SUMMARY
A study was carried out to investigate how common Cryptosporidium infections are in beef calves in Swedish suckler herds and to explore which species and subtypes that occur. We further aimed at identifying factors associated with shedding of Cryptosporidium oocysts in this type of calf management. The study was conducted in two regions in Sweden and included 30 herds. Faecal samples were collected from calves younger than 3 months. A brief clinical examination was done and a questionnaire was used to collect data on management routines. Faeces were cleaned and concentrated and oocysts identified by epifuorescence microscopy. Cryptosporidium positive samples were analyzed at the 18S rRNA and GP60 genes to determine species and Cryptosporidium parvum subtype, respectively. Logistic regression was used to identify factors associated with infection. Oocysts were detected in 122 (36·7%) calves from 29 (97%) herds, at 400 to 2·4 × 107 OPG. The youngest positive calves were only 1 and 2 days old. There was no association between age and Cryptosporidium infection. Cryptosporidium bovis, Cryptosporidium ryanae, C. parvum and Cryptosporidium ubiquitum were identified, with C. bovis being the major species. Two C. parvum subtypes, IIaA16G1R1 and IIdA27G1 were identified. Routines for cleaning calf pens and number of cows in calving pens were associated with infection.
Key words: Cryptosporidium, beef calves, suckler herd, diarrhea, Cryptosporidium bovis, Cryptosporidium parvum, Cryptosporidium ryanae, Cryptosporidium ubiquitum
INTRODUCTION
The protozoan genus Cryptosporidium are clinically important pathogens causing gastrointestinal disease in a variety of species, including cattle and humans (Cacciò and Widmer, 2014). The parasites are transmitted as oocysts via the faecal-oral route, either by individual-to-individual contact, by exposure to contaminated equipment or by ingestion of contaminated food or water. Cryptosporidium parvum is one of the most common causes of calf diarrhoea worldwide (Blanchard, 2012). One infected calf can shed billions of oocysts in the faeces, and thus effectively spread the infection within the herd and to the environment. Cattle are mainly infected with four Cryptosporidium species, of which three are morphologically identical but genetically different species; C. parvum, C. bovis and C. ryanae; whereas C. andersoni can be distinguished by its larger, ovoid oocysts. C. parvum has a broad host range and infects most mammals including humans and cattle whereas C. bovis and C. ryanae seem to be adapted to cattle (Robertson et al. 2014).
Bovine Cryptosporidium spp. infections are common around the world and infections are most common in preweaned calves. In studies comprising dairy calves up to 2 months of age, 5–93% of investigated calves shed oocysts and the cumulative prevalence was 100% in some herds (O'Handley et al. 1999; Santín et al. 2008). The overall picture is that there is an age related pattern in the distribution of Cryptosporidium species. C. parvum is mostly found in young calves up to 2 months of age, whereas C. bovis and C. ryanae are the most common species in older calves and young stock (Robertson et al. 2014). C. andersoni is mainly found in young stock and adult cattle (Robertson et al. 2014).
Cryptosporidium species have been shown to be ubiquitous in Swedish dairy herds; with the highest infection rates in calves, followed by young stock and cows (Silverlås et al. 2009; Silverlås & Blanco-Penedo, 2013). In the studies by Silverlås et al. (2009); and Silverlås & Blanco-Penedo (2013) C. parvum was only found in calves up to 9 weeks of age, although C. bovis was the dominating species even in this age group. This is in contrast to what has been reported from many other countries (Langkjaer et al. 2007; Plutzer & Karanis, 2007; Santín et al. 2008; Brook et al. 2009). Analysis of the GP60 locus can be used to discriminate between species specific and zoonotic C. parvum subtypes. All Swedish bovine isolates analysed so far belong to the zoonotic subtype families IIa and IId (Silverlås et al. 2010a, 2010b, 2013; Silverlås and Blanco-Penedo, 2013).
In the 1970s the number of herds with specialized beef breeds started to increase in Sweden and today approximately a third of all Swedish cows are kept in cow-calf production systems. In 2012, there were 11 375 suckler herds with an average of 17 cows per herd (Statistics Sweden, 2013). In contrast to calves in dairy herds, calves in suckler herds are kept on pasture with their dams. In recent years questions concerning the risk of spread of zoonotic pathogens like C. parvum from grazing animals to water have been frequently discussed in Sweden. In order to assess the extent of C. parvum shedding into the environment from suckler herds and the potential zoonotic implications thereof, this study was implemented. The main objectives were to investigate how common Cryptosporidium infection is in calves in Swedish suckler herds and to explore which species and subtypes that occur. We further aimed at identifying factors associated with shedding of Cryptosporidium oocysts in this type of calf management.
MATERIALS AND METHODS
Study design and sampling
The study was conducted in Halland on the western coast and Uppsala-Örebro counties in the central-eastern part of Sweden. Both regions are densely populated and have cattle herds grazing near lakes and streams. The reason for targeting these regions was that the municipalities in both regions had discussed restrictions on grazing such pastures. In total, 30 herds were recruited. According to sample size calculations, 73 herds would be needed to estimate 95% herd prevalence with 95% confidence interval (CI) assuming a perfect test. The 95% herd prevalence was based on our previous dairy herd studies (Silverlås et al. 2009; Silverlås & Blanco-Penedo, 2013). Because all calves up to 90 days of age were sampled and longitudinal studies show that all calves in infected herds shed oocysts at some stage during their first months (O'Handley et al. 1999; Santín et al. 2008), we assumed that a smaller herd sample size could be used. The study herds, 15 in each region, were a convenience sample of all suckler herds with more than 10 cows. Eligible herds in these regions were identified by Växa Sverige, an organisation owned by Swedish farmers, with knowledge about the cattle herds in the regions. In the middle region, eligible herds were also identified through a farmer network for local products. The owners were contacted by phone and by mail and those agreeing to participate were visited by a veterinarian once either during April–June 2012 (n = 24) or April–May 2013 (n = 6).
At the visit, rectal faecal samples were collected directly from all calves that were younger than 3 months. A brief clinical examination was done of the sampled animals. Samples were sent by mail when samplings were performed in the southern region and reached the laboratory within 3 days after sampling. Samples from the middle region were returned to the laboratory by car on the day of sampling. Information about the herds and management routines were collected from herd records and through interviews with the owners using a predetermined questionnaire. The information included e.g. herd size and number of animals in different age groups, purchase of animals, routines for calving and colostrum intake, cleaning of the calving area, mortality in neonatal calves and prevalence of diarrhoea and/or respiratory problems. Questions about age of calves when let out on pasture, if they were first kept in a temporary paddock, if the farm had pasture close to water and handling of calf manure were also included. All samples were collected before the pasture period had started.
Laboratory methods
Faecal consistency and colour were noted at arrival to the laboratory. One gram of each sample was cleaned and concentrated by saline-glucose flotation (Maddox-Hyttel et al. 2006). Ten μl of each cleaned sample plus 50 μl distilled water was put on teflon printed 3-well slides (Immuno-Cell, Mechelen, Belgium), dried and stained with FITC-labelled monoclonal anti-Cryptosporidium antibodies (CryptoCel IF test kit, Cellabs, Sydney, Australia). The entire wells were examined by epifluorescence microscopy and oocysts enumerated at 200 × magnification. The lower detection limit of this method is approximately 400 oocysts per gram faeces (OPG).
All samples that were Cryptosporidium positive by microscopy were further analyzed at the 18S rRNA and the 60 kDa glycoprotein (GP60) genes to determine species and C. parvum subtype, respectively. DNA was extracted from concentrated samples using a combined freeze-thawing and QIAamp DNA stool mini kit (QIAGEN, Hilden, Germany) protocol (Silverlås et al. 2010b). Samples were then subjected to nested polymerase chain reaction (PCR) protocols to amplify the 18S and GP60 genes. Amplification of the 18S gene was done essentially as described by Xiao et al. (1999, 2000). The first amplification mixture contained 1X of Platinum Taq buffer (Life Technologies), 3, 4 or 6 mm MgCl2, 200 μm of each deoxynucleoside triphosphate, 200 μm of each of primary primers, 0·16 mg/ml bovine serum albumin (BSA), 2 μl DNA solution and 1 unit of Platinum Taq polymerase in a total volume of 25 μl. After initial denaturation at 95 °C for 5 min, 35 cycles followed consisting of 94 °C for 45 s, 56 °C for 45 s and 72 °C for 60 s, and a final extension of 72 °C for 7 min. For the second amplification, 2 μl from the first reaction were added to a reaction mixture as above except containing secondary primers and MgCl2 at 3 mm concentration. The reaction condition was 95 °C for 5 min followed by 40 cycles of 94 °C for 45 s, 58 °C for 90 s and 72 °C for 60 s and then a final extension step of 72 °C for 7 min. DNA samples that failed to yield a PCR product were rerun with primers designed by (Silva et al. 2013) following the published protocol except that bovine serum albumin (BSA), 0·16 mg/ml, was added and that 3 mm MgCl2 instead of 1·5 mm was used. Amplification of the GP60 gene was done according to Chalmers et al. (2005).
Species and subtype were determined by sequencing in both directions on an ABI 3100 Genetic Analyzer (Applied Biosystems) using the internal primers and BigDye Terminator v3·1 Cycle Sequencing Kit (Applied Biosystems). CLC Main Workbench (CLC bio, Aarhus, Denmark) was used to assemble consensus sequences and manually correct mismatches. Achieved sequences were compared with sequences deposited in GenBank using Basic Local Alignment Search Tool (BLAST, NCBI http://www.ncbi.nlm.nih.gov/blast/Blast.cgi).
Statistical analysis
Data were entered into Microsoft Office Excel 2007 spreadsheets (©2006 Microsoft Corporation). Statistical analyses were performed using Stata 11 (Stata Software, StataCorp LP College Station, TX, USA). Continuous normally and non-normally distributed data were analyzed using the Mann–Whitney test. Non-continuous data were categorized and analyzed using the χ2-test. A random effects logistic regression model was built, using manual forward selection and herd as random effect, to investigate factors associated with being Cryptosporidium positive or not. Spearman rank correlations were investigated prior to modelling to avoid collinearity between factors entered into the model. Calves were considered to be clustered within herds. Variables significant at P ⩽ 0·2 in univariable random effects logistic regression were considered for building the main effects model. A flow chart of these variables was done to identify any confounders or intervening factors. Confounding was considered to be present if any variable OR changed >20% when a new variable was entered into the model. The main effects model was investigated for interactions and the fit, sensitivity and specificity and distribution of residuals.
RESULTS
The 30 herds included in the study had 10 to 99 cows (median 39) and between three and 18 calves were sampled in each herd (median 11·5). The calves were of different breeds, mainly pure bred or cross bred Angus, Charolais, Hereford, Limousine, Simmental, Holstein Friesian and Swedish Red. In total, samples were collected from 332 calves 1 to 90 days of age. One calf had poor general condition and one had tachycardia, otherwise no signs of illness were found in sampled calves.
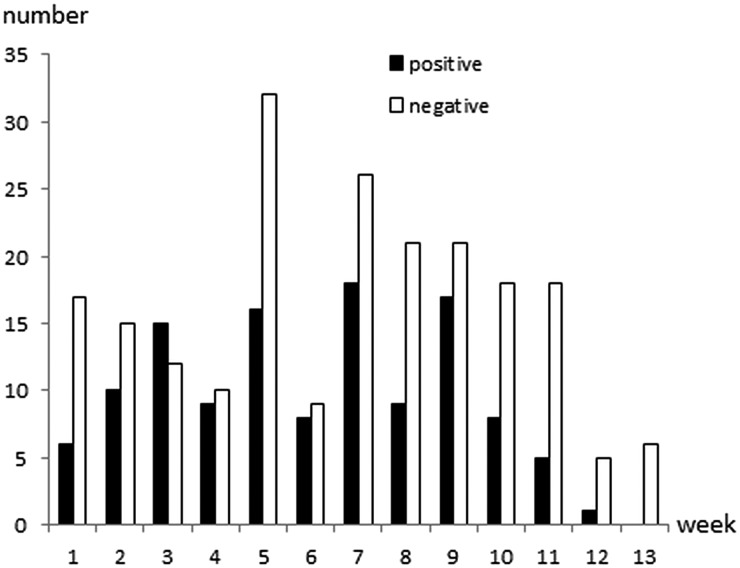
Cryptosporidium oocysts were detected in 122 (36·7%; 95% CI 31·5–42·1%) calves aged 1–81 days of age (Fig. 1). The faecal consistency was watery, loose, pasty and solid in 20, 79, 206 and 27 calves, respectively. Watery and loose stools were considered as diarrhoea, whereas pasty and solid were considered as normal consistency. There was no significant association between age, faecal consistency or colour of faeces, and Cryptosporidium infection (Table 1). Oocyst counts in the positive samples were 400 to 2·4 × 107 OPG. The two youngest positive calves were only 1 and 2 days old and they shed 1600 and 400 OPG, respectively. Calves positive for Cryptosporidium spp. were identified in 29 (97%) herds and the within-herd prevalence varied between 6·3% and 75·0% (median 42·3%). Of the sampled calves, 267 were ⩽ 62 days, and Cryptosporidium oocysts were detected in 106 (39·7%) of them.
Fig. 1.
Age and Cryptosporidium spp. oocyst shedding of 332 calves sampled in 30 Swedish suckler herds. Faecal samples were cleaned and concentrated by saline-glucose flotation, stained with FITC-labelled monoclonal anti-Cryptosporidium antibodies and examined by epifluorescence microscopy. The lower detection limit of the method is approximately 400 oocysts per gram faeces (OPG; Maddox-Hyttel et al. 2006).
Table 1.
Age, faecal consistency and faecal colour in 122 Cryptosporidium spp. positive and 210 Cryptosporidium spp. negative calves in 30 Swedish suckler herds
| Cryptosporidium spp | Test | P | ||
|---|---|---|---|---|
| Positive | Negative | |||
| Age in days | ||||
| Mean (s.d.) | 39·1 (23·0) | 43·5 (20·3) | Mann–Whitney test | 0·096 |
| Faecal consistency (number of calves) | ||||
| Normal | 89 | 144 | χ2-test | 0·400 |
| Diarrhoea | 33 | 66 | ||
| Faecal colour (number of calves) | ||||
| Brown | 95 | 172 | χ2-test | 0·372 |
| Yellow | 27 | 38 | ||
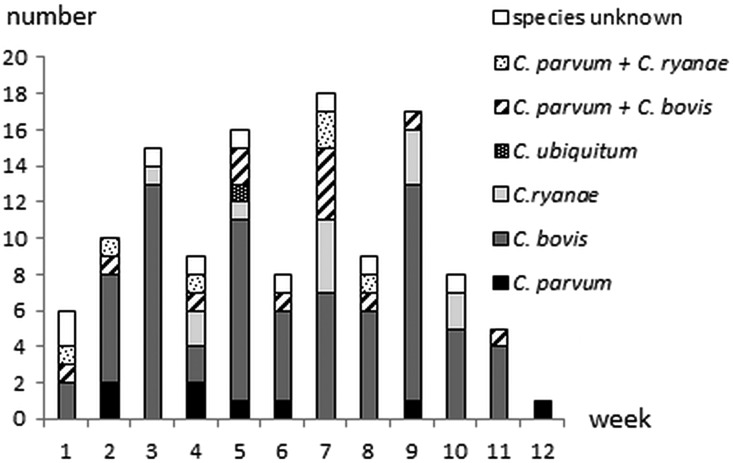
Cryptosporidium species could be determined in 113 (92·6%) of the 122 positive samples. Monoinfection with C. bovis was identified in 72 (21·7%) calves, C. ryanae in 13 (3·9%) calves, C. parvum in 8 (2·4%) calves and C. ubiquitum in 1 (0·3%) calf. In addition, mixed C. bovis/C. parvum infection was identified in 13 (3·9%) calves and mixed C. ryanae/C. parvum infection in 6 (1·8%) calves. Thus, in total C. parvum was identified in 27 of the 332 faecal samples (8·1%; 95% CI 5·4–11·6%) and in 23·9% (95% CI 16·4–32·8%) of Cryptosporidium positive samples with successful sequence results. Nine calves shedding C. parvum had watery or loose stools whereas 18 had normal faecal consistency. Species distribution per week in life is shown in Fig. 2. C. bovis, C. ryanae and C. parvum were detected in 25, 13 and 9 herds, respectively. C. bovis infection was identified in a 1-day-old and two 4-days-old calves (one of them with mixed C. bovis/C. parvum infection). Mixed C. ryanae/C. parvum infection was identified in a 7-days-old calf. There was no significant age difference for the different species (P = 0·09–0·89, Mann–Whitney test).
Fig. 2.
Cryptosporidium species distribution in 122 oocyst shedding calves in 30 Swedish suckler herds Faecal samples were cleaned and concentrated by saline-glucose flotation, stained with FITC-labelled monoclonal anti-Cryptosporidium antibodies and examined by epifluorescence microscopy. The positive samples were further analyzed at the 18S rRNA and the 60 kDa glycoprotein (GP60) genes to determine Cryptosporidium species.
Oocyst excretion ranges were 400–2·6 × 106 OPG (median 9600 OPG) for C. bovis, 800–4·4 × 105 OPG (median 4800 OPG) for C. ryanae, 400–3·8 × 106 OPG (median 1600 OPG) for C. parvum, 1·3 × 106 OPG for C. ubiquitum, 400–2·4 × 107 OPG (median 8800 OPG) for mixed C. bovis/C. parvum and 800–2·0 × 107 OPG (median 3·7 × 105 OPG) for mixed C. ryanae/C. parvum. No significant differences in OPG were found for the different species (P = 0·08–0·87, Mann–Whitney test).
GP60 analysis identified two subtypes of C. parvum: IIaA16G1R1 in 7 calves and IIdA27G1 in 16 calves with mono or mixed C. parvum infection. The subtype could not be determined for the remaining four C. parvum positive calves. Only one subtype was detected in each herd; two herds had IIaA16G1R1, five herds had IIdA27G1 and the subtype could not be determined in two herds that had only one C. parvum positive calf each.
Univariable modelling returned three variables with P ⩽ 0·2: (1) routines for cleaning calf pens, (2) number of cows in calving pens and (3) per cent of dead (for any reason) calves the season of sampling. The flow chart identified variable (3) as intervening to both (1) and (2), i.e. the outcome of (3) could be affected by the outcome of the other variables. Another problem was that all levels of (3) were not represented for all levels of (1) and (2), causing interpretation problems. Intervening variables should not be included in modelling, and thus (3) was removed. Both variable (1) and (2) could be kept in the model, which was highly significant and fit the data well (Table 2). There was no interaction between the variables. The sensitivity and specificity of the model was low (Se 0·32, Sp 0·86), and the area under the receiver operating curve (ROC) was only 0·59. One herd was identified as an outlier by residual distribution, but was kept in the model since data were properly recorded.
Table 2.
Variables significant at P ⩽ 0·2 in univariable modelling and final random effects logistic regression model of variables associated with a calf being infected with Cryptosporidium spp. at the time of sampling
| Variable | Level | Punivariable | ORfinal model | Pfinal model | 95% CI |
|---|---|---|---|---|---|
| (1) Cleaning routines for calving pens | Once a year | 0·020a | 1 | 0·0035a | |
| More often | 0·25 | 0·004 | 0·10–0·64 | ||
| (2) Number of cows in calving pen | 1 (single pen) | 0·056a | 1 | 0·0109a | |
| 5–15 | 2·5 | 0·029 | 1·10–5·89 | ||
| 20–28 | 1·2 | 0·579 | 0·57–2·69 | ||
| 30–90 | 0·47 | 0·202 | 0·15–1·49 | ||
| No data | 5·1 | 0·100 | 0·73–35·23 | ||
| (3) Per cent dead calves this year | 0 1–4 5–9 ⩾10 |
0·054a | Not included in the final model | ||
Herd was used as random effect. Walds χ2 of final model = 20·3, 5 df, P = 0·0011.
Overall P-value of variable
P = P-value, OR = Odds Ratio, CI = Confidence Interval
DISCUSSION
The present study showed that Cryptosporidium spp. are common in Swedish suckler herds. Oocysts were detected in 29 of 30 investigated herds and in 36·7% of the sampled calves. The herd prevalence was in accordance with what has been found in Swedish dairy cattle (Silverlås et al. 2009; Silverlås and Blanco-Penedo, 2013). However, the prevalence among the calves was lower than in dairy calves, where 49–52% of calves have been reported to shed oocysts (χ2-test; P = 0·002). The study on dairy calves only included calves up to 62 days of age and accordingly only beef calves up to this age were included in the comparison. This difference is consistent with findings in other studies reporting a higher prevalence in dairy calves compared with calves in suckler herds in e.g. Belgium, Canada and Zambia (Dixon et al. 2011; Geurden et al. 2006, 2007).
The most common species was C. bovis, followed by C. parvum and C. ryanae. This is in concordance with the situation in our dairy herds where C. bovis is the most prevalent species in preweaned calves (Silverlås et al. 2010b). In Sweden, C. parvum has previously only been reported in cattle younger than 9 weeks, with the main window of infection before 6 weeks of age (Silverlås et al. 2013), but in this study we detected it in two calves, 71- and 81-days-old, respectively. This could be an effect of a lower infection pressure in suckler herds resulting in calves getting infected later. In dairy herds, calves are commonly infected before they are 3 weeks old (Santín et al. 2008). In suckler herds the calves are kept with their dams on a larger area, as compared with dairy calves that are kept close together, exposing them to a lower infection pressure. A common advice to prevent Cryptosporium infection is to ensure that all newborn calves ingest an adequate amount of colostrum during their first 24 h of life (Robertson et al. 2014). In Sweden, it is a legal requirement that calves are fed colostrum within 6 h after birth. Thus, there should not be a major difference in access to colostrum between beef and dairy calves.
The maximum OPG value in C. parvum monoinfected calves was 3·8 × 106 OPG in a 12 days old calf. The other calves shed fewer oocysts and no calf older than 5 weeks shed more than 2800 OPG. The majority of calves in which C. parvum was detected had mixed infections where either C. bovis or C. ryanae was the only species detected by sequencing of the 18S rRNA locus. Presence of C. parvum was only identified when GP60 was used as an additional tool, indicating a higher infection level of the apathogenic species. Each herd was only visited once, so the maximum oocyst shedding could possibly have been higher in some individuals at other time points. Even so, together these results indicate that calves in Swedish suckler herds, where calves are usually 5–6 weeks old before turn out to pasture, are of limited importance for C. parvum oocysts shedding to the environment.
C. ubiquitum has a broad host range and wide geographic distribution and has mostly been found in sheep and humans (Fayer et al. 2010; Li et al. 2014). Cattle can be experimentally infected (Fayer et al. 2010), and natural infection with C. ubiquitum has been described once, in a 22 week old veal calf in Brittany, France (Follet et al. 2011). We report here the first finding of C. ubiquitum in cattle in Sweden. The positive sample came from a 5 week old bull calf in a herd where 11 other calves were sampled. The sequence did not contain ambiguities but was short (227 bp). PCR and sequencing of the 18S rRNA locus for this sample was thus re-run in order to confirm the result. None of the other samples from this herd contained C. ubiquitum.
The two C. parvum subtypes identified both belonged to zoonotic subtype families; IIa and IId. This is in accordance with what has been found in other European countries, where subtype family IIa is the most common in calves (Robertson et al. 2014). Subtype IIaA16G1R1 has previously been detected in calves and humans in Sweden (Insulander et al. 2013; Silverlås et al. 2010b, 2013). Recently this subtype of C. parvum caused an outbreak in Sweden through direct contact between calves and humans (Kinross et al. 2015). It is a common subtype in calves in Eastern Europe (Soba and Logar, 2008; Imre et al. 2011; Kváč et al. 2011) and has been reported from Germany, the Netherlands and Estonia (Broglia et al. 2008; Wielinga et al. 2008; Lassen et al. 2014). It has been found in pigs and lambs (Kváč et al. 2009; Imre et al. 2013), and human infection has been reported from e.g. Slovenia and Estonia (Soba and Logar, 2008; Lassen et al. 2014). The IIdA27G1 subtype has not been previously identified in Sweden and only two short sequences from calves, both from Romania, are present in GenBank (GB); IIdA27G1a (GB id KC469693) and IIdA27G1b (GB id KC469694). The post repetitive sequences of our isolates contained numerous double peaks in the chromatograms and it could not be determined whether they belonged to either of these subtypes or should have another small letter suffix. Subtype IIaA16G1R1 was the most common subtype found in calves from herds with calf diarrhoea problems (Silverlås et al. 2013). In the present study, however, none of the investigated herds reported any calf diarrhoea at the time of sampling. The distribution pattern found here, with only two subtypes in the study population and one subtype in each investigated herd, is rather uncommon and was surprising to us. A previous study from Sweden reported as many as 27 subtypes from 171 investigated C. parvum positive samples (Silverlås et al. 2013). A similar pattern with multiple subtypes has also been observed in other molecular epidemiology studies performed in Europe, although the subtype diversity varies greatly among countries (reviewed by Robertson et al. 2014). The highest GP60 diversity, besides the Swedish study, has been reported from the Netherlands, where 17 subtypes were found in 100 investigated C. parvum positive samples (Wielinga et al. 2008). It has been suggested that herd management strategies affect subtype distribution. Areas with closed herd management (few animal movements between herds) usually show a high number of subtypes in the calf population, but only one subtype in each herd (Misic & Abe, 2007; Brook et al. 2009; Soba & Logar, 2008; Silverlås et al. 2013). In contrast, fewer subtypes but two or more subtypes per herd is a common pattern in areas with animal exchange between herds (Peng et al. 2003; Trotz-Williams et al. 2006; Brook et al. 2009). The vast majority of the herds in the present study had bought animals during recent years so we expected to find the latter pattern here.
The median herd size in this study was larger than the overall median suckler herd size in Sweden at the time of the study. It is thus possible that our data is biased compared with the average Cryptosporidium spp. infection pressure and species distribution in Swedish suckler herds. Infection pressure generally increases with population size, which could mean that we have overestimated both the herd and the within-herd prevalence of Cryptosporidium spp. It is possible more herds would have come out as Cryptosporidium spp. negative if smaller herds would have been included to a larger extent. Another possible bias is that the small sample size (30 herds) may not be representative of all suckler herds. Increasing the sample size certainly increases the reliability of results but this could not be done due to lack of funding. The model identified two factors associated with Cryptosporidium infection. However, the sensitivity, specificity and prediction probability of the model was low, indicating that other important factors were missed. The reasons could be that the questionnaire was largely developed based on knowledge about routines in dairy herds, and also that even though questions were categorized farmers were allowed to give more open answers instead of just checking the most applicable answer. This led to a number of answers spanning several categories, or making it impossible to categorize answers and a number of variables could not be used for the regression analysis. There was no indication of unstable estimates due to few observations. The reason for one herd becoming an outlier in the model is most likely that this was the only herd that had not answered the question about the number of cows in calving pens, creating a unique level of that variable for those calves.
In conclusion, the results show that Cryptosporidium infection is common in Swedish suckler herds and that C. bovis is the dominant species in young calves. Approximately 8% of the calves excreted C. parvum which is consistent with earlier studies in the dairy herds. The two subtypes of C. parvum that were found both belong to zoonotic subtype families.
ACKNOWLEDGEMENTS
We thank Helena Reineck for valuable help in the laboratory.
FINANCIAL SUPPORT
This study was funded by Swedish Farmers’ Foundation for Agricultural Research (Grant number H1150199).
REFERENCES
- Blanchard P. C. (2012). Diagnostics of dairy and beef cattle diarrhea. Veterinary Clinics of North America: Food Animal Practice 28, 443–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broglia A., Reckinger S., Cacció S. M. and Nöckler K. (2008). Distribution of Cryptosporidium parvum subtypes in calves in Germany. Veterinary Parasitology 154, 8–13. [DOI] [PubMed] [Google Scholar]
- Brook E. J., Anthony Hart C., French N. P. and Christley R. M. (2009). Molecular epidemiology of Cryptosporidium subtypes in cattle in England. Veterinary Journal 179, 378–382. [DOI] [PubMed] [Google Scholar]
- Cacciò S. M. and Widmer G. (ed.) (2014). Cryptosporidium: Parasite and Disease, Springer-Verlag, Wien. [Google Scholar]
- Chalmers R. M., Ferguson C., Caccio S., Gasser R. B., Abs El-Osta Y. G., Heijnen L., Xiao L., Elwin K., Hadfield S., Sinclair M. and Stevens M. (2005). Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. International Journal for Parasitology 35, 397–410. [DOI] [PubMed] [Google Scholar]
- Dixon B., Parrington L., Cook A., Pintar K., Pollari F., Kelton D. and Farber J. (2011). The potential for zoonotic transmission of Giardia duodenalis and Cryptosporidium spp. from beef and dairy cattle in Ontario, Canada. Veterinary Parasitology 175, 20–26. [DOI] [PubMed] [Google Scholar]
- Fayer R., Santín M. and Macarisin D. (2010). Cryptosporidium ubiquitum n. sp. in animals and humans. Veterinary Parasitology 172, 23–32. [DOI] [PubMed] [Google Scholar]
- Follet J., Guyo t. K., Leruste H., Follet-Dumoulin A., Hammouma-Ghelboun O., Certad G., Dei-Cas E. and Halama P. (2011). Cryptosporidium infection in a veal calf cohort in France: molecular characterization of species in a longitudinal study. Veterinary Research 42, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurden T., Goma F. Y., Siwila J., Phiri I. G. K., Mwanza A. M., Gabriel S., Claerebout E. and Vercruysse J. (2006). Prevalence and genotyping of Cryptosporidium in three cattle husbandry systems in Zambia. Veterinary Parasitology 138, 217–222. [DOI] [PubMed] [Google Scholar]
- Geurden T., Berkvens D., Martnens C., Casaert S., Vercruysse J. and Claerebout E. (2007). Molecular epidemiology with subtype analysis of Cryptosporidium in calves in Belgium. Parasitology 134, 1981–1987. [DOI] [PubMed] [Google Scholar]
- Imre K., Lobo L. M., Matos O., Popescu C., Genchi C. and Dărăbuş G. (2011). Molecular characterisation of Cryptosporidium isolates from pre-weaned calves in Romania: Is there an actual risk of zoonotic infections? Veterinary Parasitology 181, 321–324. [DOI] [PubMed] [Google Scholar]
- Imre K., Luca C., Costache M., Sala C., Morar A., Morariu S., Ilie M. S., Imre M. and Dărăbuş G. (2013). Zoonotic Cryptosporidium parvum in Romanian newborn lambs (Ovis aries). Veterinary Parasitology 191, 119–122. [DOI] [PubMed] [Google Scholar]
- Insulander M., Silverlås C., Lebbad M., Karlsson L., Mattsson J. G. and Svenungsson B. (2013). Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiology & Infection 141, 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinross P., Beser J., Troell K., Silverlås C., Björkman C., Lebbad M., Winiecka-Krusnell J., Lindh J. and Löfdahl M. (2015). Cryptosporidium parvum infections in a cohort of veterinary students in Sweden. Epidemiology & Infection, In press, doi: 10.1017/S0950268814003318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kváč M., Sak B., Hanzlíková D., Kotilová J. and Květoňová D. (2009). Molecular characterization of Cryptosporidium isolates from pigs at slaughterhouses in South Bohemia, Czech Republic. Parasitology Research 104, 425–428. [DOI] [PubMed] [Google Scholar]
- Kváč M., Hromadová N., Květoňová D., Rost M. and Sak B. (2011). Molecular characterization of Cryptosporidium spp. in pre-weaned dairy calves in the Czech Republic: Absence of C. ryanae and management-associated distribution of C. andersoni, C. bovis and C. parvum subtypes. Veterinary Parasitology 177, 378–382. [DOI] [PubMed] [Google Scholar]
- Langkjaer R. B., Vigre H., Enemark H. L. and Maddox-Hyttel C. (2007). Molecular and phylogenetic characterization of Cryptosporidium and Giardia from pigs and cattle in Denmark. Parasitology 134, 339–350. [DOI] [PubMed] [Google Scholar]
- Lassen B., Ståhl M. and Enemark H. L. (2014). Cryptosporidiosis–an occupational risk and a disregarded disease in Estonia. Acta Veterinaria Scandinavica 56, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Xiao L., Alderisio K., Elwin K., Cebelinski E., Chalmers R., Santin M., Fayer R., Kvac M., Ryan U., Sak B., Stanko M., Guo Y., Wang L., Zhang L., Cai J., Roellig D. and Feng Y. (2014). Subtyping Cryptosporidium ubiquitum,a zoonotic pathogen emerging in humans. Emerging Infectious Diseases 20, 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox-Hyttel C., Langkjaer R. B., Enemark H. L. and Vigre H. (2006). Cryptosporidium and Giardia in different age groups of Danish cattle and pigs – occurrence and management associated risk factors. Veterinary Parasitology 141, 48–59. [DOI] [PubMed] [Google Scholar]
- Misic Z. and Abe N. (2007). Subtype analysis of Cryptosporidium parvum isolates from calves on farms around Belgrade, Serbia and Montenegro, using the 60 kDa glycoprotein gene sequences. Parasitology 134, 351–358. [DOI] [PubMed] [Google Scholar]
- O'Handley R. M., Cockwill C., McAllister T. A., Jelinski M., Morck D. W. and Olson M. E. (1999). Duration of naturally acquired giardiosis and cryptosporidiosis in dairy calves and their association with diarrhea. Journal of American Veterinary Medical Association 214, 391–396. [PubMed] [Google Scholar]
- Peng M. M., Wilson M. L., Holland R. E., Meshnick S. R., Lal A. A. and Xiao L. (2003). Genetic diversity of Cryptosporidium spp. in cattle in Michigan: implications for understanding the transmission dynamics. Parasitology Research 90, 175–180. [DOI] [PubMed] [Google Scholar]
- Plutzer J. and Karanis P. (2007). Genotype and subtype analyses of Cryptosporidium isolates from cattle in Hungary. Veterinary Parasitology 146, 357–362. [DOI] [PubMed] [Google Scholar]
- Robertson L. J., Björkman C., Axén C. and Fayer R. (2014). Cryptosporidiosis in farmed animals In Cryptosporidium: Parasite and Disease (ed. Caccio S. M. and Widmer G.), pp. 149–235. Springer-Verlag, Wien. [Google Scholar]
- Santín M., Trout J. M. and Fayer R. (2008). A longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Veterinary Parasitology 155, 15–23. [DOI] [PubMed] [Google Scholar]
- Silva S. O., Richtzenhain L. J., Barros I. N., Gomesa A. M. M. C., Silva A. V., Kozerski N. D., de Araújo Ceranto J. B., Keid L. B. and Soares R. M. (2013). A new set of primers directed to 18S rRNA gene for molecular identification of Cryptosporidium spp. and their performance in the detection and differentiation of oocysts shed by synanthropic rodents. Experimental Parasitology 135, 551–557. [DOI] [PubMed] [Google Scholar]
- Silverlås C. and Blanco-Penedo I. (2013). Cryptosporidium spp. in calves and cows from organic and conventional dairy herds. Infection and Immunity 141, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverlås C., Emanuelson U., de Verdier K. and Björkman C. (2009). Prevalence and associated management factors of Cryptosporidium shedding in 50 Swedish dairy herds. Preventive Veterinary Medicine 90, 242–253. [DOI] [PubMed] [Google Scholar]
- Silverlås C., de Verdier K., Emanuelson U., Mattsson J. G. and Björkman C. (2010a). Cryptosporidium infection in herds with and without calf diarrhoeal problems. Parasitology Research 107, 1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverlås C., Näslund K., Björkman C. and Mattsson J. G. (2010b). Molecular characterisation of Cryptosporidium isolates from Swedish dairy cattle in relation to age, diarrhoea and region. Veterinary Parasitology 169, 289–295. [DOI] [PubMed] [Google Scholar]
- Silverlås C., Bosaeus-Reineck H., Näslund K. and Björkman C. (2013). Is there a need for improved Cryptosporidium diagnostics in Swedish calves? International Journal for Parasitology 43, 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soba B. and Logar J. (2008). Genetic classification of Cryptosporidium isolates from humans and calves in Slovenia. Parasitology 135, 1263–1270. [DOI] [PubMed] [Google Scholar]
- Statistics Sweden. (2013). Yearbook of Agricultural Statistics 2013. Statistics Sweden, Agriculture Statistics Unit, Örebro. [Google Scholar]
- Trotz-Williams L. A., Martin D. S., Gatei W., Cama V., Peregrine A. S., Martin S. W., Nydam D. V., Jamieson F. and Xiao L. (2006). Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitology Research 99, 346–352. [DOI] [PubMed] [Google Scholar]
- Wielinga P. R., de Vries A., van der Goot T. H., Mank T., Mars M. H., Kortbeek L. M. and van der Giessen J. W. B. (2008). Molecular epidemiology of Cryptosporidium in humans and cattle in The Netherlands. International Journal for Parasitology 38, 809–817. [DOI] [PubMed] [Google Scholar]
- Xiao L., Escalante L., Yang C., Sulaiman I., Escalante A. A., Montali R. J., Fayer R. and Lal A. A. (1999). Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Applied and Environmental Microbiology 65, 1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Alderisio K., Limor J., Royer M. and Lal A. F. (2000). Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Applied and Environmental Microbiology 66, 5492–5498. [DOI] [PMC free article] [PubMed] [Google Scholar]