Abstract
Microtubules are not like other polymers. Whereas polymers such as F-actin will grow continuously as long as the subunit concentration is high enough, a steadily growing microtubule can suddenly shrink even when there is ample αβ-tubulin around. This remarkable behavior was discovered in 1984 when Tim Mitchison and Marc Kirschner deduced that microtubules switch from growth to shrinkage when they lose their GTP caps. Here, I review the canonical explanation of dynamic instability that was fleshed out in the years after its discovery. Many aspects of this explanation have been recently subverted, particularly those related to how GTP-tubulin forms polymers and why GTP hydrolysis disrupts them. I describe these developments and speculate on how our explanation of dynamic instability can be changed to accommodate them.
INTRODUCTION
When we observe microtubules growing under a light microscope, what catches the eye is their sudden collapse, something not observed for polymers such as F-actin. Even before the marvel of dynamic instability was directly observed (Horio and Hotani, 1986), we knew about the switching behavior of microtubules. But how was dynamic instability discovered if not by eye? We knew that αβ-tubulin binds GTP (Weisenberg et al., 1968), the nucleotide that powers its polymerization (Weisenberg, 1972). GTP hydrolysis and phosphate release converts the GTP-tubulin into GDP-tubulin, which falls off the polymer rapidly (Carlier et al., 1984). Fortunately, the hydrolysis of GTP lags behind the binding of new GTP-tubulin; this lag creates a cap of GTP-tubulin at the microtubule end (Carlier and Pantaloni, 1981). Most of this is true for F-actin and ATP, however, so why are microtubules different? Using an electron microscope, Tim Mitchison and Marc Kirschner observed that a microtubule population can dwindle even while its mean length increases (Mitchison and Kirschner, 1984a). In other words, some microtubules will shrink to nonexistence while others continue to grow. They combined this observation (and others) with the facts about GTP and deduced that individual microtubules switch from growth to shrinkage when they lose their GTP caps (Mitchison and Kirschner, 1984b). They called this switching behavior dynamic instability.
In the years that followed, an explanation for this unique switching behavior was fleshed out. When a microtubule grows, the protofilaments are straight or gently curved (0–5° bend per αβ-tubulin; Mandelkow et al., 1991; Chrétien et al., 1995). When a microtubule shrinks, however, the protofilaments curve outward steeply (12° bend per αβ-tubulin). The difference in protofilament curvature implied that the GTP-tubulins that bind to growing microtubule ends are “straight,” whereas the GDP-tubulins found in the microtubule lattice want to be “curved.” The GTP cap keeps these GDP-tubulins in shape; when the GTP cap is lost, GDP-tubulin relaxes into the curved conformation. This relaxation breaks the bonds between protofilaments, which then curve outward steeply, and the GDP-tubulin falls off the polymer rapidly. The swiftness with which the GDP-microtubule collapses is what distinguishes microtubules from F-actin and defines the phenomenon known as catastrophe.
Dynamic instability became a foundation of cell physiology; upon it we have built our explanations for how dividing cells segregate their chromosomes, how fibroblasts migrate into wounds during healing, and how neurons extend their axon and dendrites. Microtubule growth and shrinkage are essential to these processes and many more. Driving these processes are a host of microtubule-associated proteins (MAPs) that make microtubules grow faster, shrink slower, undergo catastrophe more often, and so on. To explain how MAPs drive the microtubule cytoskeleton, we first need to understand how GTP-tubulin forms polymers and why GTP hydrolysis disrupts them. But what is missing from the canonical explanation of dynamic instability? Here, I argue that recent experiments have subverted this explanation and that our best models cannot explain the diversity of MAPs that control microtubule physiology. I speculate on how a modern explanation of dynamic instability might account for the complexities of microtubule growth and catastrophe.
GROWTH
Microtubules grow when αβ-tubulin collides with the end of a protofilament and forms a noncovalent bond. These collisions occur more frequently when the tubulin concentration is higher, and thus the growth rate increases linearly with more tubulin (e.g., Walker et al., 1988). What could be simpler? Measurements of microtubule growth rates are classically fit to Oosawa's equation for polymer growth (Oosawa and Asakura, 1975), which reduces the microtubule to a single end-site that grows with a second-order on-rate constant (units of μM−1⋅s−1) and shrinks with a first-order off-rate constant (units of s−1). This “1D model” provided a simple framework for analyzing MAPs that increase microtubule growth rates, such as the microtubule polymerase XMAP215 (Gard and Kirschner, 1987).
Not every MAP can be shoehorned into the 1D model, however, because microtubules cannot be reduced to a single end-site. Rather, the bonds formed by the incoming αβ-tubulin vary depending on the context at each protofilament end. Some protofilaments extend beyond others, and this tapering creates differences in the number of lateral bonds that will form (Figure 1A). The bonds may also differ in their contact surfaces. There may be two types of lateral bond (Wang and Nogales, 2005), or both lateral and longitudinal bonds may form at different contact angles due to the outward curvature and relative flatness of the tapered end (Figure 1B; Chrétien et al., 1995). The tendency of tapered ends to curve outward is presumably caused by the intrinsic curvature found in GTP-tubulin (Ozon et al., 1997; Ayaz et al., 2012; Pecqueur et al., 2012), which defied the expectation that GTP-tubulin is straight. The tendency to curve outward will compete with the tendency of the lattice to roll up into a tube (Janosi et al., 1998). The balance of these competing tendencies creates a “sheet,” a misnomer originating from the hammered-flat appearance of microtubule ends in negative-stain electron microscopy (Simon and Salmon, 1990). As the tapered end is filled in, the microtubule end will straighten and the angles (and energies) of lateral and longitudinal bonds will change.
FIGURE 1:
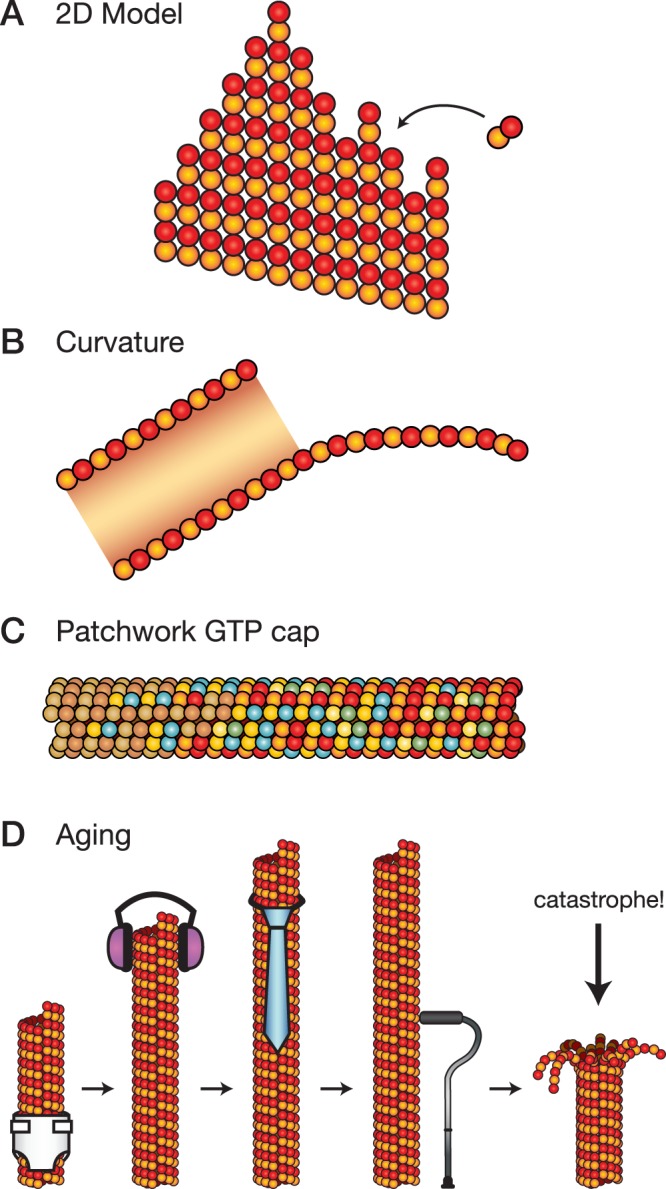
Complexities in microtubule growth and catastrophe. (A) Schematic drawing of the 2D model for microtubule growth, showing a tapered microtubule end. The number of lateral bonds that form depends on the context at each protofilament end. (B) Schematic drawing of curvature at microtubule ends, showing a cross section of a growing microtubule end and its tapered, outwardly curved, and flattened-out structure. The schematic is based on cryo–electron microscopy images in Chrétien et al. (1995). (C) Schematic drawing of a patchwork GTP cap. The different colors of β-tubulin (red, green, blue, beige) represent hypothetical nucleotide states from GTP to GDP. (D) Schematic drawing of the aging process showing a young microtubule (far left, in diaper) growing and maturing (left to right, a teenager with headphones and an adult with necktie) until it becomes old (far right, with cane). The aging process leads to catastrophe.
Accounting for these complexities has solved a few puzzles of microtubule growth. Consider that the on-rate constant from the 1D model is ∼0.1 μM−1⋅s−1 per protofilament end (Walker et al., 1988). This rate is quite slow. For comparison, globular proteins collide together with a rate constant estimated at 4 μM−1⋅s−1 (Northrup and Erickson, 1992), and G-actin polymerizes into F-actin with an on-rate constant of 12 μM−1⋅s−1 (Pollard, 1986). Presumably αβ-tubulin diffuses like any other protein of similar mass and shape. So why does tubulin bind so slowly to the ends of protofilaments? As it turns out, it doesn't. The key is to treat each protofilament end separately and determine rate constants based on context (VanBuren et al., 2005). In this “2D model,” αβ-tubulin binds to protofilament ends rapidly (at ∼4 μM−1⋅s−1) but falls off them just as rapidly when no lateral bonds are formed. The off-rate constant drops with each lateral bond until the αβ-tubulin becomes entombed by its neighbors. Computer simulations that implement this idea can reproduce microtubule growth rates (Gardner et al., 2011a) and, critically, the large fluctuations observed in the growth of individual microtubules (Schek et al., 2007).
Despite its contributions, the 2D model makes predictions that are at odds with experimental data. For example, any small reduction in the off-rate constant from a protofilament end should increase microtubule growth rates substantially. Indeed, reducing the off-rate constant seems to be the trick for XMAP215 (Brouhard et al., 2008). But paclitaxel, a chemotherapeutic that stabilizes lateral bonds (Prota et al., 2013), actually slows down microtubule growth (Zanic et al., 2013), the opposite of the predicted effect. Similarly, consider Doublecortin, a MAP involved in brain development. Doublecortin stabilizes microtubules by binding at the vertex of four αβ-tubulins (Fourniol et al., 2010) and forming cooperative assemblies (Bechstedt and Brouhard, 2012); moreover, Doublecortin binds specifically to microtubule ends (Bechstedt et al., 2014). Nonetheless, doublecortin has no effect on microtubule growth rates whatsoever (Moores et al., 2006). Something is missing. What we lack is a description of microtubule growth that accounts for the curvature of microtubule ends. Paclitaxel, for example, straightens the protofilaments at microtubule ends (Elie-Caille et al., 2007); perhaps this straightening contributes to paclitaxel's slowing of microtubule growth. The inclusion of structural phenomena in the 2D model, including the intrinsic curvature of GTP-tubulin and the variable angles of αβ-tubulin bonds, may be necessary in order to explain an assortment of MAPs and tubulin-binding drugs.
CATASTROPHE
When a microtubule is growing, the incoming α-tubulin contributes residues that complete the GTP pocket of the distal β-tubulin (Nogales et al., 1998, 1999). GTP hydrolysis is free to proceed in the penultimate αβ-tubulin, but the lag in GTP hydrolysis creates a GTP cap at microtubule ends. The size of the GTP cap was believed to be small (Voter et al., 1991), perhaps as small as a single layer of GTP-tubulin. Whatever its mean size, the cap size will fluctuate when microtubules grow due to the stochastic nature of new GTP-tubulin binding and the GTP hydrolysis reaction. The fluctuations occasionally cause the cap to disappear, exposing GDP-tubulin subunits at the microtubule end; the microtubule then falls apart “in some catastrophic manner” (Mitchison and Kirschner, 1984a).
This textbook description of the GTP cap and how it is lost has evolved significantly in recent years. The game changer has been EB1, the protein that binds with high affinity to microtubule ends. Because EB1 binds with higher affinity to GTP analogues, namely GMPCPP (Zanic et al., 2009) and GTPγS (Maurer et al., 2011), EB1's comet-shaped signal is considered a readout for (some aspect of) the GTP cap. The EB1 signal decays exponentially over hundreds of nanometers (Bieling et al., 2007), indicating that the GTP cap is not small but instead hundreds of subunits deep. However, the EB1 signal does not reach the very tip of the microtubule, suggesting that the very tip of the GTP cap differs from its “core” (Maurer et al., 2014). What could this difference be? Perhaps the very tip of the microtubule is made of GTP-tubulin proper, but core αβ-tubulins are in some other nucleotide state, such as the GDP-Pi state. Heterotrimeric G proteins, for comparison, pass through three distinct states between GTP and GDP (Sprang, 1997). EB1 binds with different affinities to the various GTP analogues (GTPγS > GDP-BeF3 >> GMPCPP; Maurer et al., 2011). These differences perhaps indicate that EB1 recognizes a novel nucleotide state—or perhaps nucleotide analogues are only analogues. In any case, the intensity of the EB1 signal drops to ∼20% of its peak intensity just before catastrophe (Maurer et al., 2014). This observation indicates that the GTP cap is lost when the density of “GTP-tubulin” drops below a threshold.
When GTP is hydrolyzed in the cap, in however many steps, αβ-tubulin undergoes a conformational change that weakens the bonds holding the lattice together. The assumption has been that lateral bonds are the ones weakened, based on images of shrinking microtubule ends in which intact protofilaments curve outward steeply (Mandelkow et al., 1991). Indeed, enhanced lateral contacts were observed in the structure of GTPγS microtubules (Maurer et al., 2012), suggesting that the GTP cap has stronger lateral bonds. In contrast, no differences in lateral bonds were observed in a comparison of the structures of GMPCPP- and GDP-microtubules (Alushin et al., 2014). The most obvious difference between the GMPCPP and GDP structures is a “compaction” of α-tubulin in the GDP state, which occurs at the contact surface of the longitudinal bond. It is not clear how the compaction of one αβ-tubulin would affect its neighbors, and the compaction may also place the lateral bonds under strain. These contrasting structures make it unclear which bonds are weakened after GTP hydrolysis. Considering that the GTP cap also overlaps the tapered, outwardly curved, and flattened-out region of a growing microtubule end, we are faced with a GTP cap that is a large region speckled with internal strains, variable bond angles, and possibly multiple nucleotide states (Figure 1C).
A patchwork GTP cap does not, however, change the fundamental idea that catastrophes occur when the GTP cap is lost due to fluctuations in growth and hydrolysis. But are these fluctuations really the cause of catastrophe? The EB1 signal grows larger when microtubules grow faster (Bieling et al., 2007), indicating that the “GTP cap” grows larger as well. Larger GTP caps should be harder to lose to fluctuations, and thus the frequency of catastrophe should drop when microtubules grow faster—for example, at higher tubulin concentrations. Such an inverse correlation has been widely assumed to hold, but the data say otherwise: catastrophes occur at similar rates across a range of αβ-tubulin concentrations. The key observation is that the probability that a microtubule undergoes catastrophe increases with time; in other words, microtubules age, and older microtubules are more susceptible to catastrophe than younger microtubules (Figure 1D; Gardner et al., 2011b). The implication is that the microtubule end remembers its past, perhaps through the accumulation of “defects” during growth (Bowne-Anderson et al., 2013) or because of increased tapering of older microtubule ends (Coombes et al., 2013).
A structural explanation of catastrophe must bring together the aging process and the reduction of the “GTP cap” to ∼20% of its size before catastrophe. Just before this drop in the EB1 signal, microtubule growth slows down (Maurer et al., 2014). The slowdown allows GTP hydrolysis events to catch up with the microtubule end and pop the cap. Presumably the aging process is predisposing the microtubule to such slowdowns. What is needed is an understanding of how different structural states of the microtubule end affect growth rates and hydrolysis rates, which might generate hypotheses about the specific nature of the aging-related defects.
CONCLUSION
Readers familiar with microtubules will have noticed that I did not discuss rescue, the process by which a shrinking microtubule is converted back into a growing one. We know too little about rescue in purified tubulin solutions to say much. A similarly mysterious process is nucleation, whether it be spontaneous nucleation or nucleation from a template. Rescue and nucleation are more complex than growth and catastrophe, which must be explained first. Such a modern explanation of dynamic instability will include the following: the intrinsic curvature of GTP-tubulin; the competing tendencies of microtubule ends to curve out and roll up; the jumble of bonds in the 2D model; the patchwork of the GTP cap; the consequences of GTP hydrolysis on bond strengths; and the aging process. The reason to develop such an explanation is, of course, to understand dynamic instability in its own right. More importantly, we will soon be able to explain a diversity of MAPs and, in so doing, the behavior of cells.
Acknowledgments
I thank S. Wolfson for shrewd editing and our many conversations on writing. I thank S. Bechstedt, S. Chaaban, C.T.J. Hsu, M. Wieczorek, and M. Zanic for comments on the manuscript. I am supported by the Canadian Institutes for Health Research (MOP-111265 and MOP-137055), the Natural Sciences and Engineering Research Council of Canada (RGPIN-2014-03791), and McGill University. I am a Canadian Institutes for Health Research New Investigator.
Abbreviation used:
- MAP
microtubule-associated protein.
Footnotes
REFERENCES
- Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P, Ye X, Huddleston P, Brautigam CA, Rice LM. A TOG:αβ-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 2012;337:857–860. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechstedt S, Brouhard GJ. Doublecortin recognizes the 13-protofilament microtubule cooperatively and tracks microtubule ends. Dev Cell. 2012;23:181–192. doi: 10.1016/j.devcel.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechstedt S, Lu K, Brouhard GJ. Doublecortin recognizes the longitudinal curvature of the microtubule end and lattice. Curr Biol. 2014;24:2366-2375. doi: 10.1016/j.cub.2014.08.039. [DOI] [PubMed] [Google Scholar]
- Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, Surrey T. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- Bowne-Anderson H, Zanic M, Kauer M, Howard J. Microtubule dynamic instability: a new model with coupled GTP hydrolysis and multistep catastrophe. Bioessays. 2013;35:452–461. doi: 10.1002/bies.201200131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Hill TL, Chen YD. Interference of GTP hydrolysis in the mechanism of microtubule assembly—an experimental study. Proc Natl Acad Sci USA. 1984;81:771–775. doi: 10.1073/pnas.81.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Pantaloni D. Kinetic analysis of guanosine 5'-triphosphate hydrolysis associated with tubulin polymerization. Biochemistry. 1981;20:1918–1924. doi: 10.1021/bi00510a030. [DOI] [PubMed] [Google Scholar]
- Chrétien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J Cell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes CE, Yamamoto A, Kenzie MR, Odde DJ, Gardner MK. Evolving tip structures can explain age-dependent microtubule catastrophe. Curr Biol. 2013;23:1342–1348. doi: 10.1016/j.cub.2013.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elie-Caille C, Severin F, Helenius J, Howard J, Muller DJ, Hyman AA. Straight GDP-tubulin protofilaments form in the presence of taxol. Curr Biol. 2007;17:1765–1770. doi: 10.1016/j.cub.2007.08.063. [DOI] [PubMed] [Google Scholar]
- Fourniol FJ, Sindelar CV, Amigues B, Clare DK, Thomas G, Perderiset M, Francis F, Houdusse A, Moores CA. Template-free 13-protofilament microtubule-MAP assembly visualized at 8 Å resolution. J Cell Biol. 2010;191:463–470. doi: 10.1083/jcb.201007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Kirschner MW. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Charlebois BD, Janosi IM, Howard J, Hunt AJ, Odde DJ. Rapid microtubule self-assembly kinetics. Cell. 2011a;146:582–592. doi: 10.1016/j.cell.2011.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 2011b;147:1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- Horio T, Hotani H. Visualization of the dynamic instability of individual microtubules by dark-field microscopy. Nature. 1986;321:605–607. doi: 10.1038/321605a0. [DOI] [PubMed] [Google Scholar]
- Janosi IM, Chrétien D, Flyvbjerg H. Modeling elastic properties of microtubule tips and walls. Eur Biophys J. 1998;27:501–513. doi: 10.1007/s002490050160. [DOI] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Bieling P, Cope J, Hoenger A, Surrey T. GTPγS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs) Proc Natl Acad Sci USA. 2011;108:3988–3993. doi: 10.1073/pnas.1014758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Cade NI, Bohner G, Gustafsson N, Boutant E, Surrey T. EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr Biol. 2014;24:372–384. doi: 10.1016/j.cub.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Microtubule assembly nucleated by isolated centrosomes. Nature. 1984a;312:232–237. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984b;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Moores CA, Perderiset M, Kappeler C, Kain S, Drummond D, Perkins SJ, Chelly J, Cross R, Houdusse A, Francis F. Distinct roles of doublecortin modulating the microtubule cytoskeleton. EMBO J. 2006;25:4448–4457. doi: 10.1038/sj.emboj.7601335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;9:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the αβ-tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Northrup SH, Erickson HP. Kinetics of protein-protein association explained by Brownian dynamics computer simulation. Proc Natl Acad Sci USA. 1992;89:3338–3342. doi: 10.1073/pnas.89.8.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa F, Asakura S. Thermodynamics of the Polymerization of Protein, London: Academic Press; 1975. [Google Scholar]
- Ozon S, Maucuer A, Sobel A. The stathmin family—molecular and biological characterization of novel mammalian proteins expressed in the nervous system. Eur J Biochem. 1997;248:794–806. doi: 10.1111/j.1432-1033.1997.t01-2-00794.x. [DOI] [PubMed] [Google Scholar]
- Pecqueur L, Duellberg C, Dreier B, Jiang Q, Wang C, Pluckthun A, Surrey T, Gigant B, Knossow M. A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end. Proc Natl Acad Sci USA. 2012;109:12011–12016. doi: 10.1073/pnas.1204129109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prota AE, Bargsten K, Zurwerra D, Field JJ, Diaz JF, Altmann KH, Steinmetz MO. Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science. 2013;339:587–590. doi: 10.1126/science.1230582. [DOI] [PubMed] [Google Scholar]
- Schek HT, Gardner MK, Cheng J, Odde DJ, Hunt AJ. Microtubule assembly dynamics at the nanoscale. Curr Biol. 2007;17:1445–1455. doi: 10.1016/j.cub.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JR, Salmon ED. The structure of microtubule ends during the elongation and shortening phases of dynamic instability examined by negative-stain electron microscopy. J Cell Sci. 1990;96:571–582. doi: 10.1242/jcs.96.4.571. [DOI] [PubMed] [Google Scholar]
- Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- VanBuren V, Cassimeris L, Odde DJ. Mechanochemical model of microtubule structure and self-assembly kinetics. Biophys J. 2005;89:2911–2926. doi: 10.1529/biophysj.105.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voter WA, O'Brien ET, Erickson HP. Dilution-induced disassembly of microtubules: relation to dynamic instability and the GTP cap. Cell Motil Cytoskeleton. 1991;18:55–62. doi: 10.1002/cm.970180106. [DOI] [PubMed] [Google Scholar]
- Walker RA, O'Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP, Salmon ED. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberg RC. Microtubule formation in vitro in solutions containing low calcium concentrations. Science. 1972;177:1104–1105. doi: 10.1126/science.177.4054.1104. [DOI] [PubMed] [Google Scholar]
- Weisenberg RC, Borisy GG, Taylor EW. The colchicine-binding protein of mammalian brain and its relation to microtubules. Biochemistry. 1968;7:4466–4479. doi: 10.1021/bi00852a043. [DOI] [PubMed] [Google Scholar]
- Zanic M, Stear JH, Hyman AA, Howard J. EB1 recognizes the nucleotide state of tubulin in the microtubule lattice. PLoS One. 2009;4:e7585. doi: 10.1371/journal.pone.0007585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M, Widlund PO, Hyman AA, Howard J. Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat Cell Biol. 2013;15:688–693. doi: 10.1038/ncb2744. [DOI] [PubMed] [Google Scholar]


