Abstract
end-1 and end-3 are GATA transcription factors important for specifying endoderm cell fate in Caenorhabditis elegans. Deletion of both factors together results in larval arrest, 0% survival and a fate change in the endoderm-specifying E lineage. Individual deletions of either factor, however, result in the development of viable, fertile adults, with 100% of worms developing to adults for end-1(−) and 95% for end-3(−). We sought to quantify the variable phenotypes seen in both deletions using automated cell lineaging. We quantified defects in cell lifetime, cell movement and division axis in end-3(−) embryos, while quantifying perturbations in downstream reporter gene expression in strains with homozygous deletions for either gene, showing that each deletion leads to a unique profile of downstream perturbations in gene expression and cellular phenotypes with a high correlation between early and late defects. Combining observations in both cellular and gene expression defects we found that misaligned divisions at the E2 stage resulted in ectopic expression of the Notch target ref-1 in end-3(−) embryos. Using a maximum likelihood phylogenetic approach we found end-1 and end-3 split to form two distinct clades within the Caenorhabditis lineage with distinct DNA-binding structures. These results indicate that end-1 and end-3 have each evolved into genes with unique functions during endoderm development, that end-3(−) embryos have a delay in the onset of E lineage cell fate and that end-1 has only a partially penetrant ability to activate E lineage fate.
Keywords: C. elegans, Endoderm, GATA factors, Gene expression, Cell fate, Cell migration, Gastrulation
Introduction
In Caenorhabditis elegans the progenitor E cell blastomere arises at the 8-cell stage and gives rise to a single tissue type, the intestine. Its process of organogenesis has been characterized from the E blastomere through embryonic development and to adulthood (Leung et al., 1999). Along with other transcription factors, end-1 and end-3 are essential for E lineage cell fate and with med-1 and med-2 are the first zygotically expressed transcription factors in the E lineage (Maduro et al., 2007; Raj et al., 2010). During the period of end-1 and end-3 expression, the E lineage goes from being developmentally plastic to fixed in its developmental fate (Yuzyuk et al., 2009). This fixation culminates in the activation of the GATA factor elt-2, which is essential for E lineage development and maintains E lineage cell fate throughout the rest of development and adulthood (Fukushige et al., 1998, 1999; McGhee et al., 2007, 2009).
Both end-1 and end-3 are GATA transcription factors. The GATA transcription factor gene family members are distinguished by their ability to bind the simple consensus DNA motif WGATAR (McGhee et al., 2007). This DNA motif is bound by the highly conserved DNA-binding motif CXXC(X)17–19CXXC. Of the ten GATA factors encoded by the C. elegans genome, end-1 and end-3 are more closely related to each other than to any of the other factors, suggesting that they arose directly through gene duplication and originally shared functions (Gillis et al., 2008). However, they have both been conserved at least since the C. briggsae and C. elegans split; a period of at least 40 million years (Cutter, 2008; Maduro et al., 2005a), indicating that each factor likely acquired distinct functions (Force et al., 1999). Genetic investigations support the hypothesis that the genes have both redundant and distinct functions. In support of redundant function, deletion of either factor individually has little impact on viability with almost all embryos developing into fertile adults: 95% of end-3(−) and 100% of end-1(−) embryos developing to adulthood (Maduro et al., 2005a). Also, ectopic expression of either factor through heat shock-driven transgenic arrays is sufficient to convert other embryonic cells to an endoderm fate (Maduro et al., 2005a; Zhu et al., 1998).
In support of distinct functions, end-1 and end-3 have distinct expression patterns. Expression of end-3 in the parent of the E cell persists for one cell cycle while end-1 expression begins later in E and persists for more than two cell cycles (Baugh et al., 2003; Maduro et al., 2007; Raj et al., 2010; Zhu et al., 1997). Along with the increase in lethality, deletions of end-3 but not end-1 led to a variable decrease in downstream elt-2 expression (Raj et al., 2010). Lee et al. (2006) found a delay in gastrulation for both end-3(−) and end-1(−), but the end-3(−) phenotype was more severe. Finally, Maduro et al. (2005b) found that deletions in end-3 can lead to an increase or decrease of the total number of E lineage cells as seen by an elt-2 reporter gene. They also showed that in a pop-1(RNAi) background end-3(−) embryos failed to express end-1 leading the authors to suggest that end-3 provides some input into end-1 activation. Thus end-1 and end-3 appear to retain essential functions in common but also to have acquired distinct roles that confer selective advantage which has led to their retention.
In order to better understand the contributions of the end-1 and end-3 transcription factors to E-lineage cell fate we quantified the phenotypic consequences of individual deletions of end-1 and end-3 during C. elegans development. We examined four key aspects of development using 4D imaging and automated cell tracking; cell lifespan, cell movement, cell division axis and gene expression. Analyzing more than 200 embryos using 4D imaging and automated tracking allowed us to quantify minor and variable phenotypes greater than previously attempted. We determined phylogenies for the end-1 and end-3 genes in five sequenced Caenorhabditis species using Pristionchus pacificus as an outgroup. This phylogenetic analysis revealed a canonical end-1 and end-3 GATA domain. From these results we draw three conclusions; that end-1 and end-3 have evolved into unique genes with specific function during C. elegans development, that end-3(−) embryos have a delay in the onset of E lineage fate and that end-1 has a partially penetrant ability to activate E lineage fate.
Results
E lineage phenotypes of end-1 and end-3 mutations
Early E lineage development is characterized by three events: a lengthening of the cell cycle through the introduction of a G2 phase beginning in the E2 cells, gastrulation of Ea and Ep from the posterior surface into the central part of the embryo, and left-right divisions of Ea and Ep. We used 4D movies to quantify the effects of end-1(−), end-3(−) homozygous mutations and of end-1(−)/+, end-3(−)/+double heterozygotes on these E phenotypes.
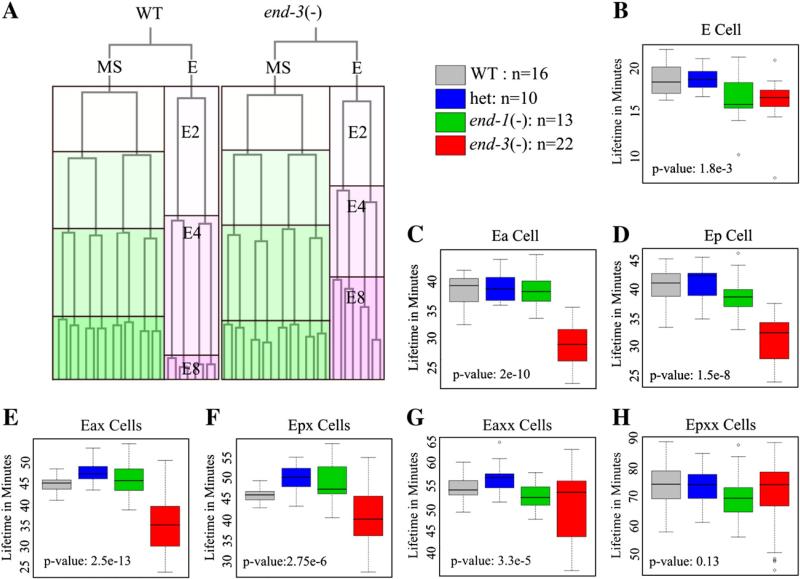
The cell lifetime of the E founder cell in wild type is only slightly longer than that of its MS sister cell, but the lifetimes of its E2 and E4 descendants are more than 150% longer of the comparable MS descendants (Fig. 1A, Sulston et al., 1983). In contrast, the lifetimes of the E2 and E4 cells in end-3(−) embryos are significantly shorter and more variable than in wild type (Figs. 1C–F, Maduro et al., 2005a). Only in the E8 stage of end-3(−) embryos are average lifetimes extended comparable to wild type, but they remain more variable (Figs. 1G and H). However, no significant differences in lifetimes were observed for end-1(−) embryos or for end-1(−)/+, end-3(−)/+double heterozygotes.
Fig. 1.
Deletion of end-3 leads to an acceleration of early E lineage cell divisions relative to WT. (A) Example of typical E lineage acceleration showing E lineage divisions relative to their sister lineage MS for wild type (left) and end-3(−) (right). Each box represents one cell lifetime with MS colored in green and E colored in pink. (B–H) The average cell lifetime for E lineage cells of end-3(−) (red), WT (gray), end-1(−) (green) and heterozygous deletion (blue) embryos. Each boxplot represents the cell lifetimes for a particular cell or set of cells. The total number of series used is shown after the color key. (B) Cell lifetimes for the E cell. (C and D) Cell lifetimes for the daughters of E: Ea and Ep. (E and F) Cell lifetimes for the daughters of Ea and Ep. (G and H) Cell lifetimes for the granddaughters of Ea and Ep.
To determine if the early shortened life spans in end-3(−) embryos might lead to extra cell divisions, we followed the E lineage further in embryogenesis. In wild type, only 4 of the 16 Exxxx cells undergo an additional round of division, yielding 20 total E progeny by the end of embryogenesis. One or more extra E16 divisions occurred in half the end-3(−) embryos (11/22) to produce more than 20 cells. The majority of these extra divisions (8/11) were in cells derived from the anterior daughter of E, Ea. Interestingly, the lineage derived from Ea also has a faster rate of cell division in end-3(−) embryos compared with that of its posterior sister, Ep (data not shown). This is consistent with results of Maduro et al. (2007) which found that end-1 expression was decreased in Ea cells.
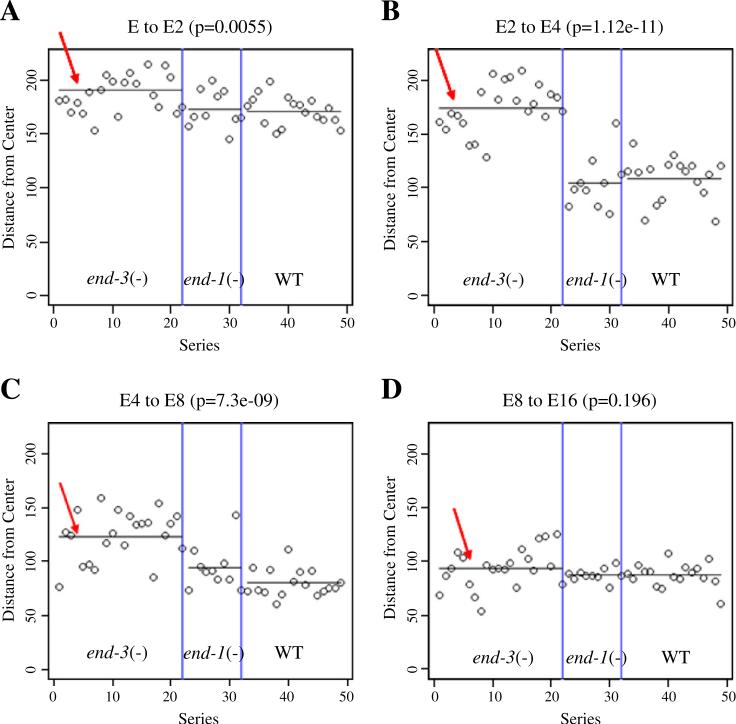
Relative to wild type, end-3(−) embryos also had a significant delay in migration of the E progeny to the center of the embryo (Fig. 2, Lee et al., 2006). To control for the shortened E life spans in end-3(−) embryos we normalized time relative to the MS lineage and total cell number. Already by the end of the E cell lifetime, when end-3 expression is just beginning, end-3(−) embryos were detectably different in position compared to wild type. This difference became very apparent by the end of the E2/E4 boundary and persisted through the E4/E8 boundary (Figs. 2B and C). However, by the E8/E16 boundary, the E lineage cells of end-3(−) animals had moved centrally, equivalent to wild type (Fig. 2D). We saw no significant difference in the behavior of end-1(−) embryos and end-1(−)/+, end-3(−)/+double heterozygotes compared to wild type.
Fig. 2.
Deletion of end-3 leads to a delay in gastrulation of the E lineage. Progress toward gastrulation was determined by taking the center of mass for a set of cells and measuring its distance from the center of the embryo. Each dot represents a single embryo. Measurements were taken (A) one time point prior to the division of E to Ex, (B) one time point prior to the beginning of the division of Ex to Exx (C) one time point prior to the beginning of the Exx to Exxx division and (D) one time point prior to the beginning of the Exxx to Exxxx division. The average distance from the center of the embryo is shown with a line for each strain (red arrow). P-values were calculated on the difference between the average distances of E-derived cells for the end-3(−) embryos as compared to WT, time of development was normalized to wild type for all embryos.
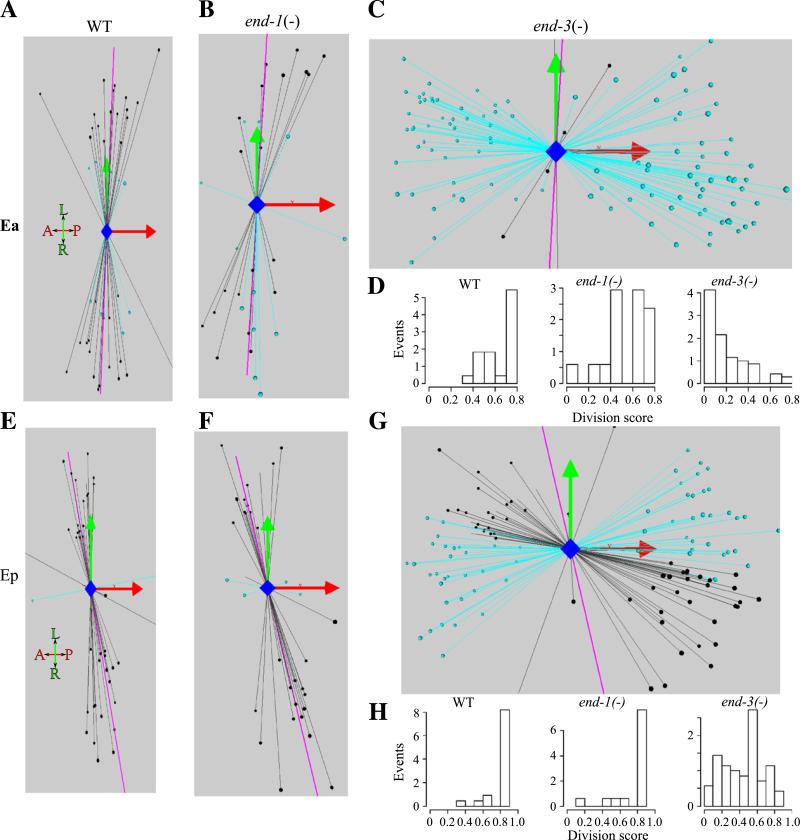
Since the division of Ex cells in wild type is left–right rather than the more usual anterior–posterior division and occurs after the Ex cells have gastrulated, we asked whether this orientation is altered in end-3(−) embryos. We developed methods to characterize the division axis and its variation and applied them to all cells up to the 350-cell stage (including the E8 to E16 division) in 25 wild type embryos. For all divisions in wild type, our scoring system yields a mean value of 0.92, indicating that the division axis in wild type is highly reproducible for most divisions. (Under the scoring system, divisions that align precisely with the average axis score 1 and cells that divide orthogonally to the average axis score a 0.) In contrast, the left–right divisions of Ea and Ep in wild type have average scores of 0.63 and 0.8 respectively and represent two of the more variable divisions of the embryo through 350 cells.
We examined the division orientation of Ea and Ep for both end-3(−) and end-1(−) embryos (Fig. 3). Compared to the wild type axis, the Ea division for end-3(−) embryos had an average score of 0.22 with most divisions aligned more closely with the AP axis rather than the LR axis (Fig. 3C). The Ep change for end-3(−) embryos was less dramatic but still shifted toward the AP axis, with a score of 0.46 (Fig. 3F). These extreme shifts in division axis prevented us from directly recognizing the Exl and Exr daughters. Instead, we chose to designate the anterior daughter as the Exl cell for comparative purposes. These designations have the advantage that they preserve some of the positional characteristics of the named cell later in development. A more minor alteration was observed in end-1(−) embryos where the average Ea cell division was 0.46, with a slight shift on average toward the AP axis (Fig. 3B). In end-1(−) embryos the division axis of Ep cells was not significantly different from wild type (Fig. 3E). The subsequent divisions in both end-3(−) and end-1(−) embryos had division axes similar to wild type, e.g., for E4 divisions the scores were 0.88 and 0.79 respectively (wild type score for E4 was 0.88).
Fig. 3.
Deletion of end-3 leads to a highly penetrant and severe defect in the orientation of the division of the E2 cells. Each panel shows the division axis for a group of E2 cells in either WT, end-1(−) or end-3(−) embryos. The AP and LR axes are denoted by the red and the green arrows respectively with the DV axis shown in a blue line that is coming out of the panel. The average WT division axis is shown as a purple line in all panels. Divisions that are determined to be typical within a standard measure of variance are shown as black lines, while those divisions determined to be atypical are shown with light blue lines. (A–C) 3D representation of the Ea division in WT, end-1(−) and end-3(−) embryos respectively. (E–G) 3D representation of the Ep division in WT, end-1(−) and end-3(−) embryos respectively. (D and H) Each division score is plotted for WT, end-1(−) and end-3(−) in bar plots for both the Ea and Ep divisions respectively. All embryos have been normalized for orientation and size.
The phenotypic effects seen in end-3(−) embryos were strikingly similar to the defects of gad-1(−) embryos (Knight and Wood, 1998), isolated in a search for gastrulation defective mutants. In gad-1(−) embryos “the E cells divide early with abnormal spindle orientations and fail to migrate into the embryo, and no subsequent gastrulation movements occur.” But in addition to these early phenotypes, gad-1(−) embryos eventually arrest at the lima bean stage. To determine if the gad-1 defects might be in part mediated by end-3 or end-1, we examined gad-1(RNAi) embryos for alterations in end-3 or end-1 expression, using integrated end-1::RFP and end-3::RFP promoter fusions. While gad-1 (RNAi) blocked gastrulation and produced altered axes of Ex divisions as expected, there was no significant difference in the onset of end-3 or end-1 signals in the treated embryos compared to wild type. These results suggest that gad-1 is not the main activator of end-3 or end-1 but may act downstream. The gut granules observed later in surface cells of gad-1 arrested embryos (Knight and Wood, 1998) might reflect wild type activation of end-3 and end-1 and E-lineage fate followed by a subsequent failure downstream.
The observed phenotypes when end-3 function is absent are consistent with a variably delayed developmental program in the E lineage and indicate that end-1 has a more limited ability to activate E lineage cell fate in the absence of end-3 activity. Nonetheless, end-1 function appears to eventually allow most end-3(−) embryos to complete gut development. The distinct effects of end-3 might arise either because of its earlier expression (Raj et al., 2010), because of different affinities for various target genes compared to end-1, because end-3 plays some role in activation of end-1 or because of some combination of all three.
E lineage-specific reporter expression is abnormal in end-1− and end-3 − embryos
The persistence of end-1 and end-3 paralogs in the Caenorhabditis species as well as their distinctive phenotypic effects suggests that they evolved to regulate different target genes in gut development. To look directly for expression differences between the two genes, we assayed nine transcription factor reporter genes in end-1(−) and end-3(−) embryos. Five were known to have roles in E lineage specification (end-1, end-3, elt-7, elt-2 and ref-1; Neves et al., 2007; Sommermann et al., 2010) whereas four were selected based on evidence of expression in the E lineage (tlp-1, pax-3, pha-4 and nhr-57; Murray et al., in preparation). Six of these reporters are expressed throughout the E lineage (end-1, end-3, elt-7, elt-2, pha-4 and nhr-57) and three are expressed in specific subsets of E lineage cells (ref-1, pax-3 and tlp-1). Five of the genes are also expressed in other cells in the embryo (ref-1, pax-3, tlp-1, pha-4 and nhr-57).
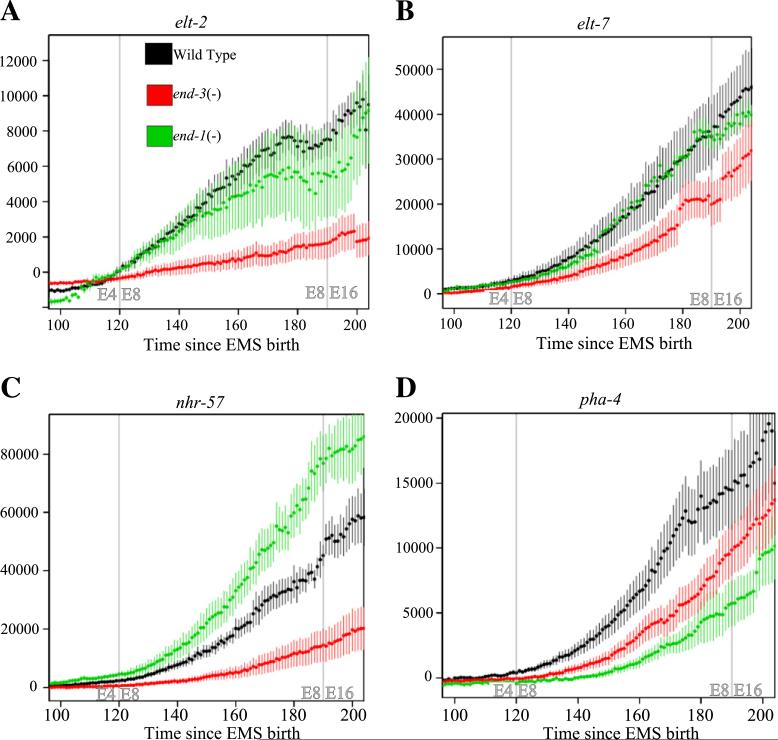
In end-3(−) embryos four of the E ubiquitous reporters show reduced expression in the E lineage and in no other lineage, starting at the beginning of the E8 stage (Fig. 4). For two of these reporters, elt-7 and pha-4, expression differences were slight and may reflect only a delay in onset (Figs. 4B and D). However, two other reporters, elt-2 and nhr-57, had substantially decreased overall signal throughout development (Fig. 4A and C). In three out of ten end-3(−) embryos there was a complete lack of elt-2 reporter expression even at the 350 cell stage and reporter expression in no more than one E lineage cell by the 500 cell stage. These three embryos all had an extra round of cell division and showed the most extreme E lineage phenotypes. Since elt-2 is necessary and sufficient for the intestinal cell fate, the lack of reporter expression would suggest a severe defect in gut development. Thus, despite the overall increase in E derived cells, there can be a reduced number of elt-2 expressing cells, consistent with results of Maduro et al. (2007).
Fig 4.
Alteration in transcription factor expression for end-3(−) and end-1(−) embryos. Deletion of end-3 leads to decreases in the expression of (A) elt-2, (B) elt-7, (C) nhr-57 and (D) pha-4 while deletion of end-1 leads to decreases only in elt-2 and pha-4 expression. Each graph shows the expression of fluorescent reporters in the E lineage for WT (black), end-1(−) (green) and end-3(−) (red) embryos. Single dots represent the average fluorescent intensity of each reporter for each treatment at a given point in development, colored lines are two standard error. Gray lines represent the beginning of the E4 to E8 and the E8 to E16 divisions in wild type.
The end-1(−) embryos had a more complex series of phenotypes with these four reporters. Only one reporter, pha-4, had a decrease in expression but this was more pronounced than in end-3(−) embryos (Fig. 4D). On the other hand, end-1(−) embryos produced an increase of nhr-57 reporter expression (Fig. 4C). We saw no changes in elt-2 and elt-7 reporter expression relative to wild type in end-1(−) embryos (Figs. 4A and B). These differences in effects are consistent with distinct roles for end-1 and end-3 in E lineage gene activation. We saw no change in the average level of expression for end-1 or end-3 reporters in either deletion (data not shown); however we did see a small number of embryos with a significant reduction of expression relative to WT, consistent with recent observations by Raj et al. (2010).
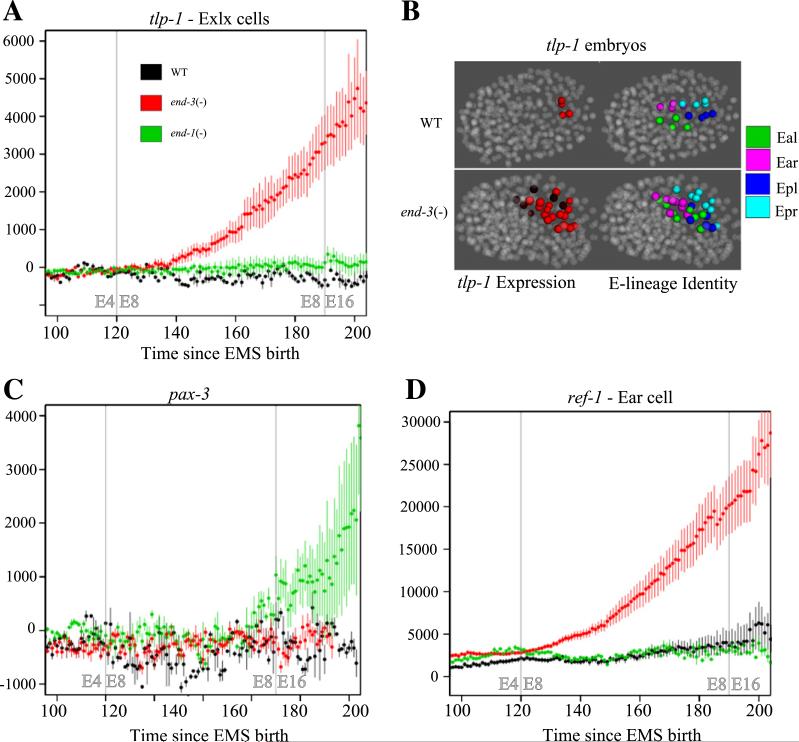
We next examined two of the three genes expressed in specific subsets of E lineage cells. tlp-1 is involved in asymmetrical cell fate of the T-cells during larval development and is responsive to WNT signaling (Kagoshima et al., 2005; Yoda et al., 2005; Zhao et al., 2002). Expression of the tlp-1 reporter is restricted in wild type to the daughters of the two most posterior E8 cells, Eplp and Eprp (Fig. 5B). This reporter expression begins just prior to the E8 to E16 division and is stronger in the Eprp-derived cells. In end-1(−) embryos there is increased reporter expression in these cells as well as an earlier onset of tlp-1 reporter expression (data not shown). Associated with this increase is a low level of ectopic reporter expression in the Epla cell. More strikingly, however, there is widespread ectopic tlp-1 reporter expression in end-3(−) embryos (Figs. 5A and B). Expression was seen for each of the E8 cells in at least one imaged embryo (Fig. 5B). On average five cells of the E8 cells expressed tlp-1 in end-3(−) embryos at the E8 stage as compared with two in wild type embryos. The Exxp posterior daughters showed expression more frequently, with 3.2 out of 4 posterior daughters showing expression as compared with 1.6 anterior daughters (Exxa) showing expression on average. The extent of ectopic expression is again associated with the more severe end-3(−) phenotypes. The ectopic tlp-1 reporter expression was similar to tlp-1 reporter expression in the C lineage in terms of onset, level and posterior bias. pax-3, like tlp-1, is expressed in the posterior-most cells in the E lineage in wild type (Earp and Eprp). The reporter pax-3 had decreased expression in end-3(−) embryos in the Exxx cells, but in end-1(−) embryos an ectopic increase in the Ealp cells was seen (Fig. 5C).
Fig. 5.
Alteration in transcription factor expression for end-3(−) and end-1(−) embryos. end-3(−) embryos have increased expression of (A) tlp-1 and (D) ref-1 in a specific subset of cells while end-1(−) has increased expression of (C) pax-3 in the Earp cell. (A–C,D) Each graph shows the expression of fluorescent reporters in the E lineage in WT, end-1(−) and end-3(−) embryos. All coloring is the same as Fig. 4. (B) 3D representation of tlp-1 expression in the E lineage for WT and end-3(−) embryos. A typical WT embryo is shown above and an end-3(−) embryo below. On the left is a 3D representation of tlp-1 expression at a similar point in development for WT (top) and end-3(−) (bottom) embryos. Higher expression is denoted by cells by red intensity. On the right is a 3D representation of the same time point, this time with specific sublineages of E colored according to their identity with Eal = green, Ear = pink, Epl = blue and Epr = cyan.
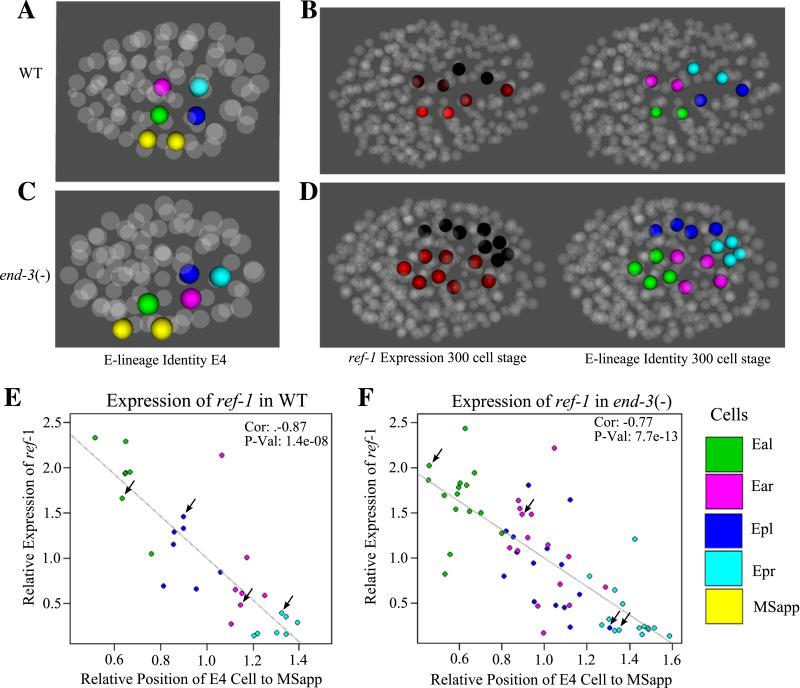
Atypical E2 division axis in end-3 − embryos correlates with ectopic ref-1 reporter expression
In the most severely affected end-3(−) embryos, the abnormal A–P orientation of the Ex division can result in both daughters of Ea lying more proximal to MSapp than the daughters of Ep (Fig. 6C). In wild type, Eal and Epl are adjacent to MSapp and MSapa and receive a Notch signal from them (Neves et al., 2007) that results in increased reporter expression of ref-1 in the Eal and Epl cells. This expression leads to a cascade of intercellular signaling within the E lineage that produces the gut-twist seen at the E16 stage (Hermann et al., 2000). These events are critical for proper gut function later in development. In end-3(−) embryos as a result of the abnormal proximity to MSapp, ref-1 reporter expression can be increased in Ear daughters compared to wild type and in turn ref-1 reporter expression in Epl daughters can be decreased. These levels are negatively correlated with the distance of the cell from MSapp (Fig. 6F). In end-3(−) embryos the expression of ref-1 had a partially penetrant increase in the daughters of the Ear cell associated with a corresponding decrease in the daughters of the Epl cells (Figs. 5D and 6D). We hypothesize that atypical divisions at the Ex stage in end-3(−) embryos result in this ectopic expression. When we examined the ref-1 reporter expression in end-3(−) embryos one cell cycle after the Exx stage it still correlated with proximity to MSapp, but Ear and Epl now have overlapping levels of expression and proximity. Further, those Ear cells with the higher ref-1 reporter expression had often assumed the position normally occupied by Epl daughters in the gut by the Exxx stage (Fig. 6D). Compared to wild type embryos there was an overall increase in ref-1 reporter expression in both end-1(−) and end-3(−) embryos (data not shown). That an Ea daughter had assumed the location and ref-1 reporter expression associated with an Ep daughter indicates a switch in cell fate.
Fig. 6.
Mislocalization of the E4 cells in end-3(−) embryos leads to an improper increase of ref-1 expression in Ear daughters along with a corresponding decrease in Epl daughters. (A–D) 3D representation of ref-1 expression in the E lineage for WT and end-3(−) embryos. (A and C) Position of E4 cells relative to the Notch-signaling cells MSapp and MSapa in WT and end-3(−) embryos. (B and D) Expression one cell cycle later (left) along with the corresponding cell identities (right). Higher ref-1 expression is shown by the level of red intensity. Cell identities (right) Eal = red, Ear = pink, Epl = blue, Epr = cyan and MSap = yellow. (E and F) Expression of ref-1 in E8 cells as a function of E4 proximity to MSapp. Cell identity is colored according to the legend in the bottom right of the figure. Expression and proximity are plotted as a ratio of the average for the E cells of individual embryos. WT is shown on the left and end-3(−) on the right. Arrows indicate cells from embryos shown in A and B.
Because of the graded effect of MSapp on ref-1 reporter expression in the mutant embryos, we reexamined ref-1 expression in WT embryos. We found strong reporter expression in Eal and Epl cells but also weak reporter expression in both right daughters of Ex cells, with the level of expression correlated with the proximity of the cells to the MSapp cells (Fig. 6E). In wild type embryos Epl is further from the MSapp cell than Eal, and has a significantly lower level of ref-1 reporter expression (p-value=6.2e−5). In addition Ear and Epr are yet more distant from MSapp and have still lower expression. Plotting ref-1 reporter expression versus distance from MSapp produces a high negative correlation (Fig. 6E). In fact, the Exx daughters from different animals can be grouped into four distinct clusters by their distance from MSapp and their relative expression level. These results are consistent with images from Neves and Priess (2005). This correlation is also seen for MSapa, but is less significant (data not shown).
Evolution of end-1 and end-3 suggests a structural basis for distinct function
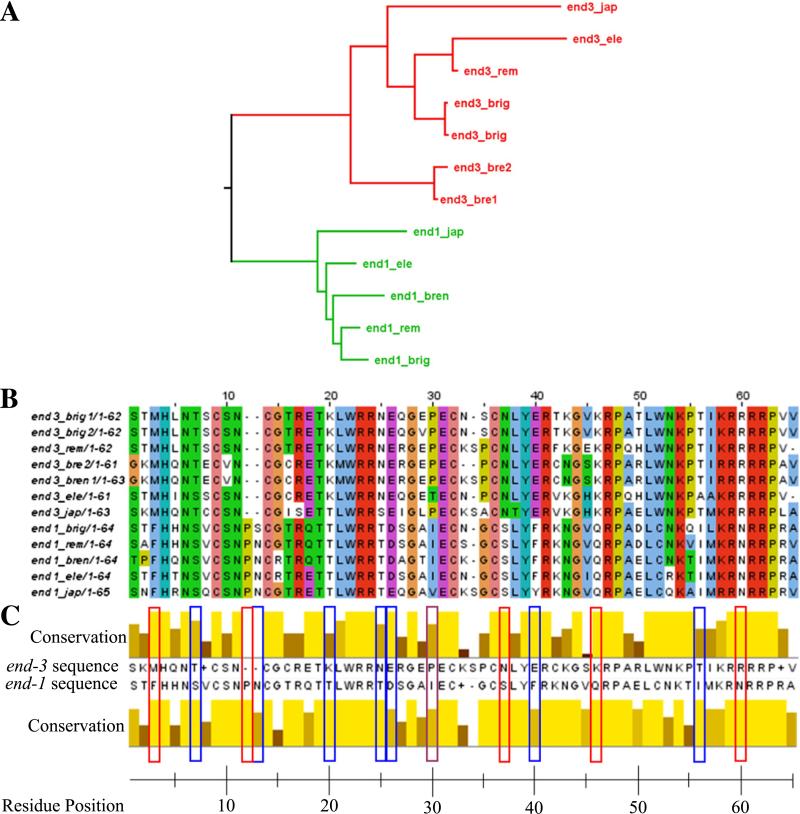
The distinct phenotypic effects of end-1 and end-3 deletions and their different effects on gene expression patterns in the E lineage demonstrate that the two genes have acquired distinct functions since their duplication. To gain additional support for this and to provide insight into the underlying molecular basis for these distinct functions we examined the phylogenetic relationship of the genes in Caenorhabditis species and the possible effects of substitutions on protein structure (Fig. 7). Using a maximum likelihood method (Guindon and Gascuel, 2003) we developed a gene phylogeny for end-1 and end-3 based on the highly conserved GATA domain. The phylogeny shows that the duplication that gave rise to the end-1 and end-3 genes appears to have occurred after the split from P. pacificus but before the split from C. japonica. end-1 and end-3 form separate clades, with one end-1-like gene in each of the five sequenced Caenorhabditis species but with one or two end-3-like genes in each species. The distinct clades are consistent with the hypothesis that the two factors have acquired specific functions within the Caenorhabditis species. We also aligned all the genes across their entire length and determined the end-1/end-3-specific phylogeny using the same maximum likelihood method. This phylogenetic tree had identical clade structure with the tree generated using just the GATA domain (Fig. 7A). These results are similar to those found by Maduro et al. (2005a) and Gillis et al. (2008), but reveal exactly when end-1 and end-3 duplicated. Further, the tree groups the end-1 and end-3 clades together with elt-3 as an outgroup. Further, we find that the elt-3 clade diverged after the split from P. pacificus, indicating elt-3 may have arisen as the result of an earlier end-3 duplication.
Fig. 7.
end-1 and end-3 have unique phylogenies and GATA domain structures. (A) By using a maximum likelihood phylogenetic analysis end-1 and end-3 were found to have diverged into two distinct clades. Branch lengths represent total amino acid substitution rate for each domain protein. (B) Alignment of all the end-1 and end-3 genes found in Caenorhabditis, residues colored according to Clustal standard colors. (C) Consensus GATA domain for end-1 and end-3 as determined using Clustal. Highlighted in red boxes are those residues that are divergent between the end-1 end-3 clades, but with perfect conservation within the two clades. Those residues highlighted in blue boxes have a single difference in one clade and those residues highlighted in purple boxes have one difference in each clade.
We next examined the structure of the DNA-binding domain of both end-1 and end-3 and found they have evolved clade-specific features. Although many key residues are conserved between the two DNA-binding domains, more than half the consensus sequence is divergent. By examining those residues that are different between the clades but contain a single substitution or less within a clade, thirteen residues stand out as specific for either end-1 or end-3 (Fig. 7C). For six of these sites end-3 has the same residue as the most closely related P. pacificus GATA factor, indicating a conservation of the ancestral allele, while at only one site end-1 has the same residue as P. pacificus. Most striking are the two amino acids inserted at positions 12 and 13 in end-1. This insertion is not present in any other GATA factor from fungi to human (Lowry and Atchley, 2000). Further, the amino acid identity for this insertion is perfectly conserved for all end-1 genes for one residue and only one substitution is found in the other, indicating that the insertion is under functional constraint throughout the Caenorhabditis lineage. Three other examples of clade-specific substitution stand out for their potential impact on protein function. First, the site at amino acid twenty, with a consensus threonine for end-3 and a lysine for end-1, interacts with the thymine of the GATA DNA motif (Vonderfecht et al., 2008). Second, the amino acid at site forty six, with a lysine in end-3 and a glutamate in end-1, is within a motif that is important for protein– protein interaction. Finally, the region from forty-three to fifty-six, which has four conserved differences, interacts extensively with the DNA backbone. None of these three clade-specific examples is conserved in P. pacificus, indicating both end-1 and end-3 have diverged in their abilities to bind DNA from their potential ancestral form. The clade-specific substitutions in these several regions have the potential for substantial impact on function and indicate potential targets for mutational studies into the function of end-1 and end-3.
Discussion
We quantified cell lifetimes, cell movements, division axis and downstream reporter expression in order to understand the specific roles of the highly related GATA factors end-1 and end-3 during E lineage development. The more severe phenotypes seen in end-3(−) embryos suggests that this gene has retained the critical ancestral function of the end-1/3 progenitor gene. In contrast the mild phenotypes in end-1(−) embryos suggest that it has evolved novel or derived functions. This is supported by the distinct abilities each gene has to activate reporter genes, indicating that each has diverged to activate two distinct, but overlapping E lineage regulatory pathways. The distinct and separate clades of end-1 and end-3 within the Caenorhabditis lineage and the unique structure of each within the GATA DNA binding domain further support separate function. We hypothesize this ability to activate distinct regulatory pathways confers robustness during the development of the E lineage and explains why these genes have been preserved throughout the Caenorhabditis lineage.
There are three potential reasons for the specific differences between end-1 and end-3: expression timing, unique DNA binding motifs and total expression levels. Raj et al. (2010) and Maduro et al. (2005a) have shown that end-3 expression begins late during the E cell lifetime and persists until mid-way through the Ex cell lifetimes. Yuzyuk et al. (2009) have shown that during this time in development most cells, including the E lineage, lose their plasticity and become fixed in developmental cell fate. In contrast, end-1 expression does not begin until later and does not reach peak expression until the Exx lifetimes. So while end-3 expression peaks during the period of cell fate fixation in the E lineage, end-1 does not get activated until after E lineage has normally become fixed in its cell fate. In the absence of proper E fate activation the E lineage reverts to a C lineage-like fate (Owraghi et al., 2010). Therefore, we suggest that in the absence of end-3 the E lineage cells begin to adopt a C-like fate early in development, but once end-1 reaches peak expression the E lineage cell fate is eventually activated. There is also evidence from pop-1 (RNAi) experiments that end-3 may regulate some end-1 expression, which might contribute to this more severe phenotype (Maduro et al., 2007). However, Raj et al. (2010) report a small proportion of embryos have a decrease of end-1 expression in end-3(−) embryos and not until the 50 cell stage, whereas we see 100% of embryos in end-3(−) embryos with defects in movement, division axis and division timing prior to the 50 cell stage. The shortened life spans, improper gastrulation and misaligned divisions seen early in end-3(−) embryos are all a consequence of the E lineage acting like the C lineage. That we later see evidence of C lineage-like expression for tlp-1 in end-3(−) embryos would indicate that this E lineage activation remains incomplete, or that some C lineage regulatory programs remain active from the early C lineage-like activation. Possibly tlp-1 is responding to some external WNT signal, but given the lack of common location and variability in expression of those tlp-1 expressing cells it seems more likely that it is the result of some intrinsic regulatory activation.
Secondly, looking at binding specificity there are conserved differences within the DNA-binding domain of end-1 and end-3. This differential DNA-binding domain could lead to different abilities of end-1 and end-3 to bind to and then activate downstream targets. Such different abilities to activate downstream genes are seen for multiple downstream reporters in end-1(−) and end-3(−) embryos. This is perhaps most striking for elt-2/7 where end-1(−) embryos have essentially WT levels of expression for both reporters, whereas end-3(−) embryos have severe decreases in expression of both.
Finally, the total combined end-1/3 expression level seems like the least important contributor to the conservation of end-1 and end-3. Our work with the double heterozygote deletion rules out that combined expression levels somehow play a major role. That the heterozygote has the more mild phenotypes associated with end-1(−) and end-3(−) embryos would seem to indicate time and specificity are more important, but that total end-1/3 levels still play a minor role.
The graded expression of ref-1 indicates that within the E lineage ref-1 expression plays a role in differentiating anterior posterior as well as left right asymmetries. While previously ref-1 expression had been shown to differentiate the left from the right side of the E lineage, we saw a reproducible gradation of ref-1 expression from anterior to posterior, which points to ref-1 also playing a role in differentiating the anterior and posterior cells in the E lineage. That those Ear and Epl cells in end-3(−) embryos with swapped positions were also likely to swap ref-1 expression levels is a further indication that ref-1 likely plays a role in specifying differences in anterior posterior E lineage fate. Finally, the ref-1 expression and position swapping of Ear and Epl in end-3(−) embryos indicates the E lineage is still plastic and is refining its cell fate as late as the E4 stage.
The ability of the aberrant end-3(−) embryos to recover and develop into adults shows the robustness of development in C. elegans. Despite a failure to gastrulate properly, mislocalization of sublineages and the misexpression of a variety of transcription factors at least 95% of embryos develop into adults. This robustness is conferred in part by the ability of end-1 to act in a parallel pathway of gene expression and development which corrects for the early defects and solidifies cell fate. This parallel pathway is essential for robust development and helps explain why end-1 and end-3 have both been conserved throughout the Caenorhabditis lineage.
Methods
Strains
RB1331 (end-3(ok1448) V), VC271 (end-1(ok558) V) designated end-3(−) and end-1(−) respectively.
| Reporter | Lineage marker (WT) |
Lineage marker (mutants) |
WT strain |
end-3(–) strain |
end-1(–) strain |
|---|---|---|---|---|---|
| pha-4(4.1 kb)::H1-Wcherry; unc-119(+) | stIs10024;zuIs178 | stIs10026 | RW10381 | RW10379 | RW10380 |
| tlp-1::H1-Wcherry; unc-119(+) | stIs10024; zuIs178 | stIs10026 | RW10681 | RW11111 | RW11131 |
| nhr-57::H1-Wcherry; unc-119(+) | stIs10024; zuIs178 | stIs10026 | RW10520 | RW11113 | RW11128 |
| pax-3::H1-Wcherry; unc-119(+) | stIs10024; zuIs178 | stIs10026 | RW11114 | RW11114 | RW11133 |
| elt-7::H1-Wcherry; unc-119(+) | stIs10024; zuIs178 | stIs10026 | RW10131 | RW11110 | RW11129 |
| ref-1::H1-Wcherry; unc-119(+) | stIs10024; zuIs178 | stIs10026 | RW10752 | RW11127 | RW11130 |
| end-3::H1-Wcherry; unc-119(+) | stIs10024; zuIs178 | stIs10026 | RW10378 | RW11109 | RW11134 |
| end-1::H1-Wcherry; unc-119(+) | stIs10024; zuIs178 | stIs10026 | RW10385 | RW11115 | RW11131 |
| elt-2::TY1 EGFP 3xFLAG; unc-119(+) | stIs10116; itIs37 | stIs10116; itIs37 | RW10714 | RW11128 | RW11132 |
Heterozygous strain construction
RW10379 males were crossed into hermaphrodites homozygous for end-1(−) and pha-4::H1-Wcherry. We used GFP as a marker of successful mating of the end-3 deletion to the end-1 deletion. We used DIC imaging and manual lineaging prior to the onset of GFP expression at the 26-cell stage.
Imaging/lineaging
All embryos were imaged and lineaged according to the protocol outlined in Bao et al., 2006; Murray et al., 2006 and Murray et al., 2008. 3D representations of embryos were made using the in-house program AceTree (Boyle et al., 2006).
Expression analysis
We calculated expression using red pixel intensity in nuclei from imaged embryos. For red intensity we calculated intensity of pixels per nuclei by assuming each nuclei measured 30 by 30 pixels total. To calculate total red we added red intensity from 0 to 255 for each pixel across every pixel in the 30 by 30 grid. To account for background red intensity we calculated local background red intensity from non-expressing cells and subtracted from each nuclei.
Expression onset
To determine the time fluorescence first appeared (onset), we used the initial fifty time-points to calculate a linear model describing how background expression levels (noise) change with time. No detectable expression occurs during these time points. Values for the first fifty values are subtracted from the model and the variance around the line is calculated. Candidate onset times are then tested against this same line. Onset time is determined by measuring if the expression deviates from the model by more than the variance calculated during the first fifty time points. Once a value that varies from the fitted line by more than the variance is found a Bartlett test of variance is used to determine if the difference is significant. We determined to have occurred when the Bartlett test was significant to a p-value of 0.0001.
Cell lifetime normalization
We lineaged MS and ABalpa to the 350-cell-stage for controls. We plotted division-times for ABalpa and MS against a single WT standard. Once plotted, we fitted a slope against each plot. We used the average slope for the MS and ABalpa as a correction factor for each series.
Movement
For each series we calculated a center of mass for the E derived cells for each time point. We chose time points just prior to the division of the anterior-most E cell at the four division time-points during E development. We determined the center of the embryo by examining the embryo at the 350-cell stage and taking a set of cells known to lie on the exterior of the embryo. We extrapolated this center back to the time point of interest. We calculated the distance the E derived cells from the middle of the embryo for their center of mass. In those embryos with accelerated E lineage divisions we used total cell number as an estimate for when Ea normally divide.
Axis
Typical division axis was defined using twenty-two benchmark imaged series. We defined the division axis score as the dot product of the observed axis vector to the expected axis vector after normalizing both vectors to have magnitudes of one.
Proximity and ref-1 expression
We calculated the average proximity to the MSapp cell for all four E4 cells for the entirety of their lifespan. We determined the average distance of the E4 cells from the MSapp cell for each individual embryo. Distances of the individual cells were then calculated in terms of a ratio to this average. We did this to control for embryo size and compression. Similarly we calculated average ref-1 expression for all E8 cells, one cell cycle later, and expression levels calculated in terms of a ratio to this expression level. We did this to control for variations in fluorescent detection. Individual cells were then plotted in terms of each of these ratios. Correlation coefficients and p-values were calculated using a Pearson's product-moment correlation using the R statistics package.
GATA gene discovery, Clustal alignment and ML trees
We systematically BLASTed all ten C. elegans canonical annotated GATA factors along with their C. briggsae orthologs against the genomes of the sequenced Caenorhabditis species: C. brenneri, C. remanei and C. japonica along with P. pacificus for use as an outgroup (Altschul et al., 1990). We aligned the highly conserved GATA domains using ClustalW (Thompson et al., 2002). Once aligned, we determined a phylogeny based on maximum likelihood using the phyml package. We determined this phylogeny using the standard settings as follows: assume zero invariable sites, 6 rate categories, a gamma parameter of 0.5 and optimized branch lengths and topology. We calculated one hundred bootstraps.
We determined those genes most closely related to end-1 and end-3 and aligned them across the entire length of the gene. We BLASTed each genomic region using the full length C. elegans end-1 and end-3 genes. We hand annotated the entire gene using those segments with an E value less than 0.01 with predicted splice junction as a guide. Once all of the potential end-1 and end-3 genes were annotated we re-BLASTed against the entire set of sequenced Caenorhabditis (Altschul et al., 1990).
We aligned the Caenorhabditis end-1 and end-3 genes in an effort to determine amino acid substitution rates. We aligned the end-1 and end-3 sets of genes separately to their respective ortholog sets. We then aligned these two alignments against each other. We did a further alignment to the elt-3 genes, which were used as an outgroup. Having aligned these genes, we calculated a second maximum likelihood tree using the same parameters to determine the substitution rate per amino acid residue (Anisimova and Gascuel, 2006; Guindon et al., 2005).
References
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Bao Z, Murray J, Boyle T, Ooi S, Sandel M, Waterston R. Automated cell lineage tracing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2707–2712. doi: 10.1073/pnas.0511111103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim AK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130(5):889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- Boyle T, Bao Z, Murray J, Araya C, Waterston R. AceTree: a tool for visual analysis of Caenorhabditis elegans embryogenesis. BMC Bioinforma. 2006;7:275. doi: 10.1186/1471-2105-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 2008;25(4):778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Hawkins M, McGhee J. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- Fukushige T, Hendzel M, Bazett-Jones D, McGhee J. Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11883–11888. doi: 10.1073/pnas.96.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis WQ, Bowerman BA, Schneider SQ. The evolution of protostome GATA factors: molecular phylogenetics, synteny, and intron/exon structure reveal orthologous relationships. BMC Evol. Biol. 2008;8:112–127. doi: 10.1186/1471-2148-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann G, Leung B, Priess J. Left–right asymmetry in C. elegans intestine organogenesis involves a LIN-12/Notch signaling pathway. Development. 2000;127:3429–3440. doi: 10.1242/dev.127.16.3429. [DOI] [PubMed] [Google Scholar]
- Kagoshima H, Sawa H, Mitani S, Bürglin T, Shigesada K, Kohara Y. The C. elegans RUNX transcription factor RNT-1/MAB-2 is required for asymmetrical cell division of the T blast cell. Dev. Biol. 2005;287:262–273. doi: 10.1016/j.ydbio.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Knight JK, Wood WB. Gastrulation initiation in Caenorhabditis elegans requires the function of gad-1, which encodes a protein with WD repeats. Dev. Biol. 1998;198(2):253–265. [PubMed] [Google Scholar]
- Lee J, Marston D, Walston T, Hardin J, Halberstadt A, Goldstein B. Wnt/ Frizzled signaling controls C. elegans gastrulation by activating actomyosin contractility. Curr. Biol. 2006;16:1986–1997. doi: 10.1016/j.cub.2006.08.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung B, Hermann G, Priess J. Organogenesis of the Caenorhabditis elegans intestine. Dev. Biol. 1999;216:114–134. doi: 10.1006/dbio.1999.9471. [DOI] [PubMed] [Google Scholar]
- Lowry J, Atchley W. Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding domain. J. Mol. Evol. 2000;50:103–115. doi: 10.1007/s002399910012. [DOI] [PubMed] [Google Scholar]
- Maduro M, Hill R, Heid P, Newman-Smith E, Zhu J, Priess J, Rothman J. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev. Biol. 2005a;284:509–522. doi: 10.1016/j.ydbio.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Maduro M, Kasmir J, Zhu J, Rothman J. The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development. Dev. Biol. 2005b;285:510–523. doi: 10.1016/j.ydbio.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Maduro M, Broitman-Maduro G, Mengarelli I, Rothman J. Maternal deployment of the embryonic SKN-1–NMED-1,2 cell specification pathway in C. elegans. Dev. Biol. 2007;301:590–601. doi: 10.1016/j.ydbio.2006.08.029. [DOI] [PubMed] [Google Scholar]
- McGhee J, Sleumer M, Bilenky M, Wong K, McKay S, Goszczynski B, Tian H, Krich N, Khattra J, Holt R, Baillie D, Kohara Y, Marra M, Jones S, Moerman D, Robertson A. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev. Biol. 2007;302:627–645. doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- McGhee J, Fukushige T, Krause M, Minnema S, Goszczynski B, Gaudet J, Kohara Y, Bossinger O, Zhao Y, Khattra J, Hirst M, Jones S, Marra M, Ruzanov P, Warner A, Zapf R, Moerman D, Kalb J. ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev. Biol. 2009;327:551–565. doi: 10.1016/j.ydbio.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J, Bao Z, Boyle T, Waterston R. The lineaging of fluorescently-labeled Caenorhabditis elegans embryos with StarryNite and AceTree. Nat. Protoc. 2006;1:1468–1476. doi: 10.1038/nprot.2006.222. [DOI] [PubMed] [Google Scholar]
- Murray J, Bao Z, Boyle T, Boeck M, Mericle B, Nicholas T, Zhao Z, Sandel M, Waterston R. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat. Methods. 2008;5:703–709. doi: 10.1038/nmeth.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J, Boyle T, Preston E, Vafeados D, Mericle B, Zhao Z, Bao Z, Boeck M, Waterston R. Gene expression in the C. elegans embryo: lineage motifs and multidimensional specification. doi: 10.1101/gr.131920.111. in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves A, Priess J. The REF-1 family of bHLH transcription factors pattern C. elegans embryos through Notch-dependent and Notch-independent pathways. Dev. Cell. 2005;8:867–879. doi: 10.1016/j.devcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Neves A, English K, Priess J. Notch-GATA synergy promotes endoderm-specific expression of ref-1 in C. elegans. Development. 2007;134:4459–4468. doi: 10.1242/dev.008680. [DOI] [PubMed] [Google Scholar]
- Owraghi M, Broitman-Maduro G, Luu T, Roberson H, Maduro M. Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network. Dev. Biol. 2010;340:209–221. doi: 10.1016/j.ydbio.2009.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Rifkin S, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermann EM, Strohmaier KR, Maduro MF, Rothman JH. Endoderm development in Caenorhabditis elegans: the synergistic action of ELT-2 and -7 mediates the specification→differentiation transition. Dev. Biol. 2010;347:154–166. doi: 10.1016/j.ydbio.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J, Schierenberg E, White J, Thomson J. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. Multiple Sequence Alignment Using ClustalW and ClustalX. Current Protocols in Bioinformatics. 2002:2.3.1–2.3.22. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- Vonderfecht TR, Schroyer DC, Schenck BL, McDonough VM, Pikaart MJ. Substitution of DNA-contacting amino acids with functional variants in the Gata-1 zinc finger: a structurally and phylogenetically guided mutagenesis. Biochem. Biophys. Res. Commun. 2008;369(4):1052–1056. doi: 10.1016/j.bbrc.2008.02.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda A, Kouike H, Okano H, Sawa H. Components of the transcriptional mediator complex are required for asymmetric cell division in C. elegans. Development. 2005;132:1885–1893. doi: 10.1242/dev.01776. [DOI] [PubMed] [Google Scholar]
- Yuzyuk T, Fakhouri THI, Kiefer J, Mango SE. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev. Cell. 2009;16(5):699–710. doi: 10.1016/j.devcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Fitch D, Herman M. TLP-1 is an asymmetric cell fate determinant that responds to Wnt signals and controls male tail tip morphogenesis in C. elegans. Development. 2002;129:1497–1508. doi: 10.1242/dev.129.6.1497. [DOI] [PubMed] [Google Scholar]
- Zhu J, Hill R, Heid P, Fukuyama M, Sugimoto A, Priess J, Rothman J. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 1997;11:2883–2896. doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Fukushige T, McGhee J, Rothman J. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev. 1998;12:3809–3814. doi: 10.1101/gad.12.24.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]