Abstract
Normal placentation relies on an efficient maternal adaptation to pregnancy. Within the decidua, natural killer (NK) cells and dendritic cells (DC) have a critical role in modulating angiogenesis and decidualization associated with pregnancy. However, the contribution of these immune cells to the placentation process and subsequently fetal development remains largely elusive. Using two different mouse models, we here show that optimal placentation and fetal development is sensitive to disturbances in NK cell relative abundance at the fetal–maternal interface. Depletion of NK cells during early gestation compromises the placentation process by causing alteration in placental function and structure. Embryos derived from NK-depleted dams suffer from intrauterine growth restriction (IUGR), a phenomenon that continued to be evident in the offspring on post-natal day 4. Further, we demonstrate that IUGR was accompanied by an overall reduction of global DNA methylation levels and epigenetic changes in the methylation of specific hepatic gene promoters. Thus, temporary changes within the NK cell pool during early gestation influence placental development and function, subsequently affecting hepatic gene methylation and fetal metabolism.
Maternal adaptations to pregnancy are critical during mammalian development. Cumulative evidence suggests that immune-competent cells in the developing decidua are important for placental development and embryo survival. For instance, the cross-talk between natural killer (NK) cells and dendritic cells (DC) during early pregnancy has emerged as a key regulator of angiogenesis and decidualization,1, 2 both of which are critical events for normal placentation. In this context, we have shown that NK cells prevent the development of alloimmunity during normal pregnancy by modulating the immune-regulatory functions of DC during early gestation.2, 3 Furthermore, unbalanced relative NK cell abundance (i.e., upon in vivo NK cell depletion following fms-like tyrosine kinase 3 ligand (FL)-mediated DC expansion) upregulated the expression of anti-angiogenic growth factors, suggesting that NK cells are required for early vascular responses associated with pregnancy. Indeed, NK cells regulate the remodeling of maternal spiral arteries, transforming them from low capacity high resistance into high capacity low resistance vessels.4 As for DC, these cells not only influence maternal immune tolerance to fetal alloantigens,3 but also coordinate pregnancy-associated vascular adaptations including vessel permeability and blood flow to the implantation site via the CXCL12/CXCR4 pathway.1, 5
The placenta is the key organ supporting the survival and growth of the fetus during gestation. It constitutes an interface between the maternal and fetal circulations, allowing nutrient uptake, waste elimination and metabolic/gas exchange.6 In mice, placental development begins early in the blastocyst (embryonic day E3.5) and continues to show dramatic changes throughout gestation. The mouse placenta consists of three layers: the labyrinth (comprising highly branched villi that maximize nutrient transport), the junction zone (containing trophoblast giant cells and spongiotrophoblast cells that invade the decidua and maternal vessels) and the maternal decidua (including immune and decidual cells and the maternal vasculature).7 Inadequate differentiation of these layers leads to detrimental changes in placental architecture, which affect the supply of nutrients or oxygen to the fetus. Faulty placental development has been implicated in the pathophysiology of several human pregnancy disorders, including preeclampsia, intrauterine growth restriction (IUGR) and preterm labor.7 In this context, it is likely that the fetus adapts to the pathological situation by modifying its energy metabolism in order to sustain development and continue pregnancy.
Changes in fetal metabolism are thought to occur via epigenetic modification of mitochondrial gene expression patterns, causing changes in mitochondrial number or function.8, 9 Indeed, animal models of IUGR have shown a reduction in mitochondrial DNA (mtDNA) content, accompanied by reduced expression of mtDNA encoded genes and epigenetic changes in fetal growth factors (e.g., insulin-like growth factor I (IgfI) and IgfII). In addition, many studies have suggested a role for nuclear receptors in the observed fetal metabolic adaptations toward adverse conditions in utero. These include receptors and associated coactivators, such as the peroxisome proliferator-activated receptor alpha (Pparα), peroxisome proliferator-activated receptor gamma coactivator 1 alpha (Pgc1α), hepatocyte nuclear factor 4 alpha (Hnf4α) and the glucocorticoid receptor (Nr3c1).8 During adaptation to restricted fetal development, modifications to nuclear receptors act to optimize growth and differentiation. In a process called fetal programming, these modifications can persist through adulthood possibly contributing to the observed increase in late-onset disorders at an adult age.10, 11
As the cross-talk between NK cells and DC influences both decidualization and angiogenesis, in this work we analyzed how in vivo manipulation of relative DC–NK cell abundance within the decidua impacts the placentation process and subsequently fetal development. We show that although manipulation of the relative abundance of DC and NK cells in early gestation does not seem to compromise fetal survival, both placental function and structure are specifically altered compared with control mice. Using a candidate gene approach, we demonstrate that these structural changes in the placenta are associated with epigenetic changes in the methylation of specific hepatic gene promoters in pups principally after NK cell depletion. The gene-specific changes were further accompanied by an overall reduction of global DNA methylation levels. Our data confirm that temporary changes within the NK and/or the DC pool during early gestation influence placental development and function, subsequently affecting hepatic gene methylation and fetal metabolism.
Results
Temporary aNK in utero affects fetal and post-natal body weight
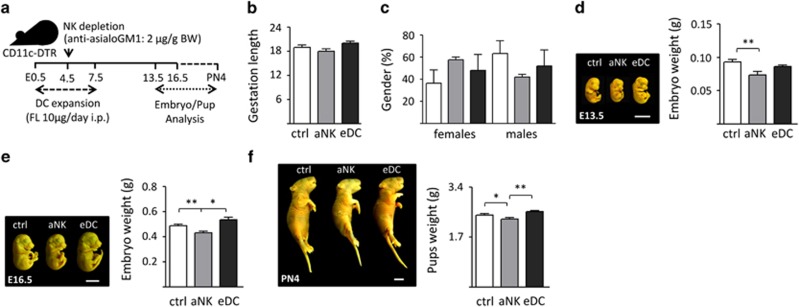
We recently established a mouse model in which DC expansion (eDC) or NK cell ablation (aNK) during early pregnancy provoked changes in the decidualization and angiogenesis processes associated with implantation, but did not affect fetal survival.2 In the eDC model, we treated CD11c.DTR female mice, whose pregnancy outcome parameters are comparable with C57BL/6 dams,12 during early pregnancy with Fms-related tyrosine kinase 3 ligand (FL) for 5 days ensuring a three-fold expansion of DC population within the decidua.2 For aNK, an antibody-mediated depletion strategy was chosen over a mouse strain with a genetical NK cell deficiency, because we pursued temporal ablation of NK cells during our experiment over a permanent NK cell deficiency.2 Of note, the antibody-mediated strategy-specific-depleted NK cells without altering other cell subsets (i.e., granulocytes; Supplementary Figure 1). In order to determine whether temporary changes in NK cell or DC relative abundance during early gestation could have an impact on fetal growth, we characterized the offspring of aNK and eDC mice from E13.5 through post-natal day (PN)-4, and compared them with control mice (Figure 1a). In these experiments, the length of gestation and gender distribution of the offspring were not altered upon aNK and eDC treatments with respect to the control group (Figures 1b and c). However, fetuses carried by aNK females appeared to be smaller compared with eDC and control female mice on E13.5 and E16.5 (Figures 1d and e). In addition, we noted that pups delivered by aNK female mice showed a significant decrease in body weight compared with control and eDC mice on PN4 (Figure 1f), suggesting that aNK influences fetal and post-natal body mass.
Figure 1.
Effects of NK cell ablation and DC expansion on pregnancy progression and fetal development. (a) Experimental design: pregnant CD11c-DTR females were injected i.p. with anti-asialo GM1 for NK ablation on E4.5 or with 10 μg FL for DC expansion from E0.5 to 7.5, as described in methods. Fetuses (E13.5 and 16.5) or pups (post-natal day 4, PN4) were analyzed. (b) Pregnancy progression following NK cell depletion or DC expansion in female mice. Mean length of gestation (days) did not vary among the different groups analyzed. (c) Gender distribution of offspring from aNK and eDC female mice. No differences were observed in the percentage of male and female pups with respect to controls. (d–f) Left panels: morphometric characterization of fetal development in aNK and eDC female mice. The figure shows representative examples of fetuses (E13.5 and E16.5) and pups (PN4) obtained from control, aNK and eDC dams. Scale bar=0.5 cm. Analysis of fetal development following NK cell depletion or DC expansion in female mice. Fetuses carried by aNK or eDC females on E13.5 and 16.5 showed no differences in Theiler stage of development with respect to control mice. (d–f) Right panels: mean body weights resistered in the offspring following NK depletion or DC expansion during early pregnancy. Reduction of body weight was detected in E13.5, E16.5 fetuses carried by aNK dams, which persists until PN4 with respect to weights rescorded in control and eDC mice. In all figures, data shown are mean±S.E.M. derived from six to eight mice per group or 48–56 embryos/pups per group. Differences are denoted as *P<0.05 and **P<0.001, as analyzed by ANOVA followed by Tukey post-hoc test
Decreased body weights in the progeny correlate with structural changes and impaired placental function
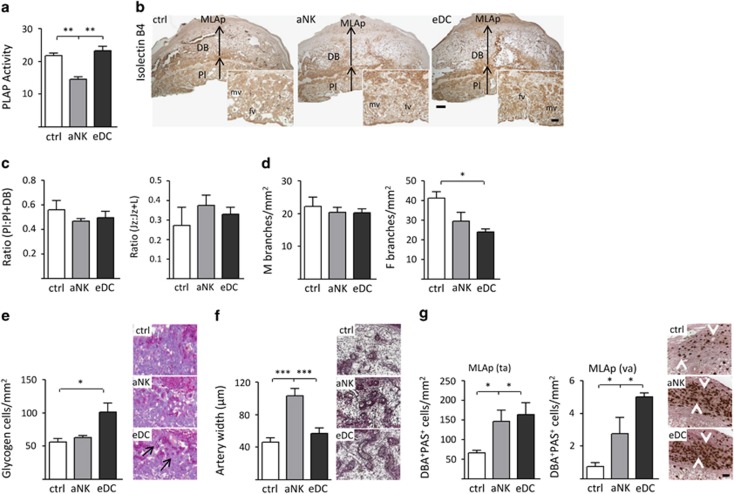
As uterine NK cells have been associated with the modulation of placental physiology during early and mid-pregnancy,13, 14 our next aim was to characterize structural changes in placental development. It was previously described that increased synthesis of placental alkaline phosphatase (PLAP), which facilitates the transfer of nutrients across the cell membrane to the developing fetus, corresponds to the increased nutritional demand of the fetus during growth.15, 16 While control and eDC placentas showed no overt differences, aNK mice displayed lower levels of PLAP activity (Figure 2a). However, the proportional depth of the placental disc (Pl) including the labyrinth (L) and junctional zone (Jz), relative to the decidua, was similar between the analyzed groups (Figures 2b and c). Further morphometric analysis revealed that aNK placentas had a similar fractional area occupied by the Jz compared with control and eDC mice. To investigate differences in placental vascularity we focused on the labyrinth zone, which consists of cells of trophectodermal and mesodermal origin that together undergo branching morphogenesis resulting in a large surface area for nutrient and gas exchange between the mother and fetus. Interestingly, upon analysis of the isolectin B4 (IB4) staining, a marker that highlights the extracellular matrix surrounding fetal blood vessels,17 eDC placentas exhibited a significant decrease in the number of embryonic vascular branches when compared with control mice (Figure 2d, right panel). Of note, the density of maternal branches did not show differences between the analyzed groups (Figure 2d, left panel). In addition, histological examination of the junctional layer showed an increased number of periodic acid-schiff (PAS)-positive vacuolated cells (glycogen cells) in eDC mice (Figure 2e). Morphological changes of the maternal spiral arteries (e.g., increased diameter and a concomitant loss of mural smooth muscle cells) occur during mid gestation in order to increase blood flow to the placental bed.6 Trichrome staining showed that although control implantation sites had dilated, represented by thin-walled arteries in the central, proximal region of the decidua basalis at E10.5 (Figure 2f), the vessels in this region of aNK placentas had narrower lumens and a cuffed appearance, indicative of a thickened arterial wall (Figure 2f). Analysis of the presence of PAS+DBA+ granulated NK cells, which facilitate dilation of spiral arteries,14 showed similar abundances and distribution patterns (e.g., tissue-associated (ta) and vascular-associated (va)) in the decidua basalis in all groups analyzed (Figure 2g). In the mesometrial lymphoid aggregate of pregnancy , however, aNK and eDC mice showed increased numbers of both va and ta NK cells.
Figure 2.
Manipulation of the relative NK cell–DC abundance during early pregnancy alters placental development and function. (a) NK cell depletion alters placental function, as noted by decreased placental alkaline phosphatase (PLAP) activity on E10.5 with respect to control and eDC mice. (b) Histological analysis of Isolectin B4 (IB4) stained uterine sections obtained during E10.5. DB, decidua basalis; Jz, junctional zone; MLAp, mesometrial lymphoid aggregate of pregnancy; PI, placenta. (c) Placental depth related to the decidua (as noted by the PI:PI+DB ratio) and the fractional area occupied by the Jz were similar in all groups analyzed. (d) Left panel; density of maternal vascular (IB4−). Right panel: fetal vascular density of labyrinth, as noted by the presence of IB4+ vessels. (e) Representative examples of E10.5 uterine sections stained with PAS. Arrows highlight glycogen vacuolated cells associated with the Jz. (f) Spiral artery remodeling, as analyzed on trichrome-stained sections on E10.5. Insets show that spiral arteries in dNK mice remain undilated, as noted by artery width (μm). (g) Accumulation of mature NK cells (PAS+DBA+) in the MLAp. Number of both tissue- (ta) and vascular-associated (va) NK cells was significantly increased in dNK and eDC mice with respect to control females. In all figures, data shown are mean±S.E.M. derived from six to eight mice per group and significant difference are noted as *P<0.0 and ***P<0.001 according to the Mann–Whitney U-test. Scale bars=100 and 50 μm
Epigenetic regulation within the developing and post-natal liver is affected by in utero manipulation of NK cell–DC relative abundance
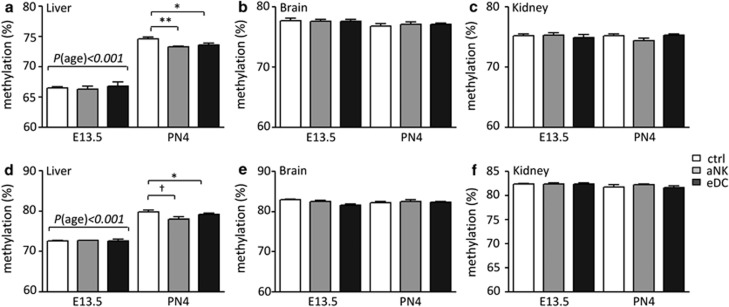
To determine whether changes in placental structure upon NK depletion and DC expansion were associated with epigenetic dysregulation, we first determined the global DNA methylation levels for fetal liver, brain and kidney by LUminometric Methylation Assay (LUMA) (Figures 3a–c). DNA methylation levels in the fetal liver strongly depended on the gestational age, where an average 10% increase in methylation levels was observed on PN4 compared with E13.5. However, no significant differences were observed between the control, aNK and eDC groups in the fetal liver on E13.5. Interestingly, we observed a significant change in the methylation levels in the livers derived from aNK and eDC dams on PN4 (Figure 3a). In addition, global methylation in both the fetal brain and kidney did not significantly differ between fetuses carried by control, aNK and eDC dams (Figures 3b and c). The global methylation levels as determined by LUMA in these organs appeared to be independent of the gestational age of the fetal mice, as no differences were observed between E13.5 and PN4 (Figures 3b and c).
Figure 3.
Global methylation levels as determined by LUMA and pyrosequencing of the LINE-1 promoter after bisulfite conversion. Data are shown for both embryonic day 13.5 (E13.5) and post-natal day 4 (PN4). (a) LUMA-Liver, (b) LUMA-Brain, (c) LUMA-Kidney, (d) LINE-Liver, (e) LINE-Brain and (f) LINE-Kidney. White bars show global methylation within the control group, gray bars from the NK-depleted group and the black bars from the DC expanded group. Data represent mean±S.D.-derived from 10 to 15 embryos/pups per group. Significant differences are noted as *P<0.05 and **P<0.01 according to ANOVA followed by Tukey post-hoc test
As a second approach to determine global methylation levels, we developed a LINE-1 pyrosequencing assay. The methylation of five individual CpG's within the LINE-1 promoter was analyzed after bisulfite conversion and amplification by PCR (Supplementary Table 1). As shown in Figure 3d, LINE-1 methylation in the liver was higher at PN4 compared with E13.5. In addition, on PN4 LINE-1 methylation patterns were lower in the aNK- and eDC-derived pups compared with the controls (Figure 3d, Supplementary Figure 1). The brain and kidney LINE-1 methylation analysis showed similar patterns between the experimental groups on E13.5 and PN4 (Figures 3e and f).
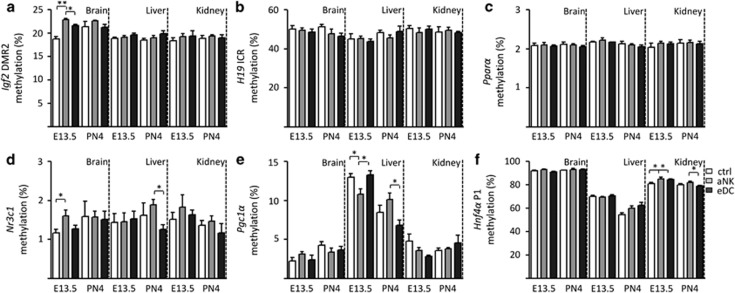
In order to evaluate the effect of NK–DC treatments on organ- and gene-specific changes in epigenetic regulation, we next focused on the methylation levels of several candidate gene promoters (Supplementary Table 2). As a first step, we measured methylation of the insulin-like-growth-factor 2 (Igf2) locus (Igf2/H19), a classical example of imprinting. DNA methylation was similar between the experimental groups in all organs or time points for both the Igf2 Differentially Methylated Region 2 (Igf2-DMR2, Figure 4a) and the H19 Imprinting Control Region (H19 ICR, Figure 4b). However, the Igf2-DMR2 in the fetal brain at E13.5 derived from fetuses carried by aNK dams showed higher methylation levels compared with control and eDC dams. In addition, we examined nuclear receptors (e.g., promoters of the glucocorticoid receptor: nuclear receptor subfamily 3, group C, member 1 (Nr3c1), Pparα, Pgc1α and Hnf4α). Overall, very low levels of methylation were found within the CpG island-associated promoters of both Nr3c1 and Pparα (Figures 4c and d). Nr3c1 showed subtle but significantly higher methylation levels in the fetal brain on E13.5 and in the liver on PN4 in offspring-derived from aNK dams (Figure 4d), while methylation levels of the Pparα promoter were not different for the three organs and two time points analyzed (Figure 4c).
Figure 4.
Promoter-specific DNA methylation. (a) Igf2-DMR2, (b) H19 ICR, (c) Pparα, (d) Nr3c1, (e) Pgc1α and (f) Hnf4α P1 were analyzed in the brain, liver and kidney. Data are shown for both embryonic day 13.5 (E13.5) and post-natal day 4 (PN4). White bars show promoter-specific DNA methylation within the control group, gray bars from the NK-depleted group and the black bars from the brain, liver and kidney tissues derived from the DC expanded group. Data represent mean±S.D. derived from 10 to 15 embryos/pups per group. Significant differences are noted as *P<0.05 and **P<0.01 according to ANOVA followed by Tukey post-hoc test
The Pgc1a promoter showed a remarkable pattern in the fetal liver, where on E13.5 the aNK group displayed significantly decreased methylation levels compared with control and eDC groups. However, on PN4 the liver tissue derived from aNK dams showed a significant increase in the methylation levels in comparison to eDC group (Figure 4e). Next, we analyzed the methylation levels of the Hnf4a promoter, in which we found a significant upregulation in the kidney of the aNK group on E13.5 and PN4 in comparison with control and eDC mice (Figure 4f).
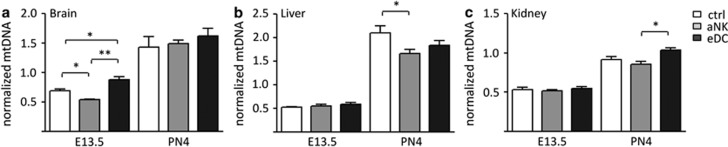
Finally, the relative mtDNA copy number was measured by qPCR. In the brain, significant differences were detected on E13.5 but disappeared with the overall increase in copy number toward PN4 (Figure 5a). However, in liver and kidney tissue we found that the overall mtDNA copy number was increased between the post-natal groups (Figures 5b and c), indicating differential regulation in different organs analyzed.
Figure 5.
Relative mtDNA content. (a) brain, (b) liver and (c) kidney. White bars show global methylation within the control group, gray bars from the NK-depleted group and the black bars from the tissue-derived DC expanded group. Data are shown for both embryonic day 13.5 (E13.5) and post-natal day 4(PN4). Data represent mean±S.D. derived from 10 to 15 embryos/pups per group. Significant differences are noted as *P<0.05 and **P<0.01 according to ANOVA followed by Tukey post-hoc test
Discussion
The placenta is a transitory organ that is essential for the development of healthy offspring. DC and NK cells within the decidua influence processes such as tolerance to alloantigens, angiogenesis and decidualization, which could have an impact on placental development. In this study, we provide clear evidence that alterations of NK cell and DC relative abundance during early pregnancy modify the placentation process and these modifications are associated with epigenetic changes in the offspring. Overall, aNK had a stronger effect on placental dysfunction than DC expansion. Depleted NK dams had narrower arterial lumens and diminished PLAP activity, which could influence nutrient delivery and contribute to the growth restriction observed in the offspring. Changes observed in aNK females, mostly in the architecture of the spiral artery walls within the decidua basalis, agree well with the classical role ascribed to NK cells in modulating placental physiology during early and mid-pregnancy.13, 14 However, a striking observation from this study is that eDC placentas displayed similar alterations. Like aNK mice, eDC females had an increase in accumulation of mature NK cells in the MLAp; indicating that similar mechanisms based on NK cell reconstitution (i.e., increased recruitment and/or maturation of precursors in the deciduas) are triggered in both groups to sustain placental function. Importantly, the similarities regarding arterial remodeling observed between eDC and aNK females highlight the notion that DC-derived signals shape the uterine milieu at the onset of implantation, establishing a critical window that determines NK cell functions later in gestation.
The fact that eDC placentas had a significant decrease in fetal vascular branch density further suggests that fms-like tyrosine kinase 3 ligand (FL) axis could be implicated in the regulation of placental morphogenesis. This is an intriguing hypothesis that awaits further investigation; especially considering previous reports on placental expression of the FL receptor in the labyrinth region.18, 19 In this context, the labyrinth layer is the main site of contact between fetal and maternal circulation, consisting of a highly branched fetal vascular network in close apposition to trophoblast-lined maternal sinusoids. Labyrinth formation involves complex reciprocal interactions between trophoblasts and incipient vessels.20 However, it is important to note that offspring-derived from eDC did not suffer from IUGR, suggesting that the changes in fetal branch density do not affect the nutritional status upon FL treatment.
Abnormal placental function is often associated with IUGR, a relatively common problem and a cause of significant perinatal morbidity and mortality.9 Recently, IUGR has been associated with epigenetic changes, due to diminished nutrient supply and/or stress responses in the offspring.21 Epigenetic modifications mainly consist of DNA methylation and histone modification, both of which are able to regulate gene expression. Some of these changes are likely heritable, that is, they are maintained through multiple cell divisions and thereby contribute to the increased risk for chronic disease (Developmental Origins of Health and Disease hypothesis). In our study, we demonstrated that the placental dysfunction seen in offspring derived from aNK dams was associated with differential DNA methylation. Genome-wide methylation levels were found to be significantly reduced in the PN4 liver of offspring derived from aNK dams, most probably reflecting an indirect contribution of maternal NK cells to epigenetic programming of the offspring. Thus, NK cells would contribute to shaping the intrauterine milieu by virtue of their role as major regulators of maternal vascular development and remodeling, a physiological process that will ensure adequate nutrient and oxygen supply to the developing fetus. In contrast, we observed no differences in general DNA methylation in the brain and kidneys. The absence of differential methylation in the brain may be explained by counter regulatory mechanisms, which spare the brain tissue from metabolic changes in the rest of the body.22 In addition, the kidney seems to be more susceptible to maternal undernutrition23 with high impact on kidney development and nephron numbers, possibly involving gene-specific epigenetic changes rather than global DNA methylation levels.
The vast majority of mammalian DNA methylation occurs at CpG dinucleotides,24 concentrated in genomic regions called CpG islands. Such regulatory regions are often unmethylated within gene promoters of actively transcribed housekeeping genes and tumor suppressor genes,25 whereas CpG islands of silent genes are predominantly methylated.26 Our studies in gene-specific DNA methylation demonstrated that offspring-derived from aNK dams display an increased methylation. We found high methylation of Igf2-DMR2 and glucocorticoid receptor (Nr3c1) in the PN4 brain and altered methylation patterns in Pgc1a in liver and Hnf4a in the kidney. Hnf4a is expressed in the liver and kidney (proximal tubules) and previous studies have shown epigenetic regulation of this locus during fetal development.27, 28 As methylation changes were present on both E13.5 and PN4, our observations may be related to long-term epigenetic adaptations. Interestingly, the promoter region of Hnf4a near the transcription start site is suggested to behave like an organ-specific differentially methylated region (T-DMR) and found to be hypomethylated in the liver,29 a finding which was supported by our data.
Similarly, the non-CpG island-associated promoter of the Pgc1a showed distinct organ-specific methylation patterns, where overall methylation levels were higher in the fetal liver compared with brain and kidney. This also points toward developmental regulation in the fetal liver between E13.5 and PN4 that seems to be disturbed in the aNK-derived offspring. Although we do not have an explanation for these organ-specific effects, it is tempting to speculate that either some loci are more vulnerable to environmental factors than others, or that specifically regulated mechanisms, such as preferential perfusion of the brain or reduced blood flow to the liver during placental dysfunction act on the epigenome. The brain, which is preferentially perfused as a fetal adaptive mechanism to shortage of supplies during poor placental function, is thus largely protected from detrimental effects of poor placental function, whereas the liver, in this adaptive mechanism, is underperfused.30 In addition, it is unclear to what extent the observed organ- and developmental-specific effects on methylation in our model may be related to underlying differences in cell-type populations, as it has become increasingly clear that different cell-type populations within a tissue share a unique epigenetic signature.30
Finally, changes in mitochondrial DNA copy number occurred between the control, aNK and eDC offspring. The brain was affected early in fetuses derived from aNK and eDC dams, but brain mitochondrial copy number increased during development and reached the same level in all groups analyzed. This might be indicative of an intrinsic regulation of copy numbers in the brain, which is spared from the detrimental effects of placental dysfunction. In the liver and kidney, the differences occurred at PN4, which may be related to the compensatory underperfusion of these organs. Together, our findings indicate that the fetal epigenome is indeed affected by maternal depletion of NK cells, whereas the eDC mice are comparable with control animals. Furthermore, genome-wide epigenetic profiling of the fetal liver has suggested that its development extends to several weeks postnatally and is accompanied by large changes in associated promoter methylation and gene expression.31, 32 Although it is well established that environmental cues can also directly alter the epigenetic state of genes, a large body of evidence suggests that (detrimental) environmental cues during critical periods of organ development can leave specific permanent epigenetic marks that extend well into adulthood. Our data indicate that placental dysfunction due to NK cell temporary ablation may indeed disturb long-term epigenetic regulation at least in the liver and kidney, while the brain is spared. Hence, future research needs to address whether this leads to long-term metabolic changes.
Materials and Methods
Mice
Five- to six-week-old C57BL/6 CD11c.DTR female mice, which express the diphtheria toxin receptor (DTR) under the control of the CD11c promoter (described by Jung et al.33), were purchased from Jaxmice (Sacramento, CA, USA) and maintained in our animal facility with a 12L/12D cycle. The presence of a vaginal plug after cohabitation of CD11c.DTR females (H2b) with Balb/c males (H2d) was denoted as day E0.5 of semiallogeneic pregnancy. Animal care and experimental procedures were performed according to the state authority for Animal Use in Research and Education. Expansion of DC or depletion of NK cells was performed as previously described.2 Briefly, temporary depletion of NK cells was achieved by injecting i.p. anti-asialo GM1 Ab (WAKO, Dusseldorf, Germany, Cat. no. 986–10001, 2 μg/g BW) on E4.5. Control CD11c.DTR females received i.p. injection of PBS supplemented with rabbit normal serum (2 μg/g BW). eDC was performed by treating pregnant female mice on E0.5 with one daily i.p. injection of human recombinant (FL: BioX cell, Cologne, Germany, Cat. no. BE0098, 10 μg/mouse/day) for 7 consecutive days. On E10.5, E13.5 and E16.5, mice from the respective groups (n=5-6) were narcotized, killed and uterine tissue from the implantation sites were processed for histological analysis. In addition, fetuses (E13.5 and E16.5) and pups (post-natal day 4, PN4) were obtained from control, aNK and eDC female mice and fixed in Bouin's solution and subsequently cleared in 70% ethanol for body weight measurement and Theiler stage analysis.34
Histological analysis
Paraffin-embedded uterine samples from E10.5 were cut into 4-μm-thick sections and stained with hematoxylin-eosin (H&E), PAS and Masson's trichrome according to standard procedures. Samples were evaluated for alkaline phosphatase activity using our standard protocol. Briefly, slides were rehydrated and washed in TBS 0.05% Tween. For staining, slides were coated with a Fast Red Violet–Naphthol AS-BI phosphate mixture and incubated in the dark for 10 min at room temperature. The slides were rinsed in TBS 0.05% Tween and mounted with Kaiser's aqueous mounting media. Negative controls were established by incubating the slides in Dual Endogenous Enzyme Block (S2003, Dako, Hamburg, Germany) for 30 min at room temperature prior to the staining step. Tissue sections were examined using a light microscope (Axiophot, Zeiss, Oberkochen, Germany) and photographs were taken with an Axio Cam HRc (Zeiss). Photo documentation was performed using the digital image analysis system Spot advanced software, version 8.6 (Visitron Systems GmbH, Puchheim, Germany).
Isolectin B4 staining
Staining of 8 μm sections was performed by washing in TBS, followed by blocking of endogenous peroxidase through incubation with 3% hydrogenperoxide (H2O2) in methanol for 30 min at room temperature. After incubation with 2% normal serum for 20 min, primary Ab against IB4 (1 : 200, Sigma-Aldrich, Munich, Germany) was incubated over night at 4 °C. The slides were then washed and incubated with HRP-conjugated secondary Abs (Jackson ImmunoResearch, Suffolk, England) for 1 h at room temperature. The signal was detected by incubating at room temperature with a 0.2 mg/ml diaminobenzidine (Dako Cytomation, Hamburg, Germany) in 0.05% H2O2 substrate solution. After washing, nuclei were counterstained with 0.1% Mayer's hematoxylin followed by a standard dehydration procedure and mounting in Vitro-Clud medium (R Langenbrinck, Baden-Württemberg, Germany). Negative controls with irrelevant IgG showed no specific immunoreactivity.
DBA lectin/PAS dual staining
After deparaffinization and rehydratation, the paraffin-embedded sections were stained using the protocol previously described.35
Fetal development analysis
Embryos/fetuses were fixed with Bouin's solution for 2–24 h, then washed with 70% ethanol several times (daily, until Bouin was cleared) and stored in 70% ethanol at room temperature. Body weight was recorded after fixation. Developmental stages were determined as previously described (http://www.emouseatlas.org/emap/home.html).
DNA isolation
Fetal tissues (brain, kidneys and liver) were homogenized using a Tissue Lyser LT (Qiagen, Hilden, Germany) in standard lysis buffer containing RNAse A and Proteinase K. The samples were subsequently extracted two times with 25 : 24 : 1 chloroform/phenol/isoamylalcohol, pH 8.0 (Ultrapure, Fluka, BioChemika, Buchs, Switzerland), precipitated with isopropanol and washed using 70% ethanol. All samples were characterized on a Nanodrop2000c (ThermoScientific, Pittsburgh, PA, USA) for purity and concentration of the genomic DNA. Integrity and purity were confirmed on a 0.8% agarose gel.
LUMA
LUMA was used to measure global levels of DNA methylation by restriction digestion using the methylation sensitive HpaII/MspI isoschizomers and quantification of the resulting overhangs by pyrosequencing.36, 37 Individual samples were digested with both HpaII+EcoRI and MspI+EcoRI, (Fermentas) as separate reactions, where the EcoRI functions as an internal control. In total, 500 ng of genomic DNA was used for the individual restrictions, incubated for 4 h at 37 °C. Lambda DNA and in vitro CpG methyltransferase (M.SssI; New England Biolabs) treated Lambda DNA were used as controls. Samples were run on a Pyromark Q24 platform (Qiagen, KJ Venlo, The Netherlands) in duplicate. Peak heights were normalized to EcoRI to calculate global methylation levels as previously described.36, 37
Pyrosequencing of gene-specific promoters and LINE-1
As a second approach for the determination of global methylation levels, we developed a pyrosequencing assay targeting the mouse LINE-1 repetitive element promoter. The LINE-1 promoter consists of four repeats, is present as thousands of individual copies on both the human and mouse genomes and may function as a representative of the entire genome.38, 39, 40 In brief, the blastn algorithm (with low complexity filtering off) was used to retrieve a list of LINE-1 promoter elements located in the mouse genome and a ClustalX alignment was made.41 Bisulfite-specific primers were developed using the Pyromark Assay design 2.0 software (Qiagen). The primers amplify a reasonably conserved region of the mouse LINE-1 promoter containing five CpGs in total (see Supplementary Table 1 for a detailed description including sequences and schemes). To identify common single-nucleotide polymorphisms that could interfere with the pyrosequencing assay itself, the PCR products were cloned and sequenced (Baseclear, Leiden, The Netherlands). From these sequence data, an A/T SNP was identified that could lead to out-of-frame sequencing; this was added as a non-CpG SNP to the sequence to analyze (AQ-mode). Genomic DNA (200 ng) was bisulfite-converted using the EZ DNA methylation gold kit, (Zymo Research, Leiden, The Netherlands) according to the manufacturer's protocol. LINE-1 was amplified by HotStar Mastermix (Qiagen) (see Supplementary Table 1 for the PCR conditions). The PCR product was purified using streptavidin Sepharose HP beads and hybridization of the sequencing primer with the biotinylated PCR product was performed as described in the PyroMark Q24 vacuum workstation guide (Qiagen). Gene-specific assays were amplified using a universal primer design as described previously.42 Finally, the Pyromark Q24 software (Qiagen) was used to determine the methylation percentage of the individual CpG positions analyzed. Primer sequences and standard curves for assay validation are shown in Supplementary Figure 2.
Statistical analysis
The number of animals included in each experimental group was indicated accordingly. Data are presented as mean±S.D. from two or three replicate experiments. Statistical significance was determined using ANOVA followed by Tukey's test, with a P-value of less than 0.05 being considered as significant.
Acknowledgments
We thank Dr. Jolita Ciapaite (University Medical Center Groningen) for providing the primers for mitochondrial copy number and P Moschansky (SMB's laboratory) for her excellent technical assistance in generating this work. This work was supported by Deutsche Forschungsgemeinschaft (DFG) grant BL1115/1-1 to SMB and the Dutch Heart Foundation (grant number 2004T4801); The Netherlands Organization for Health Research and Development (grant number 91211053) to TP, NF and GB received a doctoral fellowship from Charité and the German Academic Exchange Service (Deutscher Akademischer Austauschdienst, DAAD) respectively. IT-G was financed by the Ministerio de Educación y Ciencia (Spain) through a Post-doctoral fellowship.
Glossary
- NK
natural killer cells
- DC
dendritic cells
- IUGR
intrauterine growth restriction
- FL
fms-like tyrosine kinase 3 ligand
- mtDNA
mitochondrial DNA
- PN
post-natal day
- Pl
placenta
- Jz
junctional zone
- MLAp
mesometrial lymphoid aggregate of pregnancy
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Author contributions
TP and SMB designed research; NF, MVZ, GB, IT-G and SMB performed research; NF, MVZ, GB and IT-G analyzed data; MLC, MR and SAS gave input on writing the manuscript; TP and SMB wrote the manuscript.
Edited by R De Maria
Supplementary Material
References
- Barrientos G, Tirado-González I, Freitag N, Kobelt P, Moschansky P, Klapp BF, et al. CXCR4(+) dendritic cells promote angiogenesis during embryo implantation in mice. Angiogenesis. 2013;16:417–427. doi: 10.1007/s10456-012-9325-6. [DOI] [PubMed] [Google Scholar]
- Gonzalez IT, Barrientos G, Freitag N, Otto T, Thijssen VL, Moschansky P, et al. Uterine NK cells are critical in shaping DC immunogenic functions compatible with pregnancy progression. PLoS ONE. 2012;7:e46755. doi: 10.1371/journal.pone.0046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blois SM, Kammerer U, Alba Soto C, Tometten MC, Shaikly V, Barrientos G, et al. Dendritic cells: key to fetal tolerance. Biol Reprod. 2007;77:590–598. doi: 10.1095/biolreprod.107.060632. [DOI] [PubMed] [Google Scholar]
- Croy BA, Zhang J, Tayade C, Colucci F, Yadi H, Yamada AT. Analysis of uterine natural killer cells in mice. Methods Mol Biol. 2010;612:465–503. doi: 10.1007/978-1-60761-362-6_31. [DOI] [PubMed] [Google Scholar]
- Plaks V, Birnberg T, Berkutzki T, Sela S, BenYashar A, Kalchenko V, et al. Uterine DCs are crucial for decidua formation during embryo implantation in mice. J Clin Invest. 2008;118:3954–3965. doi: 10.1172/JCI36682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson SL, Lu Y, Whiteley KJ, Holmyard D, Hemberger M, Pfarrer C, et al. Interactions between trophoblast cells and the maternal and fetal circulation in the mouse placenta. Dev Biol. 2002;250:358–373. doi: 10.1016/s0012-1606(02)90773-6. [DOI] [PubMed] [Google Scholar]
- Watson ED, Cross JC. Development of structures and transport functions in the mouse placenta. Physiology (Bethesda) 2005;20:180–193. doi: 10.1152/physiol.00001.2005. [DOI] [PubMed] [Google Scholar]
- Inbar-Feigenberg M, Choufani S, Butcher DT, Roifman M, Weksberg R. Basic concepts of epigenetics. Fertil Steril. 2013;99:607–615. doi: 10.1016/j.fertnstert.2013.01.117. [DOI] [PubMed] [Google Scholar]
- Roberts JM, Hubel CA. The two stage model of preeclampsia: variations on the theme. Placenta. 2009;30 Suppl A:S32–S37. doi: 10.1016/j.placenta.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmann S, Bettkober M, Zelmer A, Seeger K, Faigle M, Eltzschig HK, et al. Hypoxia upregulates the histone demethylase JMJD1A via HIF-1. Biochem Biophys Res Commun. 2008;372:892–897. doi: 10.1016/j.bbrc.2008.05.150. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natll Acad Sci USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey G, Frank P, Shaikly V, Barrientos G, Cordo-Russo R, Ringel F, et al. In vivo dendritic cell depletion reduces breeding efficiency, affecting implantation and early placental development in mice. J Mol Med (Berl ) 2008;86:999–1011. doi: 10.1007/s00109-008-0379-2. [DOI] [PubMed] [Google Scholar]
- Chakraborty D, Rumi MA, Konno T, Soares MJ. Natural killer cells direct hemochorial placentation by regulating hypoxia-inducible factor dependent trophoblast lineage decisions. Proc Natl Acad Sc USA. 2011;108:16295–16300. doi: 10.1073/pnas.1109478108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen Z, Smith GN, Croy BA. Natural killer cell-triggered vascular transformation: maternal care before birth. Cell Mol Immunol. 2011;8:1–11. doi: 10.1038/cmi.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Seo H, Mano H, Furuhashi M, Goto S, Tomoda Y, et al. Expression of human placenta alkaline phosphatase in placenta during pregnancy. Placenta. 1990;11:319–327. doi: 10.1016/s0143-4004(05)80223-1. [DOI] [PubMed] [Google Scholar]
- Onyesom I, Opajobi AO, Uzuegbu UE, Oriero D, Mordi J, Awhin PE, et al. Relationship between placental alkaline phosphatase activity and cord blood glucose, albumin and neonatal birth weight at term. Invest Clin. 2009;50:491–495. [PubMed] [Google Scholar]
- Ohlsson R, Falck P, Hellström M, Lindahl P, Boström H, Franklin G, et al. PDGFB regulates the development of the labyrinthine layer of the mouse fetal placenta. Dev Biol. 1999;212:124–136. doi: 10.1006/dbio.1999.9306. [DOI] [PubMed] [Google Scholar]
- deLapeyriere O, Naquet P, Planche J, Marchetto S, Rottapel R, Gambarelli D, et al. Expression of Flt3 tyrosine kinase receptor gene in mouse hematopoietic and nervous tissues. Differentiation. 1995;58:351–359. doi: 10.1046/j.1432-0436.1995.5850351.x. [DOI] [PubMed] [Google Scholar]
- Hannum C, Culpepper J, Campbell D, McClanahan T, Zurawski S, Bazan JF, et al. Ligand for FLT3/FLK2 receptor tyrosine kinase regulates growth of haematopoietic stem cells and is encoded by variant RNAs. Nature. 1994;368:643–648. doi: 10.1038/368643a0. [DOI] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Sookoian S, Gianotti TF, Burgueno AL, Pirola CJ. Fetal metabolic programming and epigenetic modifications: a systems biology approach. Pediatr Res. 2013;73:531–542. doi: 10.1038/pr.2013.2. [DOI] [PubMed] [Google Scholar]
- Scherjon SA, Smolders-DeHaas H, Kok JH, Zondervan HA. The "brain-sparing" effect: antenatal cerebral Doppler findings in relation to neurologic outcome in very preterm infants. Am J Obstet Gynecol. 1993;169:169–175. doi: 10.1016/0002-9378(93)90156-d. [DOI] [PubMed] [Google Scholar]
- Huxley RR, Lloyd BB, Goldacre M, Neil HA. Nutritional research in World War 2: the Oxford Nutrition Survey and its research potential 50 years later. Br J Nutr. 2000;84:247–251. doi: 10.1017/s0007114500001483. [DOI] [PubMed] [Google Scholar]
- Ibrahim AE, Thorne NP, Baird K, Barbosa-Morais NL, Tavaré S, Collins VP, et al. MMASS: an optimized array-based method for assessing CpG island methylation. Nucleic Acids Res. 2006;34:e136. doi: 10.1093/nar/gkl551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Costello JF, Plass C. Methylation matters. J Med Genet. 2001;38:285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandovici I, Smith NH, Nitert MD, Ackers-Johnson M, Uribe-Lewis S, Ito Y, et al. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc Natl Acad Sci USA. 2011;108:5449–5454. doi: 10.1073/pnas.1019007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souren NY, Lutsik P, Gasparoni G, Tierling S, Gries J, Riemenschneider M, et al. Adult monozygotic twins discordant for intra-uterine growth have indistinguishable genome-wide DNA methylation profiles. Genome Biol. 2013;14:R44. doi: 10.1186/gb-2013-14-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi S, Hirabayashi K, Sato S, Li W, Takahashi Y, Hirakawa T, et al. DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res. 2008;18:1969–1978. doi: 10.1101/gr.074070.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen G, Hanson M, Kiserud T, Crozier S, Inskip H, Godfrey KM. Fetal liver-sparing cardiovascular adaptations linked to mother's slimness and diet. Circ Res. 2005;96:12–14. doi: 10.1161/01.RES.0000152391.45273.A2. [DOI] [PubMed] [Google Scholar]
- Waterland RA. Epigenetic epidemiology of obesity: application of epigenomic technology. Nutr Rev. 2008;66 Suppl 1:S21–S23. doi: 10.1111/j.1753-4887.2008.00060.x. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Kellermayer R, Rached MT, Tatevian N, Gomes MV, Zhang J, et al. Epigenomic profiling indicates a role for DNA methylation in early postnatal liver development. Hum Mol Genet. 2009;18:3026–3038. doi: 10.1093/hmg/ddp241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Unutmaz D, Wong P, Sano G, De los Santos K, Sparwasser T, et al. In vivo depletion of CD11c(+) dendritic cells abrogates priming of CD8(+) T cells by exogenous cell-associated antigens. Immunity. 2002;17:211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos G, Tirado-González I, Freitag N, Kobelt P, Moschansky P, Klapp BF, et al. CXCR4(+) Dendritic cells promote angiogenesis during embryo implantation in mice. Angiogenesis. 2012;16:417–427. doi: 10.1007/s10456-012-9325-6. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Yamada AT, Croy BA. DBA-lectin Reactivity Defines Natural Killer Cells that have Homed to Mouse Decidua. Placenta. 2009;30:968–973. doi: 10.1016/j.placenta.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Karimi M, Johansson S, Ekstrom TJ. Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics. 2006;1:45–48. doi: 10.4161/epi.1.1.2587. [DOI] [PubMed] [Google Scholar]
- Karimi M, Johansson S, Stach D, Corcoran M, Grandér D, Schalling M, et al. LUMA (LUminometric Methylation Assay)—a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006;312:1989–1995. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- Han JS, Boeke JD. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression. Bioessays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.