Endosomal sorting requires consecutive steps of membrane remodeling and fusion in the course of endosomal maturation. Tracing of cargo relative to machinery reveals similar temporal localization of ESCRT and endosomal fusion machinery, which precedes the retromer complex. However, blocking fusion with the vacuole does not impair maturation.
Abstract
Organelles of the endolysosomal system undergo multiple fission and fusion events to combine sorting of selected proteins to the vacuole with endosomal recycling. This sorting requires a consecutive remodeling of the organelle surface in the course of endosomal maturation. Here we dissect the remodeling and fusion machinery on endosomes during the process of endocytosis. We traced selected GFP-tagged endosomal proteins relative to exogenously added fluorescently labeled α-factor on its way from the plasma membrane to the vacuole. Our data reveal that the machinery of endosomal fusion and ESCRT proteins has similar temporal localization on endosomes, whereas they precede the retromer cargo recognition complex. Neither deletion of retromer nor the fusion machinery with the vacuole affects this maturation process, although the kinetics seems to be delayed due to ESCRT deletion. Of importance, in strains lacking the active Rab7-like Ypt7 or the vacuolar SNARE fusion machinery, α-factor still proceeds to late endosomes with the same kinetics. This indicates that endosomal maturation is mainly controlled by the early endosomal fusion and remodeling machinery but not the downstream Rab Ypt7 or the SNARE machinery. Our data thus provide important further understanding of endosomal biogenesis in the context of cargo sorting.
INTRODUCTION
Organelles in the endomembrane system dynamically associate with a variety of proteins to mediate membrane fission and fusion events during protein trafficking. The association of proteins with their particular target membrane depends on the lipid composition, phosphorylation of specific phosphoinositides, and other peripheral and integral membrane proteins. The surface of these organelles is, however, not static but undergoes remodeling to function in membrane tubulation or membrane fusion (Huotari and Helenius, 2011). Within the endocytic pathway, such remodeling processes are particularly dramatic at the early endosome/late endosome transition. Early endosomes contain multiple tubular extensions, start to form intraluminal vesicles (ILVs), and eventually convert into late endosomes during a process termed endosomal maturation.
The machinery of membrane remodeling is closely linked to the Rab GTPases Rab5 at the early endosome and Rab7 at the late endosome. Rabs are switch-like proteins with C-terminal prenyl anchors. In their GDP form, Rabs are kept soluble in the cytosol by binding to the GDP dissociation inhibitor (GDI). Membrane-bound guanine nucleotide exchange factors (GEFs) recruit the Rab-GDP and trigger nucleotide exchange to the active Rab-GTP, which can then bind to effector proteins such as fusion factors or recycling protein complexes (Gerondopoulos et al., 2012; Barr, 2013; Blumer et al., 2013; Cabrera and Ungermann, 2013).
The process of endocytosis is critical for the down-regulation of plasma membrane proteins, which is generally initiated by ligand binding or in response to quality control measures. Membrane proteins are subsequently packaged into endocytic vesicles, which fuse with early endosomes in a Rab5-dependent reaction (Zeigerer et al., 2012). In yeast, this process requires the homologous Vps21 protein, the tethering protein Vac1, and the soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) fusion machinery, including the Pep12 syntaxin-like SNARE (Horazdovsky et al., 1994; Gerrard et al., 2000; Peterson and Emr, 2001; Cabrera et al., 2013). In addition, Vps21-GTP interacts with the hexameric class C core vacuole/endosome tethering (CORVET) complex to promote endosome–endosome fusion (Peplowska et al., 2007; Balderhaar et al., 2013; Cabrera et al., 2013). Early endosomes then mature to late endosomes. In this context, endosomal sorting complexes required for transport (ESCRT) proteins promote the sorting of membrane proteins into ILVs (Henne et al., 2011) and thus profit from multiple endosome–endosome fusion events (Zeigerer et al., 2012), by which additional surface is generated for this process. Consequently, endosomes are converted into more rounded structures. In parallel, cargo receptors that transport lysosomal hydrolase from the Golgi to the endosome release their cargo and are collected by the retromer complex. This complex consists of the cargo recognition complex (CRC), consisting of Vps35-Vps26 and Vps29, and the dimeric sorting nexin (SNX) complex, Vps5-Vps17 (Seaman, 2012). It is believed that recruitment and activation of the Rab7-like Ypt7 then triggers the formation of endosomal tubules (Rojas et al., 2008; Seaman et al., 2009; Balderhaar et al., 2010; Liu et al., 2012). The recruitment of Ypt7 depends on its GEF complex, Mon1-Ccz1 (Nordmann et al., 2010; Cabrera and Ungermann, 2013; Cabrera et al., 2014), which functions similarly in higher eukaryotes (Kinchen and Ravichandran, 2010; Poteryaev et al., 2010; Yousefian et al., 2013; Cui et al., 2014; Singh et al., 2014). It is likely that activation of Ypt7 is connected to the amount of Vps21 on early endosomes, suggesting coordinated turnover from one Rab GTPase to the subsequent one (del Conte-Zerial et al., 2008; Barr, 2013).
Even though earlier studies analyzed the morphology of endosomes during cargo uptake (Prescianotto-Baschong and Riezman, 1998, 2002; Griffith and Reggiori, 2009), the relative function of the machinery involved in endosomal maturation has not been analyzed in detail. We therefore set out to correlate the spatiotemporal distribution of this machinery with actively transported cargo. Our colocalization data and mutant analyses reveal that endosomal protein complexes function in similar time windows on endosomes but do not sense the absence of the final Rab GTPase Ypt7 or the fusion machinery.
RESULTS
Trafficking of α-factor through the endocytic pathway
To determine the relative localization of proteins involved in endosomal tubulation, fission, and fusion relative to incoming and transported cargo and thus monitor endosomal maturation during this process, we used the α-factor receptor Ste2 in yeast. This receptor resides in the plasma membrane and binds α-factor, a small peptide pheromone that is secreted by yeast cells with mating type α (Jenness et al., 1983; Burkholder and Hartwell, 1985; Blumer et al., 1988). Binding of the peptide to the receptor triggers its uptake into endocytic vesicles and subsequent trafficking via endosomes to the lytic vacuole for degradation (Singer and Riezman, 1990; Schandel and Jenness, 1994). The ligand was labeled at a central lysine residue with a linker connected to Cy5/DY647 (Figure 1A) to trace the molecule in the fluorescence microscope without losing its binding capacity toward the receptor (Toshima et al., 2006; Figure 1A). To analyze protein localization relative to incoming cargo, we first cooled cells to 4°C to block endocytosis, followed by addition of labeled α-factor. After removal of unbound ligand, cells were resuspended in medium at 23°C to allow endocytosis and subsequent passage through the endocytic pathway (Figure 1B). When cells were heated to 23°C, the probe was readily taken up and was present in intracellular puncta after 5 min. Further incubation then resulted in labeling of the vacuolar lumen, which became first visible after 15 min (Figure 1C, white arrows). After 40 min, all α-factor was quantitatively localized to the vacuolar lumen (Figure 1C), in agreement with previous studies (Toshima et al., 2006, 2014). Uptake of α-factor required expression of the Ste2 receptor (Figure 1D), which was endocytosed with its bound ligand (Figure 1E), leaving behind the likely unbound receptor at the plasma membrane (Figure 1E). Furthermore, we compared uptake of α-factor relative to another plasma membrane transporter, Mup1, which is endocytosed upon methionine addition (Menant et al., 2006), and observed colocalization of both during passage through the endocytic pathway (Figure 1F, white arrows). The stronger plasma membrane staining of Mup1 is likely due to a continuous uptake of this protein compared with the rapid uptake of Ste2 when α-factor is bound. Our combined data indicate that labeled α-factor migrates with the same kinetics and characteristics as other cargo through the endosomal pathway.
FIGURE 1:
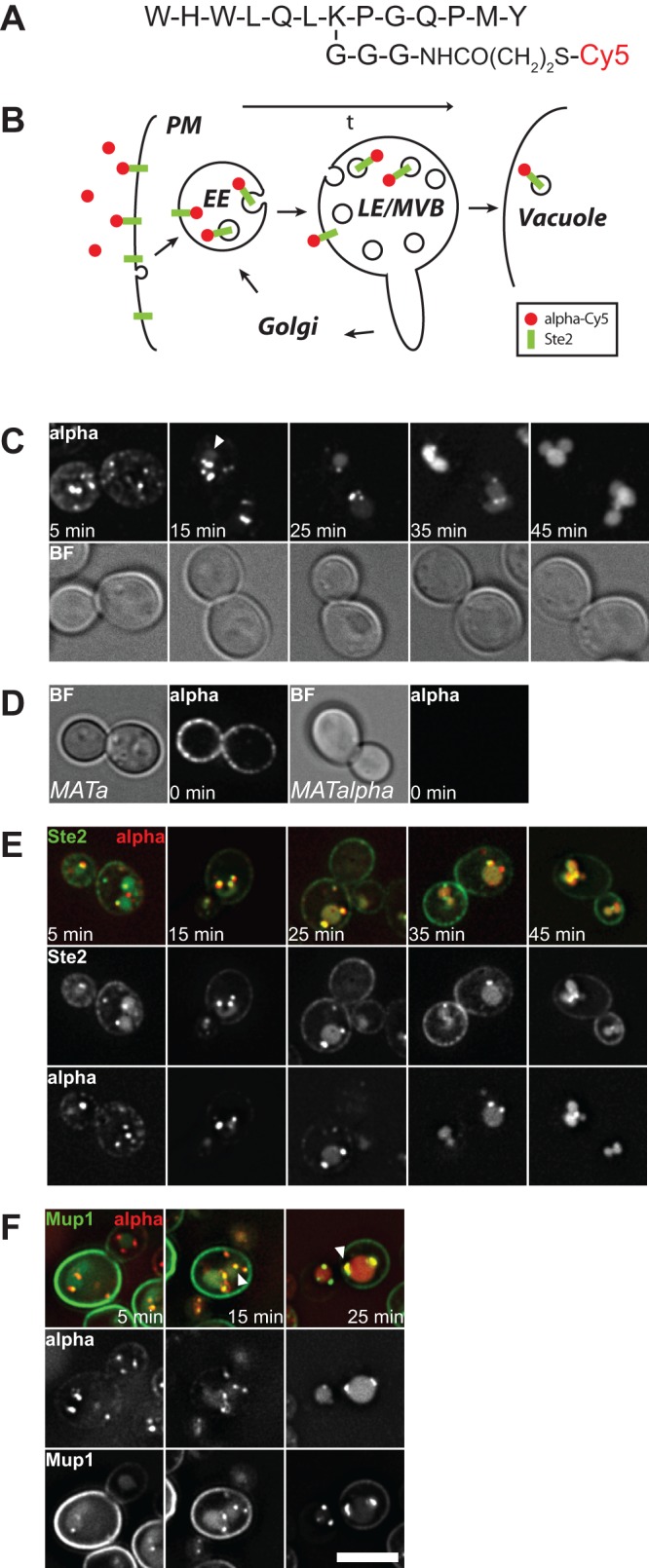
Dynamics of α-factor trafficking through the endocytic pathway. (A) Schematic representation of α-factor peptide sequence including a linker at the central lysine for dye labeling. (B) Model of α-factor and Ste2 trafficking to analyze the endocytic pathway. (C) Wild-type cells were grown in synthetic medium supplemented with amino acids to logarithmic phase, cooled to 4°C to block endocytosis, treated with fluorescent α-factor, washed, and resuspended in medium at 23°C to allow uptake of the probe. Indicated time points refer to the time interval after cells were heated to 23°C. White arrows indicate first appearance of vacuolar Cy5 staining. (D) Uptake of α-factor depends on cell mating type. Cells were treated as in C and analyzed in the microscope directly after washing at 4°C. (E) α-Factor and its receptor Ste2 colocalize during passage through the endocytic pathway. Cells expressing endogenously tagged Ste2-GFP were treated as in C and analyzed in the fluorescence microscope. (F) Mup1 and α-Cy5 are transported through the same endosomes. Cells were grown in synthetic medium without methionine to accumulate endogenously expressed Mup1-3xmCherry at the plasma membrane. Cells were then treated as in C and resuspended in medium containing 1 mM methionine at 23°C to allow endocytosis and trigger Mup1 uptake. White arrows indicate Mup1 and α-Cy5 colocalizing endosomes. BF, bright field; scale bar, 5 μm.
Analysis of the spatiotemporal protein localization during endosomal transport
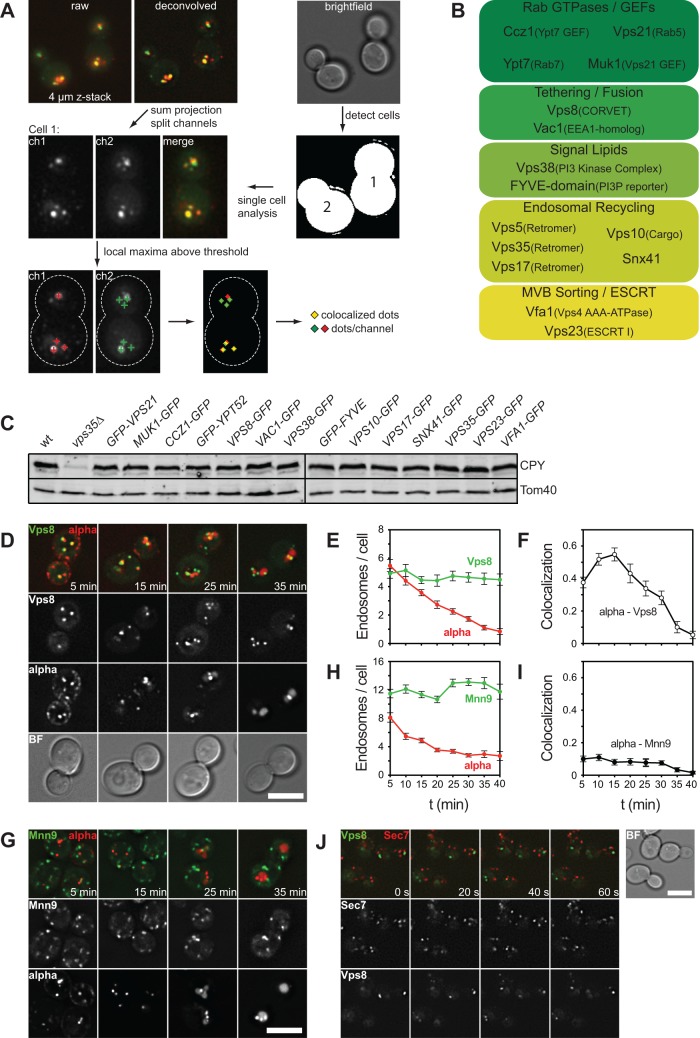
To investigate the spatiotemporal membrane localization of proteins involved in endosomal biogenesis during maturation, we chromosomally tagged one gene at a time with green fluorescent protein (GFP) and followed colocalization with Cy5-labeled α-factor during uptake of the probe. To measure colocalization in a quantitative and unbiased manner, we developed a tool for ImageJ to measure colocalization (Figure 2A; for details, see Materials and Methods). To analyze all endosomes in a cell at a certain time, we took z-stacks of GFP and Cy5 in 5-min intervals after cells had been heated to 23°C and then used our tool to determine the number of colocalizing structures per cell by segmentation of endosomes in each channel. Structures that showed substantial overlap between the channels were scored as “colocalized.” Structures that were >800 nm were removed to exclude vacuolar signals. For each endosomal protein, we counted on average 120 cells/time point to reduce effects due to experimental variations.
FIGURE 2:
Spatiotemporal analysis of protein (co)localization on endosomes. (A) Workflow to measure colocalization of α-factor on endosomes. We acquired and deconvolved 4-μm z-stacks with 400-nm spacing of the fluorescence channels. A bright-field image of the same image section was used to segment single yeast cells. A sum projection of the deconvolved image stack was used to detect local intensity maxima in each channel in every cell to colocalize and quantify the respective dots per cell (see Materials and Methods). (B) Schematic representation of 16 proteins chosen for analysis of the endocytic pathway, categorized by function. (C) All tagged constructs are functional. Trafficking of CPY was monitored by analysis of internal CPY content using antibodies against CPY and Tom40 as loading control on whole-cell extracts. (D) α-Factor uptake assay in cells expressing endogenously tagged VPS8-GFP. Cells were analyzed as in Figure 1C. (E, F) Quantification of α-Cy5 uptake assay shown in D. Endosome number per cell (E) and colocalization of endosomes in the Cy5 and GFP channels (F) were quantified using the ImageJ plug-in described in A. Colocalization represents the fraction of Cy5-labeled α-factor endosomes per cell that are colocalized with the GFP channel or (colocalized endosomes per cell)/(Cy5-positive endosomes per cell) at the respective time points of the assay. (G) α-Factor uptake assay was carried out as in D in cells expressing endogenous Mnn9-GFP. (H, I) Quantification of uptake assay shown in G as in E and F. (J) Analysis of dynamic colocalization of endosome and Golgi markers. Cells expressing endogenous Vps8-GFP and Sec7-mCherry were grown as described for the α-uptake assay and analyzed by time-lapse microscopy in 20-s intervals. Graphs show mean values and SEM. BF, bright field; scale bars, 5 μm.
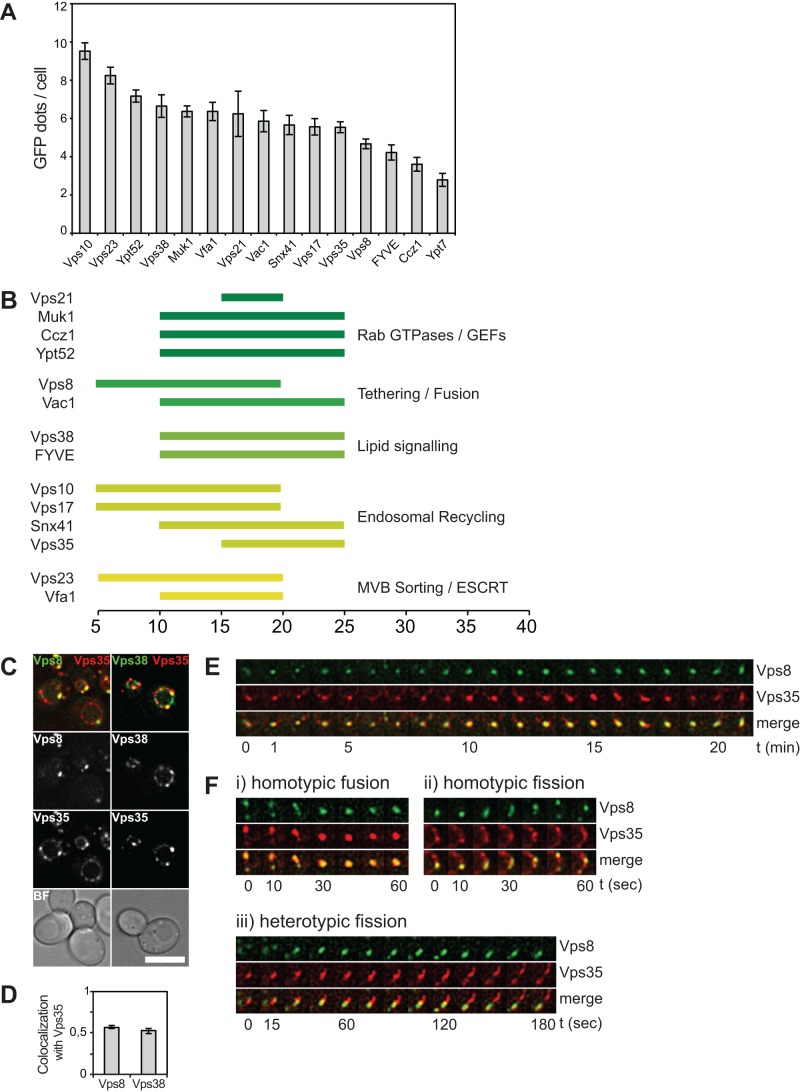
For our analysis of endosomal maturation, we selected representative proteins for specific fusion and fission reactions at endosomes. These included the Rabs Vps21 and Ypt7 and respective GEFs, the interacting tethering proteins CORVET and Vac1, proteins involved in lipid signaling, such as the Vps38 subunit of the phosphoinositol-3-kinase Vps34, a FYVE probe to label phosphoinositide-3-phosphate (PI3P) on endosomes, and subunits of the retromer complex, as well as ESCRT proteins (Figure 2B). Furthermore, we verified that all of the tagged constructs remained functional by quantitative measurement of carboxypeptidase Y (CPY) transport to the vacuole. Any vps mutant results in a block of vacuolar transport and subsequent secretion from the cells (Figure 2C).
Our analysis results in three important pieces of information. First, we monitor the number of endosomes (as marked by the GFP-tagged protein) over time. Second, we observe the time it takes for α-factor to reach the vacuole, observed by decreasing number of α-factor– positive structures at later time points. This may change in certain mutants or during stages of starvation. Finally, we can derive the degree of colocalization of α-factor endosomes with GFP-tagged proteins over time and thus determine the likely activity of the tagged protein relative to the monitored cargo. This may allow us to monitor sequential localization of endosomal proteins.
To test our tool in combination with the α-factor uptake assay, we monitored its colocalization with the endosomal CORVET subunit Vps8 over time (Figure 2, D–F, and Supplemental Movie S1). Vps8-GFP is a reproducible endosomal marker and is found in ∼5 endosomal dots/cell during the entire experiment (Figure 2E). During the course of the experiment, α-factor accumulated in dot-like structures already at 5 min, which we counted as endosomes. This amount decreased over time, in agreement with initial endosomal fusion and subsequent uptake into the vacuole (Figure 2, D–F). Figure 2F shows the degree of overlap between Vps8- and α-factor–positive structures. This graph indicates the peak of colocalization at 15 min, which then steadily decreased as α-factor migrated to the vacuole. The peak is the results of multiple assays and is therefore a reliable measure of endosomal localization during cargo transport.
To control specificity of our assay, we monitored α-factor relative to the Golgi protein Mnn9, which was found in at least 12 dots/cell but did not colocalize significantly with α-factor, unlike Vps8 (Figure 2, G–I). Furthermore, we did not observe any overlap between the Golgi marker Sec7, labeled with mCherry, and Vps8-GFP, even over shorter time periods (Figure 2J).
Early endosomal proteins behave similarly during cargo uptake
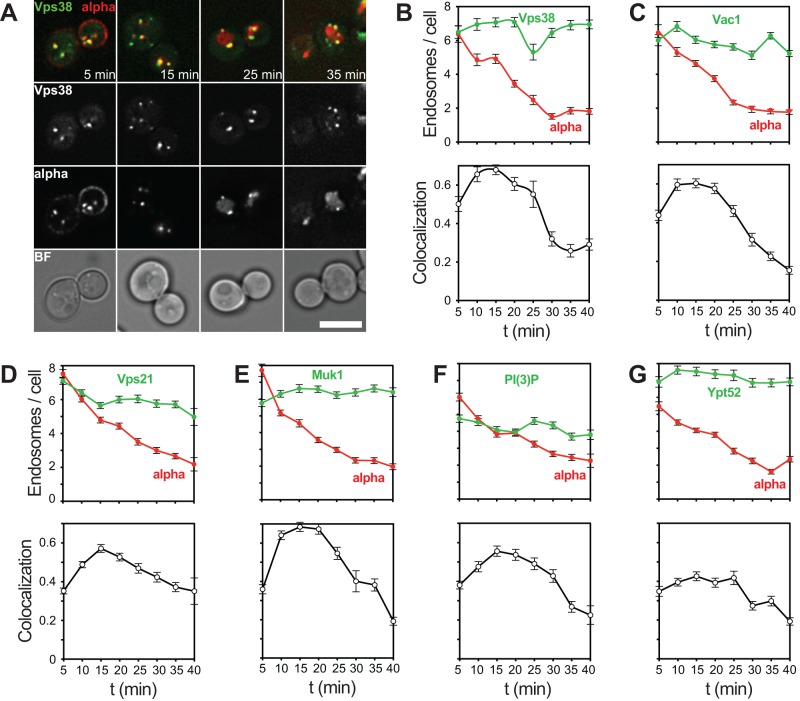
We then began with our analysis of the endosomal pathway by focusing on putative early endosomal proteins. Vps38 is part of the phosphoinositide 3-kinase complex. Like Vps8, Vps38 was present in approximately six endosomes and had maximal colocalization at 15 min with the Cy5-labeled α-factor (Figure 3, A and B). Despite some experimental variation, similar results were obtained for the Vac1 tethering protein (Figure 3C), Vps21 (Figure 3D), its GEF Muk1 (Figure 3E), the GFP-tagged FYVE domain, which monitors PI3P levels in cells (Figure 3F), and the Rab5 homologue Ypt52 (Figure 3G). These data indicate that these endosomal proteins reside on similar endosomal compartments at early stages of endocytic transport.
FIGURE 3:
Analysis of proteins involved in early endosomal fusion and PI3P generation. (A) α-Factor uptake assay in cells expressing endogenously tagged Vps38-GFP. Experiment was carried out as described in Figure 1C (see Materials and Methods). (B) Quantification of uptake assay shown in A. (C–G) Quantification of α-factor uptake assay in cells expressing GFP-tagged Vac1, Muk1, Vps21, Ypt52, and FYVE domain. Scale bars, 5 μm.
CORVET mediates fusion of early endosomes
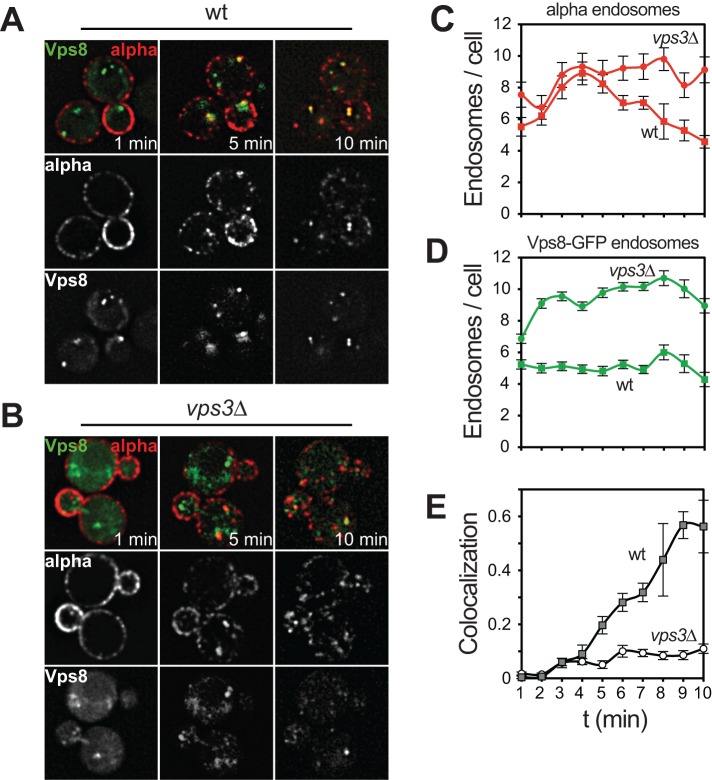
We next asked whether we could dissect the role of early endosomal proteins and obtain insights into early endosomal fusion processes. For this, we monitored the formation of α-factor–positive endosomes directly after endocytosis. After 1 min of the uptake assay, α-factor is completely associated with the plasma membrane, followed by an increase in internal vesicles per cell until 5 min (Figure 4, A and C) and increased colocalization with Vps8 (Figure 4E). Beyond 5 min, these structures coalesce into fewer structures, probably due to fusion of endocytic vesicles to endosomes or endosome–endosome fusion. To test this hypothesis, we asked whether loss of the endosomal CORVET complex would affect this early stage. CORVET has two Rab-specific subunits, Vps3 and Vps8 (Peplowska et al., 2007; Plemel et al., 2011; Epp and Ungermann, 2013). In vps3∆ cells, Vps8 is still associated with the remaining four subunits and can thus be used as an endosomal marker (Markgraf et al., 2009; Ostrowicz et al., 2010). In vps3∆ strains, we observed a similar initial increase in α-factor–positive endosomes as in wild type until 4 min (Figure 4, B and C). However, unlike in wild type, the number of α-factor–positive endosomes remained high in vps3∆ cells (Figure 4C). Moreover, even though the number of Vps8-positive endosomes was higher in vps3∆ cells (Figure 4D), the colocalization of Vps8-GFP and α-factor increased over time in wild-type cells but remained low in vps3∆ cells (Figure 4E). These data indicate that CORVET is required to deliver endocytic cargo to Vps8-positive endosomes, probably by mediating specific fusion reactions.
FIGURE 4:
Analysis of the first 10 min of the endocytic pathway. (A, B) α-Factor uptake assay in cells expressing Vps8-GFP in the wild type (A) or vps3∆ mutant (B). Experiment was carried out as in Figure 1C, but cells were directly spotted on cover slides for microscopy after washing in cold medium without incubation at 23°C. The same image section was analyzed for 10 min. (C–E) Quantification of the experiment in A and B as in Figure 2. Graphs show mean values ± SEM. Scale bars, 5 μm.
Analysis of endosomal recycling during maturation
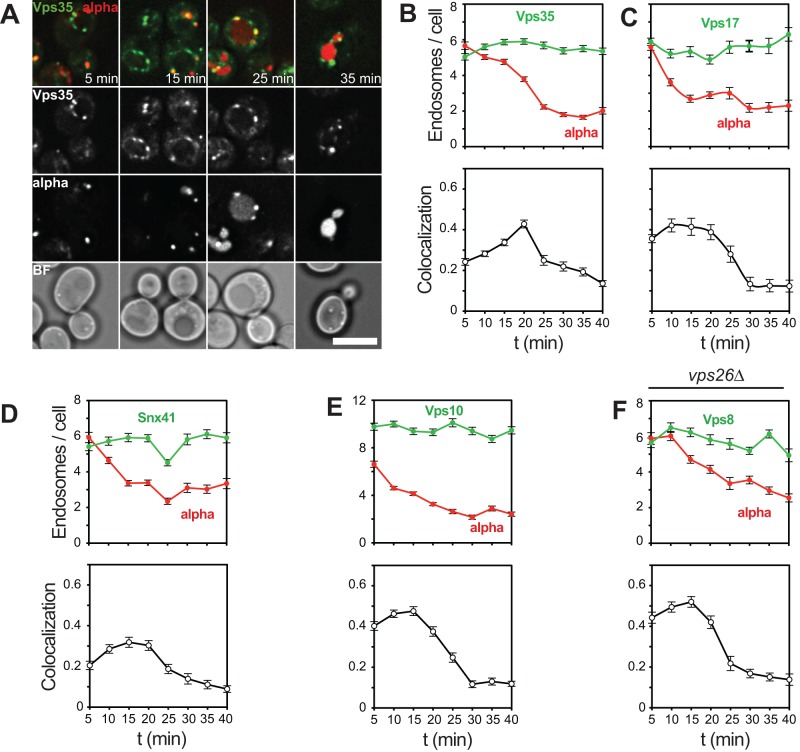
In a next set of experiments, we analyzed proteins involved in endosomal membrane recycling relative to α-factor trafficking to the vacuole. Considering that recycling should follow the initial delivery of other hydrolases to the endosome, such as CPY, we expected a delayed overall localization of these proteins relative to early fusion factors. We thus analyzed Vps35 as a central subunit of the CRC part of the retromer complex. Unlike the previously analyzed fusion factors, Vps35 had a clear colocalization peak that was shifted to 20 min (Figure 5, A and B). In contrast, Vps17 as part of the SNX BAR complex of retromer and Snx41 both peaked at 15 min, indicating that their function may be required earlier (Figure 5, C and D). We next analyzed Vps10, which transports cargo such as CPY from the Golgi to the endosome and is subsequently recycled by the retromer complex (Cooper and Stevens, 1996; Seaman et al., 1997). To reveal the cross-talk between this pathway and endocytosis, we sought the time at which the cargo receptor Vps10 would colocalize with α-factor. Vps10 was found in 12 dots/cell, in agreement with its dual localization on endosomes and the Golgi (Chi et al., 2014). Of importance, Vps10 colocalized early with α-factor, indicating that it is delivered to early endosomes (Figure 5E). Furthermore, Vps10 colocalization then decreased, probably due to constant removal of Vps10 by retromer and subsequent sorting of α-factor to vacuoles (Figure 5E).
FIGURE 5:
Mapping of proteins involved in endosomal recycling. (A) α-Factor uptake assay in cells expressing endogenous Vps35-GFP. Experiment was carried out as in Figure 1C (see Materials and Methods). (B) Quantification of assay shown in A as described in Figure 2. (C–F) Quantification of α-factor uptake assay in cells expressing GFP-tagged Vps17, Snx41, and Vps10 in wild-type cells or Vps8-GFP in vps26∆ cells. (B–F) Mean values ± SEM. Scale bar, 5 μm.
We next asked whether removal of retromer would affect the kinetics of α-factor delivery along the endocytic pathway and therefore traced Vps8 and α-factor in vps26∆ cells. However, deletion of retromer did not affect the overall colocalization pattern, indicating that retromer has no major influence on processes controlling endosomal maturation and fusion (Figure 5F).
ESCRT function begins early in the yeast endocytic pathway
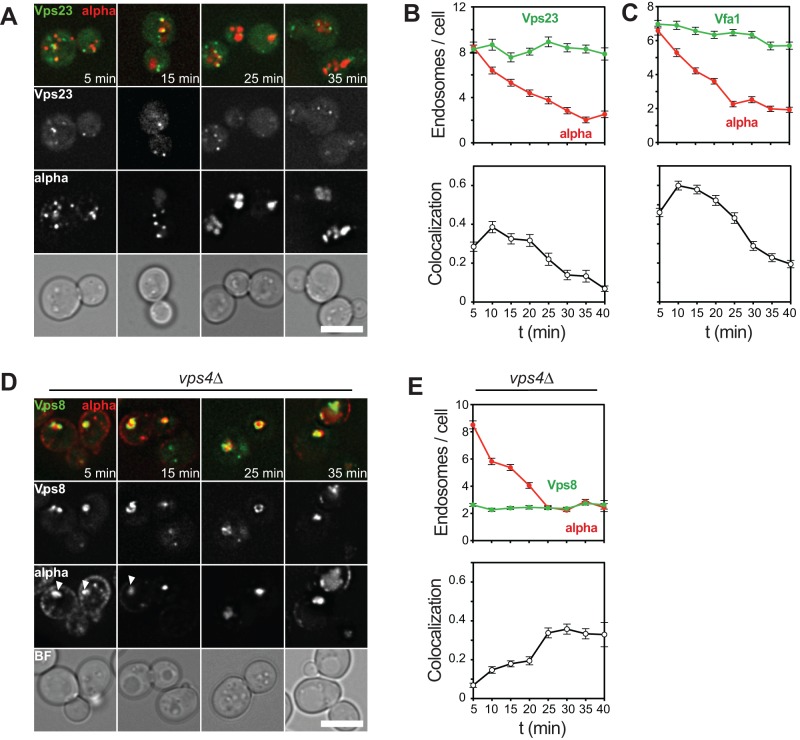
ESCRT proteins function in four distinct complexes and thus promote formation of ILVs (Henne et al., 2011). To analyze ESCRT, we selected one of the few ESCRT subunits that is not affected in function by tagging. Vps23, an ESCRT-I subunit, was monitored relative to α-factor uptake as before (Figure 6A). To our surprise, although in agreement with data from metazoan cells (Raiborg et al., 2002), Vps23 colocalized very early with α-factor and was present in up to 8 endosomal dots/cell (Figure 6, A and B). Considering that the number of α-factor–positive dots decreases over time (Figure 6B), such a function is consistent with an early role of ESCRT proteins. Another ESCRT protein, Vfa1, functions in the context of the Vps4 AAA-type ATPase (Arlt et al., 2011). Vfa1 showed similar behavior in the assay, with a peak at 10 min, indicating that Vps4 also acts early in the pathway (Figure 6C).
FIGURE 6:
Endosomal localization of proteins involved in ESCRT sorting. (A) Uptake assay of labeled α-factor in cells expressing Vps23-GFP. Experiment was carried out as described in Figure 1C. (B) Quantification of α-factor uptake assay shown in A as described in Figure 2. (C) α-Factor uptake assay and quantification of cells expressing Vfa1-GFP. (D, E) Cells expressing GFP-tagged Vps8 in vps4∆ cells were analyzed as in A and B. White arrows indicate class E compartments at early time points. Graphs show mean values ± SEM; scale bars, 5 μm.
To address the role of ESCRTs in endosomal maturation and endocytic trafficking, we deleted VPS4 from our tester strain (Figure 6D). In vps4∆ cells, endosomes collapse into an aggregated structure with lamellae-like morphology, termed the class E compartment (Babst et al., 1997), which seems to form due to increased Vps21 activity (Russell et al., 2012). These structures also accumulate Vps8, which only colocalized with α-factor efficiently at later time points, in strong contrast to the wild-type situation (Figure 6E). However, the class E compartment was already stained with α-factor after 5 min of the assay (Figure 6D, white arrows). We noticed that α-factor localized to the class E compartment even at later time points of the assay (35–40 min), which might indicate delayed transport through the pathway in this mutant.
A block in endosome–vacuole fusion does not affect endosomal maturation
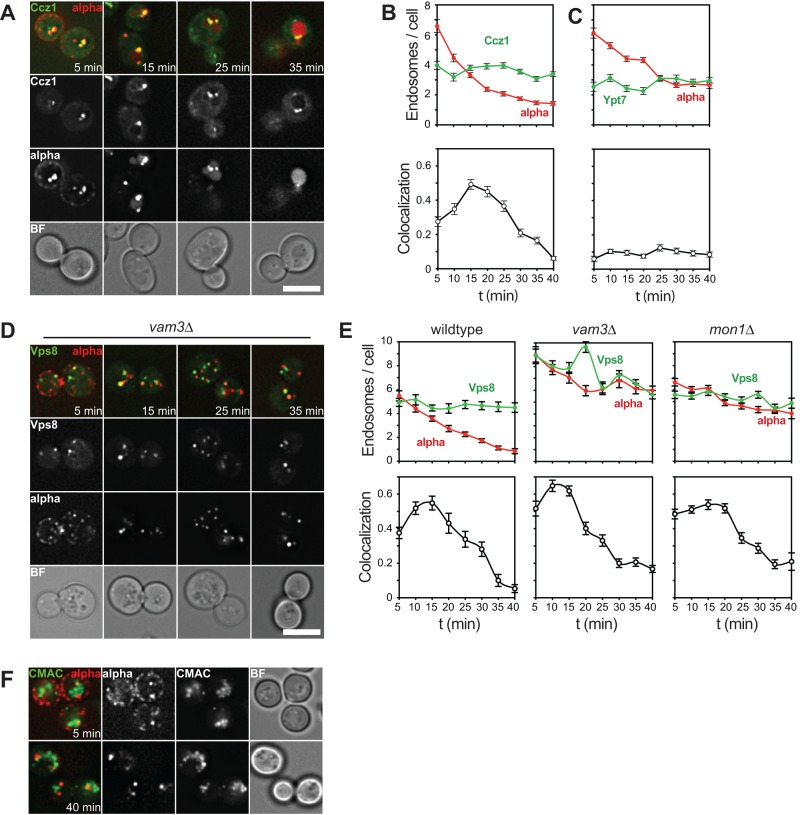
Endosomal maturation concludes with the recruitment of the fusion machinery and the fusion of endosomes with vacuoles. During this process, the Rab7-like Ypt7 protein is activated by the Mon1-Ccz1 GEF complex and can interact with both retromer and the homotypic fusion and vacuole protein sorting (HOPS) tethering complex (Balderhaar and Ungermann, 2013). We thus expected that following the GEF complex or Ypt7 would be indicative of mature late endosomes. When we monitored Ccz1-GFP relative to endocytosed α-factor, we were surprised to observe a very similar localization pattern to that of Vps8, with a peak at 15 min (Figure 7, A and B). However, its substrate, Ypt7, was hardly found on endosomes and localized predominantly to vacuole membranes, which is also reflected by the low colocalization with α-factor over time and the low Ypt7 endosome number per cell (Figure 7C). This suggests that Mon1-Ccz1 might depend on additional activating factors on endosomes to recruit and activate Ypt7.
FIGURE 7:
A block of endosome–vacuole fusion does not affect endosomal maturation. (A) α-Factor uptake assay in cells expressing Ccz1-GFP, a subunit of the Ypt7 GEF complex. (B) The experiment in A was quantified as described in Figure 2. (C) The assay described in A was carried out in cells expressing GFP-tagged Ypt7. (D) Cells expressing Vps8-GFP in a vam3∆ background were analyzed as in A. (E) Quantification of α-factor uptake assays carried out in cells expressing Vps8-GFP in the wild type (left) and vam3∆ (middle) or mon1∆ (right) mutant. (F) Uptake assay as in D with additional CMAC staining of the vacuolar lumen (see Materials and Methods). Graphs show mean values ± SEM. Scale bars, 5 μm.
We then asked whether loss of the machinery required for terminal fusion would result in altered trafficking of α-factor through the endocytic pathway. In the absence of the vacuolar SNARE Vam3, uptake of fluorescent α-factor appeared normal in the first 20 min, as reflected by normal decrease of α-factor endosomes/cell (Figure 7, D and E). However, fusion was efficiently blocked, as indicated by a plateau of 6–7 α-factor endosomes/cell at time points later than 20 min and no α-factor localizing to vacuolar structures even after 40 min of the assay (Figure 7, D and E). When we then analyzed the progression of endosomal maturation by monitoring colocalization with Vps8, we observed a colocalization pattern like that in wild-type cells (Figure 7E). Similar results were obtained in mutants lacking the GEF subunit Mon1 (Figure 7E). Vacuoles are strongly fragmented in the vam3∆ mutant, which could be detected as endosomes with our tool. We thus tested whether there is overlap of vacuoles with α-factor at 40 min of the assay by costaining of vacuoles using 7-amino-4-chloromethylcoumarin (CMAC) but did not detect any (Figure 7F). This confirms the block in endosome–vacuole fusion, in that α-factor localizes to endosomes even at later time points in this mutant. In sum, our data show that a block in fusion has no effect on upstream maturation events in our assay.
Finally, on the basis of our multiple assays, we compared the relative amounts of GFP-positive structures marked by the analyzed proteins, which we interpret as endosomes per cell (Figure 8A). Of interest, proteins that we scored as early, such as the ESCRT-I subunit Vps23, are also present in more structures (8 endosomes/cell), whereas proteins involved in late endosomal fusion, such as Ypt7 or Ccz1, are in comparably few structures (∼3). This indicates that the relative function of proteins in the endocytic pathway approximately correlates with the number of structures decorated by these proteins.
FIGURE 8:
Model of endosomal maturation in yeast. (A) Data from α-factor uptake assays were used to calculate average number of endosomes per cell with respect to analyzed endosomal proteins. Graphs show mean values ± SD. (B) Colocalization pattern of analyzed endosomal proteins. All colocalization patterns obtained with the α-factor uptake assay were normalized, and a threshold was used to show the time points with most colocalization. (C) Localization of endogenous Vps8-GFP or Vps38-GFP with Vps35 tagged with 3xmCherry. (D) Colocalization in C was analyzed as in Figure 2A. Graph shows mean values ± SEM. (E, F) Tracking of single endosomes in cells expressing Vps8-GFP and Vps35-3xmCherry. The z-stacks were acquired every 10–15 s over a time period of 40 min, followed by deconvolution, sum projection, and bleach correction using ImageJ. Endosomes were tracked manually.
Although we saw some variation in the colocalization pattern with endocytosed α-factor, most of the investigated proteins localized in a similar time frame on endosomes (Figure 8B). To this point, our analysis was based on average colocalization values with a time interval of 5 min. We wondered whether we might have missed faster protein localization dynamics on single endosomes. To investigate this, we tagged the early marker Vps8 and the late marker Vps35 with GFP or mCherry, respectively, and followed single endosomes over time with a 10- to 15-s time interval. On the basis of our colocalization assay with α-factor, we would predict that Vps8 and Vps35 would still substantially overlap on similar endosomal structures (Figure 8B). Following both endosomal markers over time, we indeed found high colocalization (∼50%) between these proteins (Figure 8, C and D). As a control, we found a similar amount of colocalization between Vps38 and Vps35, in agreement with our uptake assay (Figure 8, B–D). When we followed single endosomes, we detected endosomes containing both Vps8 and Vps35 over a time of >20 min (Figure 8E). This indicates that 1) both proteins can act simultaneously on the same structures and 2) early and late endosomes are not separate entities with separate protein profiles, at least not at this level of resolution that our analysis can provide. In addition to these long colocalization events on a single endosome, we also detected homotypic fusion and fission of endosomes with both markers and also heterotypic fission events in which Vps35 is separated from a Vps8-Vps35–positive structure (Figure 8, E and F). All of these observations were made on endosomes that were relatively immobile, since they could be traced with a time interval of 10–15 s. However, there was another population of endosomes included in our previous bulk analysis, which was much more mobile. Owing to its mobility, we were not able to follow these single endosomes even with a time interval of 2.5 s.
DISCUSSION
Based on the trafficking of an endocytosed ligand through the endocytic pathway, our work provides insights into the consecutive function of early and late factors involved in endosomal remodeling and fusion. Surprisingly, almost all endosomal factors analyzed colocalize with the endocytosed probe within a similar time window (Figure 8B). The only notable exception is the CRC subcomplex of retromer, which overall seems to come to the endosome later. Our data further show that proteins involved in endosomal fusion and ILV formation such as ESCRTs cannot be separated by our assay. This is in agreement with earlier findings on ESCRT localization in mammalian cells (Sachse et al., 2002). It also fits the idea that endosomal fusion is required to generate enough membrane for the generation of intraluminal vesicles. Finally, we demonstrate that impaired fusion of endosomes with vacuoles does not block the kinetics of α-factor through the endocytic pathway. Thus endosomal maturation does not necessarily require activation of Ypt7 or recognition by the terminal vacuole, which agrees with observation on the Rab7 GEF protein Mon1 and Ccz1 mutants in plants, where endosomal maturation also seemed unperturbed by the mutation (Singh et al., 2014). This is surprising, considering that both Mon1 and Ccz1 localized similarly to Vps8 relative to endocytosed α-factor, suggesting that recruitment of Mon1-Ccz1 does not necessarily coincide with full GEF activity. Potentially, Mon1-Ccz1 is inhibited in its activity until endosomes have reached a degree of maturation that then triggers its activity. Of interest, the Vps35 subunit of retromer is also a Ypt7 effector. According to our working model, activated Mon1-Ccz1 would recruit Ypt7, which then promotes recycling before engaging with the HOPS complex in fusion with the vacuole (Bröcker et al., 2012; Gautreau et al., 2014). How Mon1-Ccz1 activity is controlled and whether activated Ypt7 might affect the previous Rab Vps21 are then major questions that need to be addressed to understand the molecular basis of endosomal maturation. An intriguing observation of our analysis is the consecutive recruitment of retromer subcomplexes to the endosome. The SNX complex colocalized earlier with α-factor than with the CRC complex, as monitored by Vps35 (Figure 5). This situation is similar to observations made in mammalian cells, where the SNX complex localizes to Rab5-positive endosomes and subsequently recruits the CRC together with Rab7 (Rojas et al., 2008; van Weering et al., 2012). This further implies that also yeast retromer can act in separate subcomplexes, which would be in line with the CRC being part of several different recycling complexes (Kama et al., 2007; Strochlic et al., 2007; Harrison et al., 2014).
The analysis of protein localization relative to incoming labeled α-factor has been nicely used to dissect early steps in endocytosis (Toshima et al., 2006). It has also been used to monitor the localization of Vps21 relative to cargo, and these data suggested that the AP-3 pathway also intersects with endocytosis (Toshima et al., 2014). We would like to note that the analysis of class D mutants such as vps21∆, pep12∆, and vac1∆ generally interfere with the functionality of the endocytic pathway, and trafficking thus causes a very general shift in PI3P levels and protein sorting (Cowles et al., 1994; Becherer et al., 1996; Peterson et al., 1999; Tall et al., 1999; Cabrera et al., 2013). In agreement, CORVET mutants strongly impaired endosomal number and cargo trafficking (Figure 4), indicating that our assay is able to reproduce the previous proposal that CORVET acts in endosomal fusion (Cabrera et al., 2013).
One general challenge in our analysis was the consecutive action of endosomal proteins. This was not necessarily expected, as several complexes, such as CORVET and ESCRTs, are expected to coincide with function. On the other hand, retromer was separated at least partially from ESCRT-I and CORVET. However, at the single-endosome level, Vps8 and Vps35 were observed together over long time periods (Figure 8E). We also saw fusion and fission dynamics on single endosomes, which could not be further investigated. A next improvement of this analysis will therefore be single-endosome tracing, which has been hampered by the high mobility of a subpopulation of endosomes. Single-point analysis has been successfully used to investigate Golgi maturation and trans-Golgi function (Losev et al., 2006; Matsuura-Tokita et al., 2006), trans-Golgi dynamics (Daboussi et al., 2012), and early steps of endocytosis (Kaksonen et al., 2003; Toshima et al., 2006; Kukulski et al., 2012). Of note, these processes occur within short (1–5 min) time periods, whereas endosomal transport, including endosomal maturation, requires ∼15–20 min in yeast. Our data provide a starting point to also extend this analysis to substeps of endosomal maturation.
In summary, our colocalization analysis of several endosomal proteins involved in membrane remodeling and fusion highlights the interconnection between these proteins in endosomal trafficking and the independence of endosomal maturation from Rab7/Ypt7 activation.
MATERIALS AND METHODS
Yeast strains and plasmids
Genetic manipulation in the yeast Saccharomyces cerevisiae was carried out by homologous recombination of PCR-amplified cassettes as described (Janke et al., 2004). Yeast strains are listed in Table 1. The mating type of yeast strain SEY6210 was changed to MATa by insertion of pSC11, followed by looping out of the plasmid on 5-fluoroorotic acid plates, resulting in strain CUY7840. Plasmid pCU3475 (pRS406 NOP1pr-GFP-FYVE) was integrated into strain CUY7840 to express a GFP-FYVE domain from human EEA1 under control of the weak NOP1 promoter.
TABLE 1:
Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| SEY6210 | MATalpha leu2-3112 ura3-52 his3-∆200 trp-∆901 lys2-801 suc2-∆9 | Robinson et al. (1988) |
| CUY7840 | MATa leu2-3112 ura3-52 his3-∆200 trp-∆901 lys2-801 suc2-∆9 | This study |
| CUY7962 | CUY7840, MUP1::3xmCherry-hphNT1 | This study |
| CUY8119 | CUY7840, VAC1::GFP-TRP1 | This study |
| CUY8120 | CUY7840, YPT7::TRP1-PHO5pr-GFP | This study |
| CUY8121 | CUY7840, VPS35::GFP-TRP1 | This study |
| CUY8123 | CUY7840, MNN9::GFP-TRP1 | This study |
| CUY8124 | CUY7840, VPS17::GFP-HIS3 | This study |
| CUY8125 | CUY7840, SNX41::GFP-TRP1 | This study |
| CUY8127 | CUY7840, VPS23::GFP-TRP1 | This study |
| CUY8132 | CUY7840, VPS8::GFP-TRP1 | This study |
| CUY8133 | CUY7840, VFA1::GFP-TRP1 | This study |
| CUY8134 | CUY7840, CCZ1::GFP-HIS3 | This study |
| CUY8511 | CUY7840, pRS406 NOP1pr-GFP-FYVE(hEEA1)::URA3 | This study |
| CUY8512 | CUY7840, YPT52::URA3-PHO5pr-GFP | This study |
| CUY8520 | CUY7840, VPS10::GFP-TRP1 | This study |
| CUY8657 | CUY7840, STE2::GFP-TRP1 | This study |
| CUY8776 | CUY7840, VPS21::URA3-PHO5pr-GFP | This study |
| CUY8811 | CUY7840, VPS35::3xmCherry-HIS3 VPS8::GFP-TRP1 | This study |
| CUY8812 | CUY7840, VPS35::GFP-hphNT1 VPS8::3xmCherry-HIS3 | This study |
| CUY8813 | CUY7840, VPS38::GFP-TRP1 | This study |
| CUY8814 | CUY7840, MUK1::GFP-TRP1 | This study |
| CUY8835 | CUY7840, VPS35::GFP-hphNT1 vam3∆::kanMX6 | This study |
| CUY8837 | CUY7840, VPS8::GFP-TRP1 vam3∆::kanMX6 | This study |
| CUY8845 | CUY7840, VPS8::GFP-TRP1 vps3∆::natNT2 | This study |
| CUY9205 | CUY7840, SEC7::GFP-TRP1 | This study |
| CUY9210 | CUY7840, VPS8::GFP-TRP1 SEC7::mCherry-hphNT1 | This study |
| CUY9337 | CUY7840, VPS38::GFP-TRP1 vps3∆::hphNT1 | This study |
| CUY9410 | CUY7840, VPS38::GFP-TRP1 vam3∆::kanMX6 | This study |
| CUY9606 | CUY7840, VPS8::GFP-TRP1 vps26∆::hphNT1 | This study |
| CUY9639 | CUY7840, VPS8::GFP-TRP1 mon1∆::LEU2 | This study |
| CUY9655 | CUY7840, vps4∆::TRP1 VPS8::GFP-HIS3 | This study |
| CUY9673 | CUY7840, SLA1::GFP-TRP1 | This study |
| CUY9778 | CUY7840, VPS35::3xmCherry-HIS3 VPS38::GFP-TRP1 | This study |
Fluorescence labeling of α-factor and uptake assays
The α-factor peptide, including a propionyl-G3 linker at the central lysine residue (LifeTein, Hillsborough, NJ), was labeled with the Cy5 derivative maleimide-DY647 (Dyomics, Jena, Germany) in HBS buffer (500 mM NaCl, 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid/NaOH, 0.1 mM EDTA, pH 8) for 3 h at room temperature, followed by purification via HPLC on a C18 reversed-phase column and analysis by mass spectrometry.
For uptake assays, cells were grown in synthetic medium supplemented with amino acids and 2% glucose at 30°C for 16–20 h to logarithmic phase and an OD600 of 0.6–0.8. The cells were then washed with cold medium and kept on ice for 15 min, followed by application of 2.5 μM labeled α-factor for 15 min on ice. The cells were washed extensively with cold medium and then resuspended in medium at 23°C, which marked the starting point of the assay. The cultures were incubated at 23°C on a shaker, and different aliquots of cells were imaged at the respective time points up to 40 min after heating the cells. To analyze early steps of the endocytic pathway, cells were prepared as described and then directly applied without resuspension in warm medium to a glass slide at 23°C. To analyze Mup1-GFP and α-factor trafficking simultaneously, cells were grown in synthetic medium without methionine as described. After addition of fluorescent α-factor for 15 min, cells were washed with cold synthetic medium with 1 mM methionine and then resuspended in the same medium at 23°C to allow endocytosis.
Fluorescence microscopy
The vacuolar membrane was stained by adding 30 μM FM4-64 for 30 min, followed by washing and incubation in medium without dye for 1 h as described (Vida and Emr, 1995). Staining of vacuoles was done by the addition of 0.1 CMAC for 10 min at 30°C and subsequent washing with medium. Cells were imaged on an Olympus IX-71 inverted microscope equipped with 100× NA 1.49 and 60× NA 1.40 objectives, a sCMOS camera (PCO, Kelheim, Germany), an InsightSSI illumination system, 4′,6-diamidino-2-phenylindole, GFP, mCherry, and Cy5 filters, and SoftWoRx software (Applied Precision, Issaquah, WA). We used 4-μm z-stacks with 400-nm spacing for constrained-iterative deconvolution (SoftWoRx) and quantification.
Quantification of colocalization in yeast cells
Quantification of colocalization was performed in ImageJ (National Institutes of Health, Bethesda, MD) by a self-written graphical user interface based on ImageJ built-in routines. Briefly, single cells were segmented from bright-field images, followed by detection of endosome signals on sum projections of deconvolved image stacks. After removal of background signals, a binary image of the endosome signals was obtained by thresholding all pixels within the cell mask with the MaxEntropy threshold (Kapur et al., 1985). Signals smaller than one-third of single-endosome signals, as well as signals >800 nm, were removed from the binary mask to remove noisy background signals or large background signals from the vacuole. Afterward, signal detection and counting were conducted by calculating local maxima for each channel within the binary endosome mask (Figure 2). The output was an image containing the local maxima, which were then convolved with an endosome kernel (Airy disk of the reporter channel) to obtain a binary representation of the two channels. After logical conjunction of both representations, which results in the overlapping area, the number of colocalizing endosomes was obtained by counting the number of particles with an area >50% of the reporter Airy disk.
Supplementary Material
Acknowledgments
We thank Frederik Sündermann, Markus Babst, and Lois Weisman for helpful advice and Angela Perz for expert technical support. This work was funded by the Sonderforschungsbereich/SFB 944, Projects P8 (J.P.) and P11 (C.U.). C.U. received additional support from the Hans-Mühlenhoff Foundation.
Abbreviations used:
- BF
bright field
- CMAC
7-amino-4-chloromethylcoumarin
- CORVET
class C core vacuole/endosome tethering complex
- CPY
carboxypeptidase Y
- CRC
cargo recognition complex
- ESCRT
endosomal sorting complexes required for transport
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- HOPS
homotypic fusion and vacuole protein sorting
- ILV
intraluminal vesicle
- PI3P
phosphoinositide-3-phosphate
- SNARE
N-ethylmaleimide–sensitive factor attachment protein receptor
- SNX
sorting nexin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-08-1318) on February 5, 2015.
REFERENCES
- Arlt H, Perz A, Ungermann C. An overexpression screen in Saccharomyces cerevisiae identifies novel genes that affect endocytic protein trafficking. Traffic. 2011;12:1592–1603. doi: 10.1111/j.1600-0854.2011.01252.x. [DOI] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar HJK, Arlt H, Ostrowicz CW, Bröcker C, Sündermann F, Brandt R, Babst M, Ungermann C. The Rab GTPase Ypt7 is linked to retromer-mediated receptor recycling and fusion at the yeast late endosome. J Cell Sci. 2010;123:4085–4094. doi: 10.1242/jcs.071977. [DOI] [PubMed] [Google Scholar]
- Balderhaar HJK, Lachmann J, Yavavli E, Bröcker C, Lürick A, Ungermann C. The CORVET complex promotes tethering and fusion of Rab5/Vps21-positive membranes. Proc Natl Acad Sci USA. 2013;110:3823–3828. doi: 10.1073/pnas.1221785110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar HJK, Ungermann C. CORVET and HOPS tethering complexes—coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- Barr FA. Rab GTPases and membrane identity: causal or inconsequential. J Cell Biol. 2013;202:191–199. doi: 10.1083/jcb.201306010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer J, Rey J, Dehmelt L, Mazel T, Wu YW, Bastiaens P, Goody RS, Itzen A. RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol. 2013;200:287–300. doi: 10.1083/jcb.201209113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer KJ, Reneke JE, Thorner J. The STE2 gene product is the ligand-binding component of the alpha-factor receptor of Saccharomyces cerevisiae. J Biol Chem. 1988;263:10836–10842. [PubMed] [Google Scholar]
- Bröcker C, Kuhlee A, Gatsogiannis C, Kleine Balderhaar HJ, Hönscher C, Engelbrecht-Vandré S, Ungermann C, Raunser S. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc Natl Acad Sci USA. 2012;109:1991–1996. doi: 10.1073/pnas.1117797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder AC, Hartwell LH. The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 1985;13:8463–8475. doi: 10.1093/nar/13.23.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M, Arlt H, Epp N, Lachmann J, Griffith J, Perz A, Reggiori F, Ungermann C. Functional separation of endosomal fusion factors and the class C core vacuole/endosome tethering (CORVET) complex in endosome biogenesis. J Biol Chem. 2013;288:5166–5175. doi: 10.1074/jbc.M112.431536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M, Nordmann M, Perz A, Schmedt D, Gerondopoulos A, Barr F, Piehler J, Engelbrecht-Vandré S, Ungermann C. The Mon1-Ccz1 GEF activates the Rab7 GTPase Ypt7 via a longin-fold-Rab interface and association with PI3P-positive membranes. J Cell Sci. 2014;127:1043–1051. doi: 10.1242/jcs.140921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M, Ungermann C. Guanine nucleotide exchange factors (GEFs) have a critical but not exclusive role in organelle localization of Rab GTPases. J Biol Chem. 2013;288:28704–28712. doi: 10.1074/jbc.M113.488213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi RJ, Liu J, West M, Wang J, Odorizzi G, Burd CG. Fission of SNX-BAR-coated endosomal retrograde transport carriers is promoted by the dynamin-related protein Vps1. J Cell Biol. 2014;202:527. doi: 10.1083/jcb.201309084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Stevens T. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Emr SD, Horazdovsky BF. Mutations in the VPS45 gene, a SEC1 homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J Cell Sci. 1994;107:3449–3459. doi: 10.1242/jcs.107.12.3449. [DOI] [PubMed] [Google Scholar]
- Cui Y, Zhao Q, Gao C, Ding Y, Zeng Y, Ueda T, Nakano A, Jiang L. Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell. 2014;26:2080–2097. doi: 10.1105/tpc.114.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi L, Costaguta G, Payne GS. Phosphoinositide-mediated clathrin adaptor progression at the trans-Golgi network. Nat Cell Biol. 2012;14:239–248. doi: 10.1038/ncb2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Conte-Zerial P, Brusch L, Rink JC, Collinet C, Kalaidzidis Y, Zerial M, Deutsch A. Membrane identity and GTPase cascades regulated by toggle and cut-out switches. Mol Syst Biol. 2008;4:206. doi: 10.1038/msb.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp N, Ungermann C. The N-terminal domains of Vps3 and Vps8 are critical for localization and function of the CORVET tethering complex on endosomes. PLoS One. 2013;8:e67307. doi: 10.1371/journal.pone.0067307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau A, Oguievetskaia K, Ungermann C. Function and regulation of the endosomal fusion and fission machineries. Cold Spring Harb Perspect Biol. 2014;6:a016832. doi: 10.1101/cshperspect.a016832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondopoulos A, Langemeyer L, Liang JR, Linford A, Barr FA. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol. 2012;22:2135–2139. doi: 10.1016/j.cub.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard S, Bryant N, Stevens T. VPS21 controls entry of endocytosed and biosynthetic proteins into the yeast prevacuolar compartment. Mol Biol Cell. 2000;11:613–626. doi: 10.1091/mbc.11.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J, Reggiori F. Ultrastructural analysis of nanogold-labeled endocytic compartments of yeast Saccharomyces cerevisiae using a cryosectioning procedure. J Histochem Cytochem. 2009;57:801–809. doi: 10.1369/jhc.2009.952952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MS, Hung C-S, Liu T-T, Christiano R, Walther TC, Burd CG. A mechanism for retromer endosomal coat complex assembly with cargo. Proc Natl Acad Sci USA. 2014;111:267–272. doi: 10.1073/pnas.1316482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Horazdovsky B, Busch G, Emr S. VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J. 1994;13:1297–1309. doi: 10.1002/j.1460-2075.1994.tb06382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera M, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jenness DD, Burkholder AC, Hartwell LH. Binding of alpha-factor pheromone to yeast a cells: chemical and genetic evidence for an alpha-factor receptor. Cell. 1983;35:521–529. doi: 10.1016/0092-8674(83)90186-1. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin D. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kama R, Robinson M, Gerst JE. Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast. Mol Cell Biol. 2007;27:605–621. doi: 10.1128/MCB.00699-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur JN, Sahoo PK, Wong A. A new method for gray-level picture thresholding using the entropy of the histogram. Comput Vis Graph Image Process. 1985;29:273–285. [Google Scholar]
- Kinchen JM, Ravichandran KS. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 2010;464:778–782. doi: 10.1038/nature08853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Kaksonen M, Briggs JAG. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell. 2012;150:508–520. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- Liu T-T, Gomez TS, Sackey BK, Billadeau DD, Burd CG. Rab GTPase regulation of retromer-mediated cargo export during endosome maturation. Mol Biol Cell. 2012;23:2505–2515. doi: 10.1091/mbc.E11-11-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losev E, Reinke C, Jellen J, Strongin D, Bevis B, Glick B. Golgi maturation visualized in living yeast. Nature. 2006;441:1002–1006. doi: 10.1038/nature04717. [DOI] [PubMed] [Google Scholar]
- Markgraf DF, Ahnert F, Arlt H, Mari M, Peplowska K, Epp N, Griffith J, Reggiori F, Ungermann C. The CORVET subunit Vps8 cooperates with the Rab5 homolog Vps21 to induce clustering of late endosomal compartments. Mol Biol Cell. 2009;20:5276–5289. doi: 10.1091/mbc.E09-06-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura-Tokita K, Takeuchi M, Ichihara A, Mikuriya K, Nakano A. Live imaging of yeast Golgi cisternal maturation. Nature. 2006;441:1007–1010. doi: 10.1038/nature04737. [DOI] [PubMed] [Google Scholar]
- Menant A, Barbey R, Thomas D. Substrate-mediated remodeling of methionine transport by multiple ubiquitin-dependent mechanisms in yeast cells. EMBO J. 2006;25:4436–4447. doi: 10.1038/sj.emboj.7601330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann M, Cabrera M, Perz A, Bröcker C, Ostrowicz CW, Engelbrecht-Vandré S, Ungermann C. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol. 2010;20:1654–1659. doi: 10.1016/j.cub.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Ostrowicz CW, Bröcker C, Ahnert F, Nordmann M, Lachmann J, Peplowska K, Perz A, Auffarth K, Engelbrecht-Vandré S, Ungermann C. Defined subunit arrangement and rab interactions are required for functionality of the HOPS tethering complex. Traffic. 2010;11:1334–1346. doi: 10.1111/j.1600-0854.2010.01097.x. [DOI] [PubMed] [Google Scholar]
- Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell. 2007;12:739–750. doi: 10.1016/j.devcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Peterson MR, Burd CG, Emr SD. Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45p-dependent vesicle docking/fusion at the endosome. Curr Biol. 1999;9:159–162. doi: 10.1016/s0960-9822(99)80071-2. [DOI] [PubMed] [Google Scholar]
- Peterson MR, Emr SD. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic. 2001;2:476–486. doi: 10.1034/j.1600-0854.2001.20705.x. [DOI] [PubMed] [Google Scholar]
- Plemel RL, Lobingier BT, Brett CL, Angers CG, Nickerson DP, Paulsel A, Sprague D, Merz AJ. Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol Biol Cell. 2011;22:1353–1363. doi: 10.1091/mbc.E10-03-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141:497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Prescianotto-Baschong C, Riezman H. Morphology of the yeast endocytic pathway. Mol Biol Cell. 1998;9:173–189. doi: 10.1091/mbc.9.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescianotto-Baschong C, Riezman H. Ordering of compartments in the yeast endocytic pathway. Traffic. 2002;3:37–49. doi: 10.1034/j.1600-0854.2002.30106.x. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Biol Cell. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R, van Vlijmen T, Mardones GA, Prabhu Y, Rojas AL, Mohammed S, Heck AJ, Raposo G, van der Sluijs P, Bonifacino JS. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol. 2008;183:513–526. doi: 10.1083/jcb.200804048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell MRG, Shideler T, Nickerson DP, West M, Odorizzi G. Class E compartments form in response to ESCRT dysfunction in yeast due to hyperactivity of the Vps21 Rab GTPase. J Cell Sci. 2012;125:5208–5220. doi: 10.1242/jcs.111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse M, Urbé S, Oorschot V, Strous GJ, Klumperman J. Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol Biol Cell. 2002;13:1313–1328. doi: 10.1091/mbc.01-10-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schandel KA, Jenness DD. Direct evidence for ligand-induced internalization of the yeast alpha-factor pheromone receptor. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ. The retromer complex—endosomal protein recycling and beyond. J Cell Sci. 2012;125:4693–4702. doi: 10.1242/jcs.103440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, Harbour ME, Tattersall D, Read E, Bright N. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J Cell Sci. 2009;122:2371–2382. doi: 10.1242/jcs.048686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M, Marcusson E, Cereghino J, Emr S. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B, Riezman H. Detection of an intermediate compartment involved in transport of alpha-factor from the plasma membrane to the vacuole in yeast. J Cell Biol. 1990;110:1911–1922. doi: 10.1083/jcb.110.6.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Krüger F, Beckmann H, Brumm S, Vermeer JEM, Munnik T, Mayer U, Stierhof Y-D, Grefen C, Schumacher K, et al. Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr Biol. 2014;24:1383–1389. doi: 10.1016/j.cub.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Strochlic TI, Setty TG, Sitaram A, Burd CG. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J Cell Biol. 2007;177:115–125. doi: 10.1083/jcb.200609161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall G, Hama H, DeWald D, Horazdovsky B. The phosphatidylinositol 3-phosphate binding protein Vac1p interacts with a Rab GTPase and a Sec1p homologue to facilitate vesicle-mediated vacuolar protein sorting. Mol Biol Cell. 1999;10:1873–1889. doi: 10.1091/mbc.10.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima JY, Nishinoaki S, Sato Y, Yamamoto W, Furukawa D, Siekhaus DE, Sawaguchi A, Toshima J. Bifurcation of the endocytic pathway intoRab5-dependent and -independent transportto the vacuole. Nat Commun. 2014;5:1–11. doi: 10.1038/ncomms4498. [DOI] [PubMed] [Google Scholar]
- Toshima JY, Toshima J, Kaksonen M, Martin AC, King DS, Drubin DG. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc Natl Acad Sci USA. 2006;103:5793–5798. doi: 10.1073/pnas.0601042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weering JRT, Verkade P, Cullen PJ. SNX-BAR-mediated endosome tubulation is co-ordinated with endosome maturation. Traffic. 2012;13:94–107. doi: 10.1111/j.1600-0854.2011.01297.x. [DOI] [PubMed] [Google Scholar]
- Vida T, Emr S. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefian J, Troost T, Grawe F, Sasamura T, Fortini M, Klein T. Dmon1 controls recruitment of Rab7 to maturing endosomes in Drosophila. J Cell Sci. 2013;126:1583–1594. doi: 10.1242/jcs.114934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigerer A, Gilleron J, Bogorad RL, Marsico G, Nonaka H, Seifert S, Epstein-Barash H, Kuchimanchi S, Peng CG, Ruda VM, et al. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature. 2012;485:465–470. doi: 10.1038/nature11133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.