Abstract
A defect in the assembly of the oligosaccharide donor (Dol-PP-GlcNAc2Man9Glc3) for N-linked glycosylation causes hypoglycosylation of proteins by the oligosaccharyltransferase (OST). Mammalian cells express two OST complexes that have different catalytic subunits (STT3A or STT3B). We monitored glycosylation of proteins in asparagine-linked glycosylation 6 (ALG6) deficient cell lines that assemble Dol-PP-GlcNAc2Man9 as the largest oligosaccharide donor. Based upon pulse labeling experiments, 30–40% of STT3A-dependent glycosylation sites and 20% of STT3B-dependent sites are skipped in ALG6-congenital disorders of glycosylation fibroblasts supporting previous evidence that the STT3B complex has a relaxed preference for the fully assembled oligosaccharide donor. Glycosylation of STT3B-dependent sites was more severely reduced in the ALG6 deficient MI8-5 cell line. Protein immunoblot analysis and RT–PCR revealed that MI8-5 cells express 2-fold lower levels of STT3B than the parental Chinese hamster ovary cells. The combination of reduced expression of STT3B and the lack of the optimal Dol-PP-GlcNAc2Man9Glc3 donor synergize to cause very severe hypoglycosylation of proteins in MI8-5 cells. Thus, differences in OST subunit expression can modify the severity of hypoglycosylation displayed by cells with a primary defect in the dolichol oligosaccharide assembly pathway.
Keywords: asparagine-linked glycosylation, congenital disorders of glycosylation, lipid-linked oligosaccharide, oligosaccharyltransferase
Introduction
Asparagine-linked glycosylation is one of the most common protein modification reactions in eukaryotic cells. The oligosaccharyltransferase (OST) transfers a high mannose oligosaccharide (GlcNAc2Man9Glc3) from a lipid-linked oligosaccharide (LLO) donor onto consensus acceptor sites (N-X-T/S; X≠P) in nascent polypeptides in the lumen of the rough endoplasmic reticulum.
The biosynthetic assembly of LLO by the ALG (asparagine-linked glycosylation) pathway is initiated on the cytoplasmic face of the endoplasmic reticulum by glycosyltransferases that utilize sugar nucleotides (UDP-N-acetyl glucosamine and GDP-mannose) as the donor substrates (as reviewed by Aebi 2013). The dolichol pyrophosphate (Dol-PP)-linked oligosaccharide Dol-PP-GlcNAc2Man5 is flipped across the ER membrane, where four additional mannose residues and three glucose residues are added by glycosyltransferases that utilize Dol-P-Man and Dol-P-Glc as the saccharide donors to yield the fully assembled oligosaccharide donor (Dol-PP-(GlcNAc2Man9Glc3)).
LLO assembly intermediates that lack the terminal glucose residues are transferred by the OST at reduced rates relative to the fully assembled donor (Turco et al. 1977; Trimble et al. 1980; Karaoglu et al. 2001). Yeast mutants that lack any of the glycosyltransferases in the ALG pathway synthesize hypoglycosylated proteins (as reviewed by Burda and Aebi 1999). The ALG6 protein is the α,1-3 glucosyltransferase that adds the first of three glucose residues to the Dol-PP-GlcNAc2Man9 assembly intermediate. Yeast alg6Δ cells that accumulate Dol-PP-GlcNAc2Man9 as the largest oligosaccharide donor synthesize variants of yeast carboxypeptidase Y that on average lack one of the four oligosaccharides that are normally present on carboxypeptidase Y (Reiss et al. 1996). Mutations in human ALG pathway genes cause the majority of the currently described variants of type I congenital disorders of glycosylation (CDG) , a multisystemic disease caused by hypoglycosylation of human glycoproteins (as reviewed in Haeuptle and Hennet 2009). Sequencing of cDNAs from ALG6-CDG fibroblasts has disclosed several point mutations (e.g., A333V, S308R) that severely reduce ALG6 activity (Imbach et al. 2000; Westphal et al. 2000; Newell et al. 2003). MI8-5 cells, a temperature sensitive Chinese hamster ovary (CHO) derivative, also lack detectable ALG6 activity (Quellhorst et al. 1999; Foulquier et al. 2004), but the molecular basis of the ALG6 defect in MI8-5 cells is not known.
MI8-5 cells have proven particularly useful for the analysis of glucosylation of protein bound oligosaccharides by UDP-glucose glycoprotein glucosyltransferase (UGGT) because a protein-linked GlcNAc2Man9Glc1 glycan in MI-85 cells cannot be derived by trimming of a GlcNAc2Man9Glc3 oligosaccharide but instead is diagnostic of UGGT activity (Cacan et al. 2001; Pearse et al. 2008, 2010).
Mammalian cells express OST complexes that are composed of either STT3A or STT3B as the catalytic subunit assembled together with a shared set of accessory subunits (Kelleher et al. 2003). The two OST complexes have partially overlapping functions in N-linked glycosylation. STT3A complexes are associated with the translocation channel and mediate cotranslational glycosylation, while STT3B complexes can modify acceptor sites that have been skipped by STT3A (Ruiz-Canada et al. 2009; Shrimal, Trueman, et al. 2013). The STT3B complex can modify skipped sites cotranslationally, or posttranslocationally after the complete protein has entered the ER lumen. Kinetic analysis of the purified canine OSTs revealed that the STT3B complex has a several-fold reduced preference for the fully assembled oligosaccharide donor relative to the highly selective STT3A complex (Kelleher et al. 2003), suggesting that STT3B substrates may be less sensitive to a defect in the LLO assembly pathway. However, these kinetic experiments were conducted using purified OST complexes incorporated into phospholipid-detergent mixed micelles, so it was not clear whether the relaxed selection of LLO assembly intermediates by the STT3B complex would also occur within intact cells.
Here, we have analyzed glycosylation of a panel of glycoproteins in ALG6-CDG fibroblasts and ALG6-deficient MI8-5 cells. STT3A-dependent substrates were hypoglycosylated to a similar extent in both cell lines. Unexpectedly, STT3B substrates were more severely hypoglycosylated in MI8-5 cells than in ALG6-CDG cells. Protein immunoblot analysis revealed that MI8-5 cells express 2-fold lower levels of STT3B than parental CHO cells, indicating that the ALG6 deficiency and a reduction in STT3B content both contribute to severe hypoglycosylation of glycoproteins in MI8-5 cells.
Results
ALG6-deficient CHO and human cells
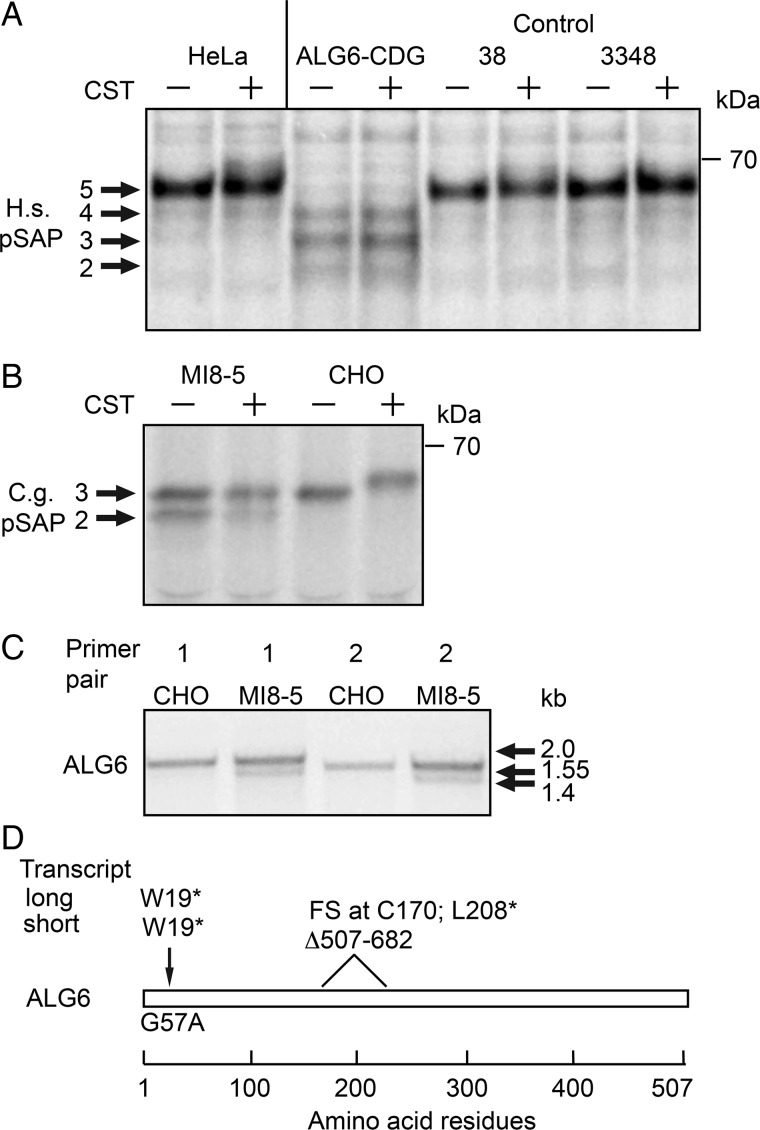
To determine whether a deficiency in LLO assembly has a differential effect upon glycosylation of glycoproteins by the STT3A and STT3B complexes, we needed cells with a severe defect in LLO biosynthesis. Our attempt to deplete ALG6 activity in HeLa cells using siRNAs was unsatisfactory as the resulting LLO pool after 72 h of siRNA treatment contained a mixture of Dol-PP-GlcNAc2Man9 and Dol-PP-GlcNAc2Man9Glc3 due to residual ALG6 activity (data not shown). As an alternative approach, we analyzed two previously described ALG6-deficient cell lines. Fibroblasts from an ALG6-CDG patient that carries the A333V mutation on the maternal ALG6 allele and the S308R and Y131H mutations on the paternal ALG6 allele were selected for this analysis (Westphal et al. 2000). The ALG6-CDG fibroblasts, control human fibroblasts (38 and 3348) and HeLa cells were pulse labeled with Tran-35S label with or without pretreatment with the glucosidase inhibitor castanospermine (Figure 1A). Glucosidases 1 and 2 sequentially remove the three glucose residues from protein bound GlcNAc2Man9Glc3 to yield GlcNAc2Man9; hence, castanospermine (CST) will block trimming of protein bound oligosaccharides produced by control human fibroblasts. Prosaposin, an endogenous human glycoprotein with five acceptor sites was immunoprecipitated and resolved by PAGE in SDS. The fully glycosylated prosaposin (pSAP) synthesized by HeLa cells or control fibroblasts migrated slightly slower when synthesized in the presence of CST due to the retention of the glucose residues on the protein-linked glycans. The ALG6-CDG fibroblasts synthesized pSAP glycoforms lacking 1-3 N-linked glycans. Importantly, CST treatment did not reduce the gel mobility of pSAP synthesized by ALG6-CDG fibroblasts consistent with the previous evidence that primary skin fibroblasts from this ALG6-CDG patient assemble Dol-PP-GlcNAc2Man9 as the largest oligosaccharide donor (Westphal et al. 2000). Endogenous CHO pSAP, which has three acceptor sites, is hypoglycosylated in MI8-5 cells (Figure 1B). CST treatment reduces the gel mobility of pSAP synthesized by CHO cells, but does not alter the gel mobility of pSAP synthesized by MI8-5 cells as reported previously (Pearse et al. 2010).
Fig. 1.
Comparison of ALG6 deficient cell lines. (A) HeLa cells, ALG6-CDG fibroblasts and control human fibroblasts (38 and 3348) or (B) MI8-5 cells and CHO cells were incubated with 1 mM castanospermine (CST) or solvent during 20 min of starvation and 10 min of labeling as indicated. Endogenous human (H.s.) or CHO (C.g.) prosaposin (pSAP) glycoforms were immunoprecipitated with anti-saposin D sera and resolved by SDS–PAGE. The indicated protein molecular weights shown in all figures are of prestained molecular-weight markers electrophoresed on all gels. (C) Agarose gel electrophoresis of the ALG6 coding sequence from CHO and MI8-5 cells after amplification by RT-PCR using two primer pairs. (D) Schematic representation of the Alg6 coding sequence in MI8-5 cells. The G57A mutation introduces a stop codon into both ALG6 mRNAs. The short mRNA transcript lacking exon 8 has a frameshift (FS) mutation at codon 170 followed by a nonsense mutation at codon 208.
The molecular basis of the ALG6 deficiency in MI8-5 cells was unknown. RT-PCR amplification of the ALG6 mRNA from MI8-5 cells revealed the presence of two mRNAs, one of which migrated more rapidly than the ALG6 mRNA from the parental CHO cells (Figure 1C). Sequencing of ALG6 cDNAs showed that both the long and short mRNAs have a G57A mutation that replaces W19 with a stop codon (Figure 1D). In addition, the shorter mRNA has an internal 175 bp deletion corresponding to the absence of exon 8. Thus, both copies of the ALG6 gene in MI8-5 cells encode null alleles consistent with the previous evidence that glucosylated LLO is not synthesized by MI8-5 cells (Quellhorst et al. 1999; Foulquier et al. 2004).
Protein N-glycosylation in ALG6-deficient cell lines
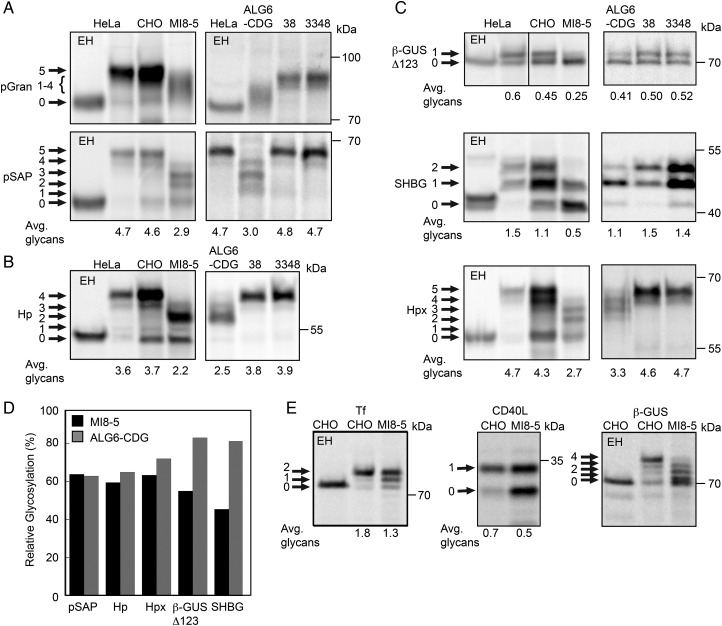
Having confirmed that MI8-5 cells and ALG6-CDG fibroblasts are deficient in ALG6 glucosyltransferase activity, we next asked whether this deficiency had a similar impact upon glycosylation of a panel of human glycoproteins. We have identified acceptor sites in certain glycoproteins that are particularly sensitive to depletion of the STT3A or STT3B complexes in HeLa cells. First, we analyzed glycosylation of human pSAP and progranulin (pGran), which are cotranslationally glycosylated by the STT3A isoform of the OST complex (Ruiz-Canada et al. 2009; Shrimal, Ng, et al. 2013). CHO and MI8-5 cells were transformed with expression plasmids for human pSAP or progranulin to allow a direct comparison of the same glycoprotein substrates since CHO pSAP and pGran have fewer NXT/S sites than human pSAP and pGran. Brief pulse labeling or pulse-chase labeling periods were used for all radiolabeling experiments to insure that the observed glycoform patterns for the substrates are not altered by Golgi localized oligosaccharide processing reactions or by ER-associated protein degradation. The labeling periods for each glycoprotein substrate were optimized in previous publications (Ruiz-Canada et al. 2009; Shrimal and Gilmore 2013; Shrimal, Ng, et al. 2013; Shrimal, Trueman, et al. 2013). Human pGran, the precursor for granulins A–G, has five glycosylation sites. Granulin was hypoglycosylated in both ALG6-deficient cell lines (Figure 2A). However, we were not able to quantitate the extent of hypoglycosylation due to the diffuse migration of the granulin glycoforms. Human pSAP was hypoglycosylated by MI8-5 cells and ALG6-CDG to a similar extent as indicated by the similar decrease in average glycans per chain (Figure 2A). The surprisingly low sequence identity (66%) between CHO pSAP and human pSAP, and the absence of two glycosylation sites in CHO pSAP likely explains why glycosylation of human pSAP is more severely reduced in MI8-5 cells relative to CHO cells. Haptoglobin (Hp), a secreted protein with four glycosylation sites, is primarily glycosylated by a cotranslational STT3A-dependent pathway (Shrimal and Gilmore 2013). As observed for pSAP, glycosylation of Hp was reduced in both ALG6-deficient cell lines (Figure 2B). Based upon analysis of three STT3A-dependent substrates (pSAP, pGran and Hp) in both ALG6-deficient cell lines, we estimate that the mammalian STT3A complex skips 30–40% of sequons when cells lack the fully assembled oligosaccharide donor (Figure 2D).
Fig. 2.
Hypoglycosylation of proteins in ALG6 deficient cell lines. Human (A–D) and Chinese hamster derived (A–E) cell lines were transfected with expression vectors for the following human glycoproteins: (A) progranulin (pGran-DDKHis) and prosaposin (pSAP-DDKHis); (B) haptoglobin (Hp-DDKHis); or (C) a β-glucuronidase derivative with a single C-terminal glycosylation site (β-GUSΔ123-MycDDK), sex hormone-binding globulin (SHBG) or hemopexin (Hpx-DDKHis); (E) transferrin (Tf-DDKHis), CD40 ligand (CD40L-Myc) or wild-type β-glucuronidase (β-GUS-MycDDK). The cells were pulse-labeled or pulse-chase labeled as described in Materials and Methods. The glycoprotein substrates were precipitated with anti-DDK, anti-Myc or anti-SHBG sera and then resolved by SDS–PAGE. Endogenous progranulin and prosaposin were immunoprecipitated from fibroblasts and HeLa cells using anti-pSAP and anti-pGRAN sera. EH designates treatment with endoglycosidase H. Quantified values below gel lanes (A–C, E) are the average number of glycans per protein chain from two or more experiments. (D) The relative glycosylation efficiency (%) of substrates that were assayed in both ALG6-deficient cell lines was calculated by averaging the glycosylation efficiency of the mutant cell line relative to the relevant wild-type control cell line.
We expected that STT3B-dependent substrates would be less sensitive to the loss of ALG6 activity because the purified STT3A complex displays a stronger preference for fully assembled LLO than the STT3B complex when assayed in vitro using synthetic peptides as acceptor substrates (Kelleher et al. 2003). The wild-type and ALG6-deficient cells were transformed with a β-glucuronidase derivative (β-GUSΔ123) or sex hormone-binding globulin (SHBG), two secreted proteins that have one or two extreme C-terminal STT3B-dependent glycosylation sites, respectively (Shrimal, Trueman, et al. 2013). Although wild-type human fibroblasts and CHO cells glycosylate β-GUSΔ123 with similar efficiency, MI8-5 cells had a more severe defect in glycosylation of β-GUSΔ123 than the ALG6-CDG cells (Figure 2C). These differences in relative glycosylation efficiency by the two ALG6-deficient cell lines were quite reproducible. The N380RS site in SHBG is modified with roughly 50% efficiency in HeLa cells resulting in the glycoform doublet (Shrimal, Trueman, et al. 2013). MI8-5 cells also had a more severe defect in glycosylation of SHBG than the ALG6-CDG cells. Based upon analysis of SHBG and β-GUSΔ123, 20% of STT3B-dependent sites were skipped in ALG6-CDG cells relative to the control human fibroblasts, consistent with the hypothesis that glycosylation of acceptor sites by STT3B is less sensitive to a defect in LLO assembly. Conversely, >50% of STT3B sites were skipped in MI8-5 cells relative to the parental CHO cell line (Figure 2D). Thus, ALG6-CDG and MI8-5 cells yielded conflicting results concerning the ability of the STT3B complex to utilize a non-optimal donor substrate. Hemopexin (Hpx) has an extreme C-terminal STT3B-dependent site and one internal site that is partially STT3B dependent. As observed for SHBG and β-GUSΔ123, glycosylation of Hpx was more defective in MI8-5 cells than in ALG6-CDG cells (Figure 2C and D).
Three additional substrates were analyzed to validate the extent of protein Hypoglycosylation in MI8-5 cells (Figure 2E). Serum transferrin (Tf), the standard diagnostic marker for protein hypoglycosylation by CDG-1 patients (Freeze and Aebi 2005), is an STT3A-dependent substrate (Shrimal, Ng, et al. 2013; Shrimal, Trueman, et al. 2013). In addition to fully glycosylated Tf, MI8-5 cells synthesized Tf variants lacking one or both glycans. CD40L is a type 2 integral membrane proteins that shows mild (10–20%) hypoglycosylation in STT3B depleted cells (Shrimal, Trueman, et al. 2013). Compared with the parental CHO cells, glycosylation of CD40L was reduced by 30% in MI8-5 cells (Figure 2E). Glycosylation of the three internal sites in β-GUS is not reduced in HeLa cells unless both STT3A and STT3B are depleted (Shrimal, Trueman, et al. 2013). Accurate quantification of the extent of β-GUS hypoglycosylation by MI8-5 cells was not feasible due to the close spacing of the major glycoforms that lack 3 and 4 glycans (Figure 2E). Analysis of these three additional glycoproteins revealed that MI8-5 cells have a severe defect in protein N-glycosylation that effects all substrates tested regardless of whether the acceptor sites are primarily glycosylated by STT3A or STT3B.
MI8-5 cells assemble low levels of the STT3B complex
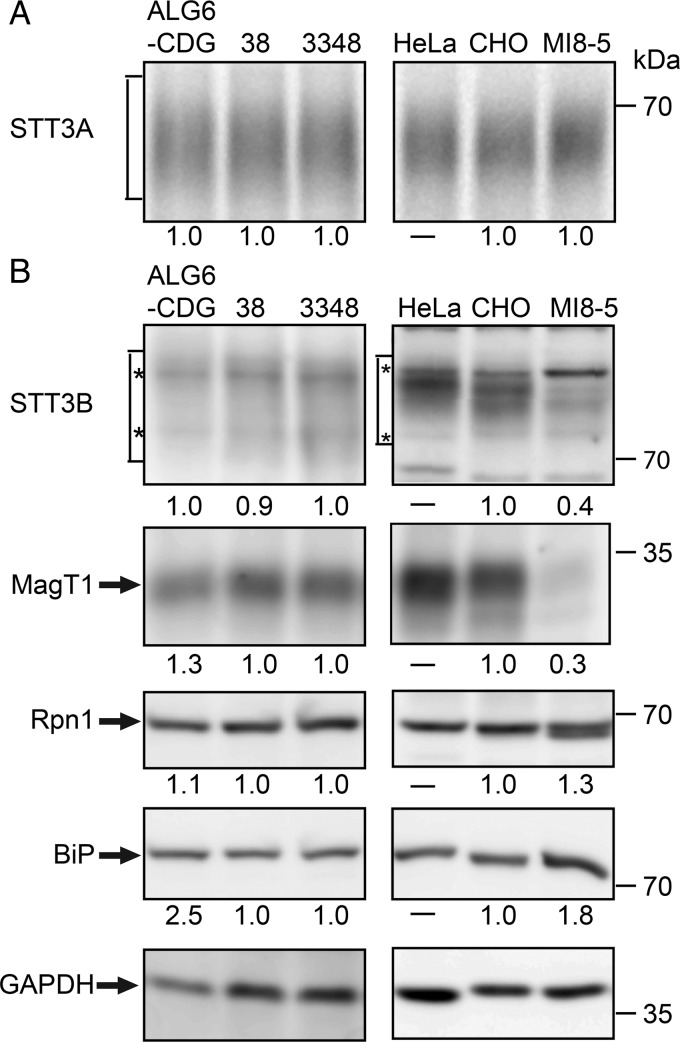
The unexpected difference in glycosylation of STT3B-dependent substrates in the two ALG6-deficient cell lines prompted further analysis of the glycosylation machinery in these cells. STT3A levels were compared by immunoprecipitation of STT3A from cells that were pulse labeled for 50 min (Figure 3A). Pulse labeling was used for this analysis because nonspecific background bands in the vicinity of STT3A made accurate quantitation of immunoblots difficult. Previously, we had found that a 50 min pulse labeling period allowed us to detect a reduction in stable expression of STT3A in fibroblasts from a STT3A-CDG patient (Shrimal, Ng, et al. 2013). There were no obvious difference in STT3A levels between the ALG6-deficient cell lines and the cell-type-matched controls. Expression levels shown below gel images for the ALG6-CDG and MI8-5 cells were normalized relative to control fibroblasts and CHO cells, respectively.
Fig. 3.
Expression of OST subunits in ALG6-deficient cell lines. (A) STT3A was immunoprecipitated from total cell extracts of cells that were pulse-labeled for 50 min with Tran-35S label. Equal quantities of 35S-labeled protein were used for each immunoprecipitation. (B) Protein immunoblot detection of STT3B, MagT1, ribophorin 1 (Rpn1), the ER chaperone BiP and GAPDH. Seventy-five micrograms of protein were electrophoresed in each lane. The asterisks designate background bands that migrate in the vicinity of STT3B. Expression of the OST subunits and BiP were adjusted relative to the gel loading control (GAPDH), and then normalized to the relevant wild-type control (3348 fibroblasts or CHO cells) to compare the relative expression of OST subunits in the ALG6-deficient cell lines. Quantified values below gel lanes are for the displayed image that is representative of two or more experiments.
Total cell extracts prepared from wild-type and mutant cells were resolved by SDS–PAGE for protein immunoblot analysis using antibodies raised against OST subunits (Figure 3B). HeLa cell extracts were electrophoresed adjacent to CHO and MI8-5 cell extracts to provide a protein mobility marker. Although the diffuse migration of STT3B makes accurate quantification challenging, the immunoblot signal for STT3B was roughly 2-fold lower in MI8-5 cells than in CHO cells. To insure that the observed reduction in STT3B was not explained by a technical problem in cell extract preparation, we used the soluble protein glyceraldehyde 3′ phosphate dehydrogenase (GAPDH) as a protein loading control, and adjusted the calculated expression levels accordingly. The cellular levels of MagT1, a subunit of the STT3B complex, are reduced in HeLa cells that have been treated with a STT3B-specific siRNA and in STT3B-CDG fibroblasts that express low levels of STT3B (Cherepanova et al. 2014). Protein immunoblotting revealed that the MagT1 content of MI8-5 cells correlates nicely with the reduction in STT3B content (Figure 3B). Presumably, MagT1 that is not assembled into the STT3B complex is unstable. Low expression of STT3B and MagT1 in MI8-5 cells is not a general consequence of an ALG6 deficiency, as the ALG6-CDG cells have normal levels of both proteins. Ribophorin I, which is a subunit of both the STT3A and STT3B complexes, migrates as a doublet in MI8-5 cells. There was no reduction in ribophorin I content in the ALG6-CDG cells, but instead we observed a slight increase in ribophorin I levels in MI8-5 cells. Hypoglycosylation of proteins in the ER causes an induction of the unfolded protein response (UPR) as detected by increased expression of lumenal chaperones including BiP (Schroder and Kaufman 2005). BiP levels are elevated in ALG6-CDG cells relative to control human fibroblasts (Figure 3B) consistent with chronic activation of the UPR pathway. We observed a smaller increase in BiP levels in MI8-5 cells, in agreement with a previous report that ER chaperone levels show little or no induction in MI8-5 cells despite splicing of the XBP-1 mRNA by IRE1 (Pearse et al. 2008).
The Cricetulus griseus (Chinese hamster) STT3B sequence in the current NCBI database (XP 003496314.1) predicts a protein of 751 residues, which is considerably shorter than other mammalian STT3B proteins (822–826 residues). Residues 31–751 of the C. griseus STT3B sequence are >99% identical to residues 104–824 of Mesocricetus auratus (golden hamster) STT3B. Residues 1–30 of the current C. griseus STT3B sequence are spurious, bearing no homology to any segment of the STT3B protein. The missing protein sequence (∼103 residues) of CHO STT3B is encoded by exon 1, which is located 43 kB upstream of exon 2 in the human STT3B gene. This region of the C. griseus genome is missing from the most recent genome assembly. As our anti-STT3B antibody was raised against the N-terminus of human STT3B, we can exclude the possibility that CHO STT3B initiates at an internal methionine residue.
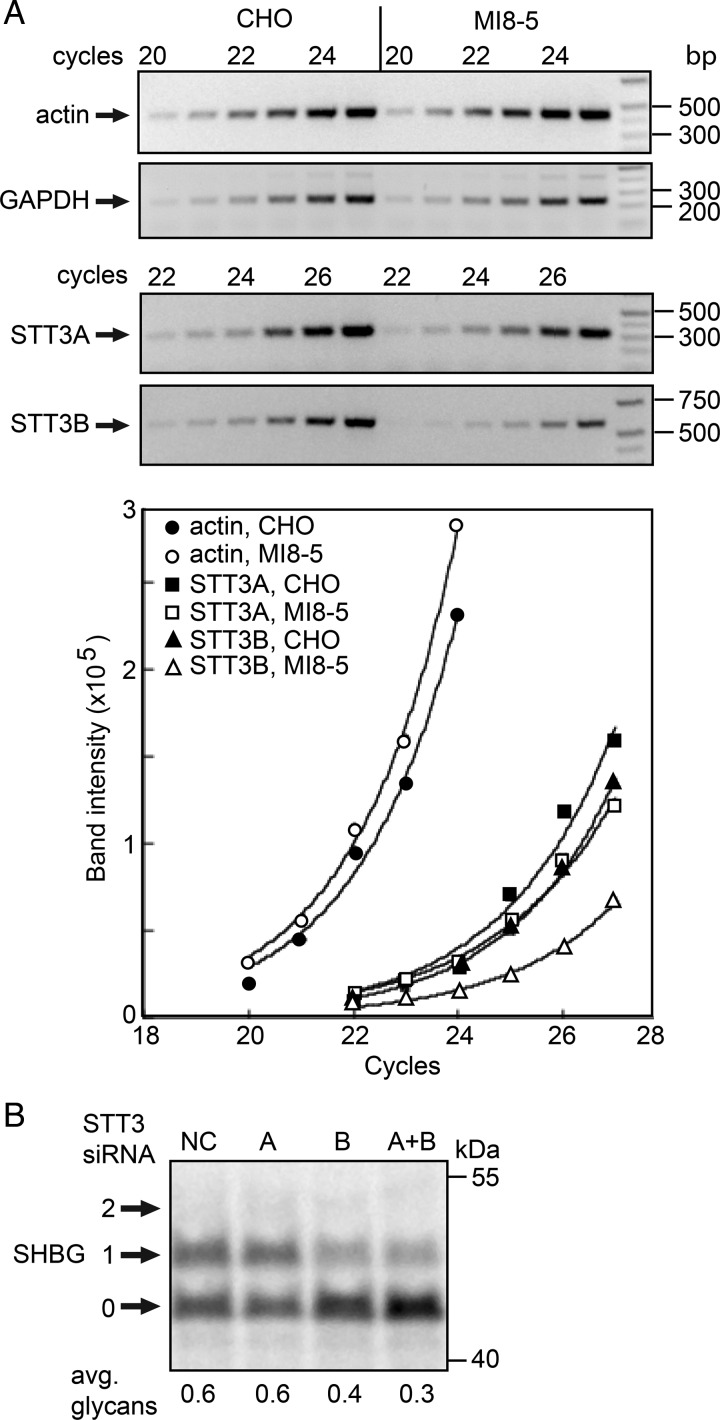
Sequencing of CHO and MI8-5-derived STT3B cDNAs disclosed no differences in the mRNA sequence encoding residues 104–824 of STT3B, hence a point mutation in this region cannot account for a reduction in STT3B expression. Our attempts to amplify the 5′ coding segment of CHO STT3B mRNA using primers designed using the M. auratus STT3B sequence were unsuccessful due to the high GC content of the 5′ segment of the STT3B mRNA. We next asked whether MI8-5 cells contain reduced amounts of mRNA encoding STT3B relative to STT3A and control mRNAs using semi-quantitative RT-PCR (Figure 4A). RT-PCR amplification profiles for actin, GAPDH and STT3A mRNAs were nearly identical in CHO and MI8-5 cells indicating that the specific content of these control mRNAs was similar. RT-PCR amplification of STT3B mRNA from MI8-5 cells showed an obvious lag relative to CHO cells indicating that the reduction in protein expression correlates with a reduction in STT3B mRNA content in MI8-5 cells.
Fig. 4.
MI8-5 cells express low amounts of active STT3B. (A) Actin, GAPDH, STT3A and STT3B expression in CHO and MI8-5 cells was examined by semi-quantitative RT-PCR. Amplification products collected after six sequential cycles (20–25 or 22–27) were resolved on agarose gels and stained with ethidium bromide. Images of the PCR amplification products were scanned and fit to the equation I = abN, where N = cycle number. (B) MI8-5 cells were treated for 48 h with a negative control siRNA (NC) or siRNAs specific for STT3A or STT3B. The cells were then transfected with an SHBG expression vector and pulse-labeled 24 h later. SHBG glycoforms (0–2 glycans) were immunoprecipitated with anti-SHBG and resolved by SDS–PAGE.
Previously, we reported that extreme C-terminal glycosylation sites like those in SHBG are almost exclusively modified by the STT3B complex (Shrimal, Trueman, et al. 2013). We next asked whether the STT3B protein detected in MI8-5 cells is active and responsible for the residual glycosylation of SHBG. Treatment of MI8-5 cells with an STT3B-specific siRNA caused a further reduction in SHBG glycosylation (Figure 4B), while treatment of MI8-5 cells with an STT3A siRNA did not alter SHBG glycosylation unless STT3A and STT3B were simultaneously depleted. Together, these three lines of evidence indicate that the MI8-5 cells express low levels of active STT3B, likely due to a defect in transcription or splicing of the STT3B mRNA.
Partial restoration of glycosylation activity in MI8-5 cells
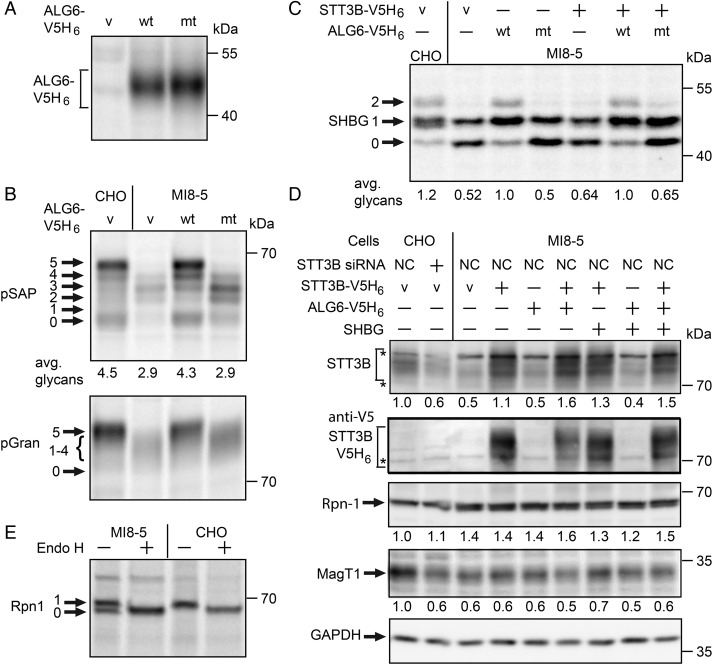
To better understand how the block in donor oligosaccharide assembly contributes to hypoglycosylation of proteins, we transfected MI8-5 cells with a wild-type or a mutant version (A333V S308R) of an ALG6-V5H6 expression vector. ALG6-V5H6 expression was detected by immunoprecipitation of the pulse-labeled protein with anti-V5 sera (Figure 5A). The tagged ALG6 construct was necessary, as commercially available ALG6 antibodies recognize a 58 kDa HeLa cell protein, but do not recognize ALG6-V5H6, which migrates as a diffuse 46 kDa protein due to the presence of multiple membrane spanning segments.
Fig. 5.
Restoration of N-glycosylation activity in ALG6-deficient cells. (A–D) CHO or MI8-5 cells were transfected with the indicated combinations of the following expression vectors: empty vector (v); wild type (wt) or mutant (mt) ALG6-V5H6, STT3B-V5H6, or glycoprotein substrates (pSAP, pGran or SHBG). (A) ALG6 was immunoprecipitated with anti-V5 sera from total cell extracts of pulse-labeled MI8-5 cells using equal quantities of 35S-labeled protein for each immunoprecipitation. (B) Glycosylation of substrates in CHO or MI8-5 cells expressing wild-type or mutant ALG6-V5H6 was assayed by a pulse-labeling procedure. All proteins were immunoprecipitated with anti-DDK sera. (C) Glycosylation of SHBG was evaluated by pulse-labeling 48 h after transfection with the protein expression vectors. (D) Cells were treated for 24 h with a negative control siRNA (NC) or siRNA specific for STT3B as indicated prior to transfection with the protein expression plasmids. Total cell extracts were prepared 48 h after transfection. Equal amounts of total proteins (75 µg) were resolved by SDS–PAGE for protein immunoblotting. Values beneath gel lanes were normalized relative to the GAPDH intensity, and then expressed relative to the parental cell line (CHO cells). The asterisks designate background bands that migrate in the vicinity of STT3B. Quantified values below gel lanes (B, C) are the average of results obtained in two or more experiments. (E). Ribophorin I was immunoprecipitated from pulse-labeled (25 min) CHO and MI8-5 cells, and treated with endoglycosidase H as indicated prior to electrophoresis.
Expression of wild-type ALG6, but not the ALG6 mutant, restored glycosylation of the two STT3A-dependent substrates, pSAP and progranulin (Figure 5B). To further explore the impact of reduced STT3B expression on N-glycosylation in MI8-5 cells, we transfected cells with combinations of expression vectors for STT3B-V5H6, ALG6-V5H6 and the substrate SHBG. After 48 h of culture, the transfected cells were pulse labeled to assay glycosylation of SHBG (Figure 5C). Overexpression of wild-type ALG6-V5H6 in MI8-5 cells was relatively effective at restoring glycosylation of SHBG. Apparently, the low STT3B content is less critical when the fully assembled oligosaccharide donor is present. Nonetheless, the low STT3B content of MI8-5 cells is a contributing factor to the hypoglycosylation of proteins in MI8-5 cells. In contrast to ALG6 expression, overexpression of STT3B-V5His6 caused only a minor improvement in glycosylation of SHBG either in the presence or absence of ALG6 expression (Figure 5C).
Total cell extracts of the transfected cells were analyzed by protein immunoblotting to determine why expression of the STT3B-V5H6 construct was relatively ineffective in restoring glycosylation of SHBG. Expression of STT3B-V5H6 in MI8-5 cells could be detected using an antibody to the V5 epitope tag, and by a marked increase in the intensity of the immunoblot signal for STT3B. As the STT3B protein must be incorporated into a hetero-heptameric complex to be functional, we determined whether overexpression of STT3B was accompanied by increased stable expression of MagT1. After normalization relative to GAPDH, overexpression of STT3B-V5H6 was not accompanied by an increase in the immunoblot signal for MagT1 (Figure 5D). When CHO cells are treated with the STT3B-specific siRNA, the immunoblot signals for STT3B and MagT1 were both reduced to levels that are similar to that observed for MI8-5 cells (Figure 5D), thereby confirming that stable MagT1 expression correlates with STT3B expression. Cotransfection of STT3B-V5H6 with or without ALG6-V5H6 and a plasmid encoding an assay substrate (SHBG) also resulted in enhanced STT3B expression without recovery of MagT1 content. Overexpression of Alg6 did not increase the expression of STT3B or MagT1, providing additional evidence that the reduction in the STT3B complex in MI8-5 cells is not an indirect consequence of the ALG6 deficiency.
Overexpression of STT3B-V5H6 or ALG6-V5H6 in MI8-5 cells did not eliminate the ribophorin I doublet (Figure 5D). Digestion of CHO and MI8-5 cell extracts with endoglycosidase H indicated that the ribophorin I doublet is explained by incomplete modification of the single glycosylation site in ribophorin I (Figure 5E). Thus, ribophorin I is another example of a protein that is hypoglycosylated in MI8-5 cells but not in ALG6-CDG cells.
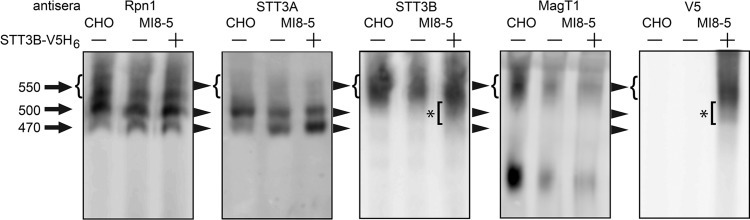
The data presented in Figure 5 raised the possibility that assembly of the STT3B complex is impaired in MI8-5 cells despite overexpression of STT3B-V5H6. Blue native gel electrophoresis (BN-PAGE) can be used to resolve the mammalian OST into three complexes with apparent molecular weights of 470, 500 and 550 kDa (Roboti and High 2012a). The three OST complexes were detected when digitonin extracts of CHO and MI8-5 cells were resolved by BN-PAGE using antibodies to the shared subunit ribophorin I (Figure 6). The two smaller complexes (470 and 500 kDa) contain STT3A as a catalytic subunit and differ with respect to the absence or presence of KCP2 (Roboti and High 2012a). The more diffusely migrating 550 kDa complex contains STT3B as the catalytic subunit. CHO and MI8-5 cells have similar total levels of the two STT3A complexes (Figure 6). As the ratio between 470 and 500 kDa complexes varies between experiments for HeLa cells (Roboti and High 2012b), the 470 kDa complex might arise by dissociation of KCP2 during sample preparation for BN-PAGE. MI8-5 cells contain less of the 550 kDa STT3B complex (Figure 6). When digitonin extracts from MI8-5 cells that express STT3B-V5H6 were analyzed, we observed that the V5-tagged STT3B protein was incorporated into the 550 kDa complex as well as a more rapidly migrating complex of roughly 500 kDa (Figure 6, bracket marked by an asterisk). Consistent with native coimmunoprecipitation experiments (Cherepanova et al. 2014), MagT1 is a subunit of the 550 kDa STT3B complex. MI8-5 cells incorporate reduced amounts of MagT1 into the 550 kDa STT3B complex even when STT3B-V5H6 is overexpressed. The 500 kDa STT3B complex observed upon overexpression of STT3B-V5H6 appears to lack MagT1. Taken together, these results indicate that the failure of STT3B-V5H6 to substantially enhance glycosylation of SHBG is explained by a scant increase in fully assembled STT3B complexes.
Fig. 6.
Assembly of the STT3B complex in MI8-5 cells. Digitonin extracts of CHO, MI8-5 cells or MI8-5 cells expressing STT3B-V5H6 were resolved by BN-PAGE and OST complexes detected by protein immunoblotting. The protein blots were aligned using native molecular-weight markers. The brackets marked by an asterisk indicates STT3B complexes with a lower apparent molecular weight than the 550 kDa STT3B complex present in CHO cells.
Discussion
Here, we have analyzed how STT3A- and STT3B-dependent substrates are glycosylated in cells that have a deficiency in LLO assembly. The experiments conducted with the ALG6-CDG fibroblasts indicated that STT3A substrates are more sensitive than STT3B substrates to a lesion in LLO assembly. This conclusion is in agreement with previous enzyme kinetics experiments using the purified STT3A and STT3B complexes (Kelleher et al. 2003).
Hypoglycosylation of serum Tf, as detected by isoelectric focusing, is the standard diagnostic marker for congenital disorders of glycosylation (Marquardt and Denecke 2003). Glycosylation of Tf is reduced in HeLa cells treated with STT3A siRNA, but not in cells treated with an STT3B siRNA (Shrimal, Trueman, et al. 2013), consistent with the observation that serum from an STT3B-CDG patient has a near-normal IEF pattern for serum Tf (Shrimal, Ng, et al. 2013). As an STT3A substrate, Tf glycosylation is particularly sensitive to mutations that impact LLO assembly as observed here for MI8-5 cells. Although Tf in an excellent diagnostic marker for the most common forms of CDG-1, it is an inadequate marker for detecting CDG-1 variants caused by mutations in STT3B, MagT1 and TUSC3 (Molinari et al. 2008; Shrimal, Ng, et al. 2013). Due to the reduced sensitivity of the STT3B complex to a defect in LLO assembly, the STT3B complex could potentially modify acceptor sites that have been skipped by STT3A in proteins that do not fold as rapidly as pSAP, progranulin and Tf. This may explain why glycosylation of Hpx in ALG6-CDG cells was more efficient than pSAP.
As expected, expression of wild-type ALG6 in MI8-5 cells restored normal glycosylation of the two STT3A-dependent substrates that we tested, and greatly improved glycosylation of SHBG, an STT3B-dependent substrate. Apparently, a 2-fold reduction in STT3B content is not sufficient to cause a glycosylation defect when the optimal oligosaccharide donor is present.
Although it is clear that a reduction in the STT3B complex contributes to the defect in N-linked glycosylation in MI8-5 cells, overexpression of STT3B was relatively ineffective in terms of increasing glycosylation of SHBG. Restoring wild-type levels of the STT3B complex is a more complicated process than restoring ALG6 expression because STT3B-V5H6 must assemble with a minimum of six other proteins (Rpn1, Rpn2, OST48, MagT1, DAD1 and OST4) to form an active STT3B complex. With the exception of MagT1, these non-catalytic OST subunits are also present in the STT3A complex, so STT3B-V5H6 must compete with newly synthesized STT3A for Rpn1, Rpn2, OST48, DAD1 and OST4. Using MagT1 incorporation into the 550 kDa STT3B complex as a marker for assembly of the active STT3B complex, we observed that overexpression of STT3B does not result in a restoration of wild-type levels of the STT3B complex. The oxidoreductase MagT1 is needed for glycosylation of STT3B-dependent substrates regardless of whether the acceptor site is in the vicinity of a cysteine residue (Cherepanova et al. 2014). Overexpression of STT3B in MI8-5 cells results in a mixed population of fully active STT3B complexes and inactive STT3B complexes lacking, at a minimum, MagT1. It remains unclear why overexpression of STT3B was not accompanied by a larger increase in the steady-state levels of MagT1. Previously, we observed that siRNA-mediated depletion of MagT1 does not reduce expression of STT3B (Cherepanova et al. 2014); hence, the low expression of STT3B cannot be explained by altered expression of MagT1. Sequencing of a MagT1 cDNA from MI8-5 cells disclosed no sequence differences relative to CHO MagT1 (data not shown).
MI8-5 cells were derived from CHO cells by chemical mutagenesis followed by a radiation suicide selection procedure that favored survival of cells that incorporate very low levels of mannose at 41.5°C (O'Rear et al. 1999; Quellhorst et al. 1999). Indeed, MI8-5 cells incorporate 9-fold lower levels of mannose at 41.5°C, and were later found to be inviable at the temperature that was used for the mannose incorporation step used in the initial selection procedure (Quellhorst et al. 1999). It should be noted that a 9-fold reduction in mannose incorporation at 41.5°C greatly exceeds the defect in N-linked glycosylation of proteins that we observe when MI8-5 cells are grown at 34°C. In addition to the ALG6 null mutation, MI8-5 cells express low levels of STT3B complex thereby accentuating the glycosylation defect relative to cells that lack ALG6 activity alone. An additional property of MI8-5 cells is the attenuated induction of the UPR pathway in response to hypoglycosylation of proteins (Pearse et al. 2008). A defect in the UPR signaling may help explain why MI8-5 cells are inviable at 41.5°C as these cells may be unable to cope with increased ER folding stress at the higher temperature. Despite the evidence that MI8-5 cells have a more complicated defect in glycoprotein biosynthesis and ER homeostasis than initially appreciated, this cell line remains an excellent model system to investigate glycoprotein processing, folding and ER-associated degradation (ERAD; Cacan et al. 2001; Foulquier et al. 2004; Pearse et al. 2008, 2010) provided that the severity of the glycosylation defect does not complicate the analysis.
Based upon the comparison of ALG6-CDG and MI8-5 cells, we propose that the relative expression of the OST catalytic subunits (STT3A and STT3B), isoform-specific subunits (MagT1, TUSC3, DC2 and KCP2) as well as shared subunits (Rpn-1, Rpn-2, OST48, DAD1 and OST4) will likely impact the extent of protein hypoglycosylation of proteins by CDG-1 patients that have lesions in the dolichol-linked oligosaccharide assembly pathway. This genetic interaction is similar to what has been observed in the S. cerevisiae system, where the combination of a mutation in an ALG pathway gene (e.g., alg6-1 or alg3-1) and an OST subunit gene (e.g. wbp1-1 or stt3-3) causes synthetic lethality due to extensive hypoglycosylation of proteins (Stagljar et al. 1994; Zufferey et al. 1995). There appears to be a general correlation between the extent of protein hypoglycosylation and the severity of CDG-1 symptoms caused by mutations that block a specific step in LLO assembly (Aebi and Hennet 2001). OST subunit genes should be considered as likely modifier genes that will influence the disease severity of patients with common forms of CDG-1. Cell type or tissue type specific differences in OST subunit expression could also impact the extent to which the LLO assembly defect compromises the function of specific human cells and tissues.
Materials and methods
Cell culture, siRNA transfection and protein expression
Primary skin fibroblasts from patients and controls were cultured as described previously (Eklund et al. 2005). Fibroblasts were seeded at 80% confluency in 60 mm dishes and grown for 24 h prior to transfection with expression plasmids (6 µg) using Lipofectamine LTX with PLUS reagent in Opti-MEM (GIBCO, Grand Island, NY, USA) using a protocol from the manufacturer (Life Technologies, Grand Island, NY, USA). HeLa cells (ATCC CCL-13) were cultured in 10 cm2 dishes at 37°C in DMEM (GIBCO), 10% fetal bovine serum with penicillin (100 units/mL) and streptomycin (100 µg/mL). CHO and MI8-5 cells were cultured in 10 cm2 dishes at 34°C in MEM Alpha + Glutamax (GIBCO) with 10% fetal bovine serum. Cells were seeded at 30% confluency for siRNA transfection or 80% confluency for plasmid transfection in 60 mm dishes and grown for 24 h prior to transfection with siRNA (NC—60 nM; STT3A—50 nM; STT3B—60 nM) or plasmid (8 µg) and Lipofectamine 2000 in Opti-MEM (GIBCO) using a protocol from the manufacturer (Life Technologies). Plasmid transfection was done after 48 h of siRNA transfection and cells were assayed 24 h later. The siRNAs designed to knockdown CHO STT3A and STT3B have been described previously (Malaby and Kobertz 2014). Negative control siRNA was purchased from QIAGEN.
Human pSAP and progranulin were amplified from cDNA clones (Thermo Scientific, Pittsburg, PA, USA) and inserted into the pCMV6-AC-DDK-His vector (OriGene, Rockville, MD, USA). The ALG6 coding sequence was amplified from a human cDNA clone (Origene) and inserted into pCDNA-V5H6. A common polymorphism that can cause a mild glycosylation defect (F304S, ; Westphal et al. 2002) was eliminated in the wild-type ALG6 expression construct. The A333V and S308R mutations were introduced into the ALG6 coding sequence to obtain the expression vector for the human ALG6 mutant. The SHBG coding sequence in the vector pRC/CMV (Invitrogen) was a gift from Dr. Geoffrey Hammond (Child and Family Research Institute; Vancouver British Columbia, Canada). Expression vector for β-GUS, β-GUS Δ123, Hp, Tf, CD40 ligand and Hpx have been described previously (Shrimal and Gilmore 2013; Shrimal, Trueman, et al. 2013).
Complementation of MI8-5 cells was conducted by cotransfecting cells with an expression vectors for a glycoprotein substrate (e.g., H. sapiens pSAP) and an enzyme (ALG6 and/or STT3B) to minimize the number of cells that express the glycoprotein substrate but do not express ALG6-V5H6 and/or STT3B-V5H6.
Radiolabeling and immunoprecipitation of glycoproteins
The procedures for pulse labeling and pulse-chase labeling of cells and immunoprecipitation of glycoproteins have been described (Shrimal and Gilmore 2013; Shrimal, Trueman, et al. 2013). Labeling periods were previously optimized for each substrate to allow sufficient time for posttranslocational glycosylation of the STT3B-dependent substrates, without having the glycoform pattern perturbed by glycan processing reactions in the Golgi or by ERAD-mediated degradation. The pulse and chase intervals were as follows: (i) pSAP and β-GUSΔ123, 10 min pulse; (ii) pGran, 15 min pulse; (iii) Hp, SHBG and Hpx, 10 min pulse–10 min chase; (iv) Tf, CD40L and β-GUS, 5 min pulse–20 min chase. The glycoprotein substrates were precipitated with anti-DDK, anti-Myc, anti-SapD, anti-Gran or anti-SHBG sera and then resolved by SDS–PAGE. As indicated, immunoprecipitated proteins were digested with endoglycosidase H (New England Biolabs, Ipswich, MA, USA).
Dry gels were exposed to a phosphor screen (Fujifilm, Tokyo, Japan) and scanned in Typhoon FLA 9000 (GE Healthcare, Pittsburgh, PA, USA) and quantified using AlphaEase FC (Santa Clara, CA, USA) to determine the average number of glycans per protein. The average number of glycans/protein was calculated as the sum of the fractional contribution of each glycoform multiplied by the number of glycans/glycoform. Low amounts of nonglycosylated pSAP, Hp and Hpx were detected in transfected CHO and MI8-5 cells. These nonglycosylated proteins (pSAP, Hp and Hpx) were not used to calculate the average number of glycans/protein as the 0-glycan form corresponds to a non-translocated precursor.
Antibodies were obtained from the following sources: anti-DDK (anti-FLAG; Sigma, St. Louis, MO, USA), anti-SHBG (R&D Systems, Minneapolis, MN, USA), anti-granulin (R&D Systems), anti-V5 (Life Technologies), anti-Myc (Santa Cruz Biotechnology, Inc. Dallas, TX, USA) and anti-SapD (gift from Prof. Konrad Sandhoff, University of Bonn).
Blue native gel electrophoresis
The OST complexes were resolved by BN-PAGE as described (Shibatani et al. 2005). Briefly, the BN-PAGE resolving gel was cast as a 6–13% polyacrylamide gradient in 50 mM Bis–Tris–HCl (pH 7.0) and 500 mM aminocaproic acid and overlaid with a 4% stacking gel. Cells were lysed at 4°C by a 30-min incubation with 20 mM Tris–Cl (pH 7.6), 150 mM NaCl, 5 mM Mg(OAc)2, 3 mM MnCl2, 2% digitonin, DNase I and 1× protease inhibitor cocktail. Cell lysates were clarified by centrifugation (10 min at 13,000 rpm). A supernatant fraction containing 200 µg of protein was mixed with 15% glycerol, 2 mM DTT and 1/40 volume of 5% Coomassie Blue G250 in 500 mM 6-aminohexanoic acid and loaded on the gel. The gels were run at 4°C for 16 h at 70 V with a cathode buffer of 0.02% Coomassie Blue G250 in 50 mM Tricine and 15 mM Bis–Tris, pH 7.0, and an anode buffer of 50 mM Bis–Tris, pH 7.0. After 16 h the cathode buffer was changed to contain 0.002% Coomassie Blue G250 and electrophoresis was continued at 500 V until the dye front reached the end of gel. A native protein gel marker (Life Technologies) was electrophoresed on gels to obtain apparent molecular weights of protein complexes. The proteins were transferred by wet blotting onto PVDF membranes. Membranes were destained with methanol and OST subunits were detected by protein immunoblotting.
Protein immunoblots and pulse labeling of STT3A or ALG6
Expression of STT3B, MagT1, ribophorin I, GAPDH and BiP in cells was analyzed by protein immunoblotting as described previously (Cherepanova et al. 2014). Due to the presence of non-specific background bands that are in the vicinity of STT3A on protein immunoblots, STT3A expression was evaluated by immunoprecipitation of STT3A from total cell extracts of cells that were pulse labeled for 50 min with 35S Trans-label (200 µCi/mL of media). The same labeling conditions were used to evaluate expression of ALG6-V5H6 by immunoprecipitation using anti-V5 sera.
RT-PCR
Total RNA was purified using TRIzol reagent (Life Technologies) according to the manufacturer's instructions. DNase I-treated total RNA (500 ng) was used in the RT reaction using Superscript III (Life Technologies) with either oligo dT or a gene-specific primer. Annealing was carried out at 65°C for 5 min, followed by extension at 50°C for 50 min followed by inactivation at 85°C for 5 min. 0.5 µL of this RT mix was used for a regular PCR. Amplification conditions were as follows: 94°C for 5 min; specified number of cycles or 30 cycles at 94°C for 30 s; 55°C for 30 s and 72°C for 30 s; and a final extension at 72°C for 3 min. No amplicon was observed in the absence of RT enzyme in all RT-PCR experiments. The following primer pairs were used: ALG6-1F 5′GCTGCTAATTGCAGAGATGGTGATATTGC and ALG6-1R 5′GCTTTGGTCCATACACAATTTGTATAGATAAC; ALG6-2F 5′ATGGAAAACTGGTACTGGATGACACTTGTGG and ALG6-2R 5′GCTTTGGTCCATACACAATTTGTATAGATAAC; STT3A-F 5′CAAGTTTCTTTGGATGGTCCGGATTGGAGG and STT3A-R 5′CGATTATCCAGGTCCTTTACCTTGTATATCC; STT3B-F 5′CAAAATACAGATGAACATGCCCGGGTCATG and STT3B-R 5′TGACTCGAGGCTTGTGACCTAGTGTCTCCC; actin-F 5′GAAGCTGTGCTACGTGGCCCTGGACTTC and actin-R 5′CCGGCCTCGTCGTACTCCTGCTTGGTG; GAPDH-F 5′CACCCAGAAGACTGTGGATGGCCCC and GAPDH-R 5′GCCTGCTTCACCACCTTCTTGATGTCCTC.
Funding
This work was supported by the National Institutes of Health (GM43768).
Conflict of interest statement
None declared.
Abbreviations
ALG, asparagine-linked glycosylation; β-GUS, beta-glucuronidase; BN-PAGE, blue native polyacrylamide gel electrophoresis; CST, castanospermine; CDG, congenital disorders of glycosylation; CHO, Chinese hamster ovary; Dol, dolichol; ERAD, ER-associated degradation; GAPDH, glyceraldehyde 3′ phosphate dehydrogenase; Hp, haptoglobin; Hpx, hemopexin; LLO, lipid-linked oligosaccharide; OST, oligosaccharyltransferase; pGran, progranulin; pSAP, prosaposin; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; SHBG, sex hormone-binding globulin; Tf, transferrin; UGGT, UDP-glucose glycoprotein glucosyltransferase; UPR, unfolded protein response pathway.
Acknowledgements
We thank Dr. Sharon Krag (Johns Hopkins University) and Dr. Hudson Freeze (Sanford-Burnham Medical Research Institute) for providing MI8-5 cells and human skin primary fibroblasts (ALG6-CDG and healthy control fibroblasts), respectively.
References
- Aebi M. 2013. N-linked protein glycosylation in the ER. Biochim Biophys Acta. 1833:2430–2437. [DOI] [PubMed] [Google Scholar]
- Aebi M, Hennet T. 2001. Congenital disorders of glycosylation: Genetic model systems lead the way. Trends Cell Biol. 11:136–141. [DOI] [PubMed] [Google Scholar]
- Burda P, Aebi M. 1999. The dolichol pathway of N-linked glycosylation. Biochim Biophys Acta. 1426:239–257. [DOI] [PubMed] [Google Scholar]
- Cacan R, Duvet S, Labiau O, Verbert A, Krag SS. 2001. Monoglucosylated oligomannosides are released during the degradation process of newly synthesized glycoproteins. J Biol Chem. 276:22307–22312. [DOI] [PubMed] [Google Scholar]
- Cherepanova NA, Shrimal S, Gilmore R. 2014. Oxidoreductase activity is necessary for N-glycosylation of cysteine-proximal acceptor sites in glycoproteins. J Cell Biol. 206:525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund EA, Newell JW, Sun L, Seo NS, Alper G, Willert J, Freeze HH. 2005. Molecular and clinical description of the first US patients with congenital disorder of glycosylation Ig. Mol Genet Metab. 84:25–31. [DOI] [PubMed] [Google Scholar]
- Foulquier F, Duvet S, Klein A, Mir AM, Chirat F, Cacan R. 2004. Endoplasmic reticulum-associated degradation of glycoproteins bearing Man5GlcNAc2 and Man9GlcNAc2 species in the MI8–5 CHO cell line. Eur J Biochem. 271:398–404. [DOI] [PubMed] [Google Scholar]
- Freeze HH, Aebi M. 2005. Altered glycan structures: The molecular basis of congenital disorders of glycosylation. Curr Opin Struct Biol. 15:490–498. [DOI] [PubMed] [Google Scholar]
- Haeuptle MA, Hennet T. 2009. Congenital disorders of glycosylation: An update on defects affecting the biosynthesis of dolichol-linked oligosaccharides. Hum Mutat. 30:1628–1641. [DOI] [PubMed] [Google Scholar]
- Imbach T, Grunewald S, Schenk B, Burda P, Schollen E, Wevers RA, Jaeken J, de Klerk JB, Berger EG, Matthijs G, et al. 2000. Multi-allelic origin of congenital disorder of glycosylation (CDG)-Ic. Hum Genet. 106:538–545. [DOI] [PubMed] [Google Scholar]
- Karaoglu D, Kelleher DJ, Gilmore R. 2001. Allosteric regulation provides a molecular mechanism for preferential utilization of the fully assembled dolichol-linked oligosaccharide by the yeast oligosaccharyltransferase. Biochemistry (Mosc). 40:12193–12206. [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Karaoglu D, Mandon EC, Gilmore R. 2003. Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol Cell. 12:101–111. [DOI] [PubMed] [Google Scholar]
- Malaby HL, Kobertz WR. 2014. The middle x residue influences cotranslational N-glycosylation consensus site skipping. Biochemistry (Mosc). 53:4884–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T, Denecke J. 2003. Congenital disorders of glycosylation: Review of their molecular bases, clinical presentations and specific therapies. Eur J Pediatr. 162:359–379. [DOI] [PubMed] [Google Scholar]
- Molinari F, Foulquier F, Tarpey PS, Morelle W, Boissel S, Teague J, Edkins S, Futreal PA, Stratton MR, Turner G, et al. 2008. Oligosaccharyltransferase-subunit mutations in nonsyndromic mental retardation. Am J Hum Genet. 82:1150–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell JW, Seo NS, Enns GM, McCraken M, Mantovani JF, Freeze HH. 2003. Congenital disorder of glycosylation Ic in patients of Indian origin. Mol Genet Metab. 79:221–228. [DOI] [PubMed] [Google Scholar]
- O'Rear JL, Scocca JR, Walker BK, Kaiden A, Krag SS. 1999. Chinese hamster ovary cells with reduced hexokinase activity maintain normal GDP-mannose levels. J Cell Biochem. 72:56–66. [DOI] [PubMed] [Google Scholar]
- Pearse BR, Gabriel L, Wang N, Hebert DN. 2008. A cell-based reglucosylation assay demonstrates the role of GT1 in the quality control of a maturing glycoprotein. J Cell Biol. 181:309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse BR, Tamura T, Sunryd JC, Grabowski GA, Kaufman RJ, Hebert DN. 2010. The role of UDP-Glc:glycoprotein glucosyltransferase 1 in the maturation of an obligate substrate prosaposin. J Cell Biol. 189:829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quellhorst GJJ, O'Rear JL, Cacan R, Verbert A, Krag SS. 1999. Nonglucosylated oligosaccharides are transferred to protein in MI8–5 Chinese hamster ovary cells. Glycobiology. 9:65–72. [DOI] [PubMed] [Google Scholar]
- Reiss G, te Heesen S, Zimmerman J, Robbins PW, Aebi M. 1996. Isolation of the ALG6 locus of Saccharomyces cerevisiae required for glucosylation in the N-linked glycosylation pathway. Glycobiology. 6:493–498. [DOI] [PubMed] [Google Scholar]
- Roboti P, High S. 2012a. Keratinocyte-associated protein 2 is a bona fide subunit of the mammalian oligosaccharyltransferase. J Cell Sci. 125:220–232. [DOI] [PubMed] [Google Scholar]
- Roboti P, High S. 2012b. The oligosaccharyltransferase subunits OST48, DAD1 and KCP2 function as ubiquitous and selective modulators of mammalian N-glycosylation. J Cell Sci. 125:3474–3484. [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C, Kelleher DJ, Gilmore R. 2009. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell. 136:272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. 2005. ER stress and the unfolded protein response. Mutat Res. 569:29–63. [DOI] [PubMed] [Google Scholar]
- Shibatani T, David LL, McCormack AL, Frueh K, Skach WR. 2005. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry (Mosc). 44:5982–5992. [DOI] [PubMed] [Google Scholar]
- Shrimal S, Gilmore R. 2013. Glycosylation of closely spaced acceptor sites in human glycoproteins. J Cell Sci. 126:5513–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S, Ng BG, Losfeld ME, Gilmore R, Freeze HH. 2013. Mutations in STT3A and STT3B cause two congenital disorders of glycosylation. Hum Mol Genet. 22:4638–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S, Trueman SF, Gilmore R. 2013. Extreme C-terminal sites are posttranslocationally glycosylated by the STT3B isoform of the OST. J Cell Biol. 201:81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagljar I, te Heesen S, Aebi M. 1994. New phenotype of mutations deficient in glucosylation of the lipid-linked oligosaccharide: Cloning of the ALG8 locus. Proc Natl Acad Sci USA. 91:5977–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble RB, Byrd JC, Maley F. 1980. Effect of glucosylation of lipid intermediates on oligosaccharide transfer in solubilized microsomes from Saccharomyces cerevisiae. J Biol Chem. 255:11892–11895. [PubMed] [Google Scholar]
- Turco SJ, Stetson B, Robbins PW. 1977. Comparative rates of transfer of lipid-linked oligosaccharides to endogenous glycoprotein acceptors in vitro. Proc Natl Acad Sci USA. 74:4410–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal V, Kjaergaard S, Schollen E, Martens K, Grunewald S, Schwartz M, Matthijs G, Freeze HH. 2002. A frequent mild mutation in ALG6 may exacerbate the clinical severity of patients with congenital disorder of glycosylation Ia (CDG-Ia) caused by phosphomannomutase deficiency. Hum Mol Genet. 11:599–604. [DOI] [PubMed] [Google Scholar]
- Westphal V, Murch S, Kim S, Srikrishna G, Winchester B, Day R, Freeze HH. 2000. Reduced heparan sulfate accumulation in enterocytes contributes to protein-losing enteropathy in a congenital disorder of glycosylation. Am J Pathol. 157:1917–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Knauer R, Burda P, Stagljar I, te Heesen S, Lehle L, Aebi M. 1995. STT3, a highly conserved protein required for yeast oligosaccharyl transferase activity in vivo. EMBO J. 14:4949–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]