Abstract
Background:
Key challenges of biopsy-based determination of prostate cancer aggressiveness include tumour heterogeneity, biopsy-sampling error, and variations in biopsy interpretation. The resulting uncertainty in risk assessment leads to significant overtreatment, with associated costs and morbidity. We developed a performance-based strategy to identify protein biomarkers predictive of prostate cancer aggressiveness and lethality regardless of biopsy-sampling variation.
Methods:
Prostatectomy samples from a large patient cohort with long follow-up were blindly assessed by expert pathologists who identified the tissue regions with the highest and lowest Gleason grade from each patient. To simulate biopsy-sampling error, a core from a high- and a low-Gleason area from each patient sample was used to generate a ‘high' and a ‘low' tumour microarray, respectively.
Results:
Using a quantitative proteomics approach, we identified from 160 candidates 12 biomarkers that predicted prostate cancer aggressiveness (surgical Gleason and TNM stage) and lethal outcome robustly in both high- and low-Gleason areas. Conversely, a previously reported lethal outcome-predictive marker signature for prostatectomy tissue was unable to perform under circumstances of maximal sampling error.
Conclusions:
Our results have important implications for cancer biomarker discovery in general and development of a sampling error-resistant clinical biopsy test for prediction of prostate cancer aggressiveness.
Keywords: prostate cancer, biopsy, biomarkers, prognosis, sampling error, tumour heterogeneity
Prostate cancer accounts for 27% of incident cancer diagnosed in men in the USA. The American Cancer Society estimates that, nationally, 233 000 new diagnoses of prostate cancer will be made in 2014 (Siegel et al, 2014). Of all men newly diagnosed with prostate cancer, only about one in seven will progress to metastatic disease over a lifetime, whereas approximately half of them will have localised disease that has a very low risk of progression (Carter et al, 2012; Siegel et al, 2014). Despite this low risk, as many as 90% of these low-risk prostate cancer patients in the United States undergo radical treatment, usually radical prostatectomy (RP) or ablative radiation therapy (Cooperberg et al, 2010). Such treatments for low-risk patients may be excessive and often result in long-term adverse events, including urinary incontinence and erectile and bowel dysfunction (Wilt et al, 2008; Moyer and U.S. Preventative Services Task Force, 2012; Loeb et al, 2014).
Current guidance and accepted standards of care for the diagnosis and management of prostate cancer recommend the use of clinical and pathological parameters to assess the disease grade and stage on biopsy (NCCN, 2012a; NCCN, 2012b). Pathological evaluation of tissue obtained by needle biopsy is essential both to confirm a prostate cancer diagnosis and to grade the cancer. Tumour grade, as determined by the biopsy Gleason score (GS) is the most important predictor of outcome, and considered the most informative parameter for guiding management decisions. The GS is comprised of two Gleason patterns, with the more prevalent pattern specified first. The two are summed to determine the GS. According to a 2005 consensus on Gleason scoring, only three patterns (3, 4, and 5) are typically recognised on biopsy (Epstein et al, 2005). The accepted prognostic categories of GS are 3+3=6, 3+4=7, 4+3=7, 8, and 9–10. Importantly, although 3+4=7 and 4+3=7 have equivalent Gleason sums, the latter has significantly worse prognosis based on a higher amount of pattern 4 (Goodman et al, 2012; Reese et al, 2012). Approximately 80–85% of all prostate cancer biopsies have a GS of 3+3=6 or 3+4=7, representing a spectrum of cases with low to intermediate to high risk of progression (Epstein et al, 2012). Patients deemed to have indolent disease are candidates for active surveillance (AS) (Mohler et al, 2012; NCCN, 2012a; NCCN, 2012b). However, current methods of biopsy evaluation are often unable to place individual patients accurately along this spectrum (Cooperberg et al, 2010; Epstein et al, 2012).
Two recognised factors affect the accuracy of biopsy-based Gleason scoring: one is sampling variation (i.e., failing to sample the area with the highest Gleason grade), and the second is pathologist discordance in Gleason scoring (Porten et al, 2011; Corcoran et al, 2012; Epstein et al, 2012). Despite the current standard practice of multicore biopsy sampling, the most aggressive area of the tumour is frequently under-represented or over-represented (Corcoran et al, 2012; Bjurlin and Taneja, 2014). Indeed, 25–50% of cases of prostate cancer need to be either upgraded or downgraded from their initial biopsy score to a more accurate surgical GS after analysis and grading of prostatectomy tissue (Kvale et al, 2009; Davies et al, 2011; Epstein et al, 2012). Discordance between pathologists in Gleason grading can be as high as 30%, adding significantly to the difficulty of ensuring uniform and accurate prognostication (McKenney et al, 2011; Goodman et al, 2012).
Several clinical and pathological risk stratification systems have been developed to improve prediction of prostate cancer aggressiveness, including the D'Amico classification system, the Cancer of the Prostate Risk Assessment score, and the National Comprehensive Cancer Network guidelines (Cooperberg et al, 2005; D'Amico et al, 1998; NCCN, 2012a; Vellekoop et al, 2014). In addition, clinical nomogram systems such as the widely adopted systems described by Kattan and Steyerberg have been developed to predict indolent prostate cancer and prostate cancer outcome(Steyerberg et al, 2007). All such systems recognise the biopsy GS as the single most powerful variable in risk assessment. Importantly, all of the risk stratification systems used to guide clinical management depend on effective and consistent Gleason scoring and are therefore vulnerable to sampling variation and discordant scoring by pathologists.
Enhanced biopsy strategies have been proposed as one means to overcome sampling variation and errors. Among these, increasing the number or density of sampled cores might ensure more representative capture of tumour tissue. However, this could potentially increase the risk of adverse events from oversampling, and there is little evidence that it improves pathological classification (Eichler et al, 2006; Delongchamps et al, 2009). There has also been interest in novel forms of image-guided biopsy. Currently, MRI-guided biopsy appears to improve detection of aggressive cancers, but long-term studies will be needed to determine whether it can improve patient selection for AS (Robertson et al, 2013).
Using a quantitative multiplex proteomics in situ imaging system, which enables accurate biomarker measurements from the intact tumour epithelium (Shipitsin et al, 2014), we here report the identification and evaluation of 12 biomarkers that are able to predict prostate cancer aggressiveness and lethal outcome. Importantly, the markers were specifically selected to be robust to sampling error. The study was designed to simulate biased biopsy-sampling error based on coring from areas of high and low GS on prostatectomy tissue from each patient to generate ‘high' (H) and ‘low' (L) tissue microarrays (TMAs), respectively. Biomarkers were then selected based on their ability to reflect true prostate pathology and lethal outcome, regardless of whether they were measured on cores with high or low GS. This performance-based approach not only identified novel biomarkers, but also confirmed known biomarkers predictive of prostate cancer aggressiveness and lethal outcome.
Materials and methods
Reagents and antibodies
All antibodies and reagents used in this study were procured from commercially available sources as described in Supplementary Table S4. Anti-fluorescein isothiocyanate MAb–Alexa 568, anti-CK8–Alexa 488, anti-CK18–Alexa 488, anti-CK5–Alexa 555, and anti-Trim29–Alexa 555 were conjugated with Alexa dyes using the appropriate protein conjugation kits (Life Technologies, Grand Island, NY, USA).
Slide processing and staining protocol
From TMA blocks, 5-μm sections were cut, placed on Histogrip (Life Technologies)-coated slides, and processed as described previously (Supplementary Materials). Briefly, after deparaffinisation, antigen retrieval was performed in 0.05% citraconic anhydride solution for 45 min at 95 °C using a Lab Vision PT module (Thermo Scientific, Waltham, MA, USA). Staining was performed either manually or in automated fashion with an Autostainer 360 or 720 (Thermo Scientific).
The quantitative multiplex immunofluorescence (QMIF) staining procedure that combined two anti-biomarker antibodies with region-of-interest markers was performed as previously described (see Supplementary Materials and Methods).
Acquisition, processing, quality control, and annotation of FFPE prostate cancer tissue blocks
A set of FFPE human prostate cancer tissue blocks with clinical annotations and long-term patient outcome information was acquired from Folio Biosciences (Powell, OH, USA). Information about prostate cancer-specific deaths was obtained from either patient records or death certificates. Samples had been collected with appropriate institutional review board approval and all patient records were de-identified. For evaluation of candidate biomarker antibodies, FFPE human prostate cancer tissue blocks with limited clinical annotation were acquired from other commercial sources.
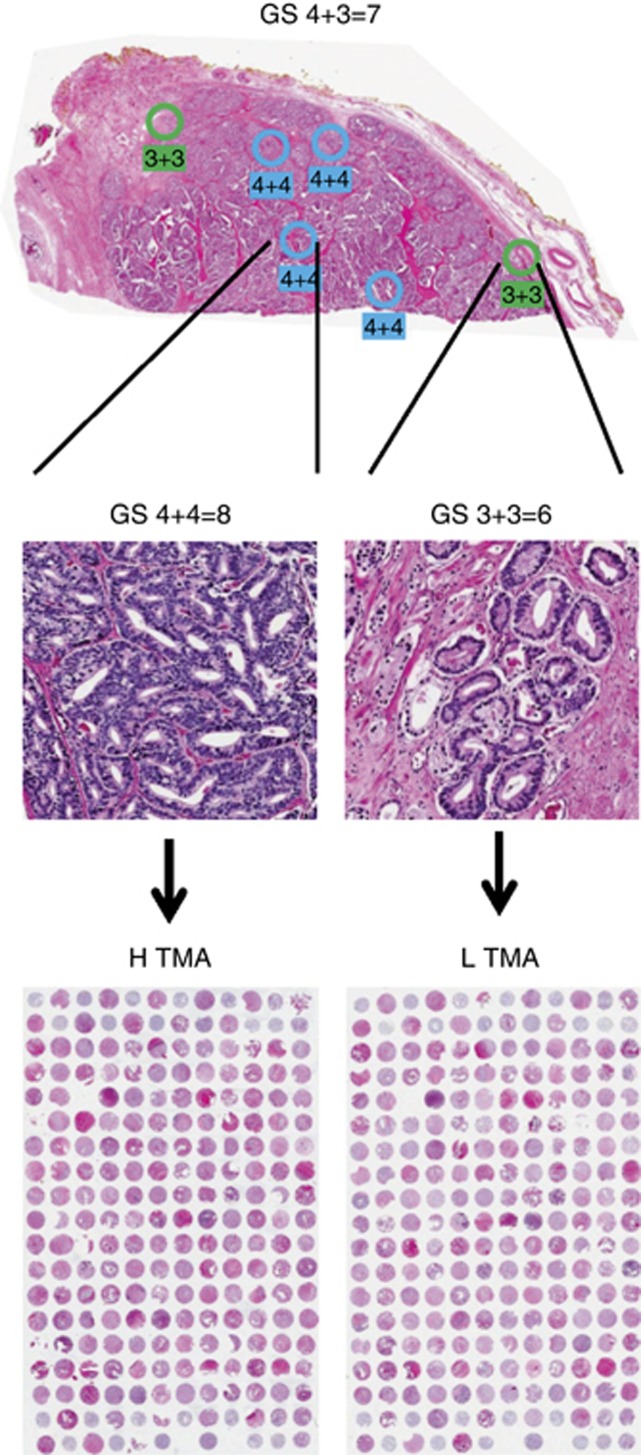
A series of 5-μm sections was cut from each FFPE block. For annotation, a 5-μm section that was the last to be cut from each FFPE block was stained with haematoxylin and eosin and scanned using a ScanScope XT system (Aperio, Buffalo Grove, IL, USA). The scanned images were remotely reviewed and annotated for GS in a blinded manner by expert clinical board-certified anatomical pathologists. Circles corresponding to 1-mm diameter cores were placed over four areas of highest and two areas of lowest Gleason patterns (see Figure 1, top).
Figure 1.
Creation of biopsy simulation tissue microarrays (TMAs). A tissue block from a prostatectomy sample was annotated with all visible Gleason patterns (top). The example shown is from a patient with an overall Gleason score (GS) of 4+3=7. As shown in a higher-magnification view (middle), patterns within the same block can be highly diverse. Two 1-mm cores were taken from each tissue block. One was taken from an area with the highest GS (4+4=8) and embedded into agarose/paraffin along with high-scoring cores from other blocks to create the H TMA (bottom left). The other was taken from an area with the lowest GS (3+3=6) and embedded into agarose/paraffin along with low-scoring cores from other blocks to create the L TMA (bottom right).
Generation of TMA blocks
TMA blocks were prepared using a modified agarose block procedure (Yan et al, 2007). See Supplementary Materials for further details.
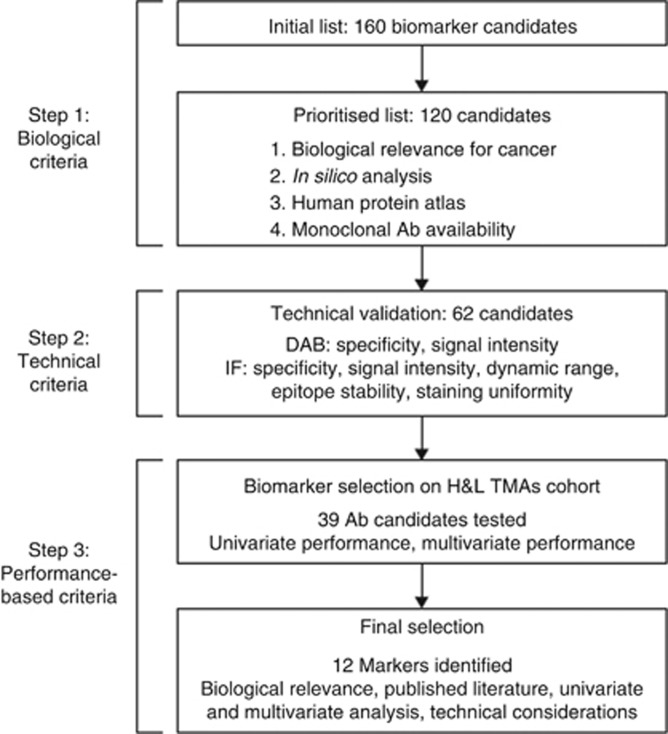
Biomarker selection
To identify biomarkers for prostate cancer aggressiveness, we developed a selection and evaluation process that could be broadly applicable across diseases and conditions. The process, shown in Figure 2, had biological, technical, performance, and validation stages. See Supplementary Methods for further details.
Figure 2.
Biomarker selection strategy. Three types of criteria were used to select 12 final biomarkers. (DAB: Ab specificity assessed based on chromogenic tissue staining with diamino benzidine (DAB); IF: Ab specificity and performance based on immunofluorescent tissue staining).
Image acquisition
Two Vectra Intelligent Slide Analysis Systems (PerkinElmer, Waltham, MA, USA) were used for automated image acquisition as described (Supplementary Materials and Methods). Multispectral images were processed into images for each separate fluorophore signal and sent for analysis with Definiens Developer script (Definiens AG, München, Germany).
Definiens automated image analysis
We developed an automated image analysis algorithm using Definiens Developer XD for tumour identification and biomarker quantification as described in Supplementary Materials.
Data stratification and end points in the analysis
Expression of 39 biomarkers was examined for correlation with tumour aggressiveness and lethality using the H and L TMAs. Disease aggressiveness was defined based on prostate pathology (aggressive disease=surgical Gleason ⩾3+4 or T3b, N+, or M+). For aggressiveness analyses, we examined marker correlation based on measurements in both L TMA samples with core Gleason ⩽3+4 and the corresponding, matched H TMA samples.
For lethal outcome analyses, we created two different sample sets: (1) all cores with an observed GS ⩽3+4; and (2) all cores.
Cohort composition
Table 1a presents the cohort composition. Only those samples that had a complete set of clinical information were included. When performing an analysis using a certain set of biomarkers, only samples with values for those markers were considered. Hence, the numbers in the table are upper bounds.
Table 1a. Clinical features of the cohort used to create L and H TMAs. A single cohort of 380 patients provided samples for the two TMAs.
| L TMA | H TMA | |
|---|---|---|
| Patients with survival and biomarker information | 360 of 380 | 363 of 380 |
| Mean age (s.d.), years | 62.2 (6.76) | 62.1 (6.83) |
| Lethal events, n (%) | 60 (16.67) | 59 (16.25) |
| Mean length of follow-up (s.d.), years | 11.55 (3.96) | 11.52 (3.98) |
|
Pathological tumour stage, n (%) | ||
| T2 | 244 (67.8) | 250 (68.9) |
| T3 | 112 (31.1) | 109 (20.0) |
| T4 | 2 (0.56) | 2 (0.55) |
| Missing | 2 (0.56) | 2 (0.55) |
|
Core Gleason score, n (%) | ||
| ⩽ 6 | 233 (64.7) | 177 (48.8) |
| 3+4 | 68 (18.9) | 98 (27.0) |
| 4+3 | 15 (4.2) | 31 (8.5) |
| 8–10 | 27 (7.4) | 47 (13) |
| Total | 343 (95) | 353 (97) |
|
Surgical Gleason score, n (%) | ||
| ⩽6 | 108 (30) | 112 (31) |
| 3+4 | 169 (47) | 138 (46) |
| 4+3 | 30 (8.3) | 30 (8.3) |
| 8–10 | 53 (15) | 53 (15) |
| Total | 360 (100) | 363 (100) |
|
Deaths stratified by surgical Gleason score, n (% of disease deaths, % of Gleason stratun) | ||
| ⩽6 | 2 (3.3, 1.9) | 3 (5.1, 2.7) |
| 3+4 | 20 (33.3, 11.8) | 17 (28.8, 10.1) |
| 4+3 | 9 (15, 30) | 10 (17, 33.3) |
| 8–10 | 29 (48.3, 54.7) | 29 (49.1, 54.7) |
| Total | 60 | 59 |
Abbreviations: H TMA=high-grade tissue microarray; L TMA=low-grade tissue microarray; TMA=tissue microarray.
For technical reasons, only 360 samples on the L TMA and 363 samples on the H TMA were usable.
Univariate analysis of aggressiveness and lethality
Our objectives for univariate analysis were two-fold: to characterise univariate behaviour as a performance assessment for potential inclusion in the final marker set, and to provide a reduced set of markers for exhaustive multivariable model exploration. See Supplementary Materials for details.
Biomarker ranking for aggressiveness via exhaustive search of multimarker models
We sought to rank the biomarkers by importance in multimarker models; 31 biomarkers, refined from the original set of 39 to improve technical performance further, were used in an exhaustive biomarker search. We considered all combinations of up to five biomarkers from the 31 biomarkers tested in the L TMA in the H and L TMA analysis. See Supplementary Materials for further details.
Biomarker ranking for lethality via exhaustive search of multimarker models
The same model-building approach done for aggressiveness was followed for the biomarker ranking for prediction of lethality. Supplementary Table S3 shows frequency of biomarker utilisation (top 5%) for lethality.
Results
Biopsy simulation
Our first goal was to develop a biopsy-sampling model to simulate and exaggerate the biopsy sample variation observed in clinical practice. For this purpose, we embedded cores from annotated prostatectomy tissue into TMAs. On the basis of centralised Gleason grading by expert urologic pathologists, a core was taken for each patient from the area with the least aggressive tumour (low GS) to generate a low-grade TMA (L TMA); in parallel, a core was taken from the area with the most aggressive tumour based on Gleason grading (high GS) to generate a high-grade TMA (H TMA) (Figure 1). Thus, we developed paired tissue TMAs with samples simulating biopsies biased in two directions, representing both more and less aggressive tumour areas from each patient.
Table 1a describes the clinical features for the multi-institution cohort of 380 patients for whom paired TMAs were prepared. Table 1b describes the subset of 301 cases with core Gleason of 3+3 or 3+4 on L TMA along with their corresponding core Gleason on H TMA and their surgical (prostatectomy) Gleason.
Table 1b. Clinical features of the cohort used to create L and H TMAs. The distribution of H TMA core Gleason scores and surgical Gleason scores among the 301 patients with L TMA core Gleason of 3+3 or 3+4.
| Core Gleason score | L TMA: n of patients | Core Gleason score | H TMA: n of patients | Surgical Gleason score | n of patients |
|---|---|---|---|---|---|
| 3+3=6 | 233 | 3+3=6 | 149 | 3+3=6 | 93 |
| 3+4=7 | 58 | 3+4=7 | 112 | ||
| ⩾ 4+3=7 | 26 | ⩾ 4+3=7 | 30 | ||
| 3+4=7 | 68 | 3+3=6 | 23 | 3+3=6 | 14 |
| 3+4=7 | 27 | 3+4=7 | 32 | ||
| ⩾4+3=7 | 18 | ⩾ 4+3=7 | 22 |
Abbreviations: H TMA=high-grade tissue microarray; L TMA=low-grade tissue microarray.
Sampling for the L TMA was specifically designed to underestimate disease severity. As shown in Tables 1a and b, 64.7% of L TMA samples had a core GS ⩽6, whereas only 30% of these L TMA samples came from patients with a surgical GS ⩽6. The probability of upgrade (Table 1b) for samples in the L TMA from cases with core GS of ⩽3+4 to a higher surgical GS was 0.64 (95% Wilson confidence interval (CI): 0.59–0.69). This probability of upgrade is higher than that seen in clinical practice (Porten et al, 2011), as expected from the sampling method and patient cohort used. Thus, by exaggerating sample variation expected in clinical practice, this biopsy simulation procedure provided a useful model to identify biomarkers that reliably predict prostate cancer aggressiveness, regardless of sample variation.
Effect of sampling error on known biomarker model performance
Prior studies have demonstrated that RP GS of seven or higher and extension of prostate cancer beyond the prostate gland are significant predictors of metastasis and prostate cancer-specific mortality (Ross et al, 2012; Shikanov and Eggener, 2012; Brimo et al, 2013). Accordingly, we defined ‘aggressive disease' based on the prostate pathology as surgical GS of at least 3+4 or pT3b (seminal vesicle invasion), N+, or M+. We tested the four-biomarker model (SMAD4, CCND1, SPP1, and PTEN) previously reported by Ding et al (2011) for its ability to predict both disease-specific death and disease aggressiveness in our sampling variation TMA cohort. Using logistic regression analysis on the patient cores in the L and H TMAs, marker coefficients and AUC were estimated on the testing set. As shown in Table 2, when measured on H TMA patient cores, the four-marker signature was able to predict disease-specific death with a median test AUC of 0.65 (95% CI of 0.59–0.74). However, when measured on L TMA, representing biased underestimation of the surgical GS, the four-marker model showed a non-significant median test AUC of 0.49 (95% CI of 0.42–0.58). Moreover, the four-marker signature was unable to predict aggressive disease in either H or L TMA (median test AUC of 0.56 (95% CI of 0.44–0.64) and of 0.56 (95% CI of 0.46–0.65), respectively). These results demonstrate the impact of sampling error on prognostic marker performance, and support the necessity of identifying alternative biomarker combinations that can predict outcomes accurately regardless of sampling variation.
Table 2. Sampling variation reduces the performance of an established lethal outcome-predictive biomarker signature.
| Markers: PTEN, SMAD4, CCND1, SPP1 | Mean AIC (2.5%, 97.5%) | Median train AUC (2.5%, 97.5%) | Median test AUC (2.5%, 97.5%) |
|---|---|---|---|
| H TMA lethal | 282.2 (275.9, 293.2) | 0.67 (0.64, 0.70) | 0.65 (0.59, 0.74) |
| L TMA lethal | 301.3 (288.6, 316.8) | 0.6 (0.58, 0.63) | 0.49 (0.42, 0.58) |
| H TMA aggressiveness | 350.1 (330.4, 367.4) | 0.62 (0.56, 0.68) | 0.56 (0.44, 0.64) |
| L TMA aggressiveness | 381.6 (353.0, 400.7) | 0.61 (0.55, 0.68) | 0.56 (0.46, 0.65) |
Abbreviations: AIC=Akaike information criterion; ROC=receiver-operating characteristic curve; TMA=tissue microarray.
The combination PTEN+SMAD4+CCND1+SPP1 has previously been shown to be prognostic for lethal outcome when measured on prostatectomy tissue. We confirmed that these markers are indeed predictive of lethal outcome when measured in the high-Gleason biopsy simulation tissue (H TMA). However, these markers are unable to predict lethality in the low-Gleason simulation biopsy (L TMA). The markers do not show statistically significant predictive performance for aggressive disease regardless of whether they were measured in high- (H TMA) or low (L TMA)-Gleason tissue areas. C statistic, area under ROC curve.
Biomarker identification
We next embarked on identifying biomarkers that would robustly predict cancer aggressiveness in spite of biopsy-sampling variation. The stepwise approach involved: (1) identification of candidate biomarkers, (2) evaluation of their biological and technical suitability, and (3) analysis of performance in H and L TMA cohorts (Figure 2).
From a search of published literature and publicly available gene expression data sets, we identified 160 biomarker candidates based on biological relevance for prostate cancer (Lapointe et al, 2004; True et al, 2006; Lapointe et al, 2007; Tomlins et al, 2007; Cheville et al, 2008; Nakagawa et al, 2008; Graff et al, 2009; Makarov et al, 2009; Pressinotti et al, 2009; Gorlov et al, 2010; Taylor et al, 2010; Chen et al, 2011; Cima et al, 2011; Ding et al, 2011; Markert et al, 2011; Ross et al, 2011; Swanson and Quinn, 2011; Ding et al, 2012; Kristiansen, 2012). We further prioritised 120 of these markers based on availability of appropriate monoclonal antibodies (MAbs) (see Supplementary Table S1 for a comprehensive biomarker candidate list). Our candidate list included well-characterised markers relevant for prostate cancer aggressiveness, such as EZH2, MTDH, FOXA1 (Hu et al, 2009; Yang and Yu, 2013; Mills, 2014), as well as the markers PTEN, SMAD4, Cyclin D1, SPP1, phospho-PRAS40-T246 (pPRAS40), and phospho-S6-Ser235/236 (pS6) previously identified as predictive of lethal outcome on prostatectomy tissue (Ding et al, 2011; Shipitsin et al, 2014).
We next procured MAbs against these 120 prioritised candidates and tested them for specificity and suitability for the QMIF assay. Candidate MAbs were further selected on the basis of signal intensity and specific immunofluorescence-staining patterns, as described (Shipitsin et al, 2014; Supplementary Materials). Candidate biomarker antibodies were selected based on signals that were more stable relative to those of epithelial markers.
Subsequently, we tested the 62 MAbs that passed the previous steps and determined their dynamic range as well as their predictive performance. Using a small test TMA designed to represent the least aggressive areas from prostate tumours with high and low overall GSs, biomarkers were selected based on correlation of signal intensity with surgical GS. Specifically, we required minimally a three-fold difference of signals between lowest and highest expression values, in addition to the demonstrated difference in signal value distributions between nonaggressive and aggressive cases. The final 39 candidate MAbs that fulfilled these criteria were tested on the clinical cohort represented by H and L TMA blocks described above.
Univariate analysis
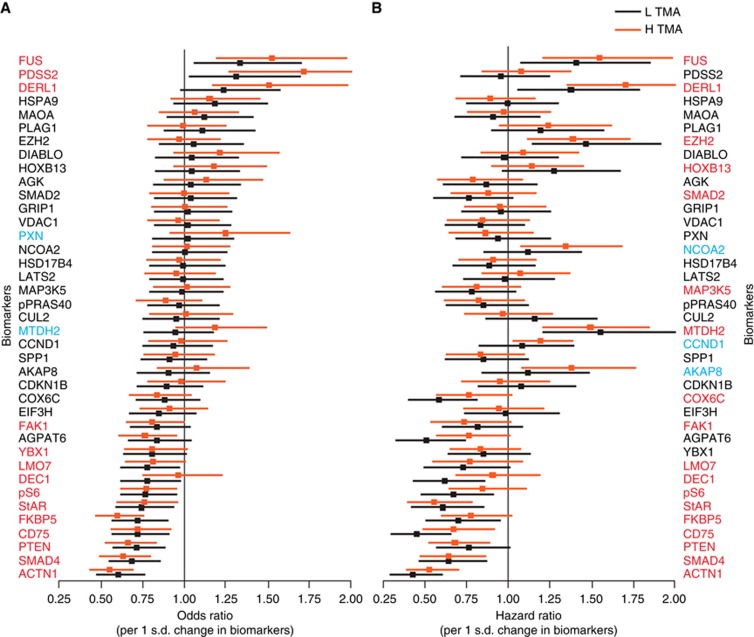
Each of the 39 biomarkers were tested for their ability to predict disease aggressiveness (surgical GS ⩾3+4 or pathological stage pT3b, and/or N+ or M+) and death from disease (survival analysis) when measured in either low- or high-Gleason areas (Figure 3). Markers shown in red text demonstrated predictive value (P<0.1) for aggressive disease or death from prostate cancer based on either an increased or decreased expression in both low- and high-Gleason areas. This result suggests that these markers are resistant to varying degrees of sampling error. There were two markers that were predictive of aggressiveness and three markers of lethal outcome when measured in high, but not in low-Gleason areas, indicating that these markers are not robust to sampling error. Conversely, no markers were identified that had predictive performance only in low, but not in high-Gleason areas. Interestingly, out of the 14 markers with significant univariate performance for aggressiveness, 12 markers also exhibited significant univariate performance for lethal outcome, consistent with a strong correlation between aggressive disease and lethal outcome. Our performance-based biomarker selection approach also confirmed correlation between lethal outcome and expression of three known prostate cancer progression markers, EZH2, HoxB13, and MTDH2, as previously reported (Hu et al, 2009; Yang and Yu, 2013; Mills, 2014).
Figure 3.
Univariate performance of 39 biomarkers measured in both low- (L TMA; black bars) and high-(H TMA; brown bars) Gleason areas for disease aggressiveness and disease-specific mortality. (A) The odds ratio (OR) for predicting severe disease pathology (aggressiveness) was calculated for each marker. Markers with an OR to the left of the vertical line are negatively correlated with the severity of the disease as assessed by pathology. Those to the right of the line are positively correlated. The markers were ranked based on OR when measured in L TMA. (B) The hazard ratio for death from disease (lethality) was calculated for each marker and plotted as described for A. Biomarkers in red indicate statistical significance at the 0.1 level in both L and H TMAs. Biomarkers in blue indicate statistical significance in only H TMA, but not L TMA. Note the large overlap of biomarkers with statistically significant univariate performance for both aggressive disease and death from disease.
Multivariate analysis: biomarker combinations predicting tumour aggressiveness
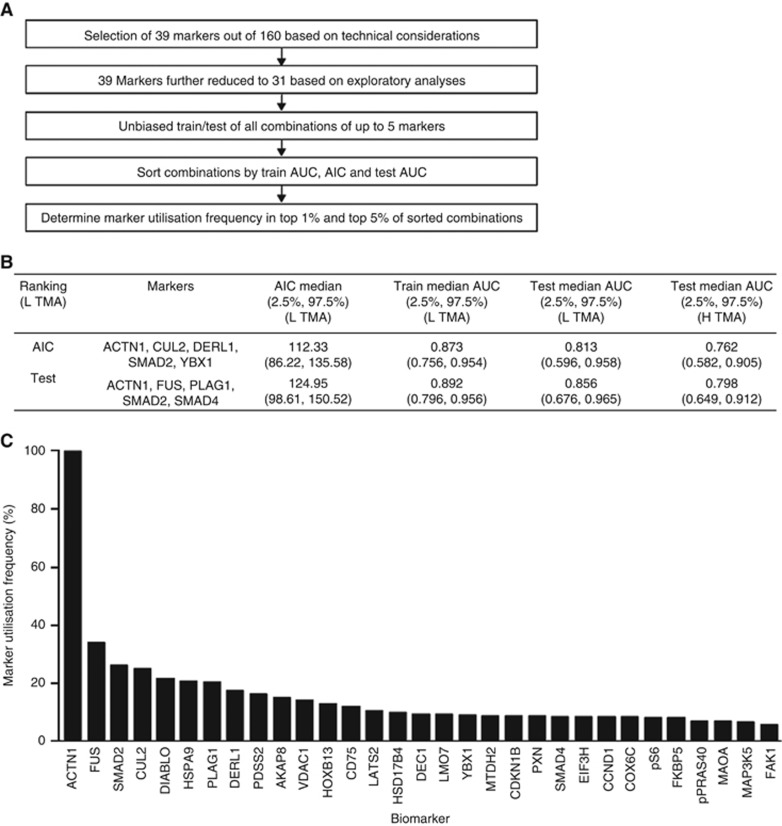
To explore the best multivariate biomarker combinations to predict disease aggressiveness, we exhaustively searched all possible models with combinations up to and including five biomarkers (Figure 4A). We focused on 31 biomarkers, further refined from the original set of 39 based on technical criteria (see Materials and Methods). Initially, an ‘extreme' model approach was used for the multivariate analysis, which included removal of ‘intermediate' samples (GS=3+4, ⩽T3a and N0) for the model building and testing. We separated patient cores in the L TMA into independent training and test sets, and tested the resulting models on both L and H TMAs for multivariate performance across sampling variation. For this purpose, we used logistic regression models to estimate biomarker coefficients using the training data set, estimated AUC from the resulting ROC in the testing set, and then repeated the process for another sampling.
Figure 4.
Performance-based biomarker selection process for disease aggressiveness. (A) The bioinformatics workflow selected the most frequently utilised biomarkers from all combinations of up to five markers from a set of 31. (B) Example of performance of top-ranked five-marker models, including comparison with training on L TMA and then testing on independent samples from L TMA and H TMA. Note that the test performances on L TMA and H TMA are consistent, with substantial overlap in confidence intervals. (C) Combinations were generated allowing a maximum of three, four, or five biomarkers. The figure shows the proteins most frequently included when five-biomarker models were used to predict aggressive disease, ranked by test.
In each case, the most frequently occurring biomarkers in the top 5 or 1% of the models, sorted by AIC (Akaike information criterion) (Lindsey and Jones, 1998) and test-set AUC, were determined. A final tally was generated encompassing ranking by test, ranking by AIC, and by both test and AIC (see Figure 4B for a representative example of a five-biomarker model ranked by AIC and test). Intriguingly, we observed a high degree of conservation of biomarker order in the top-performing biomarker models (see Figure 4C; Supplementary Table S2). The following biomarkers appeared among the top markers in at least 50% of the ranked lists: ACTN1, FUS, SMAD2, DERL1, YBX1, DEC1, pS6, HSPA9, HOXB13, PDSS2, SMAD4, and CD75. In addition, CUL2 was present in a number of highly ranked models (see Supplementary Table S2 for further details of the ranking results).
Multivariate analysis: biomarkers predicting lethal outcome
A similar modelling analysis to that applied to predict aggressiveness was performed for lethal outcome (Supplementary Table S3). Biomarkers appearing among top markers in at least 50% of the ranked lists included: MTDH2, ACTN1, COX6C, YBX1, SMAD2, DERL1, CD75, FUS, LMO7, PDSS2, FAK1, SMAD4, and DEC1 (see Supplementary Table S3 for further details of the ranking results).
Final biomarker set predictive of prostate cancer aggressiveness and lethality
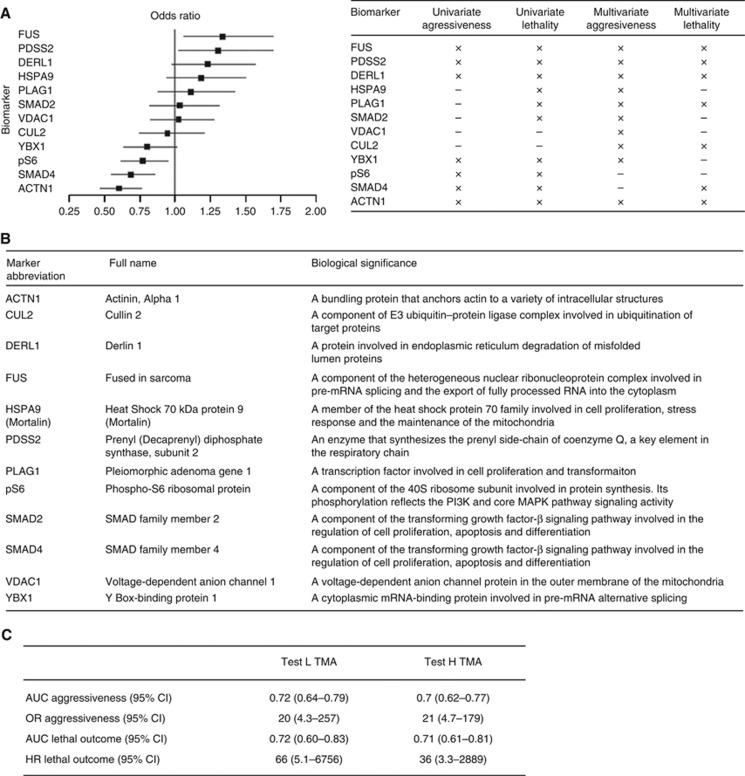
We chose a final set of 12 biomarkers based on careful integration of univariate and multivariate performance, and analytical considerations, including minimally a three-fold dynamic signal intensity range across tumour samples for all antibodies. Figure 5A shows the estimated odds ratios (ORs) associated with these 12 biomarkers for univariate prediction of aggressiveness, and Figure 5B provides a biological summary of the selected biomarkers. The final biomarker set was comprised of: FUS, PDSS2, DERL1, HSPA9, PLAG1, SMAD2, VDAC1, CUL2, YXB1, pS6, SMAD4, and ACTN1.
Figure 5.
Final biomarker set and selection criteria. (A) Twelve biomarkers were selected based on univariate performance for aggressiveness (shown as OR on left) and lethality as well as frequency of appearance in multivariate models for disease aggressiveness or lethal outcome (table on right) (B) The biomarker set comprises proteins known to function in the regulation of cell proliferation, cell survival, and metabolism. (C) A multivariate 12-marker model for disease aggressiveness was developed based on logistic regression. The resulting AUC and OR are shown. Subsequently, the risk scores generated by the aggressiveness model for all patients were correlated with lethal outcome. The resulting AUC and HR are shown.
Each of the 12 marker antibodies was rigorously validated by specificity analyses including western blotting and an immunohistochemistry assay before and after target-specific knockdown, as shown in Supplementary Figure S1.
Finally, we assessed the predictive potential of the final 12-biomarker set for both disease aggressiveness and disease-specific death on the entire patient cohort through logistic regression analyses as described (see Materials and Methods). As shown in Figure 5C, this resulted in an L TMA test AUC of 0.72 (95% CI: 0.64–0.79) and a corresponding OR for aggressive disease of 20 per unit change in risk score (95% CI: 4.3–257). To confirm the ability to generalise across sampling error, the model derived from the L TMA-training set was also tested on H TMA for prediction of aggressive disease with consistent results (Figure 5C). Without any further changes to the aggressiveness model, we examined its performance on lethal outcome prediction by correlating the aggressiveness risk scores with death from disease. Of note, we found a similar AUC for lethal outcome as for aggressiveness on both L and H TMA of 0.72 (95% CI: 0.60–0.83) and 0.71 (95% CI: 0.61–0.81), respectively. The corresponding HRs for lethal outcome on L and H TMAs were 66 per unit change (95% CI: 5.1–6756) and 36 (95% CI: 3.3–2889), respectively. We conclude that the 12 identified biomarkers are robust to sampling error and predictive of both disease aggressiveness and lethal outcome.
Discussion
There is a continuing clinical need to assess prostate cancer aggressiveness more accurately at the time of initial diagnosis as well as for follow-up of patients, both those under active patient surveillance and those receiving active treatment (Bangma and Roobol, 2012; Carter et al, 2012; Brimo et al, 2013). Currently, in men with early disease, a biopsy GS of 3+4=7 or more is one of the prognostic determinants for active treatment (NCCN, 2012a; Heidenreich et al, 2014). However, biopsy-sampling error resulting from tumour heterogeneity and discordant Gleason scoring can significantly affect the accuracy and reliability of assessing a patient's risk of cancer progression, aggressiveness, and lethality. This uncertainty in prognostication has contributed to significant overtreatment of patients with biopsies of Gleason grade 3+3 or 3+4 (Boorjian et al, 2009; Cooperberg et al, 2010; Bangma and Roobol, 2012; Epstein et al, 2012; Sandhu and Andriole, 2012; Loeb et al, 2014).
Biomarkers predictive of prostate cancer aggressiveness and lethality
In this manuscript, we describe the successful development of a performance-based strategy to identify and evaluate biomarkers predictive of prostate cancer aggressiveness and lethal outcome, even under circumstances of extreme sampling variation, an issue typically encountered during prostate biopsy taking. Using a large cohort (N=380) of annotated clinical prostatectomy samples with long-term follow-up, the areas of highest and lowest GS on each prostatectomy tissue were cored to generate paired TMAs (H and L TMAs) representing the entire cohort, thereby simulating biopsies with sampling error for each patient. Using these paired TMAs, we evaluated a large number of biomarker candidates for the ability to predict aggressive prostate pathology and lethal outcome when measured in either low- or high-grade cancer regions from each patient. We first selected biomarkers with performance against aggressiveness and lethal outcome when measured in L TMA tissue, to identify those most robust to extreme sampling error. For this purpose, we only included L TMA samples with core Gleason ⩽3+4 as clinically relevant, as biopsies with GS 4+3 or higher inevitably will be aggressive therapy candidates. Most of the identified biomarkers from univariate analyses were predictive of both disease aggressiveness and prostate cancer-specific mortality regardless of whether they were measured in L or H TMA tissue samples, and hence robust to sampling variation (Figure 3). Moreover, performance of some well-established prostate cancer biomarkers predictive of progression risk and lethal outcome (SMAD4, EZH2, MTDH2, HoxB13, and PTEN) corroborated the validity of the approach.
As part of specificity validation of our antibodies, we learned through target knockdown analyses and mass spectrometry-based protein sequencing analysis that a MAb sold as anti-DCC actually recognised the unrelated protein HSPA9, or Mortalin. We found that HSPA9 was predictive as part of multivariate models and hence was included in the final 12-marker set. Our functional analyses revealed the involvement of HSPA9 in clonogenic colony formation and cell proliferation, consistent with previously reported data (see Supplementary Figure 2 and Flachbartova and Kovacech, 2013). This further validates our unbiased, performance-based marker selection approach.
On the basis of univariate performance as well as frequency of marker appearance in multivariate models for disease aggressiveness and lethal outcome, 12 biomarkers were selected (Figure 5A). A multivariate model based on these 12 markers showed similar predictive performance for aggressiveness across tissue-sampling variation (Figure 5C). Interestingly, the risk scores generated based on the 12-marker aggressiveness model were equally predictive for the separate end point of lethal outcome across tissue-sampling variation (Figure 5C). This indicates a clear linkage between aggressive features on surgical pathology and lethality, and, importantly, validates the usage of our pathologic end point for building our biomarker panel as relevant for long-term patient outcome. We conclude that the 12 identified biomarkers are relevant for prediction of tumour behaviour and could serve as the starting point for development of a clinical, evidence-based multivariate biopsy test for evaluating prostate cancer aggressiveness as a critical aid in early decision-making steps in patient management (NCCN, 2012a; NCCN, 2012b).
Biomarkers robust to sampling error
A key objective for our study was to identify and select markers that are highly robust to sampling error. One of the key reasons for biopsy-sampling error is the heterogeneity of prostate cancer. The inability to consistently acquire tissue from the most aggressive parts of the tumour leads to frequent underestimation of tumour aggressiveness and progression risk. By coring into the highest and lowest Gleason area from each patient, we generated paired TMAs of the entire cohort study designed to simulate two biopsies from each patient, one with ‘maximal' sampling error (L TMA), and the other with minimal sampling error (H TMA). We focused on L TMAs with core Gleason ⩽3+4, as these represent the clinically relevant cases where standard of care is insufficient for accurate prognosis. We found that ∼54% of these L TMA cases were upgraded to a higher surgical GS, which is higher than observed in clinical practice (Porten et al, 2011), confirming that our approach provided a biased sampling error model.
The need for identification of biomarkers that are resistant to sampling error was underscored by examining a well-established four-marker signature based on Cyclin D1, SMAD4, PTEN, and SPP1 previously reported to be predictive of lethal outcome based on prostatectomy cohorts (Ding et al, 2011). Although we confirmed that the model was predictive for lethal outcome in H TMA, representing a situation of minimal sampling error, the model was not lethal outcome-predictive at all in our L TMA tissue cores, representing maximal sampling error (Table 2). This finding is consistent with a recent report that the four-marker signature is unable to predict lethal outcome in low-GS prostate tumours (Irshad et al, 2013).
On the basis of univariate marker analyses, we identified 14 and 18 markers with sampling error-robust performance across L and H TMA samples for disease aggressiveness and lethal outcome, respectively (markers with red font in Figure 4). Most of these univariately selected markers were predictive of both indications across sampling variation, supporting a correlation between disease aggressiveness and lethal outcome. Interestingly, although all markers that showed univariate performance for both indications on L TMA also were predictive on H TMA, two markers (PXN and MTDH2) and three markers (NCOA2, CCND1 (Cyclin D1), and AKAP8) (Sattler et al, 2000; Xu and Li, 2003; Hu et al, 2009; Ding et al, 2011; Canton and Scott, 2013) were predictive of aggressive disease and lethal outcome, respectively, only when measured on H TMA, but not on L TMA (Figure 3). This suggests that these markers are predictive primarily in situations of minimal sampling error. Indeed, all these five markers have been shown as important regulators of cellular proliferation, migration, and oncogenesis (see e.g., Sattler et al, 2000; Xu and Li, 2003; Hu et al, 2009; Ding et al, 2011; Canton and Scott, 2013). The observation that Cyclin D1 is predictive of lethal outcome only in H TMA, but not L TMA, is consistent with the finding that the four-marker signature reported by Ding et al (2011) was not predictive of lethal outcome in our L TMA tissue, as well as on low-grade prostate cancer samples (Irshad et al, 2013). The fact that no markers were predictive for aggressive disease or lethal outcome on only L TMA, but not H TMA, is interesting given that we primarily selected for markers that can predict either aggressiveness or lethal outcome in L TMA, to reflect maximal sampling error robustness. This suggests that the identified markers likely reflect field effects from more aggressive tumour regions, consistent with their similar performance in L and H TMA tissue samples.
Genetic and proteomic approaches
An extensive search to find better prostate cancer biomarkers has led to the identification of possible genetic markers for clinical risk prognostication (Lapointe et al, 2004; Lapointe et al, 2007; Tomlins et al, 2007; Cheville et al, 2008; Cuzick et al, 2011; Scher et al, 2013). However, there are conflicting results regarding the reliability of many such markers in disease prognosis. For example, although TMPRSS2–ERG gene fusions are reported to be associated with high-risk tumours, more recent studies with large cohorts report no strong correlation between these fusions and patient outcome (Gopalan et al, 2009). A multivariate gene expression-based test has recently been reported to predict metastatic disease and lethal outcome based on a conservatively managed cohort of patients from the United Kingdom (Cuzick et al, 2012), as well as biochemical recurrence after treatment in actively managed cohorts in the United States (Cooperberg et al, 2013; Bishoff et al, 2014). The influence of sampling variation on this test has yet to be determined.
The results of this study suggest that taking a proteomic approach, which measures proteins from only the tumour region of intact tissue, can possibly improve accurate risk classification at the biopsy stage. The rationale for this idea is two-fold. First, because prostate cancer is a heterogeneous, multifocal disease, biopsies frequently contain only lower-grade components, and pathologists may classify them as low-risk cancers. However, higher-grade molecular features, not reflected morphologically, have been reported to extend throughout the cancer (Boyd et al, 2012; Sowalsky et al, 2013), and therefore are measurable in seemingly lower-grade-containing biopsies. It is possible to accurately and sensitively assess such high-grade molecular features in situ from tumour regions using a proteomic approach, even in tissue samples with variable amounts of tumour and benign components. This is an advantage over gene expression-based technologies requiring tissue homogenisation, resulting in variable dilution of the higher-grade molecular features depending on the amount of intermixed benign tissue. Second, Gleason grading on biopsy is subjective, with expert pathologists disagreeing on up to 30% of cases (McKenney et al, 2011; Goodman et al, 2012). Molecular features that can be objectively measured should improve risk classification.
Limitations to the study
The fact that the biomarkers were analysed in multiple ways for univariate and multivariate performance on a single, large cohort increases the potential risk that the results are over-fitted to patients included in this study. Also, we simulated prostate cancer biopsies by taking 1-mm diameter cores from prostate tissue blocks to generate TMA blocks. Protein measurements made from cross-sections of these cores may perform differently if measured in 20 × 1-mm biopsy sections, as typically encountered in routine pathology practice.
Conclusion
There is an urgent need for a reliable and accurate prognostic test for patients with prostate cancer, given the difficulties of predicting survival outcomes for patients diagnosed with early-stage cancer and the resulting overtreatment. Further investigation of the 12 biomarkers identified in this study will enable development of an objective clinical biopsy test based on multiplex proteomics in situ imaging. The next step is to identify the optimal biomarker subset algorithm from these 12 biomarkers, and to validate this in a large clinical cohort of Gleason- and TNM-annotated biopsy samples with matched prostatectomy and pathological specimens. These studies are currently under way.
We propose that the identification strategy for protein biomarkers described herein is likely to be applicable to other tumour types, and may serve as a model for performance-based selection of biomarkers that can be used to develop prognostic or predictive tests for other tumours where histological assessment is pivotal to risk stratification and prognostication.
Acknowledgments
We would like to thank Drs Raju Kucherlapati, Ronald DePinho, Lynda Chin, Philip Kantoff, and collaborators at Metamark for insightful comments and suggestions. We thank Rowena Hughes and Winnie McFadzean from Oxford PharmaGenesis Ltd for editorial support in collating comments from authors and finalisation of the manuscript for submission. Research support: this project was funded by Metamark Genetics Inc.
DLR is a paid consultant and equity holder in Metamark. DMB and ML are paid consultants to Metamark. All other authors are paid employees of Metamark.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Bangma CH, Roobol MJ. Defining and predicting indolent and low risk prostate cancer. Crit Rev Oncol Hematol. 2012;83 (2:235–241. doi: 10.1016/j.critrevonc.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Bishoff JT, Freedland SJ, Gerber L, Tennstedt P, Reid J, Welbourn W, Graefen M, Sangale Z, Tikishvili E, Park J, Younus A, Gutin A, Lanchbury JS, Sauter G, Brawer M, Stone S, Schlomm T. Prognostic utility of the CCP score generated from biopsy in men treated with prostatectomy. J Urol. 2014;6 (14:00248–1. doi: 10.1016/j.juro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Bjurlin MA, Taneja SS. Standards for prostate biopsy. Curr Opin Urol. 2014;24 (2:155–161. doi: 10.1097/MOU.0000000000000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorjian SA, Karnes RJ, Crispen PL, Rangel LJ, Bergstralh EJ, Sebo TJ, Blute ML. The impact of discordance between biopsy and pathological Gleason scores on survival after radical prostatectomy. J Urol. 2009;181 (1:95–104. doi: 10.1016/j.juro.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Boyd LK, Mao X, Lu YJ. The complexity of prostate cancer: genomic alterations and heterogeneity. Nat Rev Urol. 2012;9 (11:652–664. doi: 10.1038/nrurol.2012.185. [DOI] [PubMed] [Google Scholar]
- Brimo F, Montironi R, Egevad L, Erbersdobler A, Lin DW, Nelson JB, Rubin MA, van der Kwast T, Amin M, Epstein JI. Contemporary grading for prostate cancer: implications for patient care. Eur Urol. 2013;63 (5:892–901. doi: 10.1016/j.eururo.2012.10.015. [DOI] [PubMed] [Google Scholar]
- Canton DA, Scott JD. Anchoring proteins encounter mitotic kinases. Cell Cycle. 2013;12 (6:863–864. doi: 10.4161/cc.24192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter HB, Partin AW, Walsh PC, Trock BJ, Veltri RW, Nelson WG, Coffey DS, Singer EA, Epstein JI. Gleason score 6 adenocarcinoma: should it be labeled as cancer. J Clin Oncol. 2012;30 (35:4294–4296. doi: 10.1200/JCO.2012.44.0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Pratt CP, Zeeman ME, Schultz N, Taylor BS, O'Neill A, Castillo-Martin M, Nowak DG, Naguib A, Grace DM, Murn J, Navin N, Atwal GS, Sander C, Gerald WL, Cordon-Cardo C, Newton AC, Carver BS, Trotman LC. Identification of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell. 2011;20 (2:173–186. doi: 10.1016/j.ccr.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheville JC, Karnes RJ, Therneau TM, Kosari F, Munz JM, Tillmans L, Basal E, Rangel LJ, Bergstralh E, Kovtun IV, Savci-Heijink CD, Klee EW, Vasmatzis G. Gene panel model predictive of outcome in men at high-risk of systemic progression and death from prostate cancer after radical retropubic prostatectomy. J Clin Oncol. 2008;26 (24:3930–3936. doi: 10.1200/JCO.2007.15.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cima I, Schiess R, Wild P, Kaelin M, Schuffler P, Lange V, Picotti P, Ossola R, Templeton A, Schubert O, Fuchs T, Leippold T, Wyler S, Zehetner J, Jochum W, Buhmann J, Cerny T, Moch H, Gillessen S, Aebersold R, Krek W. Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proc Natl Acad Sci USA. 2011;108 (8:3342–3347. doi: 10.1073/pnas.1013699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperberg MR, Broering JM, Carroll PR. Time trends and local variation in primary treatment of localized prostate cancer. J Clin Oncol. 2010;28 (7:1117–1123. doi: 10.1200/JCO.2009.26.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperberg MR, Pasta DJ, Elkin EP, Litwin MS, Latini DM, Du Chane J, Carroll PR. The University of California, San Francisco Cancer of the Prostate Risk Assessment score: a straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol. 2005;173 (6:1938–1942. doi: 10.1097/01.ju.0000158155.33890.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperberg MR, Simko JP, Cowan JE, Reid JE, Djalilvand A, Bhatnagar S, Gutin A, Lanchbury JS, Swanson GP, Stone S, Carroll PR. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol. 2013;31 (11:1428–1434. doi: 10.1200/JCO.2012.46.4396. [DOI] [PubMed] [Google Scholar]
- Corcoran NM, Hovens CM, Hong MK, Pedersen J, Casey RG, Connolly S, Peters J, Harewood L, Gleave ME, Goldenberg SL, Costello AJ. Underestimation of Gleason score at prostate biopsy reflects sampling error in lower volume tumours. BJU Int. 2012;109 (5:660–664. doi: 10.1111/j.1464-410X.2011.10543.x. [DOI] [PubMed] [Google Scholar]
- Cuzick J, Berney DM, Fisher G, Mesher D, Moller H, Reid JE, Perry M, Park J, Younus A, Gutin A, Foster CS, Scardino P, Lanchbury JS, Stone S, Transatlantic Prostate Group Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer. 2012;106 (6:1095–1099. doi: 10.1038/bjc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, Mesher D, Speights VO, Stankiewicz E, Foster CS, Moller H, Scardino P, Warren JD, Park J, Younus A, Flake DD, 2nd, Wagner S, Gutin A, Lanchbury JS, Stone S, Transatlantic Prostate Group Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12 (3:245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA, Tomaszewski JE, Renshaw AA, Kaplan I, Beard CJ, Wein A. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280 (11:969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- Davies JD, Aghazadeh MA, Phillips S, Salem S, Chang SS, Clark PE, Cookson MS, Davis R, Herrell SD, Penson DF, Smith JA, Jr., Barocas DA. Prostate size as a predictor of Gleason score upgrading in patients with low risk prostate cancer. J Urol. 2011;186 (6:2221–2227. doi: 10.1016/j.juro.2011.07.104. [DOI] [PubMed] [Google Scholar]
- Delongchamps NB, de la Roza G, Jones R, Jumbelic M, Haas GP. Saturation biopsies on autopsied prostates for detecting and characterizing prostate cancer. BJU Int. 2009;103 (1:49–54. doi: 10.1111/j.1464-410X.2008.07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, Hoshida Y, Hiller D, Hu B, Jiang S, Zheng H, Stegh AH, Scott KL, Signoretti S, Bardeesy N, Wang YA, Hill DE, Golub TR, Stampfer MJ, Wong WH, Loda M, Mucci L, Chin L, DePinho RA. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470 (7333:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Wu CJ, Jaskelioff M, Ivanova E, Kost-Alimova M, Protopopov A, Chu GC, Wang G, Lu X, Labrot ES, Hu J, Wang W, Xiao Y, Zhang H, Zhang J, Gan B, Perry SR, Jiang S, Li L, Horner JW, Wang YA, Chin L, DePinho RA. Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell. 2012;148 (5:896–907. doi: 10.1016/j.cell.2012.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler K, Hempel S, Wilby J, Myers L, Bachmann LM, Kleijnen J. Diagnostic value of systematic biopsy methods in the investigation of prostate cancer: a systematic review. J Urol. 2006;175 (5:1605–1612. doi: 10.1016/S0022-5347(05)00957-2. [DOI] [PubMed] [Google Scholar]
- Epstein JI, Allsbrook WC, Jr., Amin MB, Egevad LL. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol. 2005;29 (9:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- Epstein JI, Feng Z, Trock BJ, Pierorazio PM. Upgrading and downgrading of prostate cancer from biopsy to radical prostatectomy: incidence and predictive factors using the modified Gleason grading system and factoring in tertiary grades. Eur Urol. 2012;61 (5:1019–1024. doi: 10.1016/j.eururo.2012.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachbartova Z, Kovacech B. Mortalin - a multipotent chaperone regulating cellular processes ranging from viral infection to neurodegeneration. Acta Virol. 2013;57 (1:3–15. doi: 10.4149/av_2013_01_3. [DOI] [PubMed] [Google Scholar]
- Goodman M, Ward KC, Osunkoya AO, Datta MW, Luthringer D, Young AN, Marks K, Cohen V, Kennedy JC, Haber MJ, Amin MB. Frequency and determinants of disagreement and error in gleason scores: a population-based study of prostate cancer. Prostate. 2012;72 (13:1389–1398. doi: 10.1002/pros.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan A, Leversha MA, Satagopan JM, Zhou Q, Al-Ahmadie HA, Fine SW, Eastham JA, Scardino PT, Scher HI, Tickoo SK, Reuter VE, Gerald WL. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69 (4:1400–1406. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlov IP, Sircar K, Zhao H, Maity SN, Navone NM, Gorlova OY, Troncoso P, Pettaway CA, Byun JY, Logothetis CJ. Prioritizing genes associated with prostate cancer development. BMC Cancer. 2010;10:599. doi: 10.1186/1471-2407-10-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JR, Konicek BW, Lynch RL, Dumstorf CA, Dowless MS, McNulty AM, Parsons SH, Brail LH, Colligan BM, Koop JW, Hurst BM, Deddens JA, Neubauer BL, Stancato LF, Carter HW, Douglass LE, Carter JH. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res. 2009;69 (9:3866–3873. doi: 10.1158/0008-5472.CAN-08-3472. [DOI] [PubMed] [Google Scholar]
- Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, van der Kwast T, Mason M, Matveev V, Wiegel T, Zattoni F, Mottet N. EAU guidelines on prostate cancer. part 1: screening, diagnosis, and local treatment with curative intent-update 2013. Eur Urol. 2014;65 (1:124–137. doi: 10.1016/j.eururo.2013.09.046. [DOI] [PubMed] [Google Scholar]
- Hu G, Wei Y, Kang Y. The multifaceted role of MTDH/AEG-1 in cancer progression. Clin Cancer Res. 2009;15 (18:5615–5620. doi: 10.1158/1078-0432.CCR-09-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irshad S, Bansal M, Castillo-Martin M, Zheng T, Aytes A, Wenske S, Le Magnen C, Guarnieri P, Sumazin P, Benson MC, Shen MM, Califano A, Abate-Shen C. A molecular signature predictive of indolent prostate cancer. Sci Transl Med. 2013;5 (202:3006408. doi: 10.1126/scitranslmed.3006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen G. Diagnostic and prognostic molecular biomarkers for prostate cancer. Histopathology. 2012;60 (1:125–141. doi: 10.1111/j.1365-2559.2011.04083.x. [DOI] [PubMed] [Google Scholar]
- Kvale R, Moller B, Wahlqvist R, Fossa SD, Berner A, Busch C, Kyrdalen AE, Svindland A, Viset T, Halvorsen OJ. Concordance between Gleason scores of needle biopsies and radical prostatectomy specimens: a population-based study. BJU Int. 2009;103 (12:1647–1654. doi: 10.1111/j.1464-410X.2008.08255.x. [DOI] [PubMed] [Google Scholar]
- Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, Ferrari M, Hernandez-Boussard T, Brooks JD, Pollack JR. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67 (18:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, Ekman P, DeMarzo AM, Tibshirani R, Botstein D, Brown PO, Brooks JD, Pollack JR. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101 (3:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey JK, Jones B. Choosing among generalized linear models applied to medical data. Stat Med. 1998;17 (1:59–68. doi: 10.1002/(sici)1097-0258(19980115)17:1<59::aid-sim733>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Loeb S, Bjurlin MA, Nicholson J, Tammela TL, Penson DF, Carter HB, Carroll P, Etzioni R. Overdiagnosis and overtreatment of prostate cancer. Eur Urol. 2014;65 (6:1046–1055. doi: 10.1016/j.eururo.2013.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov DV, Loeb S, Getzenberg RH, Partin AW. Biomarkers for prostate cancer. Annu Rev Med. 2009;60:139–151. doi: 10.1146/annurev.med.60.042307.110714. [DOI] [PubMed] [Google Scholar]
- Markert EK, Mizuno H, Vazquez A, Levine AJ. Molecular classification of prostate cancer using curated expression signatures. Proc Natl Acad Sci USA. 2011;108 (52:21276–21281. doi: 10.1073/pnas.1117029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney JK, Simko J, Bonham M, True LD, Troyer D, Hawley S, Newcomb LF, Fazli L, Kunju LP, Nicolas MM, Vakar-Lopez F, Zhang X, Carroll PR, Brooks JD, Canary/Early Detection Research Network Prostate Active Surveillance Study I The potential impact of reproducibility of Gleason grading in men with early stage prostate cancer managed by active surveillance: a multi-institutional study. J Urol. 2011;186 (2:465–469. doi: 10.1016/j.juro.2011.03.115. [DOI] [PubMed] [Google Scholar]
- Mills IG. HOXB13, RFX6 and prostate cancer risk. Nat Genet. 2014;46 (2:94–95. doi: 10.1038/ng.2881. [DOI] [PubMed] [Google Scholar]
- Mohler JL, Armstrong AJ, Bahnson RR, Boston B, Busby JE, D'Amico AV, Eastham JA, Enke CA, Farrington T, Higano CS, Horwitz EM, Kantoff PW, Kawachi MH, Kuettel M, Lee RJ, MacVicar GR, Malcolm AW, Miller D, Plimack ER, Pow-Sang JM, Roach M, 3rd, Rohren E, Rosenfeld S, Srinivas S, Strope SA, Tward J, Twardowski P, Walsh PC, Ho M, Shead DA. Prostate cancer, Version 3.2012: featured updates to the NCCN guidelines. J Natl Compr Cancer Netw. 2012;10 (9:1081–1087. doi: 10.6004/jnccn.2012.0114. [DOI] [PubMed] [Google Scholar]
- Moyer and U.S. Preventative Services Task Force Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2012;157 (2:120–134. doi: 10.7326/0003-4819-157-2-201207170-00459. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kollmeyer TM, Morlan BW, Anderson SK, Bergstralh EJ, Davis BJ, Asmann YW, Klee GG, Ballman KV, Jenkins RB. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PloS One. 2008;3 (5:e2318. doi: 10.1371/journal.pone.0002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCCN 2012. NCCN Clinical Practice Guidelines in Oncology: Prostate Cancer, version 3.2012; available at: https://www.nccn.org/store/login/login.aspx?ReturnURL=http://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf (accessed 14 February 2014).
- NCCN 2012. Prostate cancer early detection, version 2.2012, available at www.NCCN.org (accessed 14 February 2014).
- Porten SP, Whitson JM, Cowan JE, Cooperberg MR, Shinohara K, Perez N, Greene KL, Meng MV, Carroll PR. Changes in prostate cancer grade on serial biopsy in men undergoing active surveillance. J Clin Oncol. 2011;29 (20:2795–2800. doi: 10.1200/JCO.2010.33.0134. [DOI] [PubMed] [Google Scholar]
- Pressinotti NC, Klocker H, Schafer G, Luu VD, Ruschhaupt M, Kuner R, Steiner E, Poustka A, Bartsch G, Sultmann H. Differential expression of apoptotic genes PDIA3 and MAP3K5 distinguishes between low- and high-risk prostate cancer. Mol Cancer. 2009;8:130. doi: 10.1186/1476-4598-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese AC, Cowan JE, Brajtbord JS, Harris CR, Carroll PR, Cooperberg MR. The quantitative Gleason score improves prostate cancer risk assessment. Cancer. 2012;118 (24:6046–6054. doi: 10.1002/cncr.27670. [DOI] [PubMed] [Google Scholar]
- Robertson NL, Emberton M, Moore CM. MRI-targeted prostate biopsy: a review of technique and results. Nat Rev Urol. 2013;10 (10:589–597. doi: 10.1038/nrurol.2013.196. [DOI] [PubMed] [Google Scholar]
- Ross AE, Marchionni L, Vuica-Ross M, Cheadle C, Fan J, Berman DM, Schaeffer EM. Gene expression pathways of high grade localized prostate cancer. Prostate. 2011;71:1568–1578. doi: 10.1002/pros.21373. [DOI] [PubMed] [Google Scholar]
- Ross HM, Kryvenko ON, Cowan JE, Simko JP, Wheeler TM, Epstein JI. Do adenocarcinomas of the prostate with Gleason score (GS) </=6 have the potential to metastasize to lymph nodes. Am J Surg Pathol. 2012;36 (9:1346–1352. doi: 10.1097/PAS.0b013e3182556dcd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu GS, Andriole GL. Overdiagnosis of prostate cancer. J Natl Cancer Inst Monogr. 2012;2012 (45:146–151. doi: 10.1093/jncimonographs/lgs031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Pisick E, Morrison PT, Salgia R. Role of the cytoskeletal protein paxillin in oncogenesis. Crit Rev Oncog. 2000;11 (1:63–76. [PubMed] [Google Scholar]
- Scher HI, Morris MJ, Larson S, Heller G. Validation and clinical utility of prostate cancer biomarkers. Nat Rev Clin Oncol. 2013;10 (4:225–234. doi: 10.1038/nrclinonc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanov S, Eggener SE. Hazard of prostate cancer specific mortality after radical prostatectomy. J Urol. 2012;187 (1:124–127. doi: 10.1016/j.juro.2011.09.044. [DOI] [PubMed] [Google Scholar]
- Shipitsin M, Small C, Giladi E, Siddiqui S, Choudhury S, Hussain S, Huang YE, Chang H, Rimm DL, Berman D, Nifong TP, Blume-Jensen P.2014. Automated quantitative multiplex immunofluorescence in situ imaging identifies phospho-S6 and phospho-PRAS40 as predictive protein biomarkers for prostate cancer lethality. Proteome Science (in press). [DOI] [PMC free article] [PubMed]
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64 (1:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- Sowalsky AG, Ye H, Bubley GJ, Balk SP. Clonal progression of prostate cancers from Gleason grade 3 to grade 4. Cancer Res. 2013;73 (3:1050–1055. doi: 10.1158/0008-5472.CAN-12-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyerberg EW, Roobol MJ, Kattan MW, van der Kwast TH, de Koning HJ, Schroder FH. Prediction of indolent prostate cancer: validation and updating of a prognostic nomogram. J Urol. 2007;177 (1:107–112. doi: 10.1016/j.juro.2006.08.068. [DOI] [PubMed] [Google Scholar]
- Swanson GP, Quinn D. Using molecular markers to help predict who will fail after radical prostatectomy. Prostate Cancer. 2011;2011:290160. doi: 10.1155/2011/290160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18 (1:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, Shah RB, Chinnaiyan AM. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39 (1:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- True L, Coleman I, Hawley S, Huang CY, Gifford D, Coleman R, Beer TM, Gelmann E, Datta M, Mostaghel E, Knudsen B, Lange P, Vessella R, Lin D, Hood L, Nelson PS. A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proc Natl Acad Sci USA. 2006;103 (29:10991–10996. doi: 10.1073/pnas.0603678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellekoop A, Loeb S, Folkvaljon Y, Stattin P. Population-based study of predictors of adverse pathology among candidates for active surveillance with Gleason 6 prostate cancer. J Urol. 2014;191 (2:350–357. doi: 10.1016/j.juro.2013.09.034. [DOI] [PubMed] [Google Scholar]
- Wilt TJ, MacDonald R, Rutks I, Shamliyan TA, Taylor BC, Kane RL. Systematic review: comparative effectiveness and harms of treatments for clinically localized prostate cancer. Ann Intern Med. 2008;148 (6:435–448. doi: 10.7326/0003-4819-148-6-200803180-00209. [DOI] [PubMed] [Google Scholar]
- Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17 (9:1681–1692. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- Yan P, Seelentag W, Bachmann A, Bosman FT. An agarose matrix facilitates sectioning of tissue microarray blocks. J Histochem Cytochem. 2007;55 (1:21–24. doi: 10.1369/jhc.6A6987.2006. [DOI] [PubMed] [Google Scholar]
- Yang YA, Yu J. EZH2, an epigenetic driver of prostate cancer. Protein Cell. 2013;4 (5:331–341. doi: 10.1007/s13238-013-2093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.