Abstract
Background:
Volatile organic compounds (VOCs) are potential biomarkers for cancer detection in breath, but it is unclear if they reflect specific mutations. To test this, we have compared human bronchial epithelial cell (HBEC) cell lines carrying the KRASV12 mutation, knockdown of TP53 or both with parental HBEC cells.
Methods:
VOC from headspace above cultured cells were collected by passive sampling and analysed by thermal desorption gas chromatography mass spectrometry (TD-GC–MS) or sensor array with discriminant factor analysis (DFA).
Results:
In TD-GC–MS analysis, individual compounds had limited ability to discriminate between cell lines, but by applying DFA analysis combinations of 20 VOCs successfully discriminated between all cell types (accuracies 80–100%, with leave-one-out cross validation). Sensor array detection DFA demonstrated the ability to discriminate samples based on their cell type for all comparisons with accuracies varying between 77% and 93%.
Conclusions:
Our results demonstrate that minimal genetic changes in bronchial airway cells lead to detectable differences in levels of specific VOCs identified by TD-GC–MS or of patterns of VOCs identified by sensor array output. From the clinical aspect, these results suggest the possibility of breath analysis for detection of minimal genetic changes for earlier diagnosis or for genetic typing of lung cancers.
Keywords: lung cancer, mutation, headspace, artificial olfactory system, GC–MS, KRAS, TP53
Lung cancer development is associated with multiple genetic and epigenetic alterations, involving both tumour suppressor genes (e.g., TP53) and specific oncogenes (e.g., KRAS), which affect cell proliferation and apoptosis (Massion and Carbone, 2003). The genetic lesions found in lung cancer are manifold, vary to some extent with the histological sub-type and can impact both on the therapeutic intervention chosen and on outcome (Osada and Takahashi, 2002; The Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine (NGM), 2013). TP53 is a tumour suppressor gene, frequently subject to both inactivating mutations and genetic loss in lung cancer (Takahashi et al, 1989); TP53 protein binds to the DNA and is involved in cell cycle regulation, induction of apoptosis and maintenance of genome integrity (Vousden and Lu, 2002). TP53 was the most commonly mutated gene in a recent genomic classification of lung cancer and found in both small cell and non-small cell lung cancers (The Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine (NGM), 2013). In the same study, KRAS had the second highest mutation incidence (The Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine (NGM), 2013); being most common in adenocarcinoma and large cell lung cancer. The human KRAS oncogene encodes a small guanosine triphosphate (GTP)-binding protein, which is located on the inner membrane and functions as a messenger for growth stimuli. Both TP53 and KRAS mutation have been implicated in response of patients to therapy. Expression of TP53 may predict good prognosis from adjuvant chemotherapy in patients with NSCLC (Bennett et al, 1993; Coate et al, 2009) and mutation of TP53 is associated with poor survival in EGFR-mutant cases (The Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine (NGM), 2013). Lung cancers carrying KRAS mutation show resistance to tyrosine kinase inhibitors (Pao et al, 2005).
The most reliable method for the detection of lung cancer genetic mutation is through DNA sequencing, which requires a good quality of tissue specimen available from biopsy (Fassina et al, 2009; Lim et al, 2009). However, this procedure is invasive, time consuming, expensive, unsuitable for non-resectable tumours and requires histopathological evaluation under the microscope (Hamilton, 2012). Additionally, DNA sequencing may delay therapy and may not be suitable for large-scale screening. Nevertheless, it is clear that a better understanding of the molecular pathogenesis of lung cancer through specific biomarkers is needed in order to improve diagnosis and treatment and by that to reduce mortality rate.
Recent work suggests that analysis of signatures based on volatile organic compounds (VOCs) may lead to novel biomarkers for lung cancer (Chen et al, 2007; Phillips et al, 2008; Dragonieri et al, 2009; Amann et al, 2011; Mazzone et al, 2011; Hakim et al, 2012; Broza and Haick, 2013). Genetic alterations associated with tumour growth may lead to VOC alterations in the microenvironment of the cell, and thus in the breath of the patient. These VOCs can be detected in a non-invasive manner, either in the headspace of the blood, saliva or cancer tissue (i.e., the air above a sample sealed in a container) or directly in the exhaled breath due to their relatively high vapour pressure (>0.1 mm Hg). The possibility of using VOCs as surrogate markers for genetic sub-types of lung cancer in situations where biopsies are unavailable for DNA sequence analysis is intriguing, but requires greater insight into the mechanism of mutation-driven VOC differences, if it is to be applied in a rational manner.
Previous studies (Smith et al, 2003; Chen et al, 2007; Filipiak et al, 2008; Sponring et al, 2009a; Barash et al, 2009; Sulé-Suso et al, 2009; Filipiak et al, 2010; Sponring et al, 2010; Peled et al, 2013) have investigated the ability of different analytical methods or nanoarrays to assign unique volatolomic fingerprints to cancer cell lines by analysing the headspace VOCs. However, while comparing multiple cancer cell lines may reflect the wide genetic diversity of lung tumour samples, the same genetic diversity is a hindrance when trying to make direct links to specific cancer-related pathways. Moreover, cancer cell lines are, by their nature, genetically unstable with high levels of aneuploidy (Ross et al, 2000). This provides the cells with some plasticity, but molecular studies may suffer from genetic drift and thus it might be more difficult to pin VOC patterns down to individual pathways.
A more tractable system to study the effect of specific cancer driver mutations on volatile release is provided by human bronchial epithelial cells (HBECs) that have been genetically manipulated (Sato et al, 2006). HBECs, immortalised through expression of telomerase and activation of CDK4, are genetically stable compared with cancer cell lines, with minimal aneuploidy (Ramirez et al, 2004). This provides a parental cell line in which the effect of additional genetic lesions can be investigated. In the derived cell line HBEC-3KT53, TP53 has been knocked down by RNAi knockdown, whereas HBEC-3KTR cell line expresses mutant KRASG12V (Sato et al, 2006). In a fourth cell line, HBEC-3KTR53, both genetic alterations were induced by simultaneous transfection (Sato et al, 2006). The three genetically altered HBEC cell lines and the vector only control (HBEC-3KT) allow us to study the effect of individual mutations on the VOC signature of lung cancer precursors and potentially model the earliest stages of lung tumorigenesis, as these lesions may be present in those at risk of subsequently developing lung cancer. TP53 and KRAS mutations are common mutations in lung cancer, sometimes occur together and have been found early in the progression of lung tumorigenesis (Massion and Carbone, 2003).
The unique volatolomic signature of the four different HBEC cell lines has been studied by means of thermal desorption combined with gas chromatography mass spectrometry (TD-GC–MS) and artificial intelligence nanoarray (Figure 1). The TD-GC–MS provides us with specific identification of the chemical composition of the VOCs, while the nanoarray is able to distinguish between the different HBEC cell lines based on the collective response of their headspace without the need for specific identification. These studies provide a proof-of-principle for the detection of different VOC patterns in closely related cells with minimal genetic alterations.
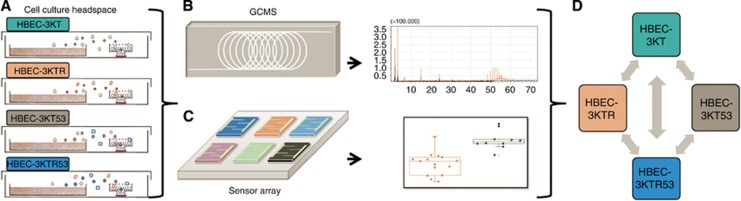
Figure 1.
In vitro studies of the unique volatolomic signatures of HBEC cell lines. (A) Open 10-cm cell culture plates with different HBEC cell lines were grown in controlled temperature and humidification atmosphere in 16-cm plates for headspace sampling on Ultra II SKC badges filled with Tenax TA. After the sorption process, headspace samples were transferred to TD-GC–MS (B) and artificial intelligence nanoarray (C) to analyse the volatolomic signature of the studied genetic changes. Analysis of TD-GC–MS and artificial intelligence nanoarray has been conducted for multiple comparisons (D).
Materials and methods
Passive collection of headspace
All HBECs (both parental and the derived HBECs) were cultured as monolayers with K-SFM (Life Technologies, Gaithersburg, MD, USA) medium containing 50 μg ml−1 bovine pituitary extract and 5 ng ml−1 EGF (Life Technologies). The parental HBEC-3KT cell line and three derived HBEC cell lines have been kindly provided to us by Professor John Minna and Professor Adi Gazdar.
For headspace sampling (Figure 1), two million cells were placed into 10-cm diameter dishes; passive VOC sampling was performed as previously described using a passive sampling badge (Ultra II, SKC Limited, Blanford Forum, UK) filled with Tenax TA sorbent (SKC Limited) placed in a 16-cm dish with the 10-cm culture (Barash et al, 2009; Barash et al, 2012; Peled et al, 2013). Tenax samples were collected following 3 days culture under standard conditions (humidified, 37 °C, 5% CO2). Control headspaces samples were also collected from plates containing only empty medium treated with the same incubation conditions. Final cell numbers were approximately 10 million per flask (no significant differences found between cell lines). Cell density control experiments were performed plating four-fold more and four-fold fewer cells.
TD-GC–MS VOC analysis
VOCs in the headspace were analysed using a TD-GC–MS QP2010 instrument (Shimadzu Corporation, Duisburg, Germany) with an SLB-5ms capillary column (with 5% phenyl methyl siloxane; 30 m length; 0.25 mm internal diameter; 0.5 μm thicknesses; from Sigma-Aldrich, Rehovot, Israel), combined with a thermal desorption (TD) system (TD20; Shimadzu Corporation). The following oven temperature profile was set: (a) 5 min at 35 °C; (b) increasing temperature until 180 °C at rate of 5 °C min−1; (c) increasing temperature until 290 °C at rate of 13.5 °C min−1; (d) 290 °C for 1 min. The GC–MS chromatograms were analysed using the GC–MS post-run analysis program (version 2.53; Shimadzu Corporation), and the compounds were tentatively identified through spectral library match (Compounds library of the National Institute of Standards and Technology, Gaithersburg, MD, USA). Quantitation was performed using area-under-curve values; subtracting relevant media-only headspace controls values (collected during the same experiment conditions).
Statistical tests were performed using SAS JMP (version 8.0, SAS Institute, Cary, NC, USA) for Wilcoxon/Kruskal–Wallis tests in search for compounds expressing differential values. Discriminant function analysis (DFA) was used as a supervised linear pattern recognition algorithm for data classification, with leave-one-out cross validation for accuracy confirmation. DFA aims to detect the most significant canonical variable such that the variance between the pre-defined classes is maximised, while the variance within each class is minimised (Ionescu et al, 2002). This way, DFA also serves as a heuristic to select the compounds (or signals) that most contribute to the separation between the study-groups. The DFA output variables (canonical variables, CVs) are obtained in manually orthogonal dimensions; thus, DFA effectively reduces the dimension of the experimental data. Furthermore, using the optimal cut-off of a receiver operating characteristic (ROC) curve based on CV1, the area under curve (ROC-AUC), sensitivities and specificities were also estimated (SPSS vers20).
VOC identification and calibration
VOCs identification and concentration were determined through external standards and calibration curves. Glass TD-tubes (Sigma-Aldrich) filled with the same tenax used for the headspace sampling were exposed to the calibration gas for 3 min at flow rate of 100 ml min−1, stimulating ∼300 ml of passive headspace sampling. A commercial permeation/diffusion tube dilution (PDTD) system (Umwelttechnik MCZ, Bad Nauheim, Germany) was used to produce gaseous standards in defined concentrations. As a carrier gas, we used purified dry nitrogen (99.9999%) from a commercial nitrogen generator (N-30, On Site Gas Systems, Newington, CT, USA). A constant gas flow of 200±1 cm3 min−1 of purified nitrogen was mixed with a constant mass flow of vaporised VOCs exiting the diffusion tube (Dynacal, VICI Metronics, Poulsbo, WA, USA) placed inside the PDTD. The mixture from the PDTD was further diluted with nitrogen to achieve the desired concentration in the range from few p.p.b. to few p.p.m. (typically two orders of magnitude). The VOC concentration could be determined by manipulating the diffusion tube diameter, the oven temperature and the vaporised VOC mass flow rate. The concentrations of VOC in TD-GC–MS for analysis of each cell line are given in Supplementary Table S1, along with the partition coefficient. Partition coefficients were calculated from the formula: λb:a=λO:W·λW:a·(a+0.3b)+λW:a(c+0.7b) (Haick et al, 2013) where λb:a is the partition coefficient blood:air, λO:W is octanol:water partition coefficient and λW:a is Henry constant at 25 °C. The values for λb:a were either calculated from the formula above or taken directly from published data (Mochalski et al, 2012; Haick et al, 2013).
Artificial intelligence nanoarray analysis
Headspace VOCs were further analysed using artificial intelligence nanoarray based on cross-reactive chemiresistors. The main two types of chemiresistors used in the nanoarray were gold nanoparticles (GNPs) and single-wall carbon nanotubes (SWCNTs) each capped with a different organic layer. The GNPs and the SWCNTs serve as the electron transfer core, while the organic layer serves as the recognition layer, allowing the adsorption of vast types of VOCs onto the sensors and thus enhancing sensitivity and specificity for a particular sensing application. Seventeen different sensors were mounted onto a custom polytetrafluoroethylene circuit board, which was then placed inside a stainless steel test chamber with a volume of 100 cm3. Full information on the synthesis and fabrication of the nanoarray is described in previous work (Barash et al, 2009; Tisch and Haick, 2010; Barash et al, 2012).
The headspace samples were heated at 270 °C for 10 min in a 750-ml stainless steel manually TD device to release the headspace VOCs. Pulses of the headspace sample from the TD chamber were then delivered by a gas sampling system into the test chamber containing the array of cross-reactive nanomaterial-based sensors (Barash et al, 2012; Peled et al, 2013). The test chamber was evacuated between exposures to release the VOCs that the sensors adsorbed with full recovery. An Agilent Multifunction switch 34980 was used to measure the resistance of all the sensors simultaneously as a function of time. Each headspace sample has been recorded for one cycle, which included 5 min of the sensors' baseline response in vacuum, followed by 5 min of VOC exposure, ending with another 5 min in vacuum. Each sensor in the array responded to all of the headspace VOCs. The collective responses from the reservoir of sensors was then analysed using the statistical pattern recognition algorithm DFA (as above).
Results
The volatolomic signature of TP53 and KRAS genetic alteration in human bronchial epithelial cell lines was studied in two steps. The first step was to analyse the cell line headspace and to identify specific VOCs associated with minimal genetic changes using a common analytical method, that is, TD-GC–MS. This TD-GC–MS analysis aimed to detect differences in individual VOC between the different HBEC cell lines and define discriminating compounds. The second step was to perform the same comparisons on independent samples using artificial intelligence nanoarray technology.
VOC quantification
Analysis of the cell headspace by TD-GC–MS identified 51 common compounds that formed a data set for multiple binary comparisons based on TD-GC–MS quantitation (area-under-curve values) relative to media-only intra-experiment controls (Supplementary Tables S1 and S2). Using a nonparametric Wilcoxon/Kruskal–Wallis test to identify differential levels (pairwise analysis between cell lines; between 12 and 40 independent cell line measurements per comparison, Table 1) at P-value of <0.05, four compounds were significantly different in at least one binary comparison (Table 1, Supplementary Table S1). The compounds are: 2-methylpropene; benzaldehyde; tridecane and 1,2,3-trimethylbenzene. For the binary comparison discriminating KRAS mutation HBEC-3KTR cells from the parental HBEC-3KT cells and for the binary comparison of the 2 KRAS-mutated cells (HBEC-3KTR and HBEC-3KTR53) compared with cells with no KRAS mutation (HBEC-3KT and HBEC-3KT53), no single compound was found to be significantly different. However, 2-methylpropene was found at higher concentrations in HBEC-3KT53 cells as compared with HBEC-3KTR53 cells, possibly indicating that the genetic background of a cell influences the VOC differences that occur with further mutation (i.e., KRAS mutation). Single compounds were able to discriminate between each comparison involving loss of TP53 (Table 1). Where discrimination was possible with single compounds, optimum cut-offs from ROC analysis gave accuracies of 77–87% (Table 1) and quantities were significantly different between cells based on mutation status (Wilcoxon P-values 0.0066–0.049).
Table 1. One compound and discriminant factor analysis model for gas chromatography mass spectrometry data.
|
One compound model |
Discriminant factor analysis model |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cell lines | n | Comparison | Compound | Accuracy (receiver operating characteristic) | Wilcoxon P-value | Compound | Accuracy (leave-one-out) | Wilcoxon P-value |
| 3KT vs 3KTR | 28 | −/+ KRAS | None | Octanal+1-methylethyl benzene+2-phenoxyethanol+1-butanol | 91% | <0.0001 | ||
| 3KT53 vs 3KTR53 | 12 | −/+KRAS | 2-Methylpropene | 86% | 0.0374 | 2,2,4-Trimethylpentane+2-methylpropene | 92% | 0.004 |
| 3KT and 3KT53 vs 3K TR and 3KTR53 | 40 | −/+KRAS | None | 2,4-Dimethylheptane+cyclohexanone+decanal+2-methylpropene | 86% | <0.0001 | ||
| 3KT vs 3KT53 | 18 | −/+TP53 | Benzaldehyde | 82% | 0.0312 | 2-Methyloctane+acetone | 83% | 0.001 |
| 3KTR vs 3KTR53 | 22 | −/+TP53 | Tridecane | 79% | 0.0325 | Ethanol+acetone+2-methylpropene | 100% | 0.0004 |
| 3KT and 3KTR vs 3KT53 and 3KTR53 | 40 | −/+TP53 | Benzaldehyde | 77% | 0.0066 | Benzaldehyde+tetradecane+2-methylpropene+2,2,4-trimethylpentane | 83% | <0.0001 |
| 3KT vs 3KTR53 | 18 | −/+KRAS and TP53 | Benzaldehyde | 82% | 0.0312 | Ethanol+2-methylpropene | 94% | 0.0007 |
| 3KTR vs 3KT53 | 22 | KRAS vs TP53 | 1,2,4-Trimethylbenzene | 87% | 0.027 | Decane+tridecane | 86% | 0.01 |
| 3KT vs 3KTR and 3KT53 and 3KTR53 | 40 | Any mutation | Benzaldehyde | 71% | 0.0417 | Ethanol+2-hydroxy benzaldehyde+benzaldehyde+decanal | 80% | 0.0002 |
Concentrations of VOCs identified in TD-GC–MS analysis are available in Supplementary Information (Supplementary Table S1) together with their partition coefficients, which can be used to estimate the concentration of the VOCs in blood and breath, as explained in the paper of Haick et al (2013). Based on this principle, discriminatory VOCs detected here would be expected to be detectable in breath samples.
VOC discriminant analysis
Performing discriminant factor analysis (DFA) on the set of 51 compounds, we identified 16 compounds in addition to the four above (Table 1) that in different combinations provided discrimination of between 72.5% and 100% accuracy with leave-one-out cross-validation between the different genetic alterations (Figure 2). Another way to look at the same data is by ROC analysis of DFA 1st canonical variable (CV1) values and comparison of CV1 values themselves. This resulted in ROC-area-under-curve (ROC-AUC) values of 0.87–1.0 and defining an optimum cut-off by ROC gave good sensitivities (70–100%) and specificities (75–92%), equivalent to accuracies of 83–100%. DFA CV1 values were significantly different between cells based on mutation status for all comparisons (Wilcoxon P-values 0.00005–0.01, Table 1, Figure 2).
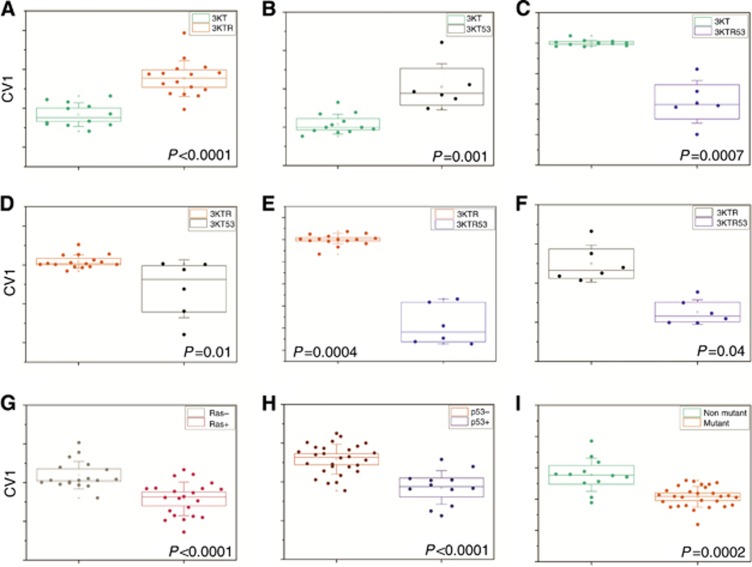
Figure 2.
DFA plot of CV values (y axis) that were calculated from the multidimensional data output obtained from the VOCs detected in the headspace of the cells by TD-GC–MS. A–C represent the CV values of the parental HBEC-3KT vs each of the mutated cell lines. D and E represent the comparison between the mutated cell line HBEC-3KTR vs the mutated cell lines HBEC-3KT53 and HBEC-3KTR53, respectively. F represents the comparison between cells carrying the TP53 knockdown but that differ in their KRAS mutation status. G–I represent the CV plots of all cell lines differing by KRAS mutation status (G), by TP53 status (H) or any mutation vs the parental non-mutated HBEC-3KT cell line (I). Each dot in the figure represents one sample.
When applying the DFA model for all four cell lines (Table 1, Figure 2) ROC curves gave an ROC-AUC value of 0.88 (95% CI 0.74–1.0, P=0.0002) for discrimination of all mutated cell lines (HBEC-3KTR, HBEC-3KT53 and HBEC-3KTR53) vs the parental HBEC-3KT cell line. In addition, the accuracy when applying leave-one-out DFA analysis was 80%, compared with 71% for the one compound (benzaldehyde) that was found to be significantly different (Table 1, Figure 2). It was also possible to discriminate cells with KRAS mutation (HBEC-3KTR and HBEC-3KTR53) compared with those without KRAS mutation (HBEC-3KT and HBEC-3KT53), irrespective of TP53 status (Table 1). Similarly, TP53 downregulation could be discriminated irrespective of KRAS status (Table 1, Figure 2).
Two interesting compounds are 2-methylpentane and 2,2,3-trimethylpentane (Table 1, Supplementary Table S1), which differ only in two methyl groups and were found to be totally depleted in two out of the four cell lines. 2-methylpentane was not found in the parental HBEC-3KT cell line and in cells expressing mutant KRASG12V; 2,2,3-trimethylpentane, on the other hand, was totally depleted in cells in which TP53 has been knocked down (HBEC-3KTR53 and HBEC-3KT53).
Artificial intelligence nanoarray
Artificial intelligence nanoarray experiments have been conducted in two phases. In the first phase, we compared the headspace signature of 15 samples from non-mutated cells (HBEC-3KT) with 12 samples from cells that express mutant KRASG12V (HBEC-3KTR). The samples were randomly selected for analysis in the nanoarray experiment. All 17 sensors in the array gave a response when exposed to the headspace samples. The signals obtained from the nanoarray response were processed and analysed by DFA to determine the features which were best able to separate the two previously defined sets of samples (based on KRAS mutation status). DFA identified three features that were able to discriminate between mutant and non-mutant cells with 100% sensitivity and specificity at the optimum cut-point (Supplementary Figure S1a). In order to address the ability of the discriminant variable to predict the RAS mutant status of cell lines based on a pre-determined DFA CV1 cut-off, data were reanalysed by randomly selecting a training set (50% of the samples, used to produce the DFA plot based on the selected features) and a test set (the remaining samples). When transposing the test sample data onto the DFA CV1 values (Supplementary Figure S1b) their RAS mutant status could be predicted. In this instance, all of the test samples had their mutation status correctly predicted by DFA leading to an accuracy of 100%. Random label shuffling validation was also performed (Supplementary Figure S1c), demonstrating that discrimination was related to mutation status and not an arbitrary ability of DFA analysis to categorise samples.
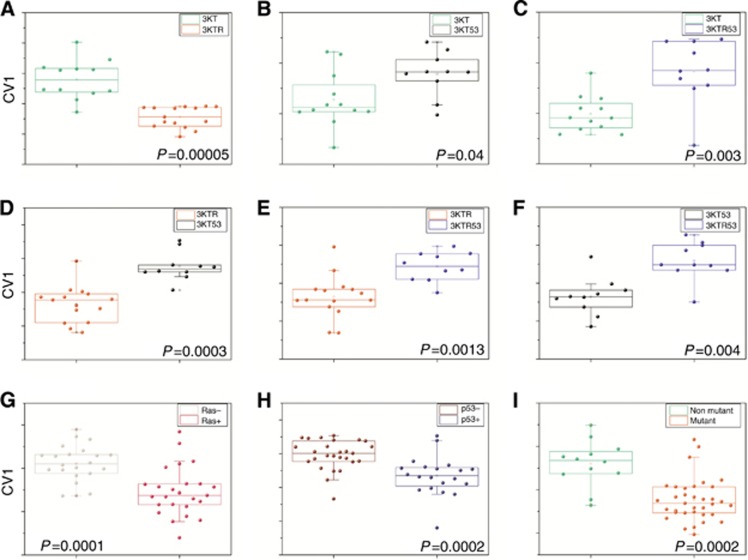
Having demonstrated the ability of the nanoarray detection to discriminate cell headspace based on the presence of a single mutation, we extended the studies to provide an independent validation of KRAS mutation detection and extend detection to other mutations (and combinations of mutations). Examples of CV1 differences are shown for each comparison in Figure 3. Nanoarray detection followed by DFA analysis proved to have significant specificity and sensitivity for: KRAS mutation (with or without additional TP53 knockdown); TP53 knockdown (with or without additional KRAS mutation); or combinations of mutations (Figure 3, Table 2). For example, we performed DFA analysis of all mutant cell lines (HBEC-3KTR, HBEC-3KT53, HBEC-3KTR53) against the parent line (HBEC-3KT) and identified four features that together gave an accuracy of 83% (in leave-one-out cross-validation analysis) (Table 2, Supplementary Table S4, Figure 3). The associated ROC curve gave an ROC-AUC of 0.87 (95% CI 0.75–0.99), P=0.0002; using the optimal cut-off the sensitivity was 70%, specificity 92% and accuracy 82%.
Figure 3.
DFA plot of CV values (y axis) that were calculated from the multidimensional data output obtained from the responses of the nanoarray to the VOCs in the headspace samples. Panels as described for Figure 2.
Table 2. Discriminant factor analysis and receiver operating characteristic analysis values of artificial intelligence nanoarray results, 2nd cohort.
|
Accuracy |
Receiver operating characteristic analysis |
Wilcoxon | ||||||
|---|---|---|---|---|---|---|---|---|
| Cell lines | n | Comparison | Leave-one-out | Leave-third-out | Sensitivity | Specificity | Area under curve (95% confidence interval) | P-value |
| 3KT vs 3KTR | 27 | −/+KRAS | 93% | 89% | 100% | 92% | 0.96 (0.88–1) | 0.00005 |
| 3KT53 vs 3KTR53 | 20 | −/+KRAS | 90% | 83% | 90% | 90% | 0.88 (0.72–1) | 0.004 |
| 3KT and 3KT53 vs 3KTR and 3KTR53 | 47 | −/+KRAS | 81% | 81% | 80% | 86% | 0.83 (0.7–0.95) | 0.0001 |
| 3KT vs 3KT53 | 22 | −/+TP53 | 77% | 64% | 80% | 75% | 0.76 (0.55–0.97) | 0.04 |
| 3KTR vs 3KTR53 | 25 | −/+TP53 | 88% | 79% | 100% | 80% | 0.88 (0.74–1) | 0.002 |
| 3KT and 3KTR vs 3KT53 and 3KTR53 | 47 | −/+TP53 | 81% | 64% | 85% | 81% | 0.82 (0.69–0.96) | 0.0002 |
| 3KT vs 3KTR53 | 22 | −/+KRAS and TP53 | 91% | 77% | 90% | 92% | 0.88 (0.69–1) | 0.003 |
| 3KTR vs 3KT53 | 25 | KRAS vs TP53 | 92% | 84% | 93% | 90% | 0.94 (0.84–1) | 0.0003 |
| 3KT vs 3KTR and 3KT53 and 3KTR53 | 47 | Any mutation | 83% | 86% | 70% | 92% | 0.87 (0.75–0.99) | 0.0002 |
Analysing the four cell lines together (e.g., HBEC-3KT and HBEC-3KT53 (no KRAS mutation) vs HBEC-3KTR and HBEC-3KTR53 (with KRAS mutation)) gave us greater sample numbers (potentially giving greater statistical power), but also allowed us to address the accuracy of sensor detection for one genetic alteration (KRAS) in the presence (or absence) of a second (TP53 depletion).
Comparison of multiple DFA models
DFA models were built using the same data, but specifying different numbers of discriminatory features (limiting to four to avoid over-fitting). While including more features in the DFA model improved accuracy (Supplementary Table S3), it was sometimes possible to achieve very good discrimination with a single sensor feature as was obtained for the second phase. For example, different features of the same single sensor (S12) had 85% accuracy in discriminating 3KTR from 3KT (ΔRmid) and 82% accuracy for 3KT vs 3KTR53 (ΔRpeak).
For any one comparison, equally good discrimination accuracy was achieved by several different sets of features (representative examples are given in Table 2 and Supplementary Table S3). Where DFA was performed with both three and four sensor features (Supplementary Table S4, Supplementary Figure S3), there was a significant correlation between CV1 values, despite sensor features being mutually exclusive. This seems to indicate that there is some inherent redundancy in the sensor arrays, which might be utilised to provide added robustness. It is unclear if this is due to the same VOC being detected by different sensors, or multiple VOCs associated with mutation.
Validation of the sensor array results
In addition to demonstrating discrimination on the basis of mutation status, we wanted to test the strength of the sensor arrays when facing confounding factors. One feature of the second phase of experiments was that we performed sampling of pairs of cell lines within the same incubator with the same batch of media, whereas in the first phase the HBEC-3KT and HBEC-3KTR cells were sampled on different days. The greater sensitivity and specificity seen in the first phase may therefore partly reflect additional differences in either the environment or batches of media between experiments. Conversely the lower accuracy seen, for comparisons made between cells grown at the same time, might reflect some admixture of headspace between culture vessels (cells having been grown in culture plates that permit some diffusion, Figure 1); however, no evidence of this was seen for media-only controls included in some experiments. Media controls (media without cells incubated with passively samples within the same experiment) did not partition consistently with the cell line grown in the same experiment (data not shown).
In order to investigate the contribution to experimental variation made by different experiments, we compared multiple batches of samples produced on different days by DFA (Supplementary Figure S2a). The DFA analysis produced a CV1 that discriminated between the two different cell lines, but provided no discrimination for experiment number. Similarly, control experiments indicated that incubator position and experiment date were not confounding factors in this DFA discrimination.
Within the time frame of our experiments, there are some minor differences in cell density by the end of the experiment between cell lines, but these are not statistically significant (mean cell number at end of 3-day incubation is 10 million when starting with two million). By varying the starting number of cells four-fold up or down, it was shown that cell density can have some effect on VOC signature, in that DFA analysis of sensor array data was able to discriminate on the basis of cell numbers (Supplementary Figure S2b). However, this did not prevent us from discriminating on the basis of mutation (Supplementary Figure S2c). For this reason, while the majority of samples used using standardised cell numbers (two million cells), we did not exclude data starting with four-fold more or four-fold less.
Discussion
In principle, TD-GC–MS results show that there are detectable VOC differences between cell lines with minimal genetic differences. For example, benzaldehyde was found to be a significant compound in the volatolomic signature of both KRAS and TP53. Benzaldehyde was previously found to be decreased in NSCLC adenocarcinoma compared with SCLC and was also found in decreased levels in cell lines with squamous cell carcinoma histology as compared with adenocarcinomas (Barash et al, 2012; Peled et al, 2013). A suggested explanation of the decrease in benzaldehyde concentration is the upregulation of aldehyde dehydrogenase in lung cancer cells (Barash et al, 2012), which uses aldehydes as substrates. In a different study, we found benzaldehyde as a major factor in the headspace signatures of cell lines carrying EGFR and EML4-ALK mutation and also in lung cancer cell lines without EGFR, KRAS or EML4-ALK mutation. On cell lines having KRAS mutation, benzaldehyde was found to be totally depleted. In this study, benzaldehyde was found at lower concentrations in cells carrying KRAS mutation, TP53 knockdown or both alterations as compared with the parental HBEC-3KT cells.
The VOCs contributing to DFA discrimination are from the families of alkanes, methylated alkanes and alkenes, benzene derivatives, ketones, aldehydes and alcohols. These types of compounds were found and discussed in previous studies. Levels of VOCs as detailed in Supplementary Table S1 are in the range of few p.p.b. to tens of p.p.b. for alkanes (straight and methylated) and for alkenes. Levels of ketones and alcohols are in the range of hundreds of p.p.b. to few p.p.m. These values are similar to what was previously published by different groups (Filipiak et al, 2008; Filipiak et al, 2010). As seen in Supplementary Table S1, aldehydes levels were decreased (except for decanal) in all mutated cells as compared with parental HBEC-3KT. This may be due to upregulation of aldehyde dehydrogenase in cancer cells as explained previously.
A VOC detected here in association with TP53 downregulation (being depleted in cells with wild-type TP53), 2-methylpentane has previously been detected in NCI-H2087 cells (Sponring et al, 2009b), a lung adenocarcinoma cell line with mutated TP53. This may imply a possible biochemical pathway that involves alkane methylation in the TP53 downregulation in carcinogenesis, but far more studies are needed in order to achieve any conclusion. Another VOC being totally depleted in lung cancer cell lines associated with TP53 downregulation is 2,2,3-trimethylpentane. This compound was found also in the paper of Sponring et al (2009b) (2,3,3-trimethylpentane) but showed no significant increase or decrease as compared with medium control. This may be due to the difference in one methyl group, but further investigation is needed in order to achieve a conclusive argument.
The expected concentration of discriminant VOCs in the breath can be estimated using the partition coefficient and concentration of the VOCs in the cells headspace, as explained in the paper of Haick et al (2013). VOCs levels found here are calculated to be detectable in breath and were partly found in previous studies of breath analysis, this supports the idea that in the future, this might be utilised as a way to analyse genetic mutations through breath analysis. An example of such compound is 4-methyloctane, that was found to be increased in all mutated cell lines and was also found to be significantly increased in the breath of lung cancer patients as compared with healthy controls (Phillips et al, 2003) and in CALU-1 and NCI-H2087 lung cancer cell lines (Filipiak et al, 2008; Sponring et al, 2009b). However, in order to confirm the calculated breath concentration of the studied VOCs, further empirical studies must be conducted in order to account for the number of tumour cells, as compared with the number of healthy cells in lung cancer tissue, and for other tissue that contribute to the breath volatolome.
As the artificial intelligence nanoarray used is based on the physiochemical response to the collective VOC signal (or volatolomic signature), it is unable to discriminate which VOCs contribute to a discriminatory signal. However, it has previously been shown to provide a more accurate identification when multiple signals (features) are integrated into a single DFA CV1 value (Brereton, 1990; Ionescu et al, 2002). The inherent integration of multiple signals, that is a feature of both the nanoarrays themselves and the DFA technique, is likely a significant part of the reason for their better performance compared with TD-GC–MS, although that the sorbent material and TD-GC–MS technique employed may limit the range of VOC detected is also a factor. In the present study, nanoarray comparisons together with pattern recognition methods (i.e., DFA) demonstrated the ability to discriminate metabolic differences due to individual mutations in the presence of different genetic backgrounds based on volatolomic signature. While the greater accuracy and potential ease of use provided by sensor array-based techniques may prove a distinct advantage in the clinical setting, current research still benefits form VOC identification provided by TD-GC–MS and analogous techniques, as VOCs identified can be compared between studies and linked to metabolic processes. It is likely that the VOCs identified by TD-GC–MS and those discriminatory on sensor arrays significantly overlap, as has been demonstrated previously using synthetic mixtures of discriminatory VOC identified in lung cancer breath, including 4-methyloctane detected here (Peng et al, 2009).
The presented results of this study may, in the future, lead to the development of nanomaterial-based device for the detection of the genetic mutation profile from the headspace of a lung cancer tissue specimen obtained through biopsy, or from patients' breath. This could help guiding doctors in making medical decisions more rapidly than current molecular genetic approaches. However, it should be noted that the biochemical origin of most VOC in breath is not fully elucidated and it is possible that other organs and microbiota will also make a contribution. Although the presence of specific cancer driver mutations (RAS and TP53) apparently has an influence on the volatolomic profile of cells lines, it is unclear if that is a direct effect of these cancer signalling pathways, or accounted for by additional genetic alterations underlying the variations in metabolic phenotype. It might therefore be possible for different genetic alterations to converge on the same metabolic pathway, giving similar VOC signatures or confounding the detection of either mutation; this may partly explain the lower accuracy seen when detecting RAS mutation across multiple cancer-derived cell lines harbouring additional mutations (Peled et al, 2013).
The HBECs used in this study, despite being immortalised (through addition of CDK4 and TERT) and having up to two additional cancer-related mutations (KRAS activation and knockdown of TP53) are not malignant when introduced into mouse model systems (Sato et al, 2006). Hence they represent early, pre-cancerous stages of lung cancer progression. This raises the possibility that it may be possible to detect earlier stages of lung tumours, or pre-cancerous conditions, and identify those at higher risk of developing cancer for recruitment into screening programmes.
The genetic alterations utilised in the study affect two of the most frequently in mutated genes in lung cancer (Massion and Carbone, 2003; The Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine (NGM), 2013; Weinstein et al, 2013). While KRAS mutations are most prevalent in NSCLC of adenocarcinoma histology, TP53 mutation (usually resulting in loss of function as mimicked by the depletion in the HBEC-3KT53 and HBEC-3KTR53 cells) is common across multiple histological types; for example, in The Cancer Genome Atlas (Weinstein et al, 2013) KRAS mutations: 28% adenocarcinoma and 1% squamous cell carcinoma, TP53 mutations: 53% adenocarcinoma and 79% squamous cell carcinoma. This would support the possibility of using the discriminatory features identified here within devices for lung cancer detection, but it is likely that other genetic alterations, the physical features of the tumour and patient characteristics would modify the volatolomic signature, meaning the VOCs identified herein might not be discriminatory for all cancers with these mutations. However, that multiple mutations can be independently discriminated in cancer-derived cell lines by the same techniques has previously been demonstrated (Peled et al, 2013), showing that the presence of other mutations does not preclude determination of VOC signatures for a specific mutation. It remains to be elucidated if those discriminant signals that best detect lung cancer in the screening or diagnostic setting (which by definition should detect all cancer sub-types) can be measured at the same time as those that have the power to determine the molecular sub-type, but in principle this could be achieved by incorporation of the appropriate nanoarrays into a single device.
The advantages of cost, scale and implementation in community medicine, which are potentially available with nanoscale sensor devices, mean that along with other risk assessment tools (Cassidy et al, 2008) breath sensors may be a first-line tool to identify those who will benefit from more regular lung cancer screening. Both the ability to correlate breath VOC signatures with genetic changes in tumours and the sensitivity required for earlier diagnosis require significant further empirical evidence.
Acknowledgments
The research leading to these results has received funding from the FP7-Health Program under the LCAOS grant agreement (no. 258868).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Amann A, Corradi M, Mazzone P, Mutti A. Lung cancer biomarkers in exhaled breath. Expert Rev Mol Diagn. 2011;11 (2:207–217. doi: 10.1586/erm.10.112. [DOI] [PubMed] [Google Scholar]
- Barash O, Peled N, Hirsch FR, Haick H. Sniffing the unique "odor print" of non-small-cell lung cancer with gold nanoparticles. Small. 2009;5 (22:2618–2624. doi: 10.1002/smll.200900937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash O, Peled N, Tisch U, Bunn PA, Jr, Hirsch FR, Haick H. Classification of lung cancer histology by gold nanoparticle sensors. Nanomedicine. 2012;8 (5:580–589. doi: 10.1016/j.nano.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett WP, Colby TV, Travis WD, Borkowski A, Jones RT, Lane DP, Metcalf RA, Samet JM, Takeshima Y, Gu JR, et al. p53 protein accumulates frequently in early bronchial neoplasia. Cancer Res. 1993;53 (20:4817–4822. [PubMed] [Google Scholar]
- Brereton R. Chemometrics: Application of mathematics statistics to laboratory systems. Elis Horwood; 1990. [Google Scholar]
- Broza YY, Haick H. Nanomaterial-based sensors for detection of disease by volatile organic compounds. Nanomedicine. 2013;8 (5:785–806. doi: 10.2217/nnm.13.64. [DOI] [PubMed] [Google Scholar]
- Cassidy A, Myles JP, van Tongeren M, Page RD, Liloglou T, Duffy SW, Field JK. The LLP risk model: an individual risk prediction model for lung cancer. Br J Cancer. 2008;98 (2:270–276. doi: 10.1038/sj.bjc.6604158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu F, Wang Y, Pan Y, Lu D, Wang P, Ying K, Chen E, Zhang W. A study of the volatile organic compounds exhaled by lung cancer cells in vitro for breath diagnosis. Cancer. 2007;110 (4:835–844. doi: 10.1002/cncr.22844. [DOI] [PubMed] [Google Scholar]
- Coate LE, John T, Tsao MS, Shepherd FA. Molecular predictive and prognostic markers in non-small-cell lung cancer. Lancet Oncol. 2009;10 (10:1001–1010. doi: 10.1016/S1470-2045(09)70155-X. [DOI] [PubMed] [Google Scholar]
- Dragonieri S, Annema JT, Schot R, Van Der Schee MPC, Spanevello A, Carratú P, Resta O, Rabe KF, Sterk PJ. An electronic nose in the discrimination of patients with non-small cell lung cancer and COPD. Lung Cancer. 2009;64 (2:166–170. doi: 10.1016/j.lungcan.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Fassina A, Gazziero A, Zardo D, Corradin M, Aldighieri E, Rossi GP. Detection of EGFR and KRAS mutations on trans-thoracic needle aspiration of lung nodules by high resolution melting analysis. J Clin Pathol. 2009;62 (12:1096–1102. doi: 10.1136/jcp.2009.067587. [DOI] [PubMed] [Google Scholar]
- Filipiak W, Sponring A, Filipiak A, Ager C, Schubert J, Miekisch W, Amann A, Troppmair J. TD-GC-MS analysis of volatile metabolites of human lung cancer and normal cells in-vitro. Cancer Epidem Biomar. 2010;19:182–195. doi: 10.1158/1055-9965.EPI-09-0162. [DOI] [PubMed] [Google Scholar]
- Filipiak W, Sponring A, Mikoviny T, Ager C, Schubert J, Miekisch W, Amann A, Troppmair J. Release of volatile organic compounds (VOCs) from the lung cancer cell line CALU-1 in vitro. Cancer Cell Inter. 2008;8:17. doi: 10.1186/1475-2867-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haick H, Broza YY, Mochalski P, Ruzsanyi V, Amann A. Assessment, origin, and implementation of breath volatile cancer markers. Chem Soc Rev. 2013;43:1423–1449. doi: 10.1039/c3cs60329f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim M, Broza YY, Barash O, Peled N, Phillips M, Amann A, Haick H. Volatile organic compounds of lung cancer and possible biochemical pathways. Chem Rev. 2012;112:5949–5966. doi: 10.1021/cr300174a. [DOI] [PubMed] [Google Scholar]
- Hamilton SR. Molecular pathology. Mol Oncol. 2012;6 (2:177–181. doi: 10.1016/j.molonc.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu R, Llobet E, Vilanova X, Brezmes J, Sueiras JE, Calderer J, Correig X. Quantitative analysis of NO2 in the presence of CO using a single tungsten oxide semiconductor sensor and dynamic signal processing. Analyst. 2002;127 (9:1237–1246. doi: 10.1039/b205009a. [DOI] [PubMed] [Google Scholar]
- Lim EH, Zhang SL, Li XL, Yap WS, Howe TC, Tan BP, Lee YS, Wong D, Khoo KL, Seto KY, Tan L, Agasthian T, Koong HN, Tam J, Tan C, Caleb M, Chang A, Ng A, Tan P. Using Whole Genome Amplification (WGA) of low-volume biopsies to assess the prognostic role of EGFR, KRAS, p53, and CMET mutations in advanced-stage Non-small Cell Lung Cancer (NSCLC) J Thorac Oncol. 2009;4 (1:12–21. doi: 10.1097/JTO.0b013e3181913e28. [DOI] [PubMed] [Google Scholar]
- Massion PP, Carbone DP. The molecular basis of lung cancer: molecular abnormalities and therapeutic implications. Respir Res. 2003;4:12. doi: 10.1186/1465-9921-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone PJ, Wang XF, Xu Y, Mekhail T, Beukemann MC, Na J, Kemling JW, Suslick KS, Sasidhar M. Exhaled breath analysis with a colorimetric sensor array for the identification and characterization of lung cancer. J Thorac Oncol. 2011;7 (1:137–142. doi: 10.1097/JTO.0b013e318233d80f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochalski P, King J, Kupferthaler A, Unterkofler K, Hinterhuber H, Amann A. Human blood and plasma partition coefficients for C4-C8 n-alkanes, isoalkanes, and 1-alkenes. Int J Toxicol. 2012;31 (3:267–275. doi: 10.1177/1091581812442689. [DOI] [PubMed] [Google Scholar]
- Osada H, Takahashi T. Genetic alterations of multiple tumor suppressors and oncogenes in the carcinogenesis and progression of lung cancer. Oncogene. 2002;21 (48:7421–7434. doi: 10.1038/sj.onc.1205802. [DOI] [PubMed] [Google Scholar]
- Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2 (1:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled N, Barash O, Tisch U, Ionescu R, Broza YY, Ilouze M, Mattei J, Bunn PA, Jr, Hirsch FR, Haick H. Volatile fingerprints of cancer specific genetic mutations. Nanomedicine. 2013;9 (6:758–766. doi: 10.1016/j.nano.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Tisch U, Adams O, Hakim M, Shehada N, Broza YY, Billan S, Abdah-Bortnyak R, Kuten A, Haick H. Diagnosing lung cancer in exhaled breath using gold nanoparticles. Nat Nanotechnol. 2009;4 (10:669–673. doi: 10.1038/nnano.2009.235. [DOI] [PubMed] [Google Scholar]
- Phillips M, Altorki N, Austin JHM, Cameron RB, Cataneo RN, Kloss R, Maxfield RA, Munawar MI, Pass HI, Rashid A. Detection of lung cancer using weighted digital analysis of breath biomarkers. Clin Chim Acta. 2008;393:76–84. doi: 10.1016/j.cca.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M, Cataneo RN, Cummin ARC, Gagliardi AJ, Gleeson K, Greenberg J, Maxfield RA, Rom WN. Detection of lung cancer with volatile markers in the breath. Chest. 2003;123 (6:2115–2123. doi: 10.1378/chest.123.6.2115. [DOI] [PubMed] [Google Scholar]
- Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, Milchgrub S, Smith AL, Souza RF, Gilbey L, Zhang X, Gandia K, Vaughan MB, Wright WE, Gazdar AF, Shay JW, Minna JD. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64 (24:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24 (3:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, Ramirez RD, Sunaga N, Gazdar AF, Shay JW, Minna JD. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66 (4:2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- Smith D, Wang T, Sulé-Suso J, Spanel P, El Haj A. Quantification of acetaldehyde released by lung cancer cells in vitro using selected ion flow tube mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:845–850. doi: 10.1002/rcm.984. [DOI] [PubMed] [Google Scholar]
- Sponring A, Filipiak W, Ager C, Schubert J, Miekisch W, Amann A, Troppmair J. Analysis of volatile organic compounds (VOCs) in the headspace of NCI-H1666 lung cancer cells. Cancer Biomark. 2010;7:153–161. doi: 10.3233/CBM-2010-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponring A, Filipiak W, Mikoviny T, Ager C, Schubert J, Miekisch W, Amann A, Troppmair J. Release of volatile organic compounds from the lung cancer cell line NCI-H2087 in vitro. Anticancer Res. 2009;29:419–426. [PubMed] [Google Scholar]
- Sponring A, Filipiak W, Mikoviny T, Ager C, Schubert J, Miekisch W, Amann A, Troppmair J. Release of volatile organic compounds from the lung cancer cell line NCI-H2087 in vitro. Anticancer Res. 2009;29 (1:419–426. [PubMed] [Google Scholar]
- Sulé-Suso J, Pysanenko A, Španel P, Smith D. Quantification of acetaldehyde and carbon dioxide in the headspace of malignant and non-malignant lung cells in vitro by SIFT-MS. Analyst. 2009;134:2419–2425. doi: 10.1039/b916158a. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, Vinocour M, Levitt M, Pass H, Gazdar AF, Minna JD. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246 (4929:491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- The Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine (NGM) A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5 (209:209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisch U, Haick H. Nanomaterials for cross-reactive sensor arrays. MRS Bull. 2010;35:797–802. [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2 (8:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45 (10:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.