Abstract
Alteration of genetic make-up of the isolates and monosporidial strains of Tilletia indica causing Karnal bunt (KB) disease in wheat was analyzed using DNA markers and SDS-PAGE. The generation of new variation with different growth characteristics is not a generalized feature and is not only dependant on the original genetic make up of the base isolate/monosporidial strains but also on interaction with host. Host determinant(s) plays a significant role in the generation of variability and the effect is much pronounced in monosporidial strains with narrow genetic base as compared to broad genetic base. The most plausible explanation of genetic variation in presence of host determinant(s) are the recombination of genetic material from two different mycelial/sporidia through sexual mating as well as through para-sexual means. The morphological and development dependent variability further suggests that the variation in T. indica strains predominantly derived through the genetic rearrangements.
Keywords: Fungal variability, Genetic divergence, ISSR, Karnal bunt, RAPD
Tilletia indica (Syn. Neovosia indica) which causes Karnal bunt that affects wheat, durum wheat and triticale. The disease is sometimes called partial bunt because only part of the kernel usually is affected. The disease seldom caused any severe production losses (Gill et al., 1993). The disease always existed, causing concerns intermittently but unlike the cereal rusts it never caused serious yield reduction. But due to the reduction in grain quality this cosmetic disease of wheat became a quarantine concerns interfering with free and fair grain trade. Management of the disease has become very much crucial due to the mode of dispersal of the pathogen, survival of teliospores in soil for a long period and unavailability of tolerant wheat cultivars. Wheat show considerable variation in degree of susceptibility among cultivars and in turn T. indica also exhibits high level of genetic variability among the isolates (Mishra et al., 2001) exhibiting varying degree of virulence. The development of resistant wheat varieties with a wide genetic base claims thorough knowledge of molecular mechanism in the development of variability in T. indica.
Since the pathogen is heterothallic fungus (Fuentes-Davila and Duran, 1986), i.e. it produces haploid secondary sporidia and compatible sporidia (+ & −), when these come in contact they hybridize and cause infection. Therefore, there is continuous variation in pathogen population. Most of the earlier workers have characterized pathogenic and molecular variability based on monoteliosporic populations (Bonde et al., 1996; David and Darrel, 1996; Datta et al., 2000; Thirumalaisamy et al., 2006), which does not give a clear picture of genetic variability as teliospores are diploid. Since, secondary sporidia are infective in nature, in our lab, genetic variation among the base isolates and monosporidial strains derived from these isolates of T. indica was analyzed by morphological, growth behaviors, RAPD-ISSR based molecular polymorphism and 4 base isolates and 20 monosporidial strains were grouped in fast, moderate and slow growing culture (Seneviratne et al., 2009). However, in the present study molecular variability in base isolates and fast growing monosporidial culture populations were analyzed using RAPD-ISSR and SDS-PAGE in the presence of host factor(s).
Understanding the genetic variability in pathogen is extensively achieved through the use of genetic markers. The DNA polymorphism generated through very popular genetic markers viz. RAPD simple sequence repeats (SSRs), restricted fragment length polymorphism (RFLP) and amplified fragment length polymorphism (AFLP) have been proven to be important in producing polymorphic patterns between different individuals and was found useful in describing genetic diversity in several groups of fungi, such as: Phytophthora cactorum (Hantula et al., 1997), claviceps spp. (Tooley et al., 2000), Botryosphaeria spp. (Zhou et al., 2001), Phaeoisariopsis griseola (Mahuku et al., 2002), P. citrophthora (Cohen et al., 2003), Fusarium culmorum (Mishra et al., 2003), Rhizoctonia solani (Elbakali et al., 2003), Colletotrichum lindemuthianum (Mahuku and Riascos, 2004), Gremmeniella spp. and Phomopsis spp. (Borja et al., 2006). However, RAPD and SSRs generally require less development efforts than RFLP and AFLP and usually yield many more bands and therefore represent many more loci (Milbourne et al., 1997).
By using SDS-PAGE, we aim to know how, where, when, and what for are the several hundred thousands of individual protein species produced in a living organism, how they interact with one another and with other molecules to construct the cellular building, and how they work with each other to fit in with programmed growth and development, and to interact with their biotic environment. In the last ten years, excellent reviews and monographs on the fundamentals, concepts, applications, power, and limitations of proteomics have appeared (Cox and Mann, 2007; Cravatt et al., 2007; Han et al., 2006; Han et al., 2008; Jorrin-Novo et al., 2009; Mann 2009; Picotti et al., 2009; Schmidt et al., 2009), some of them dealing with fungal pathogens (Bhadauria et al., 2007; Kim et al., 2007; Tan et al., 2009). Fungal pathogenicity requires the coordinated regulation of multiple genes (and their protein products) involved in host recognition, spore germination, hypal penetration, appressorium formation, toxin production, and secretion. To study the infection cycle and to identify virulence factors, proteomics provides us with a powerful tool for analyzing changes in protein expression between races and stages. Despite its simplicity, SDS-PAGE can still be quite a valid technique providing relevant information, especially in the case of comparative proteomics with large numbers of samples to be compared.
Using this technique, it is possible to distinguish between isolates of different strains of T. indica and identify the genetic variation (Rai et al., 1999). The magnitude of variation in KBPN group is less (Narrow genetic base) when compared to the other groups KB3, KB9 and JK (Broad genetic base) reflecting that variability is a genetically governed process (Seneviratne et al., 2009). In order to understand natural variation among the isolates, molecular markers based studies are useful endeavor, but studying the induced morphological and development dependent variability among the isolates and strains in presence of host determinant(s) might yield good information to study the genetic mechanism(s) which causes the variability in T. indica strains. Acetone extracts from spikes emergence stage (S2) of Karnal bunt susceptible wheat variety (WH542) was found to possess growth-promotory activity of T. indica under in vitro conditions (Rana et al., 2001) and altering growth phenotypes prompted us to study their role in generation of genetic variability in T. indica. The main objectives of this work was: To test the utility of the RAPD-ISSR and SDS-PAGE method as tools to study the induction of genetic variability in presence of host determinant(s) amongst Karnal bunt isolates and monosporidial strains.
Materials and Methods
Collection of KB isolates and preparation of monosporidial strains
The mono-teliosporic culture of T. indica was prepared as described earlier (Rai et al., 2000). Three T. indica isolates KB 3, KB 9 and JK, collected from Delhi, Punjab, Jammu & Kashmir were received from Indian Agricultural Research Institute, New Delhi, India and one isolate KBPN was collected from Wheat Pathology Lab, Department of Plant Pathology, College of Agriculture, G. B. Pant University of Agriculture and Technology, Pantnagar, India. Fungal isolates and strains were cultured at Molecular Markers Laboratory, Dept. of Molecular Biology and Genetics Engineering, G. B. Pant University, Pantnagar, India on modified potato dextrose agar (PDA) and potato dextrose broth (PDB) medium in Petri plates and flasks respectively. The cultures were incubated in BOD incubator at 22±1°C under light and dark conditions. The liquid cultures were used for study the influence of host determinants on genetic variability of T. indica.
After 21 days of growth of fungus in PDA, sporidia were collected separately by decanting and germinated on the same media. Single germinating sporidia were collected microscopically and cultured as individual mono sporidial cultures. Two mono sporidial strains from KB 3 base isolate (KB3 ms-a, KB3 ms-c), seven mono sporidial strains from KB 9 base isolate (KB 9 ms-1–KB 9 ms 7), six mono sporidial strains from JK base isolate (JK ms- a and JK ms- f) and five mono sporidial strains from KBPN base isolate (KB-pn1, KB-pn3, KB-pn5, KB-pn6 and KB-pn8) were used in this study.
Preparation of plant extracts
Plant extract was prepared from spike tissues collected from susceptible (WH-542) wheat variety in boot emergence stage at anthesis (S2). 50g of wheat spikes were ground in liquid nitrogen to a fine powder using pestle and mortar. Finely ground plant tissues suspended in cold acetone the concentration of 1 g in 10 ml of acetone. The above suspension was agitated in cold condition for 5 hours and filtered through muslin cloth to remove larger debris and stored at 4°C in tightly capped bottles. Before stating the experiments acetone was evaporated at room temperature using flash evaporator or blowing hot air over the solution. Dried material obtained was re-suspended in 1/10th of the volume of sterilized distilled water and filtered through 0.22μ filter before incorporation to the culture media.
Determination of growth kinetics and detection of developmental stage dependent variation
P9, P16, P7 and KBPN isolates were cultured on solid as well as liquid media. Radial diameter measurements were taken at 3 days intervals. The sporidia were harvested from 21 days old fungal cultures by decanting method. 10 ml of sterilized distilled water was added to the plates and gently moved back and forth and water containing sporidia was decanted to sterilized Oakridge tubes and centrifuged at 4000 rpm for 10 min to get sporidial pellet. The pellets were re-suspended in minimal amount of sterilized distilled water and stored at −20°C.
Harvesting of mycelium
The growing liquid cultures of T. indica were harvested at desired time intervals. The media containing the mycelial mat of T. indica was filtered through a folded muslin cloth and washed several times in PBS (0.05 M, pH 7.2) and followed with sterilized distilled water. The wet weight of the mycelial mass was taken and the wet masses were lyophilized for 5 hours to obtain the dry weight. Dry mycelial masses were stored in −20°C.
DNA extraction and quantification
In order to isolate DNA a modified procedure Cetyltrimethyl Ammonium Bromide (CTAB) method (Murray and Thompson, 1980) was used. For DNA extraction as recommended for fungus T. indica. DNA was purified as described by Sambrook et al., 1989.
Primers used
A set of 30 decanucleotide RAPD and 19 ISSR primers were employed for PCR amplification (Seneviratne et al., 2009). One RAPD and 5 ISSR of these primers used in this present study because these primer show more polymorphism. The sequences of these primers were random and were purchased from Life Technology. The sequence and the details of primers are given in the Table 1.
Table 1.
List of primer used for PCR amplification
| S. No. | Primer code | Primer Sequence | Amplified product (bp) |
|---|---|---|---|
| 1 | ISSR-18 | ((CTC)(8)) | 500–1300 |
| 2 | ISSR-13 | (BHB (GA)(7) ) | 200–1400 |
| 3 | ISSR-17 | ((ATG)6) | 500–1400 |
| 4 | ISSR-1 | (HVH(TG)(7)) | 200–1400 |
| 5 | ISSR-19 | ((GGGTG)(3)) | 400–1400 |
| 6 | RAPD 16 | GTG AGG CGT C | 250–1600 |
PCR amplification through RAPD and ISSR Marker
PCR was carried out in 25 μl reaction mixture containing 20 ng of genomic DNA, 0.3U Taq DNA polymerase enzyme, 200 μM each dNTPs, 10 mM Tris-Hcl, 1.5 mM MgCl2 and 0.2 μM primer, 30ng from a single primer and 1 μl from each template DNA were added in each tube. The PCR amplification was achieved in a Biometra T-gradient DNA thermocycler. For RAPD amplification PCR cycle was programmed for initial denaturation of 5 min at 94°C; 37 cycles of denaturation at 94°C for 1 min, annealing at 41°C for 1 min and extension at 72°C for 2 min; followed by a final extension 72°C for 7 min. In order to perform ISSR amplification, initial denaturation of 5 min at 94°C; 45 cycles of denaturation at 94°C for 1 min, annealing at 41°C for 1 min and extension at 72°C for 2 min; followed by a final extension 72°C for 10 min. The amplified products were stored at −20°C until they were subjected for electrophoresis. PCR products were electrophoresed in 1.6% agarose gel in 0.5X TAE buffer along with 100bp DNA marker at the concentration of 250 ng/ml to identify respective bands.
Isolation and analysis of proteins
Proteins were extracted from KB isolates and it’s monosporidial strains by crushing 0.5 g mycelia with liquid nitrogen in presence of extraction buffer (50 mM Tris-base, 50 mM EDTA, 10 mM EGTA, 0.5% Triton X-100, 0.3% BME, 0.39% Ascorbic acid, 2 mM PMSF and 0.4% PVP). The samples were centrifuged at 12,000×g for 20 minute at 4°C and supernatants were collected in separate tubes and store at 4°C.
Gel electrophoresis of proteins isolated from KB isolates and it’s monosporidial strains
Protein concentration of all samples was determined by the method of Bradford (1976) using bovine serum albumin (BSA) as standard. After quantification, KB isolates and it’s monosporidial strains protein in presence and absence of host factor(s) were analyzed by using SDS-PAGE electrophoresis on 12% acrylamide gels (Laemmli et al., 1970).
Statistical Analysis
Three independent determinations of growth kinetics, detection of developmental stage dependent variation in absence and presence of host factors, were taken and mean±SE values were calculated for statistical analysis using GraphPad Prism 6.0 software.
Results and Discussion
Fungi cause the most serious diseases of plants and display greater complexity and diversity of form than other microbial pathogens. This diversity is highlighted by the extraordinary host range observed by some pathogens and the extreme pathogenic specialization of others. Variability in the pathogen T. indica has been established by using single teliospore cultures but the stability of such pathogens to be proved as pathogen is heterothallic organism, primary or secondary sporidia or hyphae as compatible mating types must fuse to form dikaryon which readily increases the chances of variation due to heterozygosity. The greater frequency of out-crossing in T. indica suggests that intraspecific and inter-specific hybridization in T. indica presumably facilitates sexual recombination and greater genetic diversity. Knowledge of diversity of the KB pathogen in the form of distinct monoteliosporic and monosporidial cultures are considered essential before effective measures for its management are developed and understood. In the present investigation, attempts have been made to study the genetic changes of isolates and monosporidial strains of T. indica grown in culture medium in the presence of host determinant(s). More emphasis has been categorically given to detect the variation using RAPD-ISSR and SDS-PAGE based molecular tools among the monosporidial strains in order to understand the mechanism of genetic polymorphism in T. indica.
Effect of host factor(s) on T. indicia on solid media
Significant variation in radial diameter was observed in all three known virulence isolates and one unknown virulence isolate studied at all three time intervals (Fig. 1A & 1B). Both fungal extracts have given a boost to the increment compare to the control. Fast growing isolate P9 and KBPN has shown higher radial diameter and the increment in radial diameter was always higher compare to the rest two (P7 and P16) fungal isolates. At the sixth day the variation in radial diameter with respect to the treatments was clearly depicted and the variation in the same at 9th day is not that apparent as two isolates (P9, KBPN and P16) have already covered the whole petri plate indicating a rapid growth in them with host extract treatment. Radial diameter variation with respect to the host factors was statistically analyzed individually with the sixth day data and it was revealed that the variation among the treatments (Control, HD 29 and WH 542) was significant (p>0.05) in all four isolates compared (P9, P16, P7 and KBPN).
Fig. 1.
(A) Influence of host factor(s) extracted from tolerant (HD 29) and susceptible (WH 542) wheat varieties on growth of Tilletia indica main isolates with different virulence levels grown on solid medium at different time interval. (B) Effect of host factor(s) on T. indica isolates with different virulence levels at 6 days of growth on solid medium.
Effect of host factor(s) on T. indicia on liquid media
All four T. indica main isolates (P7, P9, P16 and KBPN) were cultured in modified PDB liquid media in-order to detect the behaviors of fungal isolates in the liquid culture. Statistically significant variation was detected among the fungal isolates P9 and KBPN at seven days old culture with plant extracts. Most virulent strain P9 and Pantnagar isolate KBPN exhibited higher growth compare to least and moderately virulent strains (P7 and P16 respectively) as indicated in the Fig. 2. In all situations plant extracts have increased the growth of the fungus in varying degrees. Lowest growth was observed in P7 (0.48 g%) culture without host factors. Maximum growth was observed in KBPN cultured with WH 542 host extracts (2.6 g%). As it is quite evident that both type of plant extracts (HD29 and WH 542) have contributed to increase the growth of the fungus in liquid culture. But the effect of WH 542 is always higher compared to that of HD 29. Contribution of host factors to the growth increment in fungal strains were statistically analyzed individually and significant increase (P>0.05) in growth was detected in P9 culture (gm=1.42 sem=0.103 cv=12.5). KBPN cultures with host factors have exhibited highly significant variation (P>0.05) among the treatments (Control, HD 29 and WH 542) (gm=2.08, sem=0.15 and cv=12.5). Both slow growing strains (P7 and P16) have enumerated only a non significant (P>0.05) variation with respect to the treatments given (Control, HD 29 and WH 542).
Fig. 2.
Influence of host factor(s) on T. indica on growth of different isolates (P9, P16, P7 and KBPN) treated with susceptible (WH 542) and tolerance (HD 29) genotype host factor(s) having differential pathogenicity at seven days of growth in liquid medium (mycelial weight in g% on wet basis).
Determination of sporidial count
In order to detect developmental stages dependant variations, isolation and counting of sporidia on haemocytometer at 21st days were performed on the fungal cultures (P9, P16 P7 and KBPN) with or without plant host factor(s).
As indicated in the Fig. 3, a drastic reduction in sporidial count was observed in all four fungal isolates irrespective to their growth behaviors. P9 and KBPN isolates have shown the lowest sporidial count (165×106/ml and 390×106/ml) and the other two fungal isolates (P7 and P16) have shown considerably higher sporidial count in cultures without plant extracts (control) (1224.3×106/ml and 5953.3×106/ml respectively in P7 and P16). All four fungal strains cultured with plant extracts have indicated a reduction in sporidial count but the reduction with WH542 host extracts much more pronounced compared to HD 29 host extracts. Statistical analysis performed individually for each strain (P9, P16, P7 and KBPN) indicated that the variation among the treatments are highly significant at p>0.05 significant interval level. Gupta et al. (2012) Determined sporidial count of T. indica isolate grown in presence and absence of host factor(s).
Fig. 3.
Determination of sporidial count of base isolate cultures (P9, P16, P7 and KBPN) treated with susceptible (WH 542) and tolerance (HD 29) genotype host factor(s) (Number of sporidium/ml after 21 days).
Determination of alteration of genetic make-up of fungal cultures
All four main fungal strains (P9, P16, P7 and KBPN) and four fast growing monosporidial strains(JKmse, KBpn6, KB3msa and KB9ms4) representing all four groups were cultured in liquid media with Karnal bunt susceptible S2 stage wheat (WH 542) extracts at 1:10 extract to media ratio. Harvested mycelium from 14th days old cultures were subjected to genetic analysis through RAPD and ISSR markers.
Induction of genetic variability on main fungal strains (P9, P16, P7 and KBPN) with a broad genetic base
The field isolates of T. indica (P9, P16, P7 and KBPN) with a broad genetic base were cultured with host factors and subjected to genetic evaluation with RAPD markers. The banding profiles are depicted in the Fig. 4.
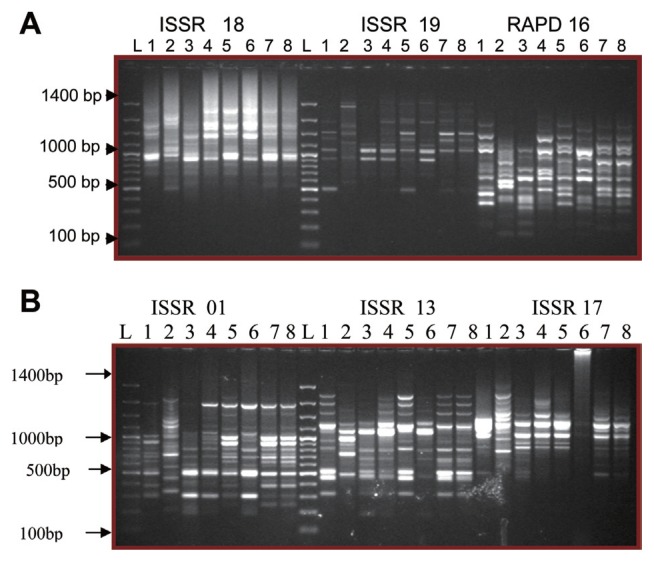
Fig. 4.
Determination of variation of genetic make-up under the influence of host factor(s) through PCR amplification of (A) 7 days and (B) 14 days fungal genome DNA with ISSR-18 and RAPD-16 primers. L, 100 bp ladder: 1, P9 (T); 2, P9 (C); 3, P16 (T); 4, P16 (C); 5, P7(T); 6, P7(C); 7, KBPN(C); 8, KBPN (T); T, Treated with host factor(s); C, Control no host factor(s).
Plant extracts treatment to fungal cultures has enhanced the genetic rearrangements and RAPD profiles indicates that the genetic rearrangements are much higher in 14th day cultures than the 7th day cultures. Fast growing fungal strains (P9 and KBPN) have always shown a clear variation in RAPD profiles irrespective to the age of the culture. RAPD primer R4 has detected genetic variation in all samples at 14 days indicating the role of host factor(s) creating a genetic rearrangement in the fungi.
Induction of genetic variability on monosporidial strains (JKmse, KBpn6, KB3msa and KB9ms4) with a narrow genetic base
The banding patterns obtained are depicted in (Fig. 5A and 5B). In most of the situation both marker systems have indicated the genetic variation between treated and untreated samples of monosporidial strains. Despite of the prominent genetic variations indicated in the other strains, KBPN group representative viz. KBpn6 always indicated less or zero genetic rearrangements compare to the other monosporidial groups tested (JK, KB3 and KB9) The results obtained under induced genetic variability studies are in accordance with the results generated through molecular characterization of fungal strains under natural conditions. The KBPN monosporidial group indicated the presence of less hot spots of recombination and the members of this group continually fail to enough generate genetic rearrangements even after the induction by the host factor(s).
Fig. 5.
Determination of genetic variation in ISSR profiles after the induction with host factor(s) using (A) ISSR-18, ISSR-19 and RAPD-16 primers and (B) ISSR-01, ISSR-13 and ISSR-17 primers. L, 100 bp ladder; 1, JKmse host factors treated(T); 2, JKmse no host factors(C); 3, KB9ms4(T); 4, KB9ms4(C); 5, KB3msa(T); 6, KB3msa(C); 7, KBpn6(T); 8, KBpn6; T, Treated with host factor(s); C, Control no host factor(s).
Influence of host factors on the alterations of mycelial proteins in Karnal Bunt (Tilletia indica) isolates with differential virulence levels
Due to various reasons, KB fungus, T. indica has emerged as the bunt fungus of choice for studies on molecular genetics, mating type, pathogenicity, growth behavior and other aspects of fungal biology. Over the last few years our appreciation of pathogenicity development mechanisms which are either constitutive or inducible, reflects the complexity of gene signaling network and the extensive diversity of pathogen as well as diverse infective mechanisms (Gupta et al., 2013). Considering all the above, in the present study, attempts were made to evaluate the natural as well as induced variability of total soluble proteins and total protein bands under the influence of host factor(s) in the development of pathogenicity of the T. indica pathogen.
Alteration of mycelia proteins in isolates of known virulence levels in the presence of host factor(s)
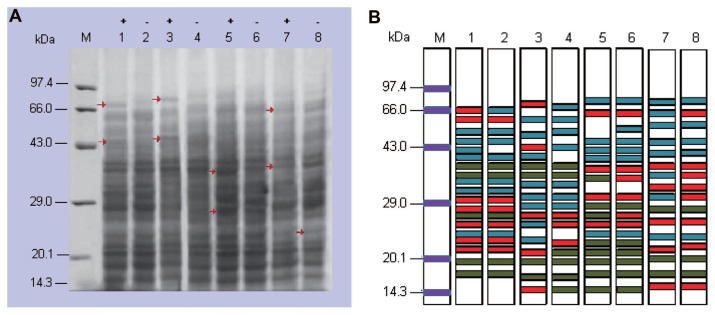
As indicated in Table 2 the drastic variation in total protein of fungal mycelia was observed among the four isolates of T. indica in the presence and absence of host factor(s). The low protein content was observed in the highly virulent (P9), moderately virulent (P16), least virulent (P7) and unknown (KBPN) isolates (3.3, 5.6, 3.6 and 4.8 mg/g of mycelia weight respectively) in absence of host factor(s) while compared with presence of host factor(s) (4.5, 6.5, 4.2 and 5.9 mg/g of mycelia weight respectively). It was interesting to see that the isolates with known virulence level exhibited very clear increase in protein concentration (36.3%, 16.2% 19.4% and 23.1% respectively) in the presence of host factor(s). Extracted proteins were subjected to protein analysis through SDS-PAGE.
Table 2.
Total soluble protein content and total number of bands in known virulence and unknown virulence isolates in presence and absence of host factor(s) at 14th day
| Fungal culture | Total Protein Concentration (mg/g of mycelia weight) | % increase | Total protein bands | Specific band/up regulated or down regulated band | Mol. Wt. (KDa) of Specific band/up regulated (↑) or down regulated (↓) band |
|---|---|---|---|---|---|
| P9 + | 4.5 | 36.3 | 18 | 2 | 51.5↑, 72.1↑ |
| P9 − | 3.3 | 18 | |||
| P16 + | 6.5 | 16.2 | 19 | 2 | 54.6↑, 74.7↑ |
| P16 − | 5.6 | 19 | |||
| P7 + | 4.2 | 19.4 | 15 | 2 | 28.4↑, 40.3↑ |
| P7 − | 3.6 | 15 | |||
| KBPN + | 5.9 | 23.1 | 16 | 3 | 23.8↑, 42.4↑, 68.9↑ |
| KBPN − | 4.8 | 16 |
The protein expression of T. indica in presence and absence of host factor(s) was observed in Fig. 6A. Some proteins were up regulated in presence of host factor(s) compared with absence of host factor(s). Two protein bands were up regulated in highly virulent (P9) isolate (51.5 and 72.1 KDa), in P16 (54.6 and 74.7 KDa) and in P7 (28.4 and 40.3 KDa). However, in KBPN isolate three protein bands were up regulated (23.8, 42.4 and 68.9 KDa).
Fig. 6.
(A) One-DE of 20 μg of mycelium protein of fast growing KB monosporidial strains at 14th day in presence and absence of host factor(s) (M-Marker; 1, P9+; 2, P9−; 3, P16+; 4, P16; 5, P7+; 6, P7−; 7, KBPN+; 8, KBPN−). This approach allowed us to observe differences in the protein band patterns in presence of host factor(s). (B) The depiction of protein profile from mycelia of fast growing KB monosporidial strains in the presence and absence of host factor(s) at 14th day (M, Marker; 1, P9+; 2, P9−; 3, P16+; 4, P16−; 5, P7+; 6, P7−; 7, KBPN+; 8, KBPN−
 Dense,
Dense,
 Medium and
Medium and
 Light.
Light.
The protein profiles (Fig. 6B) of isolates of known virulence and one Pantnagar isolate was found to produce a total of 15 to 19 bands. All the isolates were found to be variable in terms of production of proteins. The banding patterns showed that many proteins were up regulated in the presence of host factor(s) in the isolates known virulence and the size of these proteins were varied in all the isolates.
Alteration of mycelial protein in fast growing monosporidial strains in presence of host factor
As indicated in Table 3 the drastic variation in total protein of fungal mycelia was observed among the four fast growing monosporidial strains of T. indica in the presence and absence of host factor(s). The low protein content was observed in the JKMSE, KB9MS4, KB3MSA and KBPN6 (2.8, 3.9, 3.0 and 6.6 mg/g of mycelia weight respectively) in absence of host factor(s) compared to in presence of host factor(s) (4.1, 6.3, 4.0 and 8.5 mg/g of mycelia weight respectively). It was interesting to see that the fast growing monosporidial strains exhibited very clear increase in protein concentration (46.6%, 61.6%, 35.0% and 29.9% respectively) in the presence of host factor(s). Extracted proteins were subjected to protein analysis through SDS-PAGE.
Table 3.
Total soluble protein content and total number of bands in fast growing monosporidial strains grown in presence and absence of host factor(s) at 14th day
| Fungal culture | Total Protein Concentration (mg/g of mycelia) | % increase | Total protein bands at 14th Day | Specific band/up regulated or down regulated band | Mol. Wt. (KDa) of Specific band/up regulated (↑) or down regulated (↓) band |
|---|---|---|---|---|---|
| JKMSE + | 4.1 | 46.6 | 19 | 7 | 31.9↓, 35.8↓, 39.9↑, 44.8↑, 49.2↑, 78.9↓, 95.5↓ |
| JKMSE − | 2.8 | 16 | |||
| KB9MS4 + | 6.3 | 61.6 | 17 | 2 | 49.7↑, 73.9↑ |
| KB9MS4 − | 3.9 | 16 | |||
| KB3MSA + | 4.0 | 35.0 | 17 | 7 | 20.8↓, 29.8↑, 36.7↑, 38.4↓, 41.8↑, 45.5↑, 51.1↑ |
| KB3MSA − | 3.0 | 14 | |||
| KBPN6 + | 8.5 | 29.9 | 20 | 3 | 32.3↑, 43.2↑, 63.3↑ |
| KBPN6 − | 6.6 | 18 |
The protein expression in fast growing monosporidial strains of T. indica in presence and absence of host factor(s) was shown in Fig. 7A. Some proteins were up regulated in presence of host factor(s) compare with absence of host factor(s). However, some proteins were down regulated in presence of host factor(s). Three protein bands were up regulated (39.9, 44.8 and 49.2 KDa) and four protein bands were down regulated (31.9, 35.8, 78.9 and 95.5 KDa) in JKMSE strain. In KB9MS4, two protein bands were up regulated (49.7 and 73.9 KDa). Five protein bands were up regulated (29.8, 36.7, 41.8, 45.5 and 51.1 KDa) and two protein bands were down regulated (20.8 and 38.4 KDa) in KB3MSA strain. In KBPN6 strain three bands were up regulated (32.3, 43.2 and 63.3 KDa).
Fig. 7.
(A) One-DE of 20 μg of mycelium protein of fast growing KB monosporidial strains at 14th day in presence and absence of host factor(s) (M, Marker; 1, JKMSE+; 2, JKMSE−; 3, KB9MS4+; 4, KB9MS4−; 5, KB3MSA+; 6, KB3MSA−; 7, KBPN6+; 8, KBPN6−). This approach allowed us to observe differences in the protein band patterns in presence of host factor(s). (B) The depiction of protein profile from mycelia of fast growing KB monosporidial strains in the presence and absence of host factor(s) at 14th day (M-Marker; 1, JKMSE+; 2, JKMSE−; 3, KB9MS4+; 4, KB9MS4−; 5, KB3MSA+; 6, KB3MSA−; 7, KBPN6+; 8, KBPN6−).
 Dense,
Dense,
 Medium and
Medium and
 Light.
Light.
The depictions of protein profiles (Fig. 7B) of fast growing monosporidial strains were found to produce a total of 14 to 20 bands. All the strains were showed variability in terms of production of proteins. The banding patterns depict that in the presence of host factor(s) many proteins were up regulated and down regulated in the strain. The size of up regulated and down regulated proteins were varied in all the strains. Some of new proteins were also observed in the presence of host factor(s).
Performing studies in order to understand natural variation among the isolates is a useful endeavor, but studying the natural as well as induced variability among the isolates coupled with morphological and developmental stage dependent variability will give much more insights to the genetic mechanism which causes the variability in T. indica strains. Acetone extracts from spikes emergence stage (S2) of Karnal bunt susceptible wheat variety (WH-542) was found to possess growth-promotory activity of T. indica under in-vitro axenic culture condition (Rana et al., 2001) and it can be hypothesized that the enhanced growth leads to generate some genetic variability in T. indica through mitogen activated protein kinase pathways which are responsible for mating type switching (Gupta et al., 2011; Gupta et al., 2012; Gupta et al., 2013).
Since the pathogen is a heterothallic hemi-biotrophic fungus such variability is possible through recombination generated through mating between compatible mating types (Rai et al., 2000) as well as recombination originated through anastomosis and subsequent para sexual recombination (mitotic crossing over) or simply by the mutations (Nicholas, 1998). Nelson et al. (1955) inoculated a resistant wheat variety with different races of rusts, which individually failed to cause infection but in combination caused infection. It was made possible by anastomosis between the inoculated races, which resulted in the origin of a new pathogenic race. Day (1996) reported Silenins, a host factor from Silane alba appears to control entry into the parasitic developmental pathway by an effect on the mating type locus. Thus only conjugated a1 + a2 cells or a1a2 diploid cells are induced by Silenin to produce infective hyphae. The mating-type locus acts as a “master developmental switch” responding to environmental agents and controlling entry into conjugative, sporulative, or vegetative morphogenetic pathways.
Positive or promotory interactions between host and parasite may also play role in modulation of pathogenicity. Thus, susceptibility in some cases may depend on the presence of one or more plant products essential for fungal growth and development and therefore, be a positive attribute of the plant. Resistant plants would lack these products or have them in lower concentrations. Evidence for this in some plant-pathogen relationships is beginning to accumulate. Host compounds stimulate growth and virulence in Fusarium graminearum (Strange et al., 1974), Helminthosporium sorokinianum (Endo and Oertii, 1964), Botrytis cinerea (Chou and Preece, 1968) and Phoma betae (Warren, 1972). In these examples the plant product appears to act as a nutrient or fungal growth stimulant, and the fungus may be considered to be auxotrophic for this plant products. Day et al. (1981) reported an example of a host plant product which acts more like a fungal hormone and results in a highly specific redirection of morphogenesis in the fungus Ustilago violacea leading to induction of the parasitic stages previously saprophytic cells.
All four base strains (P9, P16, P7 and KB) with a broad genetic base were cultured in modified PDB with and without host factor(s) and detection in genetic variability was carried at two time intervals viz., 7th and 14th days incubation. Host extracts treatment to fungal cultures has enhanced the genetic variation probably due to rearrangement process. RAPD indicates that the genetic rearrangements are much higher at 14th day cultures than the 7th day cultures. Fast growing fungal strains (P9 and KBPN) have always shown a clear variation in RAPD profiles irrespective to the age of the culture. RAPD primer R4 has detected genetic variation in all samples at 14th days indicating the role of host factor(s) creating a genetic rearrangement in the fungi. At 7th day, ISSR 18 primer has indicated genetic rearrangements in all strains with respect to the host factor treatment and RAPD 16 primer has indicated genetic rearrangements only in P9 and KBPN strains which are fast growing compared to the others. At 14th day both primers have generated polymorphic bands in all strains with respective to the host factor(s) treatment. This indicate the genetic rearrangement is time dependent phenomenon and in the aged cultures the genetic variability is much more prominent giving much more heterogeneous genetic make-up indicating the presence of several variants in the population and may be causative factor of emergence of new fungal races for onset of disease even in resistant genotypes. Aggarwal et al. (2010) was observed genetic variability in T. indica monosporidial culture lines using 12 universal rice primers (URPs). Tripathi et al. (2011) also concluded that variation is not a generalized feature and is totally dependent on the original genetic make-up of the base isolate used for generating new monosporidial strains. This is the first report to the best of our knowledge, providing new insights about the generation of fungal diversity and clear indication of alteration of fungal development and pathogenicity in presence of host determinants.
Acknowledgments
The authors wish to acknowledge the Department of Science and Technology, Government of India for providing financial support at GB Pant University of Agriculture and Technology, Pantnagar (Grant no. SR/SO/PS-83/2005 dated 20/12/2007). The support provided by the Director, Experiment Station, and Dean, College of Basic Sciences and Humanities, GB Pant University of Agriculture and Technology, Pantnagar, is also greatly acknowledged.
References
- Aggarwal R, Tripathi A, Yadav A. Pathogenic and genetic variability in Tilletia indica monosporidial culture lines using universal rice primer-PCR. Eur J Plant Pathol. 2010;128:333–342. doi: 10.1007/s10658-010-9655-4. [DOI] [Google Scholar]
- Bhadauria V, Zhao WS, Wang LX, Zhang Y, Liu JH, Yang J, Kong LA, Peng YL. Advances in fungal proteomics. Microbiological Research. 2007;162:193–200. doi: 10.1016/j.micres.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Borja I, Solheim H, Hietala AM, Fosdal CG. Etiology and realtime polymerase chain reaction-based detection of Gremmeniella- and Phomopsis-associated disease in Norway spruce seedling. Phytopathology. 2006;96:1305–1314. doi: 10.1094/PHYTO-96-1305. [DOI] [PubMed] [Google Scholar]
- Bonde MR, Peterson GL, Fuentus-Davilo G, Auja SS, Nanda GS. Comparison of the virulence of isolates of T. indica, causal agent of Karnal bunt of wheat. Phytopath. 1996;80:1070–1074. [Google Scholar]
- Chou MC, Preece TF. The effect of pollen grains on infections caused by Botrytis cinerea Fr. Ann Appl Biol. 1968;62:11–22. doi: 10.1111/j.1744-7348.1968.tb03846.x. [DOI] [PubMed] [Google Scholar]
- Cohen S, Allasia V, Venard P, Notter S, Vernière CH, Panabières F. Intraspecific variation in Phytophthora citrophthora from citrus trees in eastern Corsica. Eur J Plant Pathol. 2003;109:791–805. doi: 10.1023/A:1026190318631. [DOI] [Google Scholar]
- Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Cravatt BF, Simon GM, Yates JR., III The biologicalimpact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- Day AW. Mating type and morphogenesis in Ustilago violacea. Bot Gaz. 1996;140:94–101. doi: 10.1086/337062. [DOI] [Google Scholar]
- Day AW, Castle AJ, Cumins JE. Regulation of parasitic development of the smut fungus, Ustilago violaceae, by extracts from host plants. Bot Gaz. 1981;142:135–146. doi: 10.1086/337203. [DOI] [Google Scholar]
- Datta R, Rajebhosale MD, Dhaliwal HS, Singh H, Ranjekar PK, Gupta VS. Intraspecific genetic variability analysis of Neovossia indica causing Karnal bunt of wheat using repetitive elements. Theoretical and Applied Genetics. 2000;100:569–575. doi: 10.1007/s001220050075. [DOI] [Google Scholar]
- David RG, Darrel JW. Using random amplified polymorphic DNA to analyse the genetic relationships and variability among three species of wheat smut (Tilletia) Botanical Bulletin of Academia Sinica. 1996;37:173–180. [Google Scholar]
- Elbakali AM, Lilja A, Hantula J, Martin M. Identification of Spanish isolates of Rhizoctonia solani from potato by anastomosis grouping, ITS-RFLP and RAMS-fingerprinting. Phytopathologia Mediterranea. 2003;42:167–176. [Google Scholar]
- Endo RM, Oertti JJ. Stimulation of fungal infection of bent grass. Nature. 1964;201:313. doi: 10.1038/201313a0. [DOI] [Google Scholar]
- Fuentus DG, Duran R. Tilletia indica: Cytology and teliospore formation in vitro and in immature kernels. Canadian J of Botany. 1986;4:1712–1719. doi: 10.1139/b86-229. [DOI] [Google Scholar]
- Gill KS, Sharma I, Aujla SS. Karnal bunt and wheat production. Punjab Agricultural University Ludhiana; 1993. p. 153. [Google Scholar]
- Gupta AK, Joshi GK, Seneviratne JM, Pandey D, Kumar A. Cloning, in silico characterization and Induction of TiKpp2 MAP kinase Kinase in Karnal bunt (Tilletia indica) under the influence of host factor(s) from wheat spikes. Mol Biol Rep. 2013;40:4967–4978. doi: 10.1007/s11033-013-2597-0. [DOI] [PubMed] [Google Scholar]
- Gupta AK, Seneviratne JM, Joshi GK, Kumar A. Induction of MAP kinase homologues during morphogenetic development and pathogenesis of Karnal bunt (Tilletia indica) under the influence of host factor(s) from wheat spikes. The Scientific World Journal. 2012 doi: 10.1100/2012/539583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta AK, Goel A, Seneviratne JM, Joshi GK, Kumar A. Molecular cloning of MAP kinase genes and in silico identification of their downstream transcription factors involved in pathogenesis of Karnal bunt (Tilletia indica) of wheat. J Proteomic Bioinform. 2011;4:160–169. [Google Scholar]
- Han X, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- Han X, Aslanian A, Yates JR., 3rd Mass spectrometry for proteomics. Curr Opin Chem Biol. 2008;12:483–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantula J, Lilja A, Parikka P. Genetic variation and host specificity of Phytophthora cactorum isolated in Europe. Mycol Res. 1997;101:565–572. doi: 10.1017/S0953756296002900. [DOI] [Google Scholar]
- Jorrín-Novo JV, Maldonado AM, Echevarría-Zomeño S, Valledor L, Castillejo MA, Curto M, Valero J, Sghaier B, Donoso G, Redondo I. Plant proteomics update 2007–2008 Second generation proteomic techniques, an appropriate experimental design, and data analysis to fulfill MIAPE standards, increase plant proteome coverage and expand biological knowledge. J Proteomics. 2009;72:285–314. doi: 10.1016/j.jprot.2009.01.026. [DOI] [PubMed] [Google Scholar]
- Kim Y, Nandakumar MP, Marten MR. Proteomics of filamentous fungi. Trends in Biotechnology. 2007;25:395–400. doi: 10.1016/j.tibtech.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Mahuku GS, Henríquez MA, Muñoz J, Buruchara R. Molecular markers dispute the existence of the Afro-Andean group of the bean angular leaf spot pathogens. Phaeoisariopsis griseola. Phytopathology. 2002;92:580–589. doi: 10.1094/PHYTO.2002.92.6.580. [DOI] [PubMed] [Google Scholar]
- Mahuku GS, Riascos JJ. Virulence and molecular diversity within Colletotrichum lindemuthianum isolates from Andean and Mesoamerican bean varieties and regions. Eur J Plant Phytol. 2004;110:253–263. doi: 10.1023/B:EJPP.0000019795.18984.74. [DOI] [Google Scholar]
- Mann M. Comparative analysis to guide quality improvements in proteomics. Nat Methods. 2009;6:717–719. doi: 10.1038/nmeth1009-717. [DOI] [PubMed] [Google Scholar]
- Mishra A, Anil Kumar, Garg GK, Sharma I. Determination of Genetic Variability among Isolates of T. indica using Random Amplified Polymorphic DNA analysis. Plant Cell Biotech Mol Biol. 2001;1:29–36. [Google Scholar]
- Mishra P, Fox RT, Culham A. Inter-simple sequence repeat and aggressiveness analysis revealed high genetic diversity, recombination and long-range dispersal in Fusarium culmorum. Annals Applied Biology. 2003;143:291–301. doi: 10.1111/j.1744-7348.2003.tb00297.x. [DOI] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbourne D, Meyer R, Bradshow JE, Baired E, Bonar N, Provan J, Powell W, Waugh R. Comparison of PCR-based marker systems for the analysis of genetic relationships in cultivated potato. Molecular Breeding. 1997;3:127–136. doi: 10.1023/A:1009633005390. [DOI] [Google Scholar]
- Nelson RR, wilcoxan RD, Cristensen JJ. Heterokaryosis as a basis for variation in Puccinia graminis Var trtici. Phytopathology. 1955;45:639–643. [Google Scholar]
- Nicholas JT. Molecular variability among fungal pathogens: Using the rice blast fungus as a case study. In: Bridge P, Couteaudier Y, Clarkson J, editors. Molecular Variability of Fungal Pathogens. 1998. pp. 1–14. [Google Scholar]
- Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai G, Kumar A, Gaur AK, Singh A, Garg GK. Stage dependent changes in protein and isozyme profiles during Growth cycle of karnal bunt (Tilletia indica) of wheat. Indian J Agric Biochem. 1999;12:59–63. [Google Scholar]
- Rai G, Kumar A, Singh A, Garg GK. Modulation of antigenicity of mycelia antigens during developmental cycle of Karnal bunt (Tilletia indica) of wheat. Indian J Experi Biol. 2000;38:488–492. [PubMed] [Google Scholar]
- Rana M, Charu A, Basant R, Anil K. Floral specificity of Karnal Bunt Infection Due to Presence of Fungal Growth-Promotory Activity in Wheat Spikes. J Plant Biol. 2001;28:283–290. [Google Scholar]
- Sambrook J, Fritch EF, Maniatis T. Molecular Cloning a laboratory manual. 2nd ed. NY: cold spring Harbor Laboratory, cold spring Harbor Laboratory Press; 1989. p. 1659. [Google Scholar]
- Schmidt A, Claassen M, Aebersold R. Directedmass spectrometry: towards hypothesis-driven proteomics. Curr Opin Chem Biol. 2009;13:510–517. doi: 10.1016/j.cbpa.2009.08.016. [DOI] [PubMed] [Google Scholar]
- Seneviratne JM, Gupta AK, Pandey D, Sharma I, Kumar A. Determination of genetic divergence based on DNA Markers amongst monosporidial strains derived from fungal isolates of Karnal bunt of wheat. Plant Pathol J. 2009;25:303–316. doi: 10.5423/PPJ.2009.25.4.303. [DOI] [Google Scholar]
- Strange RN, Majer JR, Smith Isolation and identification of choline and betaine as the two major components in anthers and wheat germ that stimulate Fusarium graminearum in vitro. Physiol Plant Pathol. 1974;4:277–290. doi: 10.1016/0048-4059(74)90015-0. [DOI] [Google Scholar]
- Tan KC, Ipcho SV, Trengove RD, Oliver RP, Solomon PS. Assessing the impact of transcriptomics, proteomics and metabolomics on fungal phytopathology. Mol Plant Pathol. 2009;10:703–715. doi: 10.1111/j.1364-3703.2009.00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Rashmi A, Anita Y. Determination of variability in monosporidial lines of Tilletia indica by RAPD analysis. Archives of Phytopathology and Plant Protection. 2011;44:1312–1321. doi: 10.1080/03235408.2010.496560. [DOI] [Google Scholar]
- Thirumalaisamy PP, Singh DV, Aggarwal R, Srivastava KD. Pathogenic variability in Tilletia indica, the casual agent of Karnal bunt of wheat. Indian Phytopathology. 2006;59:22–26. [Google Scholar]
- Tooley PW, O’Neill NR, Goley ED, Carras MM. Assessment of diversity in Claviceps africana and other Claviceps by RAM and AFLP analysis. Phytopathology. 2000;90:1126–1130. doi: 10.1094/PHYTO.2000.90.10.1126. [DOI] [PubMed] [Google Scholar]
- Warren RC. Attempts to define and mimic the effects of pollen on the development of lesions caused by Phomabetae inoculated onto sugar beet leaves. Ann Appl Biol. 1972;71:193–200. doi: 10.1111/j.1744-7348.1972.tb05082.x. [DOI] [Google Scholar]
- Zhou S, Smith DR, Stanosz GR. Differentiation of Botryosphaeria species and related anamorphic fungi using inter simple or short sequence repeat (ISSR) ingerprinting. Mycol Res. 2001;105:919–926. doi: 10.1017/S0953756201004452. [DOI] [Google Scholar]