Abstract
Background:
Prostate cancer overdiagnosis and overtreatment represents a major problem. Many men with low-grade disease on biopsy are undergraded and they harbour high-grade disease at prostatectomy with no reliable way to identify these men. We used a novel urine-based 2-gene methylation test to identify prostate cancers with aggressive features.
Methods:
Following a proof of concept study in 100 post-radical prostatectomy tissue samples, urine samples were tested from 665 men at multiple U.S. centers undergoing prostate needle biopsy for elevated prostate-specific antigen (2–10 ng ml−1). A prediction model was then developed from a combination of clinical factors and the urine-based markers. It was then prospectively tested for accurate prediction of adverse disease (surgical Gleason score ⩾7 and/or a pathological stage ⩾T3a) using urine from a separate cohort of 96 men before radical prostatectomy.
Results:
Among pre-prostatectomy men with a biopsy Gleason score <7, 41% had adverse disease of which 100% were correctly identified by the test with a negative predictive value of 100% (95% confidence interval, 86–100%).
Conclusions:
This urine-based test accurately identifies men with clinical low-risk disease who do not have adverse pathology in their prostates and would be excellent candidates for active surveillance.
Keywords: prostate cancer, risk assessment, methylation markers
Over 238 000 men are estimated to be diagnosed with prostate cancer in the United States this year, with ∼29 000 dying from the disease (American Cancer Society, 2013). Nearly 85% of these cancers will be detected by prostate-specific antigen (PSA) screening with diagnostic PSA level above 4.0 ng ml−1 (Glass et al, 2013). Two recently reported studies showed disparate findings with regard to the impact of PSA screening on prostate cancer-specific mortality, yet both showed that the single biggest risk for screening is overdetection (Andriole et al, 2009; Schröder et al, 2009). It is estimated that 10–56% of the cases of diagnosed prostate cancer would be ‘insignificant' and not have had a meaningful clinical impact if they had remained undetected/untreated (Draisma et al, 2009). In low-risk patients, the Prostate Cancer Intervention versus Observation Trial demonstrated that a survival benefit cannot be observed in radical prostatectomy vs active surveillance (AS) after 10 years of median follow-up (Wilt et al, 2012). Despite this latter finding, the vast majority of men with prostate cancer are actively treated with surgery, radiation, androgen ablation, or all the three (Cooperberg et al, 2010). This holds true even for those cancers deemed ideal for surveillance or less aggressive treatments (Barocas et al, 2008).
Overdiagnosed prostate cancers can cause a negative impact on the quality-adjusted life years following PSA screening (Heijnsdijk et al, 2012). It is difficult to assure a patient that a low-risk cancer at biopsy is truly ‘insignificant' as biopsies inherently undersample the prostate (Harnden et al, 2008; Shaw et al, 2014). Using contemporary AS protocols and standard multicore biopsy techniques, patients with no evidence of a high Gleason score cancer on biopsy still carry a significant risk of having a high-grade tumour when the prostate is removed and sampled completely (Iremashvilli et al, 2012). This is troubling as a number of clinical end points have been linked to Gleason score, including TNM stage, response to different therapies, biochemical failure, progression to metastatic disease, and survival, including prostate cancer-specific survival and overall survival (Partin et al, 1995, 2001; Kattan et al, 1998, 2003; Freedland et al, 2003; Cooperberg et al, 2005). A recent study reports that 36% of tumours with a Gleason score of 6 on biopsy are upgraded at radical prostatectomy to a Gleason score of 7 or higher (Epstein et al, 2012). A similar upgrading rate of 34% as well as an upstaging rate of 13% was also reported in both the African American and other racial groups (Jalloh et al, 2014).
Given the inherent limitations of biopsies, additional means are required to more accurately assess the tumour aggressiveness. Prostate cancer-specific methylation patterns detected in urine samples provide a potential source of additional information beyond what can be assessed by biopsy. In order to demonstrate that urine-sourced methylation patterns can discriminate between the aggressive and low-risk tumours, we developed a urine-based multiplex methylation specific PCR assay. The assay measures the copy number of β-Actin as a housekeeping gene and the methylation status of GSTP1 and APC, both of which have been shown previously to be associated with prostate cancer when detected in urine (Baden et al, 2011).
Initially, we used tissue specimens to test the ability of the assay biomarkers to discriminate Gleason scores in the primary tumour. We then use the assay to examine the methylation status of these markers in urine from men undergoing prostate cancer screening in order to develop the prediction model. The objective of the validation phase was to accurately predict the adverse disease at radical prostatectomy (surgical Gleason ⩾7 and/or pathological stage ⩾T3a) using the assay on urine specimens taken before surgery. Should the assay significantly improve risk stratification, this would improve the accuracy of identifying men who do not harbour aggressive prostate cancer and can safely avoid unnecessary therapy.
Material and methods
Patient population and sample processing
To directly measure the assay biomarkers in primary tumour tissue, a retrospective collection of 104 formalin-fixed paraffin-embedded (FFPE) radical prostatectomy tumour samples were obtained from men who underwent radical prostatectomy at the Durham VA Medical Center. After evaluating the β-Actin control, 100 of these had a valid test result. There were no restrictions on the total PSA level or any other clinical parameters.
The biopsy cohort used to train the prediction models involved a prospective trial among 18 urology clinics across the United States and included 683 men. Subjects were enrolled in this study between September 2008 and January 2009. Inclusion requirements were age between 40 and 75, total PSA levels between 2.0 and 10.0 ng ml−1, and the completion of a digital rectal examination (DRE). Among men diagnosed with prostate cancer (n=253), pathological assessments from radical prostatectomy specimens were available for 39 men.
The pre-prostatectomy cohort used for validation of the assay contains 99 male subjects from four urology clinics. Enrolment in this study commenced in September 2008 and was completed by August 2012. Subject requirements were 40–75 years of age, total PSA level between 2.0 and 10.0 ng ml−1, completion of a DRE, biopsy that confirmed prostate cancer, and scheduled to undergo a radical prostatectomy without neoadjuvant therapy.
For all the subjects in both the biopsy and pre-prostatectomy cohorts, 5–40 ml urine samples were collected following a DRE, and were processed as described previously (Baden et al, 2011). Following urine sample collection, subjects in the biopsy cohort underwent TRUS-guided needle biopsy of at least 10 cores. As such, for the biopsy cohort, urine samples were collected before biopsy. The urine samples were processed using the methylation assay. A valid assay test result was determined for 665 subjects in the biopsy cohort and 96 subjects in the pre-prostatectomy collection. A valid test result required that the amount of β-Actin in the sample was sufficient to produce an RT–PCR cycle threshold of <25 cycles.
Methylation assay
Methylation status was determined as previously described (Baden et al, 2011). Primers and Scorpion probes for two methylation markers (GSTP1, and APC) and an internal control (β-Actin) were chosen for use in a closed-tube format two-step PCR assay. Internal and external controls were used in each run; the internal control (β-Actin), to determine the adequacy of the DNA quantity and preparation, and the external control (targeted plasmid DNA), to confirm a lack of environmental contamination and to assure that reaction mixtures were functioning appropriately. Sample results that were not valid were omitted from the analyses.
Statistical analyses
The prediction model is a logistic model developed in the biopsy cohort by fitting the biopsy cohort values of β-Actin, GSTP1, APC, age, DRE, and PSA to the binary response of a biopsy Gleason score of 7 or higher vs Gleason score of 6 or lower or no cancer. Values for APC were allowed to be transformed by a restricted cubic spline as directed by the response variable, whereas β-Actin, GSTP1, PSA, and DRE were not subjected to any transformations. The calibration between the outputs of the model and the observed probabilities was evaluated using bias-corrected estimates of the risk scores vs the observed values based on nonparametric smoothers A logistic regression model using untransformed continuous PSA, age, and DRE measurements without β-Actin, GSTP1, and APC was also designed in the same manner (Harrell, 2001).
Using the prediction model scores from men in the biopsy cohort, a dichotomous cutoff was chosen to select the bottom 20% of the low-risk scores, which was found to maximise the negative predictive value (NPV) for a biopsy Gleason ⩾7. The prediction models were then applied to an independent pre-prostatectomy validation cohort (n=96) to test its ability to discriminate between adverse (surgical Gleason ⩾7 and/or stage ⩾pT3a) and low-risk (surgical Gleason 6 and stage ⩽pT2c) disease on final pathology using the pre-established cutoff from the biopsy cohort.
Bar charts and box plots were generated using MedCalc (MedCalc Software, Ostend, Belgium). Binomial confidence limits were calculated using the Wilson method in SAS software (SAS Institute Inc., Cary, NC, USA). Confidence intervals (CIs) of the prevalence-adjusted NPV were determined as outlined in Pepe, (2004). Clinical variables were plugged into a preoperative nomogram trained to predict organ confined disease (available at nomograms.org), the output of probabilities for this model were then used to make adverse disease predictions (Wang et al, 2006). Logistic modelling and all other statistical analysis was done using R (R Foundation for Statistical Computing, Vienna, Austria). P-values for group comparisons were determined by the two-sided Student's t-test. The area under the receiver operating characteristic curve (AUC) was calculated using the Wilcoxon–Mann–Whitney test statistic.
Results
Formalin-fixed paraffin-embedded tissue evaluation of the radical prostatectomy specimens
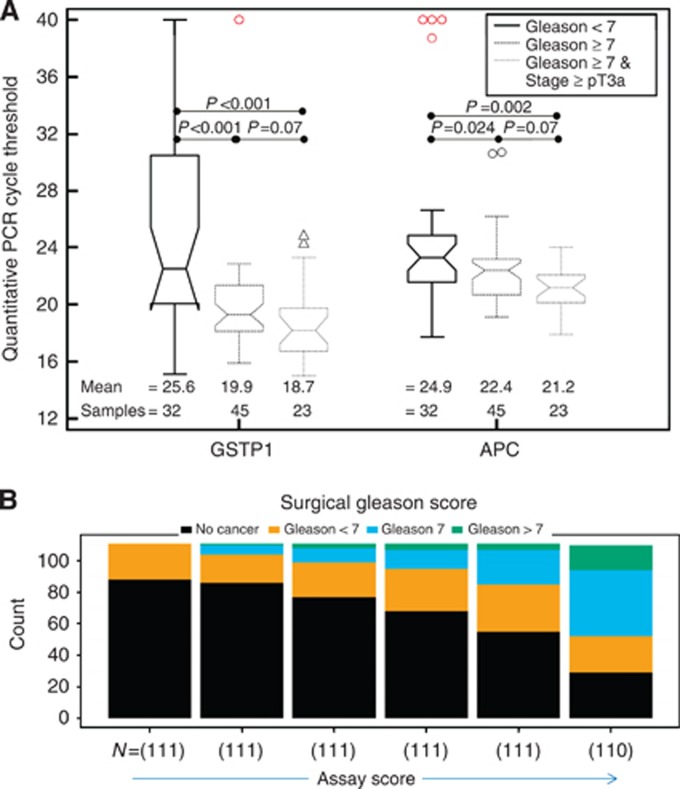
Initial assessments of the predictive power of the methylated regions of GSTP1 and APC were made by evaluating the FFPE prostate cancer tissue of 100 radical prostatectomy subjects from the Durham VA Medical Center. Clinical and pathological characteristics of the patients are described in (Table 1). GSTP1 and APC were found to have a significant increase in hypermethylation in patients with a surgical Gleason score ⩾7 compared with those with a surgical Gleason score <7 (P<0.001 and P=0.024, respectively). Furthermore, the magnitude of this difference in methylation levels increased for both the markers when the Gleason score <7 group was compared with samples that had both a Gleason score ⩾7 and a tumour stage ⩾pT3a (P<0.001 and P=0.002, respectively; Figure 1A).
Table 1. Clinicopathological characteristics of the study population.
| Subject characteristics | 100 Subjects post-operative (tissue) | 665 Subjectsbiopsy (urine) | 96 Subjectspre-prostatectomy (urine) |
|---|---|---|---|
| Number of patients (%) | |||
|
Ethnicity | |||
| Caucasian | 52 (52) | 522 (78.5) | 89 (92.7) |
| African American | 48 (48) | 96 (14.4) | 4 (4.2) |
| Hispanic | 0 (0) | 35 (5.3) | 2 (2.1) |
| Other | 0 (0) | 12 (1.8) | 1 (1) |
| Family history of prostate cancer | NA | 133 (20) | 32 (33.3) |
| Previous biopsy | NA | 151 (22.7) | 4 (4.2) |
|
DRE results | |||
| Non suspicious | NA | 589 (88.6) | 80 (83.3) |
| Suspicious | NA | 76 (11.4) | 16 (16.7) |
|
PSA value | |||
| <2.0 | 3 (3) | 1 (0.1) | 0 (0) |
| 2.0–4.0 | 17 (17) | 166 (25) | 27 (28.1) |
| 4.1–10.0 | 50 (50) | 498 (74.9) | 69 (71.9) |
| >10 | 30 (30) | 0 (0) | 0 (0) |
|
Biopsy Gleason scores | |||
| GS 6 | 56 (56) | 143 (21.5) | 64 (66.7) |
| GS 7 | 33 (33) | 91 (13.7) | 25 (26) |
| GS 8 | 9 (9) | 20 (3) | 6 (6.3) |
| GS 9 | 1 (1) | 8 (1.2) | 1 (1) |
| GS10 | 1 (1) | 1 (0.1) | 0 (0) |
|
Surgical Gleason scores | |||
| GS 5 | 4 (4) | NA | 0 (0) |
| GS 6 | 18 (18) | NA | 44 (45.8) |
| GS 7 | 68 (68) | NA | 45 (46.9) |
| GS 8 | 5 (5) | NA | 3 (3.1) |
| GS 9 | 5 (5) | NA | 4 (4.2) |
|
Surgical pathology stage | |||
| pT2a | 26 (26) | NA | 19 (19.8) |
| pT2b | 0 (0) | NA | 6 (6.3) |
| pT2c | 47 (47) | NA | 56 (58.3) |
| pT3a | 11 (11) | NA | 13 (13.5) |
| pT3b | 11 (11) | NA | 2 (2.1) |
| pT4 | 4 (4) | NA | 0 (0) |
| Percentage of patients with a positive core (median percentage of cores that are positive) |
|||
| Cancer status | 100% (NA) | 39.5% (20.4%) | 100% (33.3%) |
| Median number of cores taken (range) |
|||
| Cores tested | NA | 12 (6–35) | 12 (4–35) |
| Median years (range) |
|||
| Age | 61 (47–73) | 64 (40–75) | 61 (46–75) |
Abbreviations: DRE=digital rectal examination; NA=not available; PSA=prostate-specific antigen.
Figure 1.
The relationship between GSTP1 and APC methylation status in FFPE tissue samples and patients with a Gleason score <7, a Gleason score ⩾7, and a Gleason score ⩾7 with a pathological tumour stage of at least T3a are represented by box plots in panel A. The quantitative PCR cycle threshold values are inversely related to the degree of hypermethylation. The assay scores calculated from urine results in the biopsy cohort are shown in panel B as a stacked bar chart representing the percentage of men with a biopsy Gleason ⩾7, a biopsy Gleason <7, or no cancer. Bins were selected to contain an equivalent number of patients in each bin.
Biopsy cohort
Given the proof of principle that at least on the tissue level, GSTP1 and APC could segregate low- and high-risk tumours, we then developed a multicenter biopsy cohort to construct the prediction model. Clinicopathological characteristics for these men are shown in (Table 1). On biopsy, 263 of the 665 men with usable assay scores (40%) were diagnosed with prostate cancer and 120 (18%) had a Gleason score ⩾7. Consistent with the association of the methylation status with adverse pathology at prostatectomy in the FFPE collection, the test score from the prediction model was significantly higher in men with a Gleason score ⩾7 vs Gleason <7 or non-cancer preceding biopsy (P<0.001). As the individual's test score increased, the probability of Gleason score ⩾7 cancers increased with no appreciable increase in the risk of low-grade cancer (Figure 1B).
The prediction model trained with both the biomarkers and the clinical factors had a significantly increased AUC for predicting a Gleason score ⩾7 on biopsy compared with a prediction model using the same clinical factors without the biomarkers (0.82 vs 0.69, P<0.001) in the biopsy cohort. A cutoff was chosen as a point that maximises the NPV of the assays ability to predict a biopsy Gleason score ⩾7. At the cutoff that dichotomizes the bottom 20% of the low-risk scores there is a 5% predicted probability of having a high biopsy Gleason score. At this cutoff, 20% of the patients in the biopsy cohort are classified as low risk with an NPV of 100% and there is a <5% predicted probability of a biopsy Gleason ⩾7. The predicted probabilities of the model are well calibrated to the observed probabilities with a mean absolute error of 0.009.
Pre-prostatectomy cohort
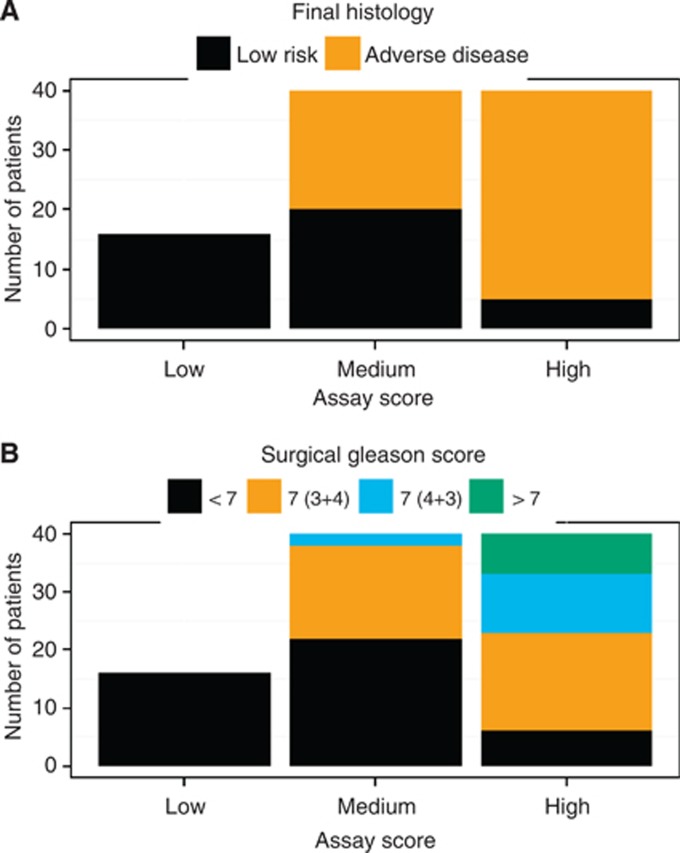
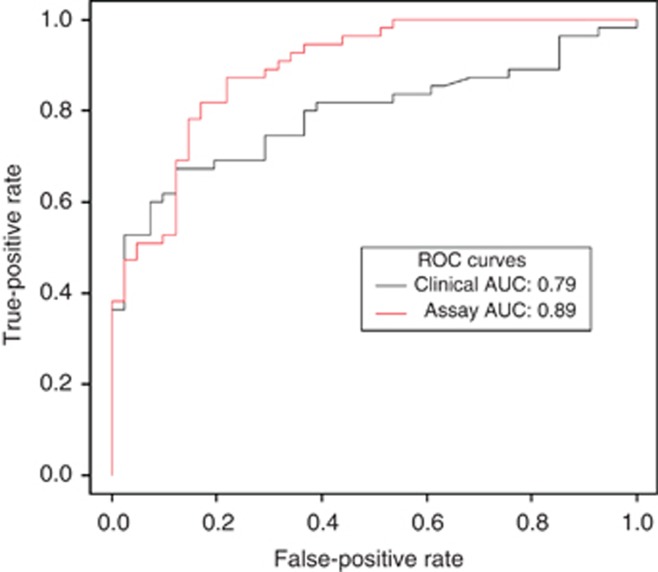
To validate the model and cutoff, we prospectively tested the urine obtained from 96 subjects undergoing radical prostatectomy. The probability of high-grade cancer and adverse pathological features increased with an individual's test score from the prediction model, (Figure 2A and B). When combined with the Gleason result from biopsy, the test score accurately stratified men with adverse disease at prostatectomy (AUC=0.89), and outperformed the clinical model alone (AUC=0.79; Figure 3). Using the 64 subjects that had a biopsy Gleason <7, a publically available preoperative nomogram designed to predict organ confined disease had an AUC of 0.73 when used to predict the adverse disease, whereas the assay model's AUC remained high at 0.83.
Figure 2.
Distribution of the urine-based assay test score in the pre-prostatectomy validation set and it association with the surgical Gleason score and adverse disease. The assay test score was divided into three bins (low, medium, and high), where the low bin represents the biopsy cohort established cutoff, and the medium and high bins were then chosen to contain an equivalent number of patients in each bin. In panel A the distribution is coloured by either the adverse or low-risk disease category, and in panel B the distribution is coloured by the surgical Gleason score.
Figure 3.
Receiver operating characteristic (ROC) curves of the assay test score for the prediction of surgical Gleason score of 7 or a pathological stage of T3A or higher.
Of the subjects with a biopsy Gleason score <7, 48% were found to have adverse disease following surgery. When the test score from the prediction model was evaluated as a dichotomous variable using the cutoff determined from the biopsy cohort with the criteria of a biopsy Gleason score ⩾7 being categorised as adverse, it correctly identified 100% of the subjects with either a surgical Gleason score of 7 or greater, or a pathological tumour stage of T3a or greater (Table 2). When used to classify subjects with a biopsy Gleason <7, the NPV of the assay for adverse disease was 100% (95% CI: 86–100%).
Table 2. Assay and clinical prediction model performance data.
|
Final histology |
||||||
|---|---|---|---|---|---|---|
| Variables | P-Values of regression coefficients | Predicted group | Gleason ⩾ 7 or stage ⩾ pT3a | Gleason <7 and stage < pT3a | AUC | Measures of performance (95% confidence interval) |
|
Biopsy Gleason grade alone | ||||||
| Biopsy Gleason Grade | NA | Biopsy Gleason ⩾7 | 29 | 3 | 0.73 | Sensitivity=53% (40–65%) |
| NPV (observed 57% prevalence)=60% (49–65%) | ||||||
| Biopsy Gleason <7 | 26 | 38 | Percentage biopsy Gleason <7=67% | |||
|
Clinical Model | ||||||
| PSA | 0.0159 | Adverse or biopsy Gleason ⩾7 | 54 | 40 | 0.79 | Sensitivity=98% (90–100%) |
| Age | <0.0001 | NPV (observed 57% prevalence)=50% (6–94%) | ||||
| DRE | 0.0005 | Low-risk and biopsy Gleason <7 | 1 | 1 | Percentage classified low-risk and biopsy Gleason <7=2% | |
|
Assay Model | ||||||
| GSTP1 | <0.0001 | Adverse or biopsy Gleason ⩾7 | 55 | 25 | 0.89 | Sensitivity=100% (95–100%) |
| APC | <0.0001 | |||||
| APC (nonlinear) | 0.0001 | NPV (observed 57% prevalence)=100% (86–100%) | ||||
| ACTB | 0.0003 | Low-risk and biopsy Gleason <7 | 0 | 16 | Percentage classified low-risk and biopsy Gleason <7=17% | |
| PSA | 0.0114 | |||||
| Age | 0.0016 | |||||
| DRE | 0.0394 | |||||
Abbreviations: AUC=area under the receiver operating characteristic curve; DRE=digital rectal examination; NPV=negative predictive value; PSA=prostate-specific antigen.
Discussion
Many diagnosed prostate cancers are slow growing or indolent. So long as the tumour remains in that state, it does not imminently threaten the health of the patient. The European Randomized study of Screening for Prostate Cancer (ERSPC) trial determined that about 30% of the detected prostate cancers remain unlikely to progress to a point where it will cause a patient's death (Schröder et al, 2012). At the same time, the ERSPC trial reported a 20% reduction in the prostate cancer mortality at 11 years from PSA screening. The Göteborg PSA Screening trial reported a 44% reduction in the prostate cancer mortality over 14 years (Hugosson et al, 2010). The resource intensive nature of PSA screening becomes clear when one looks at the number of men needed to be screened to lower the mortality rate. ERSPC reported 1.07 fewer deaths from prostate cancer per 1000 men screened. Nevertheless, it is clear from the high frequency of aggressive treatment of low-risk prostate cancer that patients and their physicians are not favouring AS or less aggressive treatments (Dall'Era et al, 2008).
To address this, a useful test not only should show significance in its ability to distinguish patients with and without adverse disease but must also be clinically actionable. A recent study looked at various nomograms via regression analysis, adjusted for the total number of biopsies, and concluded that the AUCs only ranged from 0.52–0.67, and for any progression the AUCs ranged from 0.52–0.70 (Wang et al, 2013). Although this is not a pure prediction of harbouring higher grade and/or stage from biopsy to RP, it highlights the point that nomograms that are purported to predict indolent disease per se really do not function well-enough and set the stage for use of biomarkers of whatever form. Coupled with the low-risk clinical factors, a high NPV with this methylation-based test should increase the confidence that a low-risk patient indeed has low-risk disease and provide added reassurance that AS and avoiding treatments will not negatively impact the survival. In our validation cohort of men undergoing radical prostatectomy, 25% of the subjects with grade 6 or below on biopsy and 17% of the entire cohort could have been recommended against aggressive definitive treatment following biopsy using the assay with an NPV of adverse findings at surgery of 100% (95% CI: 86%–100%) among these men.
The noninvasive nature of this assay provides an opportunity for monitoring without the risks associated with repeat biopsy. With each biopsy, there is a significantly increased risk of hospitalisation due to serious infectious and noninfectious urological complications (Loeb et al, 2013). Furthermore, a negative biopsy coupled with a PSA level of 3 ng ml−1 or higher has been estimated to cause 20% of men to experience a distressful psychological effect (Macefield et al, 2010). A negative attitude to repeat biopsy has also been reported in 20% of the subjects in the Prostate Biopsy Effects cohort nested within the Prostate Testing for Cancer and Treatment study (Rosario et al, 2012). The high NPV observed in the biopsy cohort suggests that an assay of this nature may have a place earlier in the prostate care pathway continuum, and possibly provide subjects with an elevated PSA with no history of biopsy or patients with a prior negative biopsy greater resolution of their risk, although minimising the distress of biopsy.
The shedding of cancerous cells from the prostate and exiting through the urine offers another opportunity to identify a cellular population that may otherwise have been missed, given that a needle core biopsy removes ∼1/3000th of the prostate. Using the methylation status of a series of markers secreted into the urine, the assay indicates the presence of prostate cancer with adverse features that may remain hidden from a biopsy. Hypermethylation events have been shown to correlate with an elevated risk of prostate cancer progression (Ellinger et al, 2008). Both GSTP1 and APC are hypermethylated in prostate cancer and have shown to be strongly correlated to adverse pathological features (Jerónimo et al, 2004; Zhou et al, 2004; Bastian et al, 2005; Enokida et al, 2005).
Our data provide evidence of a strong association between the presence of adverse disease and the methylation status of both APC and GSTP1 when detected in urine. We are limited by the reliance on the biopsy Gleason score to fit our prediction model because of the unavailability of post-surgical findings in this set. Although the model can classify adverse disease, the predictive probabilities from the model should not be interpreted as a calibrated probability of adverse disease. Furthermore, any direct comparisons with a validated clinical nomogram would require that the nomogram was developed using an identical definition of adverse disease. Future larger studies will be necessary to narrow the CI around the performance parameters of the assay, and to understand the impact on patient and physician decision-making.
Acknowledgments
Ortho Clinical Diagnostics provided funding for the design and conduction of the study, data collection, management, and interpretation of the data.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
References
- Andriole GL, Crawford ED, Grubb RL, III, Buys SS, Chia D, Church TR, Fouad MN, Gelmann EP, Kvale PA, Reding DJ, Weissfeld JL, Yokochi LA, O'Brien B, Clapp JD, Rathmell JM, Riley TL, Hayes RB, Kramer BS, Izmirlian G, Miller AB, Pinsky PF, Prorok PC, Gohagan JK, Berg CD, PLCO Project Team Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Cancer Society . American Cancer Society: Atlanta, GA, USA; 2013. Cancer Facts & Figures 2013. [Google Scholar]
- Baden J, Adams S, Astacio T, Jones J, Markiewicz J, Painter J, Trust C, Wang Y, Green G. Predicting prostate biopsy result in men with prostate specific antigen 2.0 to 10.0 ng/ml using an investigational prostate cancer methylation assay. J Urol. 2011;186:2101–2106. doi: 10.1016/j.juro.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Barocas DA, Cowan JE, Smith JA, Jr, Carroll PR, CaPSURE Investigators What percentage of patients with newly diagnosed carcinoma of the prostate are candidates for surveillance? An analysis of the CaPSURE database. J Urol. 2008;180:1330–1334. doi: 10.1016/j.juro.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Bastian PJ, Palapattu GS, Lin X, Yegnasubramanian S, Mangold LA, Trock B, Eisenberger MA, Partin AW, Nelson WG. Preoperative serum DNA GSTP1 CpG island hypermethylation and the risk of early prostate-specific antigen recurrence following radical prostatectomy. Clin Cancer Res. 2005;11:4037–4043. doi: 10.1158/1078-0432.CCR-04-2446. [DOI] [PubMed] [Google Scholar]
- Cooperberg MR, Broering JM, Carroll PR. Time trends and local variation in primary treatment of localized prostate cancer. J Clin Oncol. 2010;28:1117–1123. doi: 10.1200/JCO.2009.26.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperberg MR, Pasta DJ, Elkin EP, Litwin MS, Latini DM, Du Chane J, Carroll PR. The University of California, San Francisco Cancer of the Prostate Risk Assessment score: a straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol. 2005;173:1938–1942. doi: 10.1097/01.ju.0000158155.33890.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall'Era MA, Cooperberg MR, Chan JM, Davies BJ, Albertsen PC, Klotz LH, Warlick CA, Holmberg L, Bailey DE, Jr, Wallace ME, Kantoff PW, Carroll PR. Active surveillance for early-stage prostate cancer: review of the current literature. Cancer. 2008;112:1650–1659. doi: 10.1002/cncr.23373. [DOI] [PubMed] [Google Scholar]
- Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, Gulati R, Feuer E, de Koning H. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101:374–383. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger J, Bastian PJ, Jurgan T, Biermann K, Kahl P, Heukamp LC, Wernert N, Müller SC, von Ruecker A. CpG island hypermethylation at multiple gene sites in diagnosis and prognosis of prostate cancer. Urology. 2008;71:161–167. doi: 10.1016/j.urology.2007.09.056. [DOI] [PubMed] [Google Scholar]
- Enokida H, Shiina H, Urakami S, Igawa M, Ogishima T, Li LC, Kawahara M, Nakagawa M, Kane CJ, Carroll PR, Dahiya R. Multigene methylation analysis for detection and staging of prostate cancer. Clin Cancer Res. 2005;11:6582–6588. doi: 10.1158/1078-0432.CCR-05-0658. [DOI] [PubMed] [Google Scholar]
- Epstein JI, Feng Z, Trock BJ, Pierorazio PM. Upgrading and downgrading of prostate cancer from prostate to radical prostatectomy: incidence and predictive factors using the modified Gleason grading system and factoring in tertiary grades. Eur Urol. 2012;61:1019–1024. doi: 10.1016/j.eururo.2012.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedland SJ, Sutter ME, Dorey F, Aronson WJ. Defining the ideal cutpoint for determining PSA recurrence after radical prostatectomy. Urology. 2003;61:365–369. doi: 10.1016/s0090-4295(02)02268-9. [DOI] [PubMed] [Google Scholar]
- Glass AS, Cowan JE, Fuldeore MJ, Cooperberg MR, Carroll PR, Kenfield SA, Greene KL. Patient demographics, quality of life, and disease features of men with newly diagnosed prostate cancer: trends in the PSA era. Urology. 2013;82:60–65. doi: 10.1016/j.urology.2013.01.072. [DOI] [PubMed] [Google Scholar]
- Harnden P, Naylor B, Shelly MD, Clements H, Coles B, Mason M. The clinical management of patients with a small volume of prostatic cancer on biopsy: what are the risks of progression? A systematic review and meta-analysis. Cancer. 2008;112:971–981. doi: 10.1002/cncr.23277. [DOI] [PubMed] [Google Scholar]
- Harrell FE. Logistic Regression, and Survival Analysis. Springer: New York, NY, USA; 2001. Regression Modeling Strategies: with Applications to Linear Models. [Google Scholar]
- Heijnsdijk EA, Wever EM, Auvinen A, Hugosson J, Ciatto S, Nelen V, Kwiatkowski M, Villers A, Páez A, Moss SM, Zappa M, Tammela TL, Mäkinen T, Carlsson S, Korfage IJ, Essink-Bot ML, Otto SJ, Draisma G, Bangma CH, Roobol MJ, Schröder FH, de Koning HJ. Quality-of-life effects of prostate-specific antigen screening. N Engl J Med. 2012;367:595–605. doi: 10.1056/NEJMoa1201637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugosson J, Carlsson S, Aus G, Bergdahl S, Khatami A, Lodding P, Pihl CG, Stranne J, Holmberg E, Lilja H. Mortality results from the Göteborg randomized population-based prostate-cancer screening trial. Lancet Oncol. 2010;11:752–732. doi: 10.1016/S1470-2045(10)70146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iremashvilli V, Pelaez L, Manoharan M, Jorda M, Rosenberg DL, Soloway MS. Pathologic prostate cancer characteristics in patients eligible for active surveillance: a head-to-head comparison of contemporary protocols. Eur Urol. 2012;62:462–468. doi: 10.1016/j.eururo.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Jerónimo C, Henrique R, Hoque MO, Mambo E, Ribeiro FR, Varzim G, Oliveira J, Teixeira MR, Lopes C, Sidransky D. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res. 2004;10:8472–8478. doi: 10.1158/1078-0432.CCR-04-0894. [DOI] [PubMed] [Google Scholar]
- Jalloh M, Myers F, Cowan JE, Carroll PR, Cooperberg MR.2014Racial Variation in Prostate Cancer Upgrading and Upstaging Among Men with Low-risk Clinical Characteristics Eur Urole-pub ahead of print 5 April 2014; doi: 10.1016/j.eururo.2014.03.026 [DOI] [PubMed]
- Kattan MW, Eastham JA, Stapleton AM, Wheeler TM, Scardino PT. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst. 1998;90:766–771. doi: 10.1093/jnci/90.10.766. [DOI] [PubMed] [Google Scholar]
- Kattan MW, Eastham JA, Wheeler TM, Maru N, Scardino PT, Erbersdobler A, Graefen M, Huland H, Koh H, Shariat S, Slawin KM, Ohori M. Counseling men with prostate cancer: a nomogram for predicting the presence of small, moderately differentiated, confined tumors. J Urol. 2003;170:1792–1797. doi: 10.1097/01.ju.0000091806.70171.41. [DOI] [PubMed] [Google Scholar]
- Loeb S, Carter HB, Berndt SI, Ricker W, Schaeffer EM. Is repeat prostate biopsy associated with a greater risk of hospitalization? Data from SEER-Medicare. J Urol. 2013;189:867–870. doi: 10.1016/j.juro.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macefield RC, Metcalfe C, Lane JA, Donovan JL, Avery KN, Blazeby JM, Down L, Neal DE, Hamdy FC, Vedhara K, ProtecT Study Group Impact of prostate cancer testing: an evaluation of the emotional consequences of a negative biopsy result. BJC. 2010;102:1335–1340. doi: 10.1038/sj.bjc.6605648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partin AW, Piantadosi S, Sanda MG, Epstein JI, Marshall FF, Mohler JL, Brendler CB, Walsh PC, Simons JW. Selection of men at high risk for disease recurrence for experimental adjuvant therapy following radical prostatectomy. Urology. 1995;45:831–838. doi: 10.1016/S0090-4295(99)80091-0. [DOI] [PubMed] [Google Scholar]
- Partin AW, Mangold LA, Lamm DM, Walsh PC, Epstein JI, Pearson JD. Contemporary update of prostate cancer staging nomograms (Partin tables) for the new millennium. Urology. 2001;58:843–848. doi: 10.1016/s0090-4295(01)01441-8. [DOI] [PubMed] [Google Scholar]
- Pepe MS. The Statistical Evaluation of Medical Tests for Classification and Prediction. Oxford University Press: New York, NY, USA; 2004. [Google Scholar]
- Rosario DJ, Lane JA, Metcalfe C, Donovan JL, Doble A, Goodwin L, Davis M, Catto JW, Avery K, Neal DE, Hamdy FC. Short term outcomes of prostate biopsy in men tested for cancer by prostate specific antigen: prospective evaluation within ProtecT study. BMJ. 2012;344:d7894. doi: 10.1136/bmj.d7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, Denis LJ, Recker F, Berenguer A, Määttänen L, Bangma CH, Aus G, Villers A, Rebillard X, van der Kwast T, Blijenberg BG, Moss SM, de Koning HJ, Auvinen A, ERSPC Investigators Screening and prostate cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- Schröder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, Denis LJ, Recker F, Páez A, Määttänen L, Bangma CH, Aus G, Carlsson S, Villers A, Rebillard X, van der Kwast T, Kujala PM, Blijenberg BG, Stenman UH, Huber A, Taari K, Hakama M, Moss SM, de Koning HJ, Auvinen A, ERSPC Investigators Prostate-cancer mortality at 11 Years of follow-up. N Engl J Med. 2012;366:981–990. doi: 10.1056/NEJMoa1113135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw GL, Thomas BC, Dawson SN, Srivastava G, Vowler SL, Gnanapragasam VJ, Shah NC, Warren AY, Neal DE. Identification of pathologically insignificant prostate cancer is not accurate in unscreened men. BJC. 2014;110:2405–2411. doi: 10.1038/bjc.2014.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hricak H, Kattan MW, Chen HN, Scardino PT, Kuroiwa K. Prediction of organ-confined prostate cancer: incremental value of MR imaging and MR spectroscopic imaging to staging nomograms. Radiology. 2006;238:597–603. doi: 10.1148/radiol.2382041905. [DOI] [PubMed] [Google Scholar]
- Wang SY, Cowan JE, Cary KC, Chan JM, Carroll PR, Cooperberg MR. Limited ability of existing nomograms to predict outcomes in men undergoing active surveillance for prostate cancer. BJU Int. 2013;114:E18–E24. doi: 10.1111/bju.12554. [DOI] [PubMed] [Google Scholar]
- Wilt TJ, Brawer MK, Jones KM, Barry MJ, Aronson WJ, Fox S, Gingrich JR, Wei JT, Gilhooly P, Grob BM, Nsouli I, Iyer P, Cartagena R, Snider G, Roehrborn C, Sharifi R, Blank W, Pandya P, Andriole GL, Culkin D, Wheeler T, Prostate Cancer Intervention versus Observation Trial (PIVOT) Study Group Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med. 2012;367:203–213. doi: 10.1056/NEJMoa1113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Tokumaru Y, Sidransky D, Epstein JI. Quantitative GSTP1 methylation levels correlate with Gleason grade and tumor volume in prostate needle biopsies. J Urol. 2004;171:2195–2198. doi: 10.1097/01.ju.0000127728.71350.36. [DOI] [PubMed] [Google Scholar]