Abstract
Proper spatiotemporal gene expression is achieved by selective DNA binding of transcription factors in the genome. The most intriguing question is how dynamic interactions between transcription factors and their target sites contribute to gene regulation by recruiting the basal transcriptional machinery. Here we demonstrate individual binding and dissociation events of the transcription factor cAMP response element-binding protein (CREB), both in vitro and in living cells, using single-molecule imaging. Fluorescent–tagged CREB bound to its target sequence cAMP-response element (CRE) for a remarkably longer period (dissociation rate constant: 0.21 s-1) than to an unrelated sequence (2.74 s-1). Moreover, CREB resided at restricted positions in the living cell nucleus for a comparable period. These results suggest that CREB stimulates transcription by binding transiently to CRE in the time range of several seconds.
Transcription is the initial step in gene regulation in cells1. Assembly and disassembly of the transcription complex, which are governed by protein-DNA and protein-protein interactions, control spatiotemporal transcription within the nuclear space2. In particular, specific binding of a transcription factor to its target sequence is a necessary condition for recruitment of basal transcriptional machinery components such as RNA polymerase (RNAP) II and co-regulators3. To date, fluorescence recovery after photobleaching (FRAP) and fluorescence correlation spectroscopy (FCS) have demonstrated dynamic behaviour of transcription factors4,5. However, the kinetics of transcription factor binding to DNA and their spatial distributions in the nucleus remains uncertain.
CREB is a well characterized transcription factor which regulates gene expression in response to cAMP, calcium and cytokine signals in homeostasis and memory formation6,7. Biochemical and structural analyses have demonstrated that a CREB homodimer binds to a CRE in the promoter regions of its target genes6,8. Transcription could be initiated by recruitment of the co-activators CREB-binding protein (CBP)/p300 and cAMP-regulated transcriptional co-activators (CRTCs) to CREB via protein-protein interactions in response to the signals9,10. How do CREB-CRE interactions underlie transcription? How does CREB contribute to selective gene expression by being distributed spatially within the nucleus? Although CREB has been shown to be mobile in living cell nuclei11, a key issue is to quantitatively understand the spatiotemporal dynamic behavior of CREB interacting with DNA.
In the present study, we investigated real-time binding and dissociation of fluorescent-tagged CREB to CRE at the single-molecule level in vitro and in living cells. Our results show that CREB resides on the CRE sequence for a significantly longer period (more than several seconds) than on an unrelated sequence, but that DNA binding-deficient mutants do not. Dynamic behavior of CREB in living cell nuclei further demonstrates that CREB resides in fixed locations in the time range of seconds. These findings in vitro and in living cells support the view that transient and frequent binding of the transcription factor is responsible for gene regulation.
Results
Single-Molecule Imaging Reveals Real-Time Interaction between CREB and CRE in vitro
First, we directly examined interactions between CREB and CRE in vitro using single-molecule imaging12. To visualize a single molecule of CREB, HaloTag-CREB was labeled with a tetramethylrhodamine (TMR)-conjugated HaloTag ligand (TMR-CREB). We confirmed the ability of HaloTag-CREB to induce downstream transcription (Supplementary Fig. S1). TMR-CREB was loaded into a flow cell comprising two cover glasses coated with oligonucleotides containing the CRE sequence (GCACGTCA) from the human brain-derived neurotrophic factor (bdnf) locus13 (Fig. 1a). Total internal reflection fluorescence (TIRF) microscopy showed that numerous fluorescent spots of TMR-CREB remained associated with the CRE-coated cover glass for various periods (Fig. 1b and Supplementary Movie S1). Furthermore, some spots clearly exhibited two stepwise reductions in their fluorescence intensity (Fig. 1c), indicating that the CREB dimer interacting with CRE is visualized at the single-molecule level.
Figure 1.
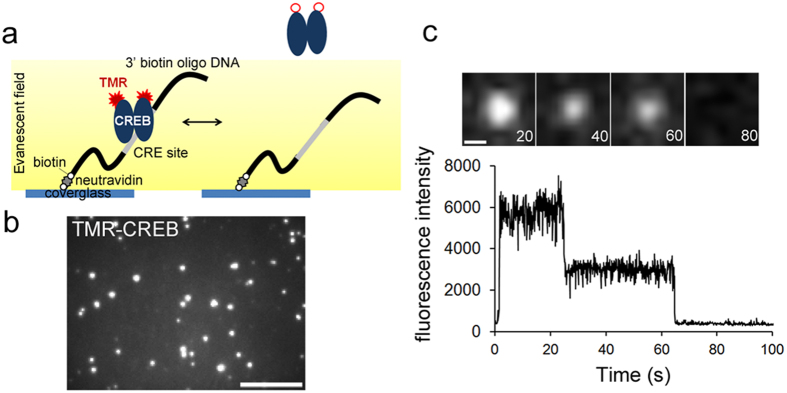
Single-molecule imaging of CREB on a DNA-coated cover glass (a) Experimental design of single-molecule imaging to visualize TMR-CREB interacting with surface-tethered DNA using TIRF microscopy. (b) TMR-CREB spots interacting with CRE were observed. TIRF images were acquired continuously at 10 frames per second for 120 s. Scale bar: 10 μm. (c) TMR-CREB spots exhibit two stepwise reductions of the fluorescence intensity. Scale bar: 0.5 μm.
CREB Binds to the CRE Sequence with Long Residence Time
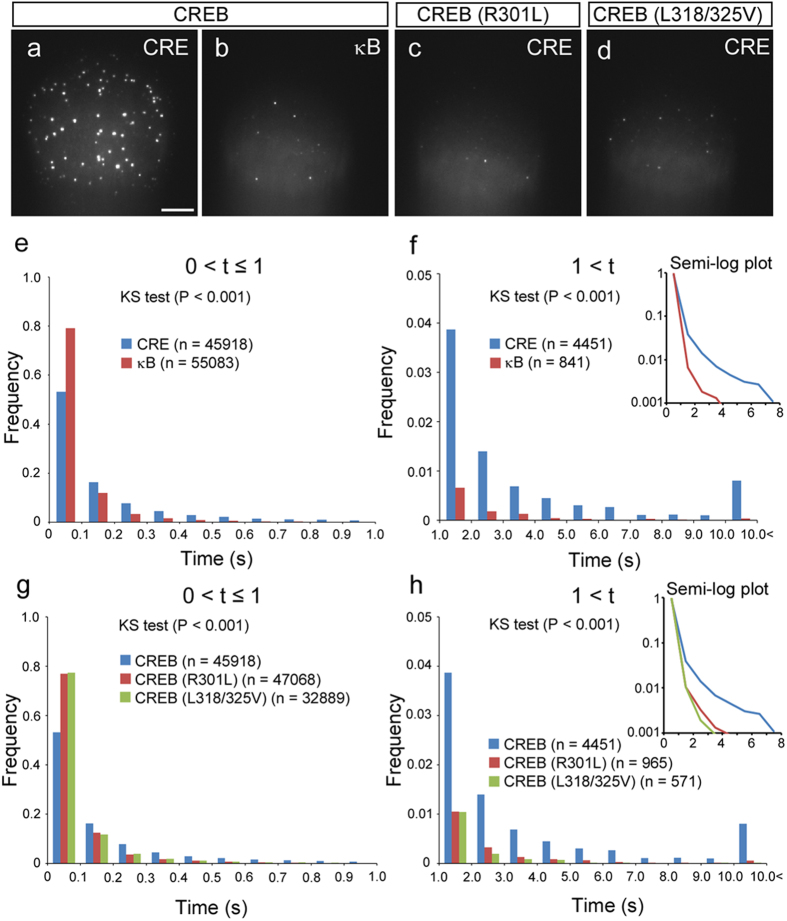
The residence times of individual CREB dimers bound to the CRE sequence were quantitated. The majority (91.8%) of TMR-CREB spots exhibited short residence times (≤1 s, Fig. 2a,e), whereas a small (8.2%) but significant fraction stayed in the same place for markedly longer periods (>1 s, Fig. 2a,f). In contrast, the fraction of TMR-CREB spots having the longer residence times was substantially lower (2.2%) on a cover glass coated with a recognition sequence for transcription factor NF-κB (the κB site sequence, GGGACTTTCC)14, indicating that TMR-CREB has a stronger affinity for the CRE sequence (Fig. 2a,b,e,f, Supplementary Fig. S2a, Supplementary Movie S2).
Figure 2.
Individual interaction between CREB and CRE in vitro. (a,b) TIRF images show individual TMR-CREB spots interacting with CRE sequence (a; the same image in Fig. 1b) and κB sequence (b). Scale bar: 10 μm. (c,d) TIRF images of TMR-mutant CREB (R301L and L318/325V) interacting with CRE sequence. (e,f) Quantitative analysis of the residence time of CREB spots interacting with CRE and κB sequences. (e) and (f) show distribution histograms for short (≤1 s) and long (>1 s) residence times of TMR-CREB, respectively. The number of TMR-CREB spots analyzed is presented in parentheses. The Kolmogorov-Smirnov (KS) test showed that the difference between the two distributions (CRE vs. κB) is significant (P < 0.001). (g,h) Quantitative analysis of the residence time of mutant CREB spots interacting with CRE sequence. (g) and (h) show distribution histograms of short (≤1 s) and long (>1 s) residence times of the spots, respectively. The number of TMR-CREB spots analyzed is presented in parentheses. The distribution of wild-type CREB is significantly different from those of the mutants (KS test, P < 0.001).
We also investigated the residence time of mutant CREB (R301L), which is unable to bind to the CRE sequence15. As expected, the population with the long residence time (1.9%) was much smaller compared to that for the intact CREB (Fig. 2c,h, Supplementary Fig. S2b, Supplementary Movie S3). Another mutant CREB (L318/325V), which cannot bind to CRE because it cannot dimerize16, also displayed a much smaller population with the long residence time (1.5%, Fig. 2d,g,h, Supplementary Fig. S2b and Supplementary Movie S4). This is consistent with previous findings from gel electrophoresis mobility shift assays15,16. These results indicate that wild-type CREB has sequence-specific binding activity with a long residence time.
The distribution histogram of the residence times of CREB bound to CRE fitted a biexponential function (Supplementary Fig. S2a). The dissociation rate constants for the short and long residences were 2.57 ± 0.33 s-1 and 0.21 ± 0.03 s-1, respectively. In contrast, the distribution histogram of the residence times of CREB on the κB sequence fitted more accurately a monoexponential function with a dissociation rate constant of 2.74 ± 0.12 s-1, which is close to the short residence in the CREB-CRE interaction (Supplementary Fig. S2a).
To test the possibility that photobleaching of TMR-CREB might have affected the rate constant, we examined the fluorescence stability of TMR-CREB in the same condition. The time constant of the photobleaching (13.8 s) was much longer than the duration of the long-residence interaction between CREB and CRE sequence (1 / 0.21 = 4.8 s) (Fig. S2a, c). Thus, it is likely that the residence times of CREB on the CRE sequence-coated substratum reflect its binding to CRE.
Single-Molecule CREB in Living Cell Nuclei
We next studied the in vivo dynamics of CREB in the nuclei of living mammalian cells at the single-molecule level17. Mouse neuroblastoma Neuro2a cells were transfected with HaloTag-CREB. To observe single-molecule TMR-CREB in the nucleus, highly inclined and laminated optical sheet (HILO) microscopy was used18 (Fig. 3a). Moreover, HaloTag-CREB was sparsely labeled by adding a low concentration of TMR-HaloTag ligand, in order to follow the dynamics of individual TMR-CREB molecules.
Figure 3.
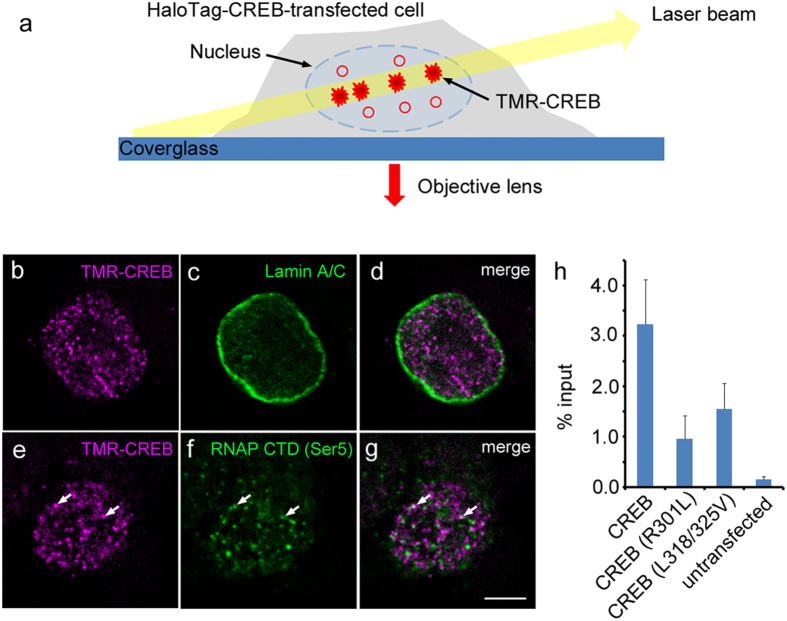
Single-molecule imaging of CREB in the nucleus. (a) Experimental design of single-molecule imaging to visualize TMR-CREB in the nucleus of living cells. A limited number of TMR-CREB molecules in the nucleus are observed with low background using HILO microscopy. (b–g) Subcellular localization of TMR-CREB in fixed HaloTag-CREB-transfected Neuro2a cells using HILO fluorescence microscopy. Scale bar: 10 μm. Immunocytochemical staining was performed with anti-lamin A/C antibody (c) anti-Ser5-phosphorylated RNAP II C-terminal domain antibody (f) and TMR-conjugated HaloTag ligand (b,e) Series of images (0.2 μm × 5 sections) were subjected to a deconvolution algorithm. Arrows indicate TMR-CREB spots colocalized with Ser5-phosphorylated RNAP II C-terminal domain. (h) ChIP and qPCR were performed to detect HaloTag-CREB residing at the CRE sequence within the c-fos promoter in cells transfected with plasmids encoding wild-type, mutant (R301L) or mutant (L318/325V) HaloTag-CREBs. Bar represents the mean ± SEM from three independent experiments.
Before live imaging, immunocytochemical analysis was performed with HILO microscopy to examine the subcellular localization of TMR-CREB in the transfected cells. A few dozen fluorescent spots of TMR-CREB were clearly observed, with low background signals (Fig. 3b,e). In addition, TMR-CREB was predominantly localized within the nucleus, surrounded by lamin A/C, a nuclear envelope component19 (Fig. 3b–d). Moreover, some of the TMR-CREB spots were found to colocalize with Ser5-phosphorylated RNAP II C-terminal domain, which suggests that a proportion of TMR-CREB molecules are present at actively transcribed loci20 (Fig. 3e–g). The average number of colocalizing spots was 2.4 ± 1.4 per optical section (n = 40 cells) (Supplementary Fig. S3).
To confirm that TMR-CREB was binding to endogenous CRE sequences in transfected cells, we performed chromatin immunoprecipitation (ChIP) assays with HaloTag affinity resin (Fig. 3h). Wild-type CREB was associated with the promoter region of c-fos as well as bdnf, each of which contains a CRE sequence8,13. In contrast, the binding fraction of both the mutant CREB (R301L) and the mutant CREB (L318/325V) apparently decreased in accordance with the observation in vitro (Fig. 2g,h).
In vivo Dynamics of CREB in the Nucleus of Living Cells
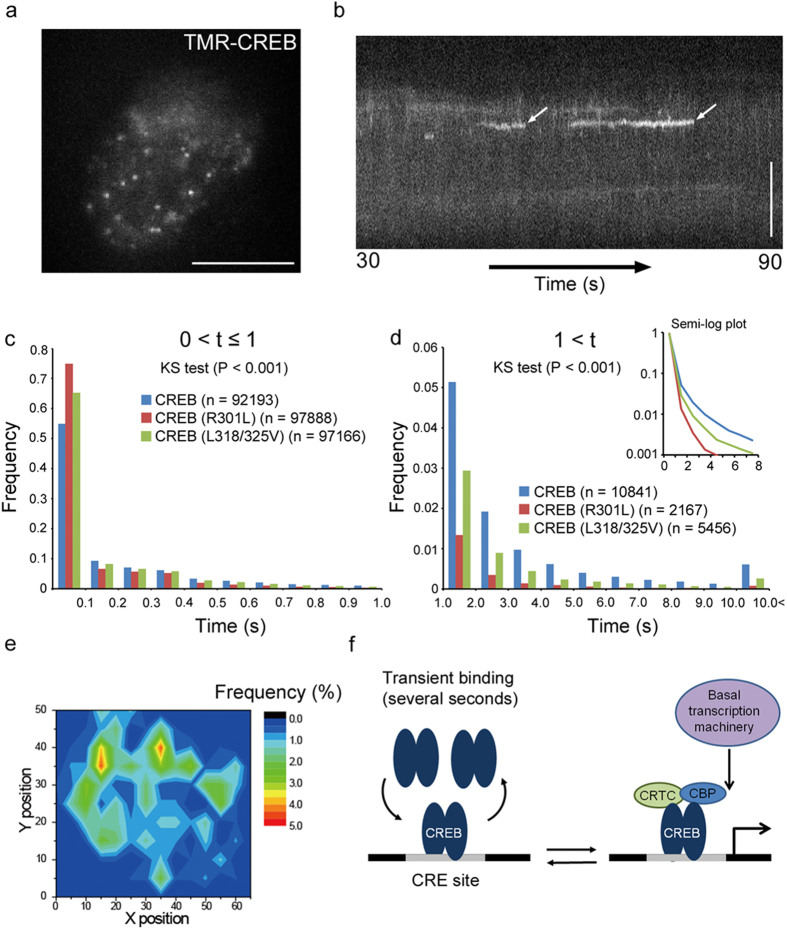
Real-time movements of TMR-CREB in living cells were investigated by HILO microscopy, and were compared to those of mutant CREB (R301L and L318/325V). If the above in vitro observations apply to the interactions of CREB with genomic DNA, TMR-CREB spots having the longer residence times (several seconds) should be detected more frequently than those of TMR-mutant CREB. Many TMR-CREB spots were clearly visible in the nuclei of living cells, with a wide range of residence times (Fig. 4a and Supplementary Movie S5) which were quantified as described above. Although the majority of both CREB (89.5%) and mutant CREB (R301L: 97.8%, L318/325V: 94.6%) spots disappeared by 1 s, the fraction with longer residence times (>1 s) was larger for wild-type CREB (10.5%) than for the mutant CREBs (R301L: 2.2%, L318/325V: 5.4%) (Fig. 4c,d, Supplementary Fig. S4, and see also Supplementary Fig. S5). The differences between wild-type and the mutant CREBs in living cells were smaller than those in vitro, suggesting that CREB may interact with nuclear proteins in addition to DNA in the nucleus. Moreover, the dissociation rate constants for TMR-CREB (3.11 ± 0.53 s-1 and 0.35 ± 0.08 s-1 for the short and long residence components, respectively) in living cells were not only different from those for the mutant CREBs but also comparable with those in vitro (Supplementary Fig. S2b, S4, Supplementary Table S1). Taken together with the findings in ChIP assay (Fig 3h), these results suggest that the fraction with the long residence time most likely corresponds to CREB specific binding to CRE. The fraction of short residence time indicates the non-specific binging to DNA. Indeed, we observed the substantial fraction of binding remained even in the mutant CREB transfected cells in ChIP assays. The subtle difference between the mutant CREB (R301L) and the mutant CREB (L318/325V) in ChIP assay also seems to reflect the residence time.
Figure 4.
Dynamics of CREB in the nucleus of living cells. (a) TMR-CREB spots were observed in the living cell nucleus by HILO fluorescence microscopy. Average of the images (for 1 s) shows individual TMR-CREB spots in the nucleus. Scale bar: 10 μm. (b) A representative kymogram (60 s) shows the long residence times of TMR-CREB spots at restricted locations (arrows). Scale bar (y-axis): 10 μm. (c,d) Distribution histograms show the short (≤1 s) and long (>1 s) residence times, respectively, of TMR-CREB and TMR-mutant CREBs in living cells. The number of spots analyzed in living cells is presented in parentheses. The distribution of wild-type CREB is significantly different from those of mutants (KS test, P < 0.001). (e) A color contour map represents spatial distribution of the frequency of appearance of TMR-CREB spots having long residence times (>1 s) in a living cell. The area covering the whole nucleus was divided into 154 equal sub-areas (1.2 × 1.2 μm). The number of TMR-CREB spots for the entire observation time (120 s) was counted in each sub-area. Twelve out of a total of 270 spots (4.4%) accumulated in two different sub-areas (hot points, orange/red). This accumulation level for the hot points is much higher than would be expected (1.8 spots, 0.7%) in a random distribution. TMR-CREB spots also tended to occur frequently in the sub-areas surrounding the hot points. (f) A model of the dynamic interaction of CREB with CRE to promote transcription initiation.
The spatial distribution of TMR-CREB spots in the nucleus was also studied. We found that TMR-CREB spots with long residence times (>1 s) appeared repeatedly at highly restricted locations in the nucleus (Fig. 4b). A location at which >4% of the long-residence-time TMR-CREB spots accumulated was defined as a hot point. As exemplified in Fig. 4e, TMR-CREB spots accumulated in two different hot points. The average number of hot points per optical section was 3.0 ± 0.6 (n = 22 cells), similar to the number of active RNAP II foci which colocalized with TMR-CREB spots (2.4 ± 1.4 per optical sections; Fig. 3g, Supplementary Fig. S3). Taken together, these results suggest that TMR-CREB interacts with genomic DNA, on presumed multiple CRE sites, in living cells, with a similar time course to that in vitro.
Discussion
The present study has shown, for the first time, a dynamic interaction between CREB and CRE at the single-molecule level. While previous DNase I footprinting and gel-shift assays had indicated a strong affinity between the two6, our findings both in vitro and in living cells revealed the existence of two modes of interaction, with a short and a long residence time. The interaction of CREB with CRE is therefore more dynamic than previously suggested21,22,23.
The short residence may represent CREB interacting weakly with non-specific DNA sequences while it searches for CRE sites in the genome. Consistent with this view, non-specific DNA binding by the lac repressor (LacI) shows very fast dissociation in living Escherichia coli cells24. Several recent studies have also demonstrated that mammalian transcription factors very transiently interact with chromatin for less than 1 s25,26,27,28,29,30 (Supplementary Fig. S5), although FRAP analysis of CREB in PC12 cells exhibits slow dissociation11. On the other hand, the long residence of CREB may reflect the specific interaction with CRE, since the long-residence fraction was almost abolished when CREB was incubated with the κB sequence (Fig. 2f). Similar residence times were obtained in living cells by single-molecule imaging (Fig. 4c,d). Therefore, we propose that CREB participates in transcription initiation by binding to the target sequence transiently in the time range of several seconds (Fig. 4f). In accordance with this view, recent single-molecule imaging studies have demonstrated that other mammalian transcription factors transiently reside in the nucleus with a similar time course of second order25,26,27,28,29,30. In particular, p53 and the glucocorticoid receptor exhibit the long residence at the target sites27.
The presence of hot points in the nucleus further implies that CREB repeatedly binds to and dissociates from active CRE sites31,32,33. CBP/p300 and CRTCs may bind efficiently only to CREB dimers residing on these CRE sites. Previous studies with tandem gene arrays have also demonstrated that transcription factors bind repeatedly at the active gene loci34,35,36,37. Thus, it is likely that the repetitive binding of transcription factors promotes transcription38,39.
Our spatial analysis also showed that a few TMR-CREB hot points were present in a single focal plane, in accordance with the number of TMR-CREB spots which colocalized with Ser5-phosphorylated RNAPII foci in fixed cells (Fig. 3g,4e, Supplementary Fig. S3). One may suppose that this number of the hot points was fewer than the total number of CRE sites in the whole genome31,32. However, it is not surprising because only a fraction of CRE sites is active in living cells33,40,41,42,43. As chromatin states and chromosome positions in the interphase nucleus are known to influence gene expression4,44, the formation of the hot point is probably limited by these chromatin components. Therefore, although further study is needed, the hot points may indicate active gene loci whose transcription is driven by the CREB-CRE interaction.
Collectively, our observations indicate that CREB contributes to transcription initiation by binding transiently to genomic CRE sites in the time range of several seconds. In addition, the present analysis using single-molecule imaging both in vitro and in living cells will enable further studies of the dynamics of transcription factors on DNA.
Methods
Plasmids
pFN21AB5414 containing HaloTag-human CREB1 cDNA was purchased from Promega. To generate the HaloTag-human CREB1 R301L and L318/325V mutant, point mutations were introduced into pFN21AB5414 by PCR-mediated site-directed mutagenesis with the mutagenic primer pairs: R301L 5’-GAAGCAGCTCGAGAGTGTCCTAGAAAGAAGAAAGAATATG-3' and its complementary oligonucleotide; L318/325V 5’-TTTAGAAAACAGAGTGGCAGTGGTTGAAAATCAAAACAAGAC AGTGATTGAGGAGCTA-3' and its complementary oligonucleotide.
DNA Coating on Cover Glasses
Cover glasses (Matsunami) were cleaned by boiling in RCA solution (6:4:1 H2O/30% NH4OH/30% H2O2) for 1 h. Poly(ethyleneimine) (PEI) and poly(acrylic acid) (PAcr) (Sigma) were dissolved at 2 mg/ml in H2O and then adjusted to pH 8.0 using either NaOH or HCl. The cleaned cover glasses were immersed in positive (+, PEI) and negative (−, PAcr) polyelectrolytes in the sequence + / wash / - / wash / + / wash for 30 min each at room temperature; each wash step involved three rinses in distilled water45. A flow cell was assembled from two cover glasses and two strips of double-sided tape as spacers. The flow cell was treated with 10 mg/ml biotinylated α-casein in PBS for 5 min and then reacted with 5 mg/ml NeutrAvidin (Life Technologies) in PBS. Biotin-labeled dsDNAs (40 nM in PBS) were adsorbed onto the cover glass surface through the avidin-biotin interaction for 5 min. Biotin-labeled 19-nt dsDNA including the CRE sequence was prepared by annealing 5’-GACAGCGCACGTCAAGGCA-biotin-3’ and its complementary oligonucleotide. Biotin-labeled 22-nt dsDNA including the κB sequence was prepared by annealing 5’-AGTTGAGGGGACTTTCCCAGGC-biotin-3’ and its complementary oligonucleotide. After each procedure, the flow cell was washed with PBS. Finally, the flow cell was treated with 10 mg/ml BSA in PBS for 5 min and then flushed with a binding buffer solution (100 mM KCl, 1 mM MgCl2, 20 mM HEPES-NaOH (pH 7.8), 0.1 M DTT, 2 mg/ml BSA, 50% sucrose). In experiments, TMR-CREB or TMR-mutant CREB in binding buffer was loaded into the flow cell. All CREB proteins were generated by the TNT Quick Coupled Transcription/Translation System (Promega) and labeled with HaloTag TMR Ligand (Promega). To determine fluorescence stability, TMR-CREB in binding buffer was loaded into a 35-mm glass-bottom culture dish (Greiner bio-one) without DNA coating.
Cell Culture
Mouse Neuro2a cells were cultured in DMEM/F12 medium (Life Technologies) supplemented with 10% fetal bovine serum (Hyclone). The cells were seeded to 80% confluence in a 4-well culture plate (Nalge Nunc) and then transfected with plasmid vectors. All plasmids used for transfection were obtained using PureLink Hipure Plasmid DNA purification kits (Life Technologies) and dissolved in PBS. Transfection with Lipofectamine2000 (Life Technologies) was performed according to the manufacturer’s instructions. After transfection, cells were cultured for 8 h and then replated on a 35-mm glass-bottom culture dish (Greiner bio-one). To label HaloTag CREB with a TMR, HaloTag CREB cDNA-transfected cells were incubated with growth medium containing 0.1 nM HaloTag TMR Ligand (Promega) for 15 min in a CO2 incubator, after which the medium was replaced.
Microscopy
An inverted microscope (Ti-E, Nikon) with oil-immersion objective (100 x, NA 1.49; Nikon) was used for all experiments. Alexa 488 and TMR were excited by 488- (20 mW; Coherent) and 561-nm (20 mW; Coherent) lasers, respectively. TIRF and HILO were used as illumination for in vitro and in vivo analysis, respectively. All fluorescence live images were obtained at 10 fps using an EM-CCD (iXon897, Andor Technology) with Solis software (Andor Technology). The images shown in the Figures are averaged representations of photos captured over 1 or 5 s. To observe the dynamics of TMR-CREB in living cells, the cell culture dish was mounted on a stage top incubator (Tokai Hit) maintained at 37 °C in an environment of humidified 5% CO2, 20% O2, and 75% N2. All images were analyzed by ImageJ software with a self-made plugin (Particle Track and Analysis). Fluorescence images of fixed cells were acquired and processed with NIS Element software linked to a deconvolution module (Nikon).
Statistical Analysis
All statistical values are presented as the mean value ± SD from at least three independent experiments. Significant differences were determined with Student’s t-test or the Kolmogorov-Smirnov test. Origin (OriginLab) and Excel (Microsoft) were used for statistical analysis and data plotting.
Immunostaining
Neuro2a cells were fixed at room temperature for 10 min in 4% paraformaldehyde/PBS. They were then permeabilized and blocked for 10 min in buffer G, composed of 5% normal goat serum (S-1000; Vector Labs) and 0.1% Triton X-100 in PBS. The cells were then incubated overnight at 4 °C with anti-lamin A/C mouse monoclonal antibody (3A6-4C11; Active Motif) at 1:1000 and anti-RNAP II C-terminal domain phospho Ser5 rabbit polyclonal antibody (Active Motif) at 1:10000 in buffer G. Primary antibodies were detected by incubation with Alexa 488-conjugated anti-mouse IgG (A11029; Invitrogen) at 1:400, or with Alexa 488-conjugated anti-rabbit IgG (A-11034; Invitrogen) at 1:400, in buffer G at room temperature for 2 h. Nuclei were stained with 0.1% 4’,6-diamidino-2-phenylindole (Sigma) in a mounting medium containing 50% glycerol and 2.3% 1,4-diazabicyclo[2.2.2]octane (Sigma) in 50 mM Tris-HCl (pH 8.0).
ChIP Assay and Quantitative real-time PCR (qPCR)
Cells were cross-linked with 1% formaldehyde in culture medium for 10 min, and lysed in lysis buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate). Cross-linked cell lysates were sonicated for 20 cycles (burst and intervals of 15 s each) with a VCX130PB (Amplitude 60%; Sonics & Materials). Sonicated samples were incubated with HaloLink Resin (Promega) for 3 h at room temperature. The resin was washed twice with nuclease-free water, once with high-salt buffer (50 mM Tris-HCl (pH 7.5), 700 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate and 1 mM EDTA), and finally three times with nuclease-free water. Samples were reverse cross-linked with reversal buffer (10 mM Tris-HCl (pH 8.0), 1 mM EDTA and 300 mM NaCl) for 6 h at 65 °C. The eluted DNA was purified using a PCR product purification kit (Promega). qPCR was performed using an ABI StepOne (Applied Biosystems). To quantify the amplified DNA, a SYBR Green PCR assay was used with KAPA SYBR FAST qPCR Kit Master Mix (KAPA Biosystems) and the primers 5'-GCGTAGAGTTGACGACAGAGC-3’ and 5'-TGGATGGACTTCCTACGTCA-3’. The obtained values were normalized to the respective input DNA.
Additional Information
How to cite this article: Sugo, N. et al. Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence. Sci. Rep. 5, 10662; doi: 10.1038/srep10662 (2015).
Supplementary Material
Acknowledgments
We thank Dr. Takashi Fujii for helpful discussions. We also thank Dr. Ian Smith for critical reading of the manuscript. This work was supported by MEXT KAKENHI on Innovative Areas “Mesoscopic Neurocircuitry” (No. 23115102) to N.Y., “Cross-talk between moving cells and microenvironment as a basis of emerging order in multicellular system” (No. 23111516), and Grant No. 20200009 to N.S. This work was also supported by JSPS KAKENHI Grant Nos. 25640035 and 15K14350 to N.S., 20300110 and 23300118 to N.Y., and by the MEXT Global COE Program.
Footnotes
Author Contributions N.S. and N.Y. conceived and designed the research project with support from T.N. and T.Y.N.S., M.M., Y.A., Y.K. and A.O. performed the experiments. N.S., M.M. and Y.A. analyzed the data. N.S. and N.Y. wrote the manuscript, which was discussed and critically edited by all co-authors.
References
- Maniatis T. & Reed R. An extensive network of coupling among gene expression machines. Nature 416, 499–506 (2002). [DOI] [PubMed] [Google Scholar]
- Ptashne M. & Gann A. Transcriptional activation by recruitment. Nature 386, 569–577 (1997). [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116, 247–257 (2004). [DOI] [PubMed] [Google Scholar]
- Hager G. L., McNally J. G. & Misteli T. Transcription dynamics. Mol. Cell 35, 741–753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelman-Ribeiro A. et al. Direct measurement of association and dissociation rates of DNA binding in live cells by fluorescence correlation spectroscopy. Biophys. J. 97, 337–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr B. & Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2, 599–609 (2001). [DOI] [PubMed] [Google Scholar]
- Lonze B. E. & Ginty D. D. Function and regulation of CREB family transcription factors in the nervous system. Neuron 35, 605–623 (2002). [DOI] [PubMed] [Google Scholar]
- Roesler W. J., Vandenbark G. R. & Hanson R. W. Cyclic AMP and the induction of eukaryotic gene transcription. J. Biol. Chem. 263, 9063–9066 (1988). [PubMed] [Google Scholar]
- Conkright M. D. et al. TORCs: transducers of regulated CREB activity. Mol Cell 12, 413–423 (2003). [DOI] [PubMed] [Google Scholar]
- Chrivia J. C. et al. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365, 855–859 (1993). [DOI] [PubMed] [Google Scholar]
- Mayr B. M., Guzman E. & Montminy M. Glutamine rich and basic region/leucine zipper (bZIP) domains stabilize cAMP-response element-binding protein (CREB) binding to chromatin. J. Biol. Chem. 280, 15103–15110 (2005). [DOI] [PubMed] [Google Scholar]
- Harada Y. et al. Single-molecule imaging of RNA polymerase-DNA interactions in real time. Biophys. J. 76, 709–715 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H. et al. Transcriptional activation of the human brain-derived neurotrophic factor gene promoter III by dopamine signaling in NT2/N neurons. J. Biol. Chem. 278, 26401–26409 (2003). [DOI] [PubMed] [Google Scholar]
- Lenardo M.J. & Baltimore D. NF-kappa B: a pleiotropic mediator of inducible and tissue-specific gene control. Cell 58, 227–229 (1989). [DOI] [PubMed] [Google Scholar]
- Walton K. M., Rehfuss R. P., Chrivia J. C., Lochner J. E. & Goodman R. H. A dominant repressor of cyclic adenosine 3',5'-monophosphate (cAMP)-regulated enhancer-binding protein activity inhibits the cAMP-mediated induction of the somatostatin promoter in vivo. Mol. Endocrinol. 6, 647–655 (1992). [DOI] [PubMed] [Google Scholar]
- Dwarki V. J., Montminy M. & Verma I. M. Both the basic region and the 'leucine zipper' domain of the cyclic AMP response element binding (CREB) protein are essential for transcriptional activation. EMBO J. 9, 225–232 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y. & Yanagida T. Single-molecule visualization in cell biology. Nat. Rev. Mol. Cell Biol. Suppl, SS1–5 (2003). [PubMed] [Google Scholar]
- Tokunaga M., Imamoto N. & Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008). [DOI] [PubMed] [Google Scholar]
- Gerace L., Blum A. & Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J. Cell Biol. 79, 546–566 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani H. P. & Greenleaf A. L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20, 2922–2936 (2006). [DOI] [PubMed] [Google Scholar]
- Benbrook D. M. & Jones N. C. Different binding specificities and transactivation of variant CRE's by CREB complexes. Nucleic Acids Res 22, 1463–1469 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock B. P. & Habener J. F. Phosphorylation of the cAMP response element binding protein CREB by cAMP-dependent protein kinase A and glycogen synthase kinase-3 alters DNA-binding affinity, conformation, and increases net charge. Biochemistry 37, 3795–3809 (1998). [DOI] [PubMed] [Google Scholar]
- Euskirchen G. et al. CREB binds to multiple loci on human chromosome 22. Mol Cell Biol 24, 3804–3814 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J., Li G. W. & Xie X. S. Probing transcription factor dynamics at the single-molecule level in a living cell. Science 316, 1191–1194 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeneweg F.L. et al. Quantitation of Glucocorticoid Receptor DNA-Binding Dynamics by Single-Molecule Microscopy and FRAP. PLoS One 9, e90532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza D., Abernathy A., Golob N., Morisaki T. & McNally J. G. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 40, e119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki T., Müller W. G., Golob N., Mazza D. & McNally J. G. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat. Commun. 5, 4456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt J. C. et al. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods 10, 421–426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speil J. et al. Activated STAT1 transcription factors conduct distinct saltatory movements in the cell nucleus. Biophys. J. 101, 2592–2600 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S. et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119, 1041–1054 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. USA 102, 4459–4464 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha-Molstad H., Keller D. M., Yochum G. S., Impey S. & Goodman R. H. Cell-type-specific binding of the transcription factor CREB to the cAMP-response element. Proc. Natl. Acad. Sci. USA 101, 13572–13577 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally J. G., Müller W. G., Walker D., Wolford R. & Hager G. L. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287, 1262–1265 (2000). [DOI] [PubMed] [Google Scholar]
- Bosisio D. et al. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. EMBO J. 25, 798–810 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayasam G. V. et al. Ligand-specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol. Cell Biol. 25, 2406–2418 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp Z. D. et al. Estrogen-receptor-alpha exchange and chromatin dynamics are ligand- and domain-dependent. J. Cell Sci. 119, 4101–4116 (2006). [DOI] [PubMed] [Google Scholar]
- Cisse I. I. et al. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341, 664–667 (2013). [DOI] [PubMed] [Google Scholar]
- Stasevich T. J. et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature 516, 272–275 (2014). [DOI] [PubMed] [Google Scholar]
- Conkright M. D. et al. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell 11, 1101–1108 (2003). [DOI] [PubMed] [Google Scholar]
- Iannello R. C. et al. Methylation-dependent silencing of the testis-specific Pdha-2 basal promoter occurs through selective targeting of an activating transcription factor/cAMP-responsive element-binding site. J. Biol. Chem. 275, 19603–19608 (2000). [DOI] [PubMed] [Google Scholar]
- Martinowich K. et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302, 890–893 (2003). [DOI] [PubMed] [Google Scholar]
- Iguchi-Ariga S. M. & Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 3, 612–619 (1989). [DOI] [PubMed] [Google Scholar]
- Lanctôt C., Cheutin T., Cremer M., Cavalli G. & Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 8, 104–115 (2007). [DOI] [PubMed] [Google Scholar]
- Kartalov E. P., Unger M. A. & Quake S. R. Polyelectrolyte surface interface for single-molecule fluorescence studies of DNA polymerase. Biotechniques 34, 505–510 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.