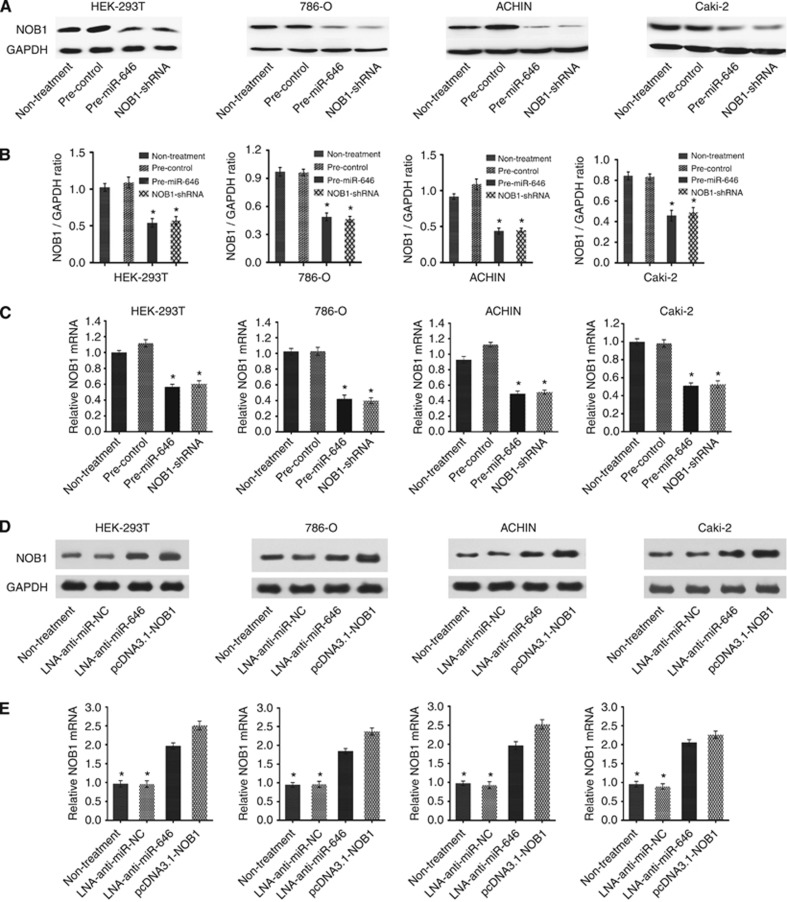
Based on our finding that NOB1 mRNA is a specific target of miR-646, we hypothesised that miR-646 might inhibit cell proliferation and arrest cell cycle progression through the downregulation of NOB1 expression. To test this assumption, 786-O, ACHN and Caki-2 cells were transiently transfected with miR-646 or miR-NC precursors. We found out that miR-646 significantly repressed the NOB1 expression, the intervention effects of which was similar to NOB1 RNA interference (Figure 2).
Figure 2.
miR-646 overexpression can downregulate NOB1 protein and mRNA expression. HEK-293T, 786-O, ACHN and Caki-2 cells were transfected with previously generated miR-646, miR-NC, NOB1-shRNA or untreated control vectors. (A and B) The results of western blot were analysed with Quantity One (n=3). miR-646 significantly repressed NOB1 protein expression in RCC cell lines. (C) The downregulation of NOB1 mRNA following miR-646 transfection in three RCC cell lines was analysed by RT–PCR (*P<0.01). (D) HEK-293T, 786-O, ACHN and Caki-2 cells were transfected with LNA-anti-miR-646, LNA-anti-miR NC, pcDNA3.1-NOB1 or untreated control vectors. The results of western blot shown miR-646 significantly increased NOB1 protein expression in RCC cell lines. (E) The upregulation of NOB1 mRNA following LNA-anti-miR-646 transfection in three RCC cell lines was analysed by RT–PCR (*P<0.01).
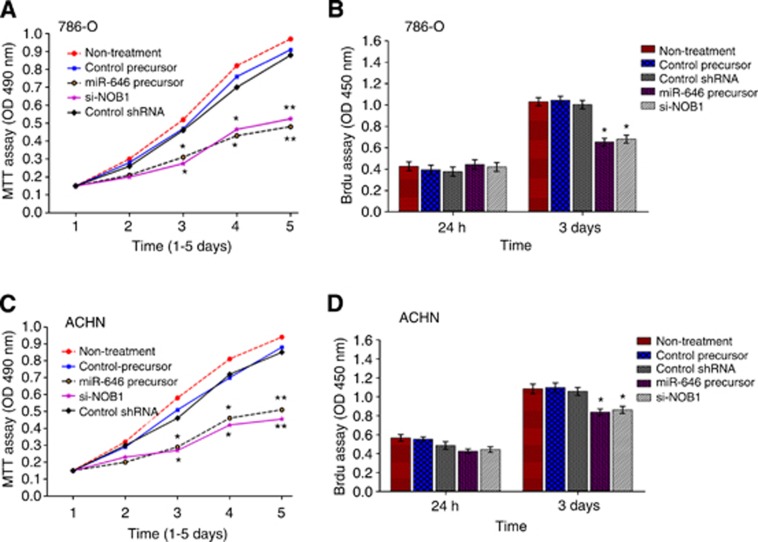
786-O and ACHN cell lines were transfected with miR-646, miR-NC, NOB1-shRNA, control shRNA or non-treated controls and were inspected using the MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H tetrazolium bromide] assay. As shown in Figure 3A and C, ectopic expression of miR-646 significantly inhibited the growth of 786-O and ACHN cell lines compared to the negative control 3 days after infection (P<0.05), and no statistically significant differences in growth rate between miR-646 overexpressing cells and NOB1-shRNA-infected cells was observed. After 5 days, the growth of 786-O cells expressing miR-646 or treated with NOB1-shRNA was decreased to 50% when compared to control cells. There were no significant differences in cell growth detected between miR-NC cells and the untreated controls (Figure 3A and C). The effect of miR-646 on the proliferation of human renal cancer cells was confirmed using a BrdU incorporation assay. Similarly, no significant difference in BrdU incorporation was observed between cells with different treatments at 24 h. However, BrdU incorporation was decreased in 786-O cells expressing miR-646 or through NOB1-shRNA treatment compared to the controls at 72 h, and similar results were observed in ACHN cells (Figure 3B and D). Overall, these data suggest that NOB1 may play a role in regulating the proliferation of human renal cancer cells.
Figure 3.
Cell viability and proliferation were detected in human renal cancer cells transfected with pre-miR-646. 786-O (A) and ACHN (C) cells were transfected with miR-646 precursor, control precursor, NOB1-shRNA, non-treatment and control shRNA for 3 days as described in the methods section before measurement of the conversion of MTT to a coloured formazan product. A statistically significant delay of cell proliferation was observed after day 3. 786-O cells (B) and ACHN cells (D) were transfected with miR-646 precursor, control precursor, NOB1-shRNA, nothing and control shRNA for 3 days as described in the methods section, and the BrdU incorporation assay was performed. BrdU incorporation was decreased in the miR-646 group and NOB1-shRNA group compared to the controls at 3 days. Significant differences between transfected cells and mock-infected cells were determined using the two-tailed Student's t-test (**P<0.01, *P<0.05).
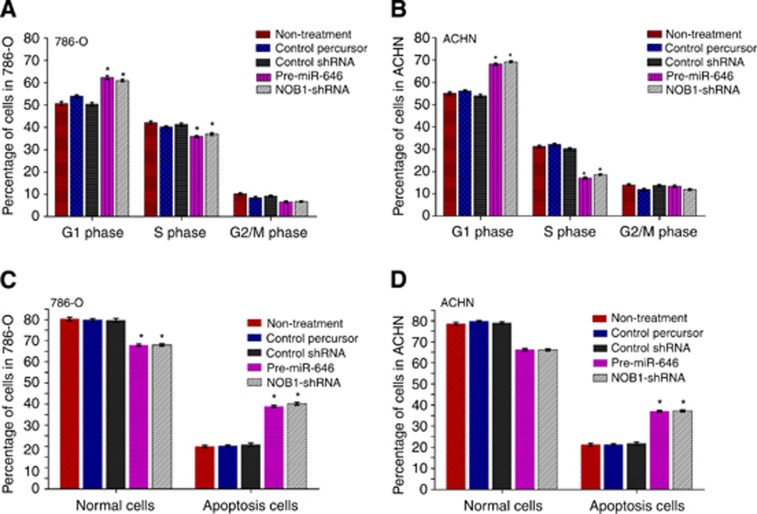
Then we examined the cell cycle distribution by FACS after transfection. Compared with miR-control cells, miR-646 overexpressing cells and NOB1 silencing cells showed a substantial decrease in the proportion of cells in S phase and an increase in the number of cells in G1 phase in 786-O and ACHN cell lines (Figure 4A and B).
Figure 4.
Effect of NOB1 and miR-646 expression on the cell cycle and apoptosis in renal cancer cells. (A and B) 786-O and ACHN cells were transfected with pre-miR-646, NOB1-shRNA, control percursor or control shRNA, respectively. After being cultured for 48 h, cells were fixed and stained with propidium iodide and cell cycle was analysed by flow cytometry. Representative analysis of three independent experiments is shown. (C and D) Quantities of the apoptosis cells with Annexin V-FITC/PI stain and flow cytometry analysis. miR-646 expression or NOB1 silencing causes G1-phase cell cycle arrest, and induced early apoptosis (Annexin V+/PI−). Three independent experiments were performed in each group (*P<0.05).
To evaluate the effect of miR-646 on apoptosis, 786-O and ACHN cells were stained with Annexin V-FITC and propidium iodide after transfection with miR-646, miR-NC, control shRNA or NOB1-shRNA for 72 h. As shown in Figure 4C and D, >80% of the untreated controls or miR-NC cells were viable, whereas only ∼68% of the miR-646 cells or NOB1 silenced cells were viable. Early apoptosis rates averaged from four experiments were 14.6% in the miR-NC group, 13.5% in the untreated control group, 28.5% in the miR-646 group, and 29.3% in the NOB1-shRNA group (P<0.01). Further, late apoptosis rates of the four groups were 8.3% with no distinct differences. We found that miR-646 induced early apoptosis in renal cancer cells.
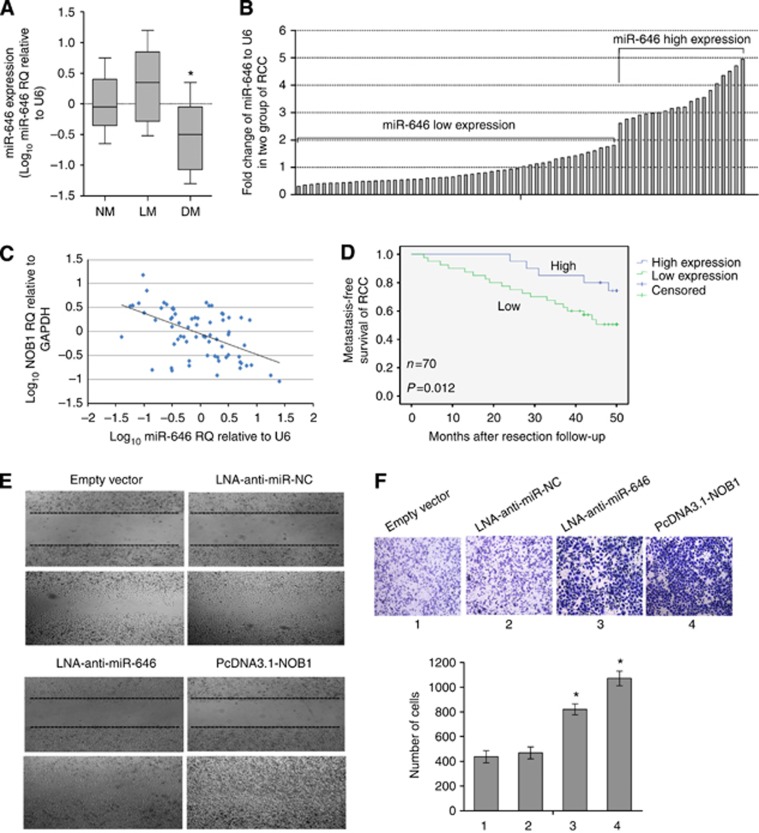
Low-level miR-646 was associated with ccRCC metastasis and shorter metastasis-free survival
We then determined whether miR-646 was associated with tumour distant metastasis. Tumour samples were divided into three groups according to their metastatic status. Non-metastatic (NM) samples were obtained from primary sites without lymphatic or distant metastases. Lymphatic metastatic (LM) samples were obtained from primary sites with lymph node metastasis, and distant metastatic (DM) samples were obtained from primary sites in the metastatic presence of distant metastases but absence of lymph node metastases. Our data showed that miR-646 expression decreased form NM and LM groups to DM group. However, there was no statistically significant difference between NM and LM groups in these variables (Figure 5A). Our findings showed that downregulated miR-646 might increase the risk of metastasis in ccRCC. To link miR-646 expression to distant metastasis, 70 patients without synchronous metastases were prospectively followed up for a 5-year observational period. The patients were divided into two groups based on relatively high or low levels of miR-646 expression at the threshold giving the lowest P value of log-rank test comparing metastasis-free survival between the two groups (Figure 5B). As shown in Figure 5C, NOB1 expression was negatively correlated with miR-646 expression in ccRCC (linear correlation analysis, r=−0.745, P<0.001). Metastases developed in 19 out of 50 cases in the low-level miR-646 group, whereas distant metastasis developed in only 2 out of 20 cases in the high-level miR-646 group during the follow-up period. Moreover, patients in the high-level miR-646 group obtained better metastasis-free survival time than that in the low-level miR-646 group (Figure 5D; log-rank test, P=0.012). Similarly, wound-healing assays were performed to investigate the motility of 786-O cell line. The pcDNA3.1-NOB1 and LNA-anti-miR-646 cells had migrated significantly faster than the empty vector and LNA-anti-miR NC cells 48 h after wound creation (Figure 5E). In addition, we used Transwell chambers precoated with Matrigel (BD Bioscience) to perform the invasion assay. As shown in Figure 5F, NOB1 enhanced the cell invasive ability by 2.75-fold relative to the vector control cells.
Figure 5.
Association of miR-646 with ccRCC distant metastasis and metastasis-free survival. (A) Real-time PCR analyses of miR-646 expression in non-metastatic (NM; n=70), lymphatic metastatic (LM; n=9) and distant metastatic (DM; n=20) ccRCC (*P<0.05 between the DM and NM groups). (B) A total of 70 cases of NM RCC were grouped into two, ‘low miR-646' (n=50) and ‘high miR-646' (n=20) according to miR-646 expression at the threshold giving the lowest P value of log-rank test comparing metastasis-free survival between the two groups. (C) NOB1 expression was negatively correlated with miR-646 expression in ccRCC (linear correlation analysis, r=−0.745, P<0.001). (D) Kaplan–Meier graph representing the probability of metastasis-free survival in ccRCC from the two groups. The status was defined as ‘occurred metastasis or not', ‘high' referred to ‘high level of miR-646 expression', and ‘low' referred to ‘low level of miR-646 expression'. (E) Inhibition of the expression of miR-646 and overexpression of NOB1promoted the migration of 786-O cells in a wound-healing assay. (F) LNA-anti-miR-646 and pcDNA3.1-NOB1 of 786-O cells exhibited more invasive potential in a transwell chamber invasion assay (*P<0.05; the P value was from a log-rank test).
miR-646 was a predictor of ccRCC distant metastasis in univariate and multivariate analyses
To evaluate the association of miR-646 with distant metastasis, a multivariate logistic regression model was constructed by considering the clinicopathological features. The features involved patients' characteristics (gender and age), tumour features (size, grade, and pT stage), NOB1 and miR-646 expression. As shown in Table 2, the covariables were transformed into binary data, and the tumour distant metastasis status was selected as the dependent variable. miR-646 expression was divided into low or high levels at the 70th percentile. The analyses indicate that miR-646 expression, NOB1, pT stage and tumour size were predictors of ccRCC distant metastasis. The risk of distant metastasis in tumours expressing low-level miR-646 was 8.2-fold higher than that in tumours expressing high-level miR-646 (P=0.017). Moreover, large tumours appeared to have more possibilities of distant metastasis than small tumours (OR=4.776, P=0.021). In univariate analysis and multivariate analysis (chi-square test), miR-646 and NOB1 expression were associated with distant metastasis (P=0.027, P=0.039 and P=0.017, P=0.048, respectively). Similarly, the tumour pT stage showed statistical significance (P=0.034 and P=0.047). Further, the grade, age and gender, were not associated with distant metastasis in this model.
Table 2. Univariate and multivariate model measured ORs and 95% CIs of RCC associated with distant and other covariates.
|
Univariate |
Multivariate |
||||
|---|---|---|---|---|---|
| Variables | DM (%) | P | OR | P | OR (95% CI) |
| Gender | 0.724 | 0.441 | 0.312 | 0.361 (0.265–1.980) | |
| Male (79) | 18 (22.8) | ||||
| Female (21) | 3 (14.3) | ||||
| Age | 0.376 | 0.801 | 0.432 | 1.459 (0.189–2.430) | |
| ⩽60 (71) | 13 (18.3) | ||||
| >60 (29) | 8 (27.6) | ||||
| Tumour size | 0.043 | 3.147 | 0.021a | 4.776 (0.476–5.832) | |
| ⩽7 (54) | 8 (14.8) | ||||
| >7 (46) | 13 (28.3) | ||||
| Grade | 0.174 | 1.756 | 0.338 | 2.249 (0.347–3.781) | |
| 1 (65) | 11 (16.9) | ||||
| 2–3 (35) | 10 (28.6) | ||||
| pT Stage | 0.034a | 1.95 | 0.047a | 3.161 (0.590–4.291) | |
| T1–T2 (65) | 7 (10.8) | ||||
| T3–T4 (35) | 14 (40.0) | ||||
| miR-646 expression | 0.027a | 5.049 | 0.017a | 5.483 (1.214–18.815) | |
| Low (70) | 19 (27.1) | ||||
| High (30) | 2 (6.7) | ||||
| NOB1 expression | 0.039a | 4.714 | 0.048a | 5.197 (2.364–20.863) | |
| Low (27) | 4 (14.8) | ||||
| High (73) | 17 (23.3) | ||||
Abbreviations: CI=confidence interval; DM=distant metastasis; OR=odds ratio; RCC=renal cell carcinoma.
Statistically significant.
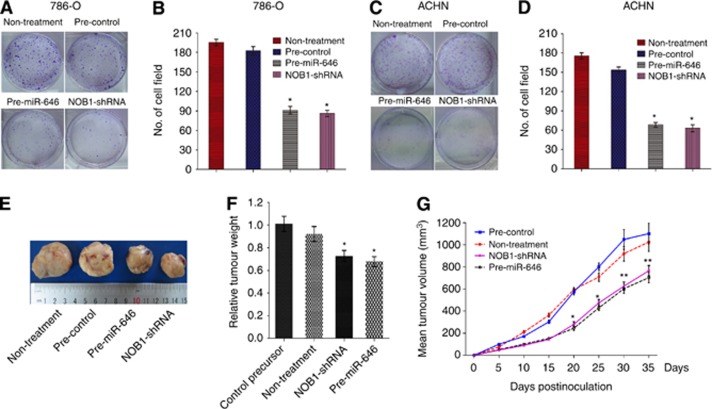
Overexpression of miR-646 inhibits carcinogenesis in human renal cancer cells
For examining the involvement of miR-646 in the tumorigenesis of human renal cancer cells in vitro, colony formation in soft agar was assessed in 786-O and ACHN cells. Similar to NOB1 silencing, overexpression of miR-646 caused a substantial reduction in colony formation in soft agar compared with the control group (P<0.05; Figure 6A–D). Further, to verify the effect of miR-646 levels on the tumorigenesis of renal cancer cells in vivo, we subcutaneously injected the genetically modified (overexpressed miR-646 or NOB1-shRNA) ACHN cells, as well as their control cells, into the forelimbs of nude mice. The tumour volume was monitored for 5 weeks. Mean tumour volume in the miR-646 group or the NOB1-shRNA group was 702.2 mm3 or 761.8 mm3, whereas tumour volumes in mice treated with saline or negative control plasmid were 973.0 mm3 or 1163.8 mm3, respectively (P<0.01) after 35 days (Figure 6E–G). The results further indicate that miR-646 overexpression could significantly inhibit the tumour formation in human renal cancer cells, suggesting that miR-646 may play a vital function in the tumorigenicity of human renal cancer cells in vitro and in vivo.
Figure 6.
The effect of miR-646 and NOB1-shRNA treatment on tumour growth in 786-O and ACHN cells and xenograft model. (A and B) Soft agar assay was performed to investigate the effect of miR-646 on tumour growth in vitro. 786-O cells had been infected with miR-646 precursor, control precursor, non-treatment or NOB1-shRNA were plated in wells coated with agar. (C and D) The same assay was performed on ACHN cells. Significant differences between transfected cells and mock-infected cells were determined using the two-tailed Student's t-test (*P<0.05). miR-646 overexpression or NOB1 silencing decreased colony formation. (E) Xenograft model was used to examine the effect of miR-646 on tumorigenesis in vivo. Tumour volume was calculated as width2 × length × 0.5 (mm3) every 5 days. miR-646 overexpression significantly inhibited tumour formation was observed. (F) Vertical bars indicate mean tumour weight at the end of the experiment. *P<0.05 as compared to the control precursor. (G) Tumour volume was evaluated periodically at the indicated days postinoculation. Data represent mean±s.d. of three independent experiments. All error bars indicated s.e.m. (**P<0.01, *P<0.05).
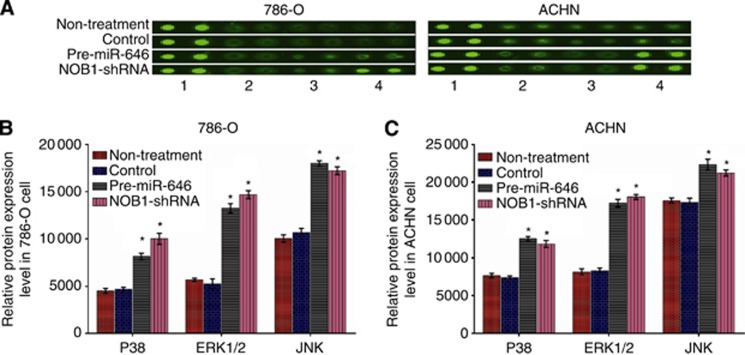
miR-646 regulates the key components of the MAPK signalling pathway
The phosphorylation of several proteins was analysed using the Phospho-Kinase Array Kit (Wayen Biotechnologies, Inc., Shanghai, China). In 786-O and ACHN cells, the phosphorylation of p38 (T180/Y182), ERK1/2 (T202/Y204, T185/Y187), and JNK (T183/Y185, T221/Y223) increased significantly after NOB1 suppression compared to the negative control (P<0.05; Figure 7).
Figure 7.
Overexpression of miR-646 transform the three key components of the MAPK pathway. (A) Cell extracts of 786-O and ACHN cells treated with pre-miR-646, NOB1-shRNA, control or nothing were incubated on a Phospho-Kinase Array and phosphorylation status was detected by subsequent incubation with anti-phosphotyrosine horseradish peroxidase. Each protein was spotted in duplicate. (1, positive control; 2, P38; 3, ERK 1/2; 4, JNK). (B and C) In 786-O and ACHN cells, the phosphorylation of all three components of MAPK pathway were significantly increased after miR-646 overexpression or NOB1-shRNA compared to the control. Values are presented as the mean±s.d. of at least three independent experiments performed in triplicate (*P<0.05).
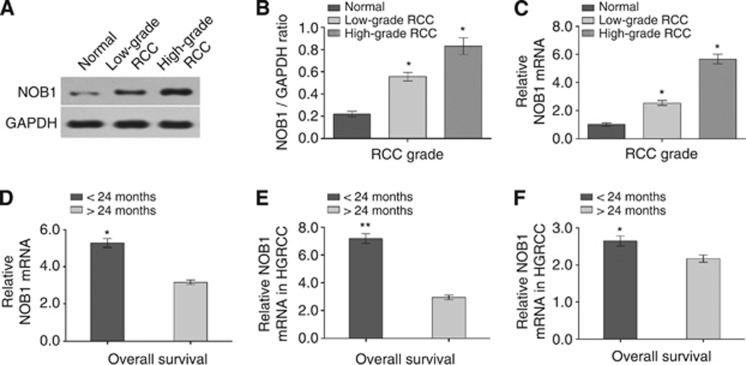
The expression of NOB1 in renal cancer tissues and overexpression correlated with poor prognosis
The expression of NOB1 in high-grade renal cancer was significantly higher than in low-grade cancer and normal tissues (P<0.001 and P=0.011, respectively). NOB1 protein expression in RCC and normal tissues were detected by western blotting as shown in Figure 8A and B. As shown in Figure 8C, the expression of NOB1 mRNA in high-grade tumour was ∼5.9-fold (P=0.013) higher, whereas in the low-grade cancer, the expression was 2.3-fold (P=0.021) higher than that of tumour-adjacent normal tissues. The difference in NOB1 expression between the high-grade cancer and low-grade cancer was statistically significant (P=0.031). We then tested the correlation between NOB1 expression and the prognosis of ccRCC patients. Among the ccRCC patients analysed, those who lived >24 months (21 patients) had lower expression of NOB1, while those who lived <24 months (36 patients) had relatively higher expression of NOB1 (P<0.01, Figure 8D). Similar results were obtained after stratification of patients into high-grade ccRCC (P<0.01; Figure 8E) and low-grade tumour (P=0.026; Figure 8F). Our results showed that higher levels of NOB1 mRNA in ccRCC patients lead to a relatively shorter survival time.
Figure 8.
The expression levels of NOB1 in different grade RCC samples. (A and B) The expression of NOB1 protein was detected by western blotting (n=3). Quantity One analysis showed that NOB1 was significantly higher in high-grade RCC samples compared with low-grade RCC (P=0.011) and normal renal samples (P<0.001). (C) Data analysis revealed that NOB1 mRNA was upregulated both high-grade RCC and low-grade RCC tissue samples (P=0.013 and P=0.021, respectively) compared with normal renal tissues by quantitative RT–PCR. (D) RCC patients who lived >2-year survival (21 patients) showed reduced NOB1 mRNA expression, whereas patients who lived <2-year survival (36 patients) showed higher NOB1 mRNA expression in spite of RCC grade (P<0.01). (E) In high-grade RCC group, those who lived >24 months (nine patients) showed NOB1 mRNA expression was downregulation, further those who lived <24 months (21 patients) showed higher NOB1 mRNA expression (P<0.01). (F) In low-grade RCC group, patients lived >24 months (11 patients) showed NOB1 mRNA expression was lower, meanwhile those who lived <24 months (13 patients) showed higher NOB1 mRNA expression (P=0.026). The differences between groups were verified by Mann–Whitney U test. Values are presented as the mean±s.d. of at least three independent experiments performed in triplicate (*P<0.05, **P<0.01).
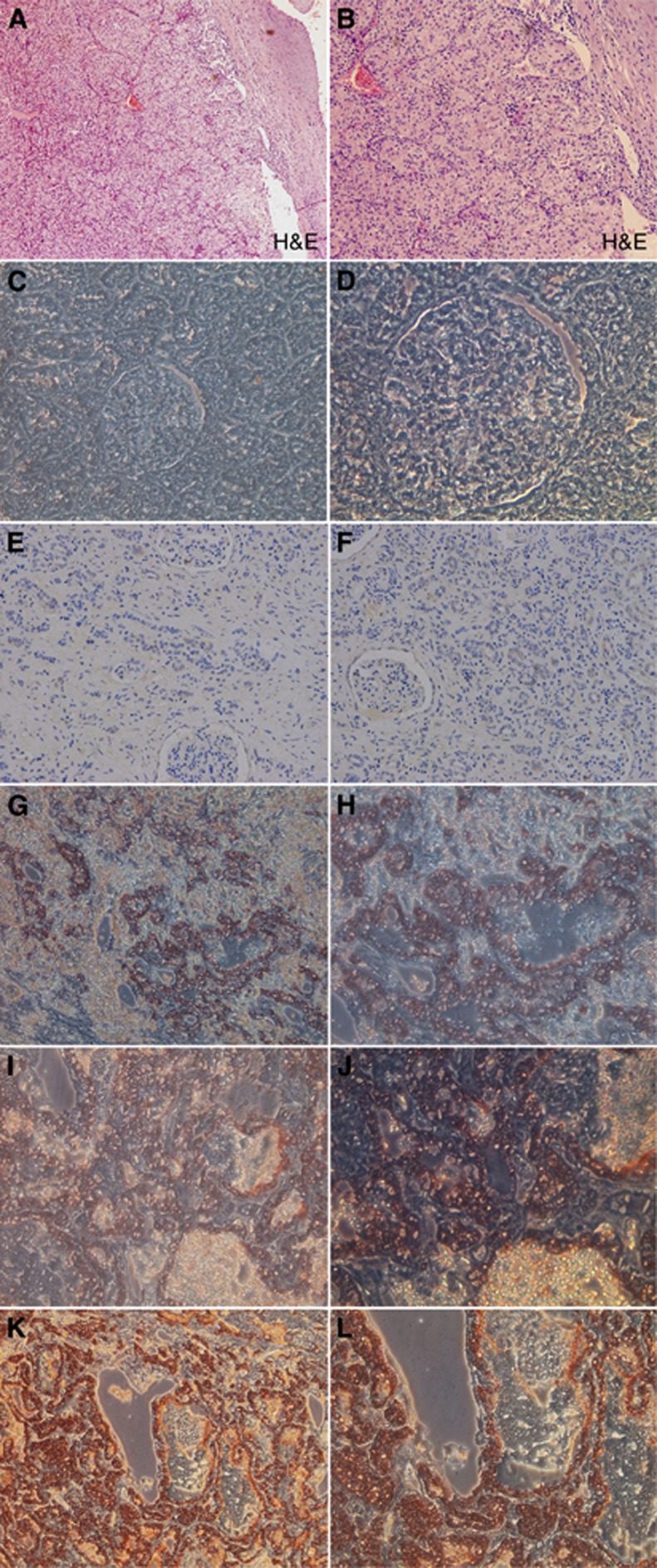
Further, immunohistochemical detection of NOB1 in renal cancer tissues (Figure 9) showed stronger staining for NOB1 in the high-grade cancer than in the low-grade cancer samples (P=0.005), whereas the tumour-adjacent normal tissues did not show significant NOB1 staining. The association between NOB1 expression and tumour grades in renal cancer strongly suggests that NOB1 might be involved in human ccRCC tumorigenesis.
Figure 9.
Immunohistochemical staining of NOB1 in RCC specimens and normal renal tissues. NOB1 immunohistochemical staining was performed on different types of RCCs and normal renal tissues, and representative images are shown. (A and B) H&E staining of RCC tissue (magnification, left × 100, right × 200). (C and D) Immunohistochemical staining of normal renal tissue, staining revealed that no significant staining was observed in normal renal tissues (magnification, left × 200, right × 400), and with the increasing of grade I–IV (E–L), renal tissue specimens expressing NOB1 protein. NOB1 staining was stronger in high-grade RCC than that in low-grade RCC.
Discussion
Small non-coding RNA has drawn more attention in the recent years due to their role in the gene transcriptional and post-transcriptional regulation. Nearly, 30% of gene expression in the human body is regulated by miRNAs (Shen et al, 2013). Recently, targeted therapies were developed to interfere with the transduction of key signalling pathways or to inhibit the function of tumour-specific molecules in malignant ccRCC (Cancer Genome Atlas Research Network, 2008; Coller et al, 2013).
MicroRNAs are unique in their ability to regulate many protein-coding genes. Increasing evidence suggests that miRNAs play an important role in the occurrence and development of many human cancers (Yi et al, 2010). The changes in miRNAs profiling are associated with almost all aspects of cancer biology such as cell proliferation, angiogenesis and invasion (Wu et al, 2012). The important roles of miRNAs in cancer suggest that they may be useful in clinical diagnosis and treatment. In the present study, we focussed on miR-646, which has been shown to suppress tumour growth in ccRCC. The downregulation of miR-646 in ccRCC was revealed to be associated with a feedback loop involving NOB1 that impaired renal cancer cell tumorigenicity.
In a previous report, NOB1, an essential protein, was found to be associated with the 26S proteasome and hence plays an essential role in universal biological processes. In this study, we showed that miR-646 inhibits tumorigenesis by blocking a novel target, NOB1, both in vitro and in vivo (Rosenzweig and Glickman, 2008). When the cell cycle of human renal cells was assessed by FACS, we observed that overexpression of miR-646 showed significant decrease in S-phase and an increase in G1-phase populations in the human renal cells, leading to a significant delay in cell proliferation. The growth inhibitory effect was observed by colony-forming and nude mouse xenograft assays, indicating that miR-646 and NOB1 are crucial for human ccRCC tumorigenesis. In addition, upregulation of NOB1 expression in human renal cancer tissue samples is related strongly to survival rate; the higher the level of NOB1, the shorter the overall survival of the patients, indicating that upregulated NOB1 plays an important role in the stages or grades of ccRCC. Our results are supported by datasets in Oncomine (www.oncomine.org). In the database, NOB1 was overexpressed in renal cancer compared to the normal kidney tissue. Also, in the dataset of French brain, NOB1 was overexpressed in human anaplastic oligodendroglioma compared to the normal brain. The results support the involvement of NOB1 in the tumorigenesis of different types of cancer.
MAPK signalling pathways can induce either cell proliferation or cell survival depending on the cell type and stimulus, the activation of the MAPK pathway has been associated with renal cancer proliferation (Salinas-Sánchez et al, 2012). The three main members that integrate the MAPK family in mammalian cells are ERK1/2, JNK and p38 MAPK, which are important in the control of cell differentiation, proliferation and apoptosis (Yao et al, 2008; Nissan et al, 2013). We hypothesised that the induction of early apoptosis by NOB1 downregulation in renal cancer cells might be related to the MAPK signalling pathway. Our results indicate that silencing of NOB1 expression increased the phosphorylation of these proteins, suggesting that the anti-tumour effect of NOB1 might be mediated by MAPK activation. NOB1 protein is a key factor linking the proteasome and cellular growth, and therefore investigation of the NOB1 function will shed some light on the mechanism of growth control by the ubiquitin–proteasome pathway (Fasanaro et al, 2010). Upregulating miR-646 or downregulating NOB1 expression can both inhibit the activation of the proteasome. Perhaps the inhibition of the proteasome leads to stabilisation of proteins that increase phosphorylation of the three key components in the MAPK pathway.
Recently, several profiling analyses reported that miRNAs were involved in tumorigenesis and metastasis in ccRCC (Ramana, 2012; Zhang et al, 2012). Interestingly, miR-15 family members were found to play critical role in pronephros development or apoptosis in RCC (Wessely et al, 2010). Moreover, miR-15 family members were downregulated in ccRCC and further downregulated in metastatic RCC (Jung et al, 2009; Heinzelmann et al, 2011). The result of our study showed that downregulation of miR-646 was correlated with high metastasis ratio in ccRCC. The aberrant expression of miR-646 protein linked to poor prognosis of patients has not been investigated in renal cancer before. Their molecular mechanisms are poorly understood and need further investigation.
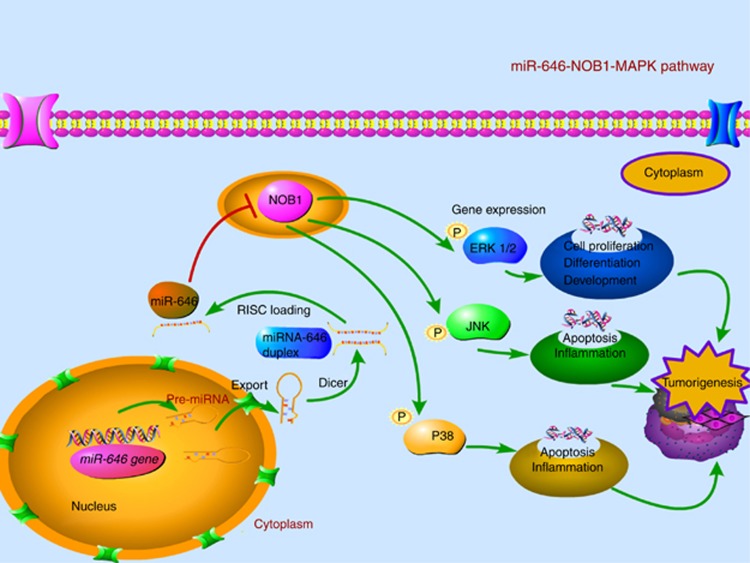
In conclusion, our studies have demonstrated that NOB1 as a novel target of miR-646. Overexpression of miR-646 decreased tumorigenesis of renal cancer cells through the modulation of MAPK pathway. Clinically, NOB1 expression might be associated with tumour grade as well as the prognosis of ccRCC. As depicted in Figure 10, the interplay among miR-646, NOB1 and the MAPK pathway scratched the surface of the terra incognita was shown. Our findings suggest that exogenous overexpression of miR-646 may be considered as a promising strategy for targeted therapies in renal cancer.
Figure 10.
Abridged general view for the interplay among miR-646, NOB1 and the MAPK pathway in ccRCC. miR-646 as a tumour suppressor by targeting NOB1, which decreased the tumorigenesis of RCC cells in vivo and in vitro through the modulation of the MAPK pathway. Overexpression of miR-646, which suppresses the expression of NOB1, activates the MAPK pathway by increasing the phosphorylation of ERK1/2, JNK and p38 MAPK, which inhibits cell proliferation and induced apoptosis in human ccRCC.
Acknowledgments
The work was partially supported by grants from the National Natural Science Foundation (No. 81000311 and No. 81270831), People's Republic of China. The funding agency had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Aguda BD. Modeling microRNA-transcription factor networks in cancer. Adv Exp Med Biol. 2013;774:149–167. doi: 10.1007/978-94-007-5590-1_9. [DOI] [PubMed] [Google Scholar]
- Ai Q, Ma X, Huang Q, Liu S, Shi T, Zhang C, Zhu M, Zhang Y, Wang B, Ni D, Li H, Zheng T, Zhang X. High-level expression of Notch1 increased the risk of metastasis in T1 stage clear cell renal cell carcinoma. PLoS One. 2012;7:e35022. doi: 10.1371/journal.pone.0035022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanaro P, Capogrossi MC, Martelli F. Regulation of the endothelial cell cycle by the ubiquitin-proteasome system. Cardiovasc Res. 2010;85:272–280. doi: 10.1093/cvr/cvp244. [DOI] [PubMed] [Google Scholar]
- Fendler A, Jung K. MicroRNAs as new diagnostic and prognostic biomarkers in urological tumors. Crit Rev Oncog. 2013;18:289–302. doi: 10.1615/critrevoncog.2013007176. [DOI] [PubMed] [Google Scholar]
- Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatsiou A, Boeckel JN, Randriamboavonjy V, Stellos K. MicroRNAs in platelet biogenesis and function: implications in vascular homeostasis and inflammation. Curr Vasc Pharmacol. 2012;10:524–531. doi: 10.2174/157016112801784611. [DOI] [PubMed] [Google Scholar]
- Gelb AB. Renal cell carcinoma: current prognostic factors. Union Internationale Contre le Cancer (UICC) and the American Joint Committee on Cancer (AJCC) Cancer. 1997;80:981–986. [PubMed] [Google Scholar]
- Gupta K, Miller JD, Li JZ, Russell MW, Charbonneau C. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat Rev. 2008;34:193–205. doi: 10.1016/j.ctrv.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Heinzelmann J, Henning B, Sanjmyatav J, Posorski N, Steiner T, Wunderlich H, Gajda MR, Junker K. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J Urol. 2011;29:367–373. doi: 10.1007/s00345-010-0633-4. [DOI] [PubMed] [Google Scholar]
- Hillman GG, Droz JP, Haas GP. Experimental animal models for the study of therapeutic approaches in renal cell carcinoma. In Vivo. 1994;8:77–80. [PubMed] [Google Scholar]
- Huang ZM, Yang J, Shen XY, Zhang XY, Meng FS, Xu JT, Zhang BF, Gao HJ. MicroRNA expression profile in non-cancerous colonic tissue associated with lymph node metastasis of colon cancer. J Dig Dis. 2009;10:188–194. doi: 10.1111/j.1751-2980.2009.00384.x. [DOI] [PubMed] [Google Scholar]
- Hutson TE, Figlin RA. Renal cell cancer. Cancer J. 2007;13:282–286. doi: 10.1097/PPO.0b013e318156fe69. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jung M, Mollenkopf HJ, Grimm C, Wagner I, Albrecht M, Waller T, Pilarsky C, Johannsen M, Stephan C, Lehrach H, Nietfeld W, Rudel T, Jung K, Kristiansen G. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med. 2009;13:3918–3928. doi: 10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller CM, Kim Y, Schmidt-Wolf IG. Targeting renal cancer with a combination of WNT inhibitors and a bi-functional peptide. Anticancer Res. 2013;33:2435–2440. [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Motzer RJ, Escudier B, Bukowski R, Rini BI, Hutson TE, Barrios CH, Lin X, Fly K, Matczak E, Gore ME. Prognostic factors for survival in 1059 patients treated with sunitinib for metastatic renal cell carcinoma. Br J Cancer. 2013;108:2470–2477. doi: 10.1038/bjc.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan MH, Rosen N, Solit DB. ERK Pathway Inhibitors: how low should we go. Cancer Discov. 2013;3:719–721. doi: 10.1158/2159-8290.CD-13-0245. [DOI] [PubMed] [Google Scholar]
- Ramana J. RCDB: Renal Cancer Gene Database. BMC Res Notes. 2012;5:246. doi: 10.1186/1756-0500-5-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R, Glickman MH. Chaperone-driven proteasome assembly. Biochem Soc Trans. 2008;36:807–812. doi: 10.1042/BST0360807. [DOI] [PubMed] [Google Scholar]
- Santarpia L, Nicoloso M, Calin GA. MicroRNAs: a complex regulatory network drives the acquisition of malignant cell phenotype. Endocr Relat Cancer. 2010;17:F51–F75. doi: 10.1677/ERC-09-0222. [DOI] [PubMed] [Google Scholar]
- Salinas-Sánchez AS, Giménez-Bachs JM, Serrano-Oviedo L, Nam Cha S, Sánchez-Prieto R. Role of mitogen-activated protein kinase (MAPK) in the sporadic renal cell carcinoma. Actas Urol Esp. 2012;36:99–103. doi: 10.1016/j.acuro.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Shen J, Stass SA, Jiang F. MicroRNAs as potential biomarkers in human solid tumors. Cancer Lett. 2013;329:125–136. doi: 10.1016/j.canlet.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Li Y, Zhou J, Xu J, Peng C, Ye F, Shen Y, Lu W, Wan X, Xie X. miR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. Am J Pathol. 2011;179:2580–2588. doi: 10.1016/j.ajpath.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM, Li Y, Nelson RA, Mu B, Onami SH, Wu JJ, Ruel NH, Wilczynski SP, Gao H, Covarrubias M, Figlin RA, Weiss LM, Wu H. Identification of a 4-microRNA signature for clear cell renal cell carcinoma metastasis and prognosis. PLoS One. 2012;7:e35661. doi: 10.1371/journal.pone.0035661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessely O, Agrawal R, Tran U. MicroRNAs in kidney development: lessons from the frog. RNA Biol. 2010;7:296–299. doi: 10.4161/rna.7.3.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YQ, Ding X, Jia YC, Huang CX, Wang YZ, Xu YH. Anti-tumor effect of beta-elemene in glioblastoma cells depends on p38 MAPK activation. Cancer Lett. 2008;264:127–134. doi: 10.1016/j.canlet.2008.01.049. [DOI] [PubMed] [Google Scholar]
- Yi Z, Fu Y, Zhao S, Zhang X, Ma C. Differential expression of miRNA patterns in renal cell carcinoma and nontumorous tissues. J Cancer Res Clin Oncol. 2010;136:855–862. doi: 10.1007/s00432-009-0726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Guo Y, Shang C, Song Y, Wu B. miR-21 downregulated TCF21 to inhibit KISS1 in renal cancer. Urology. 2012;80:1298–1302. doi: 10.1016/j.urology.2012.08.013. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ni J, Zhou G, Yuan J, Ren W, Shan Y, Tang W, Yu L, Zhao S. Cloning, expression and characterization of the human NOB1 gene. Mol Biol Rep. 2005;32:185–189. doi: 10.1007/s11033-005-3141-7. [DOI] [PubMed] [Google Scholar]