Abstract
Exosomes are virus-sized, membrane-enclosed vesicles with origins in the cellular endosomal system, but are released extracellularly. As a population, these tiny vesicles carry relatively enormous amounts of information in their protein, lipid and nucleic acid content, and the vesicles can have profound impacts on recipient cells. This review employs publically-available data combined with gene ontology applications to propose a novel concept, that exosomes transport transcriptional and translational machinery that may have direct impacts on gene expression in recipient cells. Here, we examine the previously published proteomic contents of medulloblastoma-derived exosomes, focusing on transcriptional regulators; we found that there are numerous proteins that may have potential roles in transcriptional and translational regulation with putative influence on downstream, cancer-related pathways. We expanded this search to all of the proteins in the Vesiclepedia database; using gene ontology approaches, we see that these regulatory factors are implicated in many of the processes involved in cancer initiation and progression. This information suggests that some of the effects of exosomes on recipient cells may be due to the delivery of protein factors that can directly and fundamentally change the transcriptional landscape of the cells. Within a tumor environment, this has potential to tilt the advantage towards the cancer.
Keywords: Cancer, exosomes, gene ontology, proteomics, transcriptional regulators
Cellular secretion of vesicles is vital to both normal and malignant cell function. Traditionally, cellular secretion of materials has been reserved to the classical and so-called non-classical secretory pathways. In the classical pathway nascent proteins emerging from a ribosome (usually) display an N-terminal signal sequence that targets the ribosomal complex to the endoplasmic reticulum (ER). Proteins are translated into the ER, where they may remain. Some proteins now derived from the ER but not addressed to remain there (or as proteins inserted into membranes) are targeted for the Golgi apparatus. Proteins not destined to stay in the Golgi are packaged into secretory vesicles and terminally released into the extracellular space.(1) The non-classical pathways are independent of the ER and Golgi, and three of them involve vesicular transport, including secretory lysosomes. Two of those pathways result from either direct budding of vesicles from the cell surface (yielding vesicles variously called microvesicles, ectosomes or microparticles) or a mechanism that involves the endocytic pathway, with the formation and fusion of multivesicular bodies, and the eventual exocytosis of extracellular nanovesicles, called exosomes.(1,2) Exosomes were originally regarded as a removal method for unneeded membrane proteins.(3) Recent evidence has revealed the importance of these nanovesicles beyond their role in cellular waste management, in that they are integral to protein trafficking, extracellular signaling and immunology.(4) Current research describes essential exosomal characteristics,(5) and exosome proteomics offers a glimpse into the information obtainable from these extracellular vesicles.(6) Furthermore, advancements in immunological research and drug delivery systems pose the possibility of using exosomes in the stimulation of the immune system against cancer cell antigens,(7) as well as the ability to use exosomes as drug delivery vehicles.(8)
Where Does One Find Exosomes?
Johnstone et al., studying the roles of released vesicles in reticulocyte maturation,(9) are credited with coining the term “exosomes,” which have since been found to be released from essentially every cell type.(10) Exosome secretions into culture medium have been illustrated by hematological (B-cells, T-cells, dendritic cells, mast cells and platelets) and non-hematological cells (microglia, epithelial cells, Schwann cells, adipocytes, neuronal cells, fibroblasts, germinal cells and many, many tumor types(10)). In addition, exosomes have been isolated and characterized in nearly all physiological fluids, such as plasma, serum, urine, saliva, breast milk, semen, prostatic fluid, cerebrospinal fluid, amniotic fluid, malignant effusions, pleural effusions, bronchial lavage and synovial fluid.(1)
Exosomes: Definitions and Biogenesis
Historically, extracellular vesicles have been subject to a rather ambiguous nomenclature. The derivation of exosomes is from multivesicular bodies (MVB), which are also called multivesicular endosomes (MVE).(11) Biogenesis initiates at the plasma membrane, where endocytosis of cell surface membrane materials mediates eventual molecular sorting, recycling or degradation via the lysosomal pathway. Following endocytosis and endosome formation, maturation and fusion, organelles known as late endosomes begin to display invaginations towards the inside or lumenal portion of the endosome. These bud off free into the endosomal lumen, and are called intralumenal vesicles (ILV) or intravesicular bodies (IVB); the endosome itself is now called an MVB or MVE. Notably, the membranes of the resultant ILV have undergone two membrane invaginations: one during the initial endocytosis that results in the inversion of the membrane and another invagination within the MVB that once again inverts the orientation to that of the original plasma membrane. This double inversion means that the ILV share similar transmembrane topology with the plasma membrane.(12)
The contents of ILV (“future exosomes”) depend on the membrane constituents of the MVB and the collection of cytosolic components captured or loaded into the ILV upon the intralumenal budding. Whether cargo are accidental or specific is unclear, but there are indications that some sorting signals exist. Although the exact biochemical sorting mechanisms are not elucidated, it is hypothesized or demonstrated that mono-ubiquitination, higher-order oligomerization, certain membrane anchors, members of the tetraspanin networks, and possibly protein complexes including heat shock proteins and metabolic enzymes are preferentially loaded into ILV as exosome cargo.(13) RNA-loading mechanisms are even less clear,(14) but there may be sequence motifs related to sorting.(15) microRNA loading may depend on association of members of the RNA-induced silencing complex (RISC), such as AGO2.(16) Both mRNA and miRNA found in extracellular vesicles are in different relative proportions compared to their parental cells,(17) suggesting that there may be some control over loading of RNAs.
Intralumenal vesicle content is also dictated to some extent by physical constraints. Their relatively small size is dictated by their derivation from the endosome and their lipid membrane structure.(1,10) The minimum vesicle size is determined by the lipid bilayer composition, and below a certain size the structural foundation is unstable. Lipid bilayer composition typically averages 5 nm, restricting vesicle size to approximately 30 nm.(10) Maximally, exosomes can only reach 500 nm due to their endosomal origin. Although exosome sizes can potentially range from 30–500 nm, exosomes are typically regarded as vesicles ranging from 30–100 nm.(1,10) The smaller size of exosomes, in comparison to other extracellular vesicles such as apoptotic blebs or microparticles (ranging from 100 nm to greater than 1000 nm), significantly reduces the potential vesicle capacity. Typically, the internal volume of an exosomes ranges between 20 and 90 nm3, resulting in a potential cargo volume of 4.2–380 yoctoliters.(10,18) This internal volume is comparable to the size of a ribosome complex, suggesting that exosomes could contain roughly 100 proteins and 10 000 nucleotides of nucleic acid.(10)
Intralumenal vesicle content may also have repercussions for exosome release. Endosomal sorting complex required for transport (ESCRT) protein complexes, and raft-based microdomains influencing independent assortment may play roles in the ultimate fate of ILV.(19–21) Ceramide, a lipid molecule composed of a sphingosine and a fatty acid, has been implicated in the creation of membrane microdomains and the induction of invaginations within the plasma membrane. Work by Trajkovic et al. illustrates the diminished ILV formation and exosome release in cells inhibited by sphingomyelinase, a protein integral to the cellular synthesis of ceramide.(22) Furthermore, tetraspanins, which are very prevalent on exosomes and are involved in their biosynthesis, have recently been implicated in the induction of exosomal sorting, specifically CD9 and CD82, while CD63 and CD81 are shown to bind components of the ESCRT protein complex.(23–25)
Intralumenal vesicles have several potential fates: (i) canonically, MVB fuse with the lysosome, resulting in lysosomal degradation and recycling; (ii) the vesicles may traffic to the Golgi for redistribution and recycling; (iii) “back fusion,” a circumstance where the ILV fuse back to the endosomal (MVB) limiting membrane and return their contents to the cytoplasm; and (iv) fusion of the MVB, with the plasma membrane resulting in exosomal vesicle release outside the cell.
Exosomal extracellular release occurs through fusion of the MVB with the plasma membrane. This process requires transport of the MVB to the peripheral plasma membrane, followed by docking and fusion with the plasma membrane to release of the exosomes. Fusion of MVB to the plasma membrane is highly dependent on components of the endocytic machinery, which include RAB GTPase, RAB 4, 5, 7, 11, 27, 37, cytoskeletal regulatory proteins, molecular trafficking proteins such as myosin, and membrane tethering SNARe elements.(12,23) RAB proteins are classically known to participate in the endocytic pathway and their role in exosome release is no surprise. For instance, cells with RAB35 knocked down exhibit a decline in the extracellular release of exosomes. RAB27a and b RNAi studies illustrate their vital functions in MVB docking and the subsequent release of exosomes.(23,26,27) RAB11 studies indicate a significant role in MVB release; however, the exact role of RAB11 is still under much investigation. Current understanding suggests that RAB11 proteins influence the behavior of MVB in human cells and may alter the process of exosome secretion. The significant roles of RAB proteins in MVB fusion and exosome release are well accepted, yet the exact mechanisms of RAB-influenced exosome biogenesis and release are still not fully understood. Different combinations of RAB proteins may be involved in MVB function at various stages of exosome biogenesis. The involvement of RAB proteins may be influenced by microenvironmental conditions and RAB proteins may alter the lipid and cholesterol content of MVB membranes. Alterations of MVB lipid and cholesterol motifs have structural implications for formation and release of exosomes.
The mechanism of exosome release is also influenced by cellular ion concentrations. Exosomes are known to be sensitive to changes in intracellular calcium concentrations and depolarizations by potassium concentration changes. These processes, in part, also seem to be influenced by RAB11 proteins.(28–32) The mechanisms and roles of the various proteins responsible for exosome biogenesis require further research. The understanding of these mechanisms is vital to the understanding of exosome function, significance and potential biomedical applications.
Other Extracellular Vesicles
As noted briefly above, there are other types of membrane-enclosed vesicles released by cells into the extracellular environment. Some are from specialized cells such as neurons during neurotransmitter release or other types of secretory cells, or from secretory lysosomes.(33) Apoptotic bodies are another form of EV released by cells under circumstances of cellular apoptosis (programmed cell death).(34) Other vesicles are released directly from the cell surface, and these have various names but no distinct definitions. These include terms such as microvesicles, microparticles, ectosomes, shedding vesicles, and, if from cancer cells, oncosomes.(5) One distinguishing feature of these vesicles is that they are considered to be larger than exosomes (the former typically regarded as 30–100 nm vesicles; these other EV are generally described in the 100–1000-nm range or larger). One of the problems with classifying EV is that the machinery to generate them is similar whether the vesiculation occurs at the cell surface or at the endosomal cytosolic face; thus, protein markers that can truly distinguish the different vesicle types are not obvious.(5) Differential centrifugation is also a potential method for separation of microvesicles from exosomes, but this, too, must be used with care.(35) For our purposes in this article, we will use the term “exosome” to refer to 30–100-nm diameter vesicles that are derived from the endosomal system but released extracellularly. We realize that other EV may fit some of the biochemical and biophysical parameters, but we believe this definition fits closest to accepted standards in the field.
Exosome Contents
Until recently, efforts in differentiation, isolation and characterization of these extracellular vesicles proved very challenging. Progress in isolation and characterization divulged essential defining characteristics. Notably, defining characteristics of exosomes are also related to their endosomal origins. All exosomes contain membrane transport proteins, metabolic enzymes, fusogenic proteins, tetraspanins, heat shock proteins, multivesicular body biogenesis proteins, lipid-related proteins and phospholipases.(1,10) Despite the fact that exosomes have in common these general protein families, vast diversity and variation still exists between exosomes from different cells and during cellular maturation.(10) Importantly, Carayon et al. observed changes in the protein repertoire of exosomes during erythrocyte maturation, a feature that hints at the variability of these nanovesicles as a conditionally-dependent function.(10,36,37) Besides membrane-associated proteins, exosomes contain a multitude of lipid molecules. Of these, the most common raft-associated lipids molecules are cholesterol, ceramide, sphingolipids, phosphoglycerides and fatty-acyl chains.(1,10,12,38) Thus, the lipid and protein composition of exosomes are cell-type dependent.
Although once regarded as extracellular dumps composed of irrelevant cellular content, there is clear significance to exosomal content. Exosomes contain several types of cellular material, such as proteins, bioactive lipids, metabolites and nucleic acids. Further investigations indicate disparities in exosome contents from various cells such as reticulocytes and immune cells. The variations in exosome content may indicate specific cellular exosome functions. These functions may be immunoprotective in cells such as erythrocytes, where the presence of CD55 and CD59 prevent complement-mediated attack.(28,39,40) Additional examples of cellular-specific exosome functions are observed in human immune cells, such as antigen-specific major histocompatibility complex (MHC) responses induced by B-cell exosomes in vitro. Investigational studies also indicate B-cell and dendritic cell exosome-mediated adhesion signals that are able to act at a distance.(41,42) The presence of adhesion proteins within exosomes is not limited to those from immune cells, and these proteins are found throughout many cellular-derived exosomes. Types of adhesion proteins include CD146, CD9, CD11, CD58, CD171, LGALS1, LGALS3BP, GP9, ITGB3, ITGA2B and GP1BA.(43) In addition, co-stimulatory markers such as CD80 and CD82 on antigen presenting cell-derived exosomes, CD41a and Von Willebrand factor on platelet-derived exosomes, and perforin and granzyme on cytotoxic T-cell-derived exosomes have been identified as contents of exosomes.(44–47)
Exosome content is also subject to changes in the cellular microenvironment. Changes in vesicle content have been observed during pathological disease states and chronic illnesses, as well as normal conditions.(48–50) Research into exosome cargo changes during disease may prove vital to the understanding of cellular and systemic responses to disease. Many of these studies have focused on vesicle changes during malignant disease.(51,52) Proteomic studies investigating exosomes derived from the periphery (e.g. blood or urine) of patients with cancer are significant for numerous proteins, inflammatory markers, and serum proteins and various types of nucleic acids.(53–56) Nucleic acids such as messenger RNA (mRNA) contained within cancer cell exosomes allude to the potential of distant cellular mRNA uptake and protein translation.(57,58) The presence of exosomal or other EV mRNA raises the question of whether metastatic and microenvironmental changes may be due in part to malignant cell exosome cargo contents. Investigation into this question must first proceed with research into the process of mRNA exosome cargo sorting. Current studies do not show a proportional parallel between the cellular mRNA content and the cellular derived exosomes.(17) This may indicate that mRNA exosome sorting, which is partially mediated by size and mechanism, affects the selection of exosome destined mRNA.(59,60)
Exosomes and Neoplastic Disease
Neoplastic disease progression manipulates the cancer microenvironment. One may postulate that tumor-driven microenvironment changes may allow for tumor induced-immune suppression as well as microinfiltrative disease. The metabolic relationship between rapidly proliferative cancer cells and the immediate tumor microenvironment leads to a hypoxic, acidic and nutrient-poor milieu that selects for malignant cells able to tolerate a deprived environment. Cancer cells able to survive hypoxic environments can effectively activate hypoxia-inducible factor-1 alpha (HIF1A) signaling, which results in increased expression of vascular endothelial growth factor (VEGF) and increased angiogenesis.(61) Tumor cells that tolerate increased microenvironment acidification have active proton pumps that allow for the maintenance of intracellular pH and permit normal cellular function in an acidic microenvironment.(62) Thus, the microenvironment-driven selection also predicates the survival of cancer cells that possess highly motile properties, are invasive, and can efficiently manipulate different pathways of cellular metabolism.
Cellular-mediated exosome release within the neoplastic microenvironment occurs in many cell types, such as stromal and cancer cells as well as immune cells. Exosomes released from cancer cells may significantly impact the cancer microenvironment by influencing neighboring stromal cells, immune cells and other cancer cells. Exosomes isolated from glioblastoma multiforme cells have been proposed to promote tumor growth by transporting RNA into recipient cells in the microenvironment.(17) This RNA transport results in epigenetic changes leading to novel protein translation in recipient stromal and glial cells. Other research exploring peripherally released exosomes indicates the potential for systemic mediated effects that allow for neoplastic disease progression, or, in some cases, influencing dormancy. Such examples exist in ovarian cancer, breast cancer, melanoma and colorectal cancer exosomes.(51,54,63–65) Peripherally isolated exosomes from tumor patients’ body fluids (blood, ascites fluid and urine) contained pro-oncogenic factors, like epidermal growth factor receptor (EGFR), proinflammatory cytokines, proangiogenic factors such as VEGF, and other metastatic disease promoting factors.(17,66–68) Cancer cell-derived exosomes release stromal manipulative factors, including fibroblast growth factor-2 (FGF-2), transforming growth factor beta (TGF-β), matrix metalloproteinase 2 (MMP2) and urokinase plasminogen activator.(68–71) These factors induce stromal cell changes leading to progression of malignant migration, while also altering the structural basis of the extracellular matrix (ECM). Cancer cell exosome-induced ECM remodeling may have supplementary effects, resulting in the loss of cancer cell adhesion properties that allow for neoplastic invasion.(67,68)
Cancer cell-derived exosomes also contain factors that suppress the immune system during neoplastic disease states. Tumor-mediated immunosuppression is a characteristic phenotype that is found among many types of cancers. Immunosuppression results in failed immunosurveillance/immune cytotoxicity of tumor cells and significantly increases the risk for disease progression. Mechanisms of exosome-driven immunosuppression are extensive and affect T-cells and inflammatory signaling. Tumor exosomes inhibit natural killer (NK) cells' and T cells' cytotoxic and proliferative activities via inhibition of interleukin-2 (IL-2), and also promote IL-10 and TGF-β release. Fas ligand displayed on tumor exosomes induces apoptosis.(72) Suppression of cytotoxic NK and T-cell activities has been attributed to inhibition of Jak-3-mediated pathways, suppression of cyclin D3 and significant decreases in perforin release.(73,74) The global effect of cancer-derived exosomes on the tumor microenvironment and systemic physiology poses important questions about the significance of cancer cell-derived exosomes.
Nucleic Acid Binding Proteins Within Cancer-derived Exosomes: Roles for Transcription Factors?
Cancer cell exosome proteomics concentrates on the identification/characterization of exosomal protein cargo and its implications, including the influential potential of such cargo. Overall, there has been limited attention paid to the presence of nucleic acid binding proteins found within cancer cell exosomes. Given the current focus on various RNAs as exosome cargo(75,76) it seems interesting that there are few studies regarding the roles of RNA/nucleic acid-binding proteins in exosomes (although there has been speculation concerning the release of mRNA regulatory proteins during synaptic transmissions(77)). Another concept related to that of nucleic acid-binding proteins is the idea that transcription factors may be exosome cargo constituents with the downstream effect of directed gene expression. This topic has barely creased the surface of exosome studies,(78,79) so we delved further into it with information from our own proteomic studies as well as utilization of larger comprehensive databases.
The presence of transcription factor proteins in the cargo of neoplastic cell-derived exosomes may allow for distant cell alterations in cellular transcription, resulting in altered signaling of both normal and cancer cells. The influences of transcription factors on distant cells may mediate important malignant signaling pathways that permit disease progression. Such examples may exist in the nutrient-deprived cancer microenvironment, where cancer cells’ upregulation of HIF1A, VEGF and EGFR can be transduced to nearby cells via exosomes. This microsignaling may effectively prime proximal cells for the toxic microenvironment, allowing more cells to sustain viability despite environmental nutritional depletion and acidification. These transcription factors may mediate effects observed in immune cells and stromal cells. Immune cell uptake of transcription factors may effectively suppress or alter cellular function by transcriptional dysregulation. Alterations in immune cell function may prevent neoplastic cell identification and clearance, thus allowing for neoplastic disease and inhibiting the immune system's ability to limit disease progression.
To this end, we reviewed whether transcription factors/transcriptional regulators are present within exosomes collected from tumor cell lines or from other published works, largely by proteomic identification of such proteins present in exosomes from cell lines or physiologic fluids. We also explored potential affected signaling pathways that may be influenced by transcription factors/transcriptional regulators found in exosomes.
Re-analysis of previous data
We previously published proteomic data from medulloblastoma exosomes,(73) identifying 144 proteins; of those, 14 (9.6% of the total) were categorized as “transcriptional regulators” by Ingenuity Pathway Analysis (IPA) using gene ontology-type (GO) algorithms (Table 1). Six of those (BARD1, FUBP1, HNF4A, RFC1, SMARCA1 and ZSCAN16) have direct transcriptional features; following IPA Core Analysis, the top IPA Associated Network Function was “Connective Tissue Disorders, Developmental Disorder, Hereditary Disorder” (that interactome is shown in Fig. 1). Protein interactions within the interactome include protein kinase A (PKA), BARD1, RNA polymerase, HNF4A, TP53, PSMC5, RFC1, FUBP1, SMARCA1, RNF39, KHDRBS1, ACTN2 and UBC. There are important nodes with histone H3, HNF4A, RNA Pol II, BARD1, p53 and ubiquitin C, with significant connections to cell transcription and protein degradation pathways. Notable interactome proteins/functions include: SMARCA1, involved in SWI/SNF pathways and affecting chromatin structure; major histocompatibility complex 1 expression proteins; KHDBRS1, a complex molecule that reduces Creb-binding protein (CBP)-driven transcription by competing with other transcription factors for CBP binding sites; the proteasome subunit PSMC5; the known (frequently-mutated) tumor suppressor protein, TP53, involved in many cancers; and BARD1, a protein that influences BRCA1 stability. Furthermore, RFC1 and KHDRBS1/HNF4A affect DNA repair/replication mechanisms and are involved in nuclear transcription factor expression, respectfully. In summary, medulloblastoma exosomal transcription factor protein/transcriptional regulator interactomes display significant pathway interactions with known cancer-mutated pathways such as p53 and BRCA1. Interactome pathways also have interactions with nuclear transcription factor expression, immune system MHC processing and cell cycle regulatory proteins. Mutations or alterations in any of these pathways could lead to neoplastic changes in non-cancerous cells or modulation of normal MHC I expression and immune surveillance by the host's immune system.
Table 1.
Transcriptional regulators identified in medulloblastoma exosome proteomics
| ID | Symbol | Entrez gene name | Subcell local | Function |
|---|---|---|---|---|
| P35609 | ACTN2 | Actinin, alpha 2 | Nucleus | Txn regulator |
| Q9NWB6 | ARGLU1 | Arginine and glutamate rich 1 | Nucleus | Txn regulator |
| Q99728 | BARD1 | BRCA1 associated RING domain 1 | Nucleus | Txn regulator |
| Q96AE4 | FUBP1 | Far upstream element (FUSE) binding protein 1 | Nucleus | Txn regulator |
| P41235 | HNF4A | Hepatocyte nuclear factor 4 alpha | Nucleus | Txn regulator |
| Q07666 | KHDRBS1 | KH domain containing, RNA binding, signal transduction associated 1 | Nucleus | Txn regulator |
| P62195 | PSMC5 | Proteasome (prosome, macropain) 26S subunit, ATPase, 5 | Nucleus | Txn regulator |
| P35251 | RFC1 | Replication factor C (activator 1) 1, 145 kDa | Nucleus | Txn regulator |
| P28370 | SMARCA1 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 | Nucleus | Txn regulator |
| Q16385 | SSX2 | Synovial sarcoma, X breakpoint 2 (A&B) Cancer/testis antigen 5.2, HOM-MEL-40 | Nucleus | Txn regulator |
| Q9H5I1 | SUV39H2 | Histone-lysine N-methyltransferase SUV39H2 | Nucleus | Txn regulator |
| Q9H7E2 | TDRD3 | Tudor domain containing 3 | Nucleus | Txn regulator |
| P17030 | ZNF25 | Zinc finger protein 25 | Nucleus | Txn regulator |
| Q9H4T2 | ZSCAN16 | Zinc finger and SCAN domain containing 16 | Nucleus | Txn regulator |
Proteins that were identified as transcriptional regulators (“Txn regulator”) by “Function” from(73) were tabulated according to their “ID”/Accession number (UniProt Knowledge Base, Swiss-Prot/TrEMBL, http://us.expasy.org/sprot/). “Symbol” is the Entrez gene ID with the “Entrez Gene Name” as a descriptor. “Subcell Local” refers to the subcellular localization most typically described for the protein.
Fig 1.
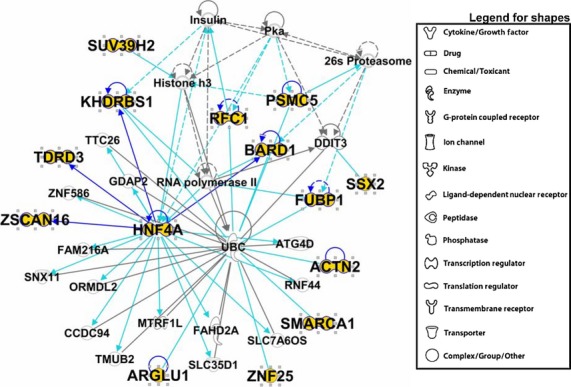
Top Ingenuity Pathway Analysis (IPA) “interactome” derived from transcriptional regulators found in medulloblastoma exosome proteomics. Proteins clustered within the Top Network/Associated Functions as derived from IPA algorithms are shown as members of this “interactome,” which was entitled “Connective Tissue Disorders, Developmental Disorder, Hereditary Disorder.” Proteins identified during that work(73) are labeled in larger bold font, with the protein symbol in gold fill. Direct connections between/among proteins are shown in solid lines; indirect interactions are shown as dashed lines (also called “edges”). Connections between proteins identified in this proteomic screen are shown in dark blue; interactions between proteins we identified and proteins not identified in our proteomics are shown in turquoise. Protein shapes are indicative of function, and that legend is to the right.
Analysis of the Vesiclepedia database
We then examined the most comprehensive exosome protein database, Vesiclepedia, a manually-curated database of exosome/extracellular vesicle proteins, lipids and RNAs (microvesicles.org), with over 43 000 protein entries currently, culled from publications and direct submissions.(80) We utilized this database as a source of overall exosome proteomic information with particular emphasis on nuclear proteins and those involved in transcription and transcriptional regulation. GO analyses were performed with, and pathways/interactomes were developed, using Ingenuity Pathway Analysis. Proteomic data were collected from Vesiclepedia (December 2013); all proteins present within the database were culled to remove duplicate proteins, as well as filtered to convert the protein/gene names to human inputs (generally the species source of the most literature information). The remaining proteins were analyzed and categorized for different protein types, allowing for interpretation of human proteins when applied to the IPA Knowledgebase. Proteins were categorized based on subcellular/extracellular localization and cellular function. Major categories derived from this analysis were cytokines, enzymes, G-protein coupled receptors, growth factors, ion channels, kinases, ligand dependent nucleic acid receptors, peptidases, phosphatases, transcription factor regulators, transcription regulators, transmembrane receptors, transporters, and proteins labeled as “other” that did not fit clearly into the categories (Table 2).
Table 2.
Ingenuity Pathway Analysis functional categories for Vesiclepedia protein compendium and total numbers per category
| Total numbers of proteins | Functional category |
|---|---|
| 1254 | “Other” |
| 728 | Enzyme |
| 325 | Transporter |
| 156 | Peptidase |
| 133 | Kinase |
| 100 | Transmembrane receptor |
| 82 | Transcription regulator |
| 55 | Phosphatase |
| 49 | Ion channel |
| 43 | Translation regulator |
| 41 | Cytokine |
| 33 | Growth factor |
| 27 | G-protein coupled receptor |
| 1 | Ligand-dependent nuclear receptor |
Ingenuity Pathway Analysis grouped the identified proteins into 14 functional subsets shown with the total numbers of proteins listed for each category.
Potential transcriptional/translational regulators in the exosome proteome
Reviewing our previous exosome proteomic data from a murine brain tumor line,(81) and from human medulloblastoma cell lines,(73) we noted a number of proteins potentially involved in transcription or translation, ranging from ribonuclear proteins to transcription factors. In these works, 10–20% of the identified proteins had some role in transcriptional and/or translational activities, suggesting the exosomes may be able to vectorally influence recipient cells not merely with RNA-type cargo, but with proteins that may govern or contribute to gene expression. We next conducted similar pathway explorations on known transcription regulators found in all the curated exosome proteomic studies. We mined the Vesiclepedia database for proteins with potential involvement in transcriptional regulation.
Proteomic data collected from the Vesiclepedia database initially revealed a total of over 13 000 proteins. This collection was filtered to remove duplicates and chemicals, and to convert non-human proteins IDs to human IDs (for better recognition in the IPA programs). This yielded a total of 3069 proteins with mapped IDs, and 3027 were available for analysis. Of these proteins, 1426 proteins are described as cytoplasmic, 383 proteins are found in the extracellular space, 346 proteins are nuclear, 627 are plasma membrane proteins and 245 are classified as “other.” Proteins in the “other” category often were structural proteins, membrane proteins with various functions, chaperones, histones and proteins with unknown activities or with difficult functions to categorize. The total protein catalog is available in Table S1.
Of the 82 proteins listed by IPA as “transcription regulators” derived from the Vesiclepedia database, (Table S2), most are multifunctional proteins that are often found in complexes with various activities in several subcellular locations. This group of proteins contains a number of known transcription factors and regulators (CREB1, NKX6-1, NRF1, PCDB1, PRDM16, PURB, RREB1, SMARCA4, STAT1, SUB1 and YBX1), a host of proteins that are histones or histone modifiers, such as histone acetylases or deacetylases and chaperones (ASH1L, CARM1, CALR, H2AFX, HDAC5, HUWE1, NIPBL, NPM1, PA2G4, RBBP7, RUVBL1, RUVBL2 and SIRT2), and binding partners of estrogen and thyroid hormone receptors (BTG1, BTG2, PHB2, PSMC5, RPL7, TRIP4 and TRIP13). IPA Core Analysis of these proteins generated the top 5 Associated Network Functions that all included “Gene Expression” as a lead component: (i) Gene Expression, Cell Cycle, Infectious Disease; (ii) Gene Expression, Cell Cycle, Cellular Assembly and Organization; (iii) Gene Expression, Cell Cycle, Cellular Development; (iv) Gene Expression, Cellular Assembly and Organization, Cellular Compromise; and (v) Gene Expression, Digestive System Development and Function, Hepatic System Development and Function. The interactome of the highest-scoring Associated Network Function (Gene Expression, Cell Cycle, Infectious Disease) is shown in Figure 2, where the extensive connectivity between that subset of factors and regulators is evident, as well as their relationships to RNA Polymerase II. Other interactomes show expected associations with histones, thyroid hormone receptors, ubiquitin and proteasomes (not shown).
Fig 2.

Top Ingenuity Pathway Analysis (IPA) interactome derived from transcriptional regulators found in the Vesiclepedia database. Proteins clustered within the Top Networks/Associated Functions as derived from IPA algorithms are shown as members of this interactome, which was entitled “Gene Expression, Cell Cycle, Infectious Disease.” Details of the symbols and edges are described in the legend for Figure 1.
Of the “Top Diseases and BioFunctions: Diseases and Disorders,” we found: Infectious Disease; Organismal Injury and Abnormalities; Renal and Urological Disease; Cancer; and Hematological Disease (P-values from 10−6 to 10−5). In “Top Diseases and BioFunctions: Molecular and Cellular Functions,” we found: Gene Expression; Cellular Growth and Proliferation; Cellular Development; Cell Death and Survival; and Cell Cycle (P-values from 10−41 to 10−7). In “Top Diseases and BioFunctions: Physiological System Development and Function,” we found: Tumor Morphology; Organismal Survival; Embryonic Development; Organismal Development; and Skeletal and Muscular System Development and Function (P-values from 10−6 to 10−4). The Top 12 Canonical Pathways are shown in Figure 3, where cancer-related signaling stands out repeatedly, as do pathways related to such signaling. These data strongly suggest that exosomes may be considered extrinsic genetic modulators not only by their nucleic acid cargo, but also by virtue of protein factors such as these transcriptional regulators that can profoundly influence recipient cell activities driven at fundamental levels.
Fig 3.
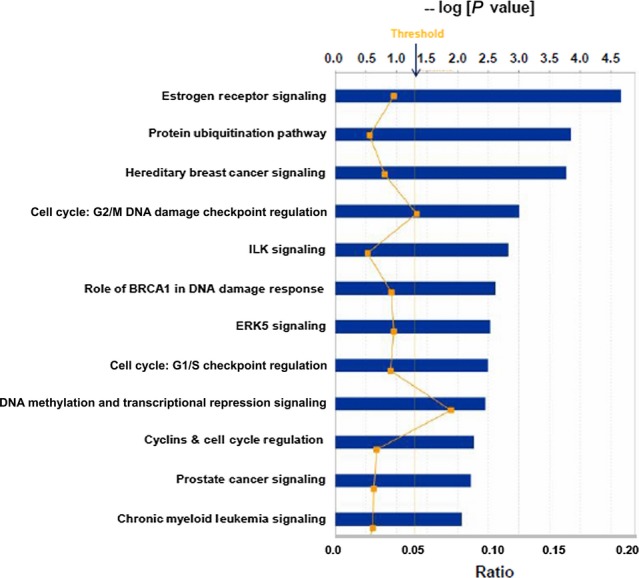
Top 12 Canonical pathways derived from Ingenuity Pathway Analysis (IPA) gene ontology analysis of transcriptional regulators found in the Vesiclepedia database. Following IPA “Core Analysis,” these 12 Canonical Pathways emerged. Graph shows category scores; “Threshold” indicates the minimum significance level (scored as –log [P-value] from Fisher's exact test, set here at 1.25). “Ratio” indicates the number of molecules from the data set that map to the pathway listed divided by the total number of molecules that map to the canonical pathway from within the IPA database.
Exosome proteomic data from our previous work (murine brain tumors and human medulloblastomas) as well as from our current unpublished datasets (not shown) suggest that transcriptional/translational regulatory factors are present within exosomes from several different cell sources. Analysis of the Vesiclepedia database confirms the presence of transcription regulatory proteins across exosomes from many different sources and experiments; we suggest that these proteins may be important for potential extrinsic modification of recipient cells. Furthermore, GO applications produce numerous pathway associations that parallel common mutated pathways seen in cancer cells, as well as landmark signaling pathways essential to cancer cell survival.
The transcription regulator interactomes from both the limited study (medulloblastoma) and the expansive study (Vesiclepedia) maintained consistent themes, such as nodes with RNA polymerase II, histone, ubiquitin and proteasome interactions. Two of the top 6 Canonical Pathways in both datasets were “Role of BRCA1 in DNA Damage Response” and “Hereditary Breast Cancer Signaling”; “Cell Cycle” and “Cellular Development” were top categories in Molecular and Cellular Functions (not shown). These common motifs suggest that there may be preferential cargo loading for transcriptional regulatory factors that may influence these downstream pathways. However, there may be skewing of the data towards tumor-related areas because perhaps as many as 40% of exosome publications are cancer studies. Nonetheless, one may argue that cancer cells employ exosomes as efficient “information packets,” possibly for the protection or maintenance of unstable proteins or proteins complexes such as transcriptional machinery(82) as well as for the abilities of exosomes to enter cells by various mechanisms.(83)
Transcription factors have the ability to modulate expression levels of specific pathways in those cells with the appropriate transcription complex cohort. Examples of potential modulated pathways from our own data may be observed in medulloblastoma cells, where the presence of transcription factors characterized in an interactome illustrates recognizable neoplastic disease pathways. These pathways include regulation of chromatin structural changes, immune system functional proteins involved in MHC class I expression, and known tumor suppressor genes. Most notable, the identification of p53 tumor suppressor gene pathway in medulloblastoma transcription factor interactomes is consistent with known p53 upstream and downstream mutations in many cancers, a central mechanism in the mutagenesis of cancer cells. Moreover, the identification of BRCA1 associated BARD1 suggests that medulloblastoma exosome cargo transcription factor proteins may have interactions with more than one tumor suppressor gene. Indeed, our work suggested that HNF4A may act as a tumor suppressor in medulloblastoma.(73) A number of the transcription regulators identified in the Vesiclepedia database were also of the suppressor/repressor category, suggesting that turning off transcription may be an important functional feature of the regulator cohort.
Another phenomenon that we did not pursue, but that did not go unnoticed, was the presence of numerous translational regulators (particularly ribosomal proteins and elongation factors) present in exosomes. DNA and RNA binding proteins could conceivably be necessary for compacting relatively large nucleic acid structures into the small interior spaces of exosomes, but the translational regulators themselves could have near-immediate impact upon entering recipient cells, driving protein production from mRNAs of exosomal or recipient cell derivation. Thus, there are several layers of potential impact from exosome–recipient cell pathways triggered by binding and/or internalization of exosomes, epigenetic changes due to transfer of nucleic acid species, as well as pathways driven (or repressed) by transcriptional/translational regulatory protein exosomal content.
The potential consequences of modulation of these pathways may be crucial to cancer development and disease progression. Although this work begins to shed light on the significance of transcription regulatory proteins in the cargo of exosomes, more research is obviously necessary to fully comprehend and characterize the important roles of such proteins, in particular the downstream driving of gene expression resulting from the transferred modulators. The presence of transcription regulatory proteins may affect multiple pathways within recipient cells, potentially altering any number of cellular outcomes. Continued research efforts to understand the effect of exosomal cargo protein is vital in the generation of new therapies, particularly in the area of predicted changes in phenotypic outcomes of recipient cells based on exosome transcriptional regulatory protein cargo.
Disclosure Statement
The authors have no conflict of interest to declare.
Supporting Information
Additional supporting information may be found in the online version of this article:
Table S1. Unique proteins identified in the Vesiclepedia database.
Table S2. Transcriptional regulators identified in the Vesiclepedia composite database.
References
- 1.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–99. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 2.Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol. 2009;10:148–55. doi: 10.1038/nrm2617. [DOI] [PubMed] [Google Scholar]
- 3.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–20. [PubMed] [Google Scholar]
- 4.Simons M, Raposo G. Exosomes-Vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–81. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raimondo F, Morosi L, Chinello C, Magni F, Pitto M. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics. 2011;11:709–20. doi: 10.1002/pmic.201000422. [DOI] [PubMed] [Google Scholar]
- 7.Tan A, De La Pena H, Seifalian AM. The application of exosomes as a nanoscale cancer vaccine. Int J Nanomedicine. 2010;5:889–900. doi: 10.2147/IJN.S13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun D, Zhuang X, Zhang S, et al. Exosomes are endogenous nanoparticles that can deliver biological information between cells. Adv Drug Deliv Rev. 2013;65:342–7. doi: 10.1016/j.addr.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Johnstone RM. The Jeanne Manery-Fisher Memorial Lecture 1991. Maturation of reticulocytes: formation of exosomes as a mechanism for shedding membrane proteins. Biochem Cell Biol. 1992;70:179–90. doi: 10.1139/o92-028. [DOI] [PubMed] [Google Scholar]
- 10.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–8. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–23. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 12.Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83:1484–94. doi: 10.1016/j.bcp.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graner MW. Brain tumor exosomes and microvesicles: Pleiotropic effects from tiny cellular surrogates. In: Garami M, editor. Molecular Targets of CNS Tumors. InTech: Rijeka, Croatia; 2012. pp. 43–78. InTech Open Access. [Google Scholar]
- 14.Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, Pegtel DM. Exosomes: fit to deliver small RNA. Commun Integr Biol. 2010;3:447–50. doi: 10.4161/cib.3.5.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batagov AO, Kuznetsov VA, Kurochkin IV. Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genomics. 2011;12(Suppl 3):S18. doi: 10.1186/1471-2164-12-S3-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–9. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 17.Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nieuwenhuysen P, Heremans K, Clauwaert J. High-pressure scattering study of Artemia salina ribosomes and polysomes. Biochim Biophys Acta. 1980;606:292–303. doi: 10.1016/0005-2787(80)90039-8. [DOI] [PubMed] [Google Scholar]
- 19.Wubbolts R, Leckie RS, Veenhuizen PT, et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278:10963–72. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 20.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–21. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Kharaziha P, Ceder S, Li Q, Panaretakis T. Tumor cell-derived exosomes: a message in a bottle. Biochim Biophys Acta. 2012;1826:103–11. doi: 10.1016/j.bbcan.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Trajkovic K, Hsu C, Chiantia S, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–7. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 23.Meckes DG, Jr, Raab-Traub N. Microvesicles and viral infection. J Virol. 2011;85:12844–54. doi: 10.1128/JVI.05853-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. Exosome release of beta-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol. 2010;190:1079–91. doi: 10.1083/jcb.201002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gan X, Gould SJ. Identification of an inhibitory budding signal that blocks the release of HIV particles and exosome/microvesicle proteins. Mol Biol Cell. 2011;22:817–30. doi: 10.1091/mbc.E10-07-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu C, Morohashi Y, Yoshimura S, et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189:223–32. doi: 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 28.Singh RK, Mizuno K, Wasmeier C, et al. Distinct and opposing roles for Rab27a/Mlph/MyoVa and Rab27b/Munc13-4 in mast cell secretion. FEBS J. 2013;280:892–903. doi: 10.1111/febs.12081. [DOI] [PubMed] [Google Scholar]
- 29.Fukuda M. Rab27 effectors, pleiotropic regulators in secretory pathways. Traffic. 2013;14:949–63. doi: 10.1111/tra.12083. [DOI] [PubMed] [Google Scholar]
- 30.Zitvogel L, Regnault A, Lozier A, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 31.Savina A, Fader CM, Damiani MT, Colombo MI. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic. 2005;6:131–43. doi: 10.1111/j.1600-0854.2004.00257.x. [DOI] [PubMed] [Google Scholar]
- 32.Clayton A, Tabi Z. Exosomes and the MICA-NKG2D system in cancer. Blood Cells Mol Dis. 2005;34:206–13. doi: 10.1016/j.bcmd.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Blott EJ, Griffiths GM. Secretory lysosomes. Nat Rev Mol Cell Biol. 2002;3:122–31. doi: 10.1038/nrm732. [DOI] [PubMed] [Google Scholar]
- 34.Wickman G, Julian L, Olson MF. How apoptotic cells aid in the removal of their own cold dead bodies. Cell Death Differ. 2012;19:735–42. doi: 10.1038/cdd.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muralidharan-Chari V, Clancy JW, Sedgwick A, D'Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci. 2010;123:1603–11. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conde-Vancells J, Rodriguez-Suarez E, Gonzalez E, et al. Candidate biomarkers in exosome-like vesicles purified from rat and mouse urine samples. Proteomics Clin Appl. 2010;4:416–25. doi: 10.1002/prca.200900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carayon K, Chaoui K, Ronzier E, et al. Proteolipidic composition of exosomes changes during reticulocyte maturation. J Biol Chem. 2011;286:34426–39. doi: 10.1074/jbc.M111.257444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89:205–12. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 39.Blanc L, De Gassart A, Geminard C, Bette-Bobillo P, Vidal M. Exosome release by reticulocytes–an integral part of the red blood cell differentiation system. Blood Cells Mol Dis. 2005;35:21–6. doi: 10.1016/j.bcmd.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Rabesandratana H, Toutant JP, Reggio H, Vidal M. Decay-accelerating factor (CD55) and membrane inhibitor of reactive lysis (CD59) are released within exosomes during In vitro maturation of reticulocytes. Blood. 1998;91:2573–80. [PubMed] [Google Scholar]
- 41.Clayton A, Turkes A, Dewitt S, Steadman R, Mason MD, Hallett MB. Adhesion and signaling by B cell-derived exosomes: the role of integrins. Faseb J. 2004;18:977–9. doi: 10.1096/fj.03-1094fje. [DOI] [PubMed] [Google Scholar]
- 42.Thery C, Duban L, Segura E, Veron P, Lantz O, Amigorena S. Indirect activation of naive CD4+ T cells by dendritic cell-derived exosomes. Nat Immunol. 2002;3:1156–62. doi: 10.1038/ni854. [DOI] [PubMed] [Google Scholar]
- 43.Little KM, Smalley DM, Harthun NL, Ley K. The plasma microparticle proteome. Semin Thromb Hemost. 2010;36:845–56. doi: 10.1055/s-0030-1267038. [DOI] [PubMed] [Google Scholar]
- 44.Segura E, Guerin C, Hogg N, Amigorena S, Thery C. CD8+ dendritic cells use LFA-1 to capture MHC-peptide complexes from exosomes in vivo. J Immunol. 2007;179:1489–96. doi: 10.4049/jimmunol.179.3.1489. [DOI] [PubMed] [Google Scholar]
- 45.Muntasell A, Berger AC, Roche PA. T cell-induced secretion of MHC class II-peptide complexes on B cell exosomes. EMBO J. 2007;26:4263–72. doi: 10.1038/sj.emboj.7601842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azevedo LC, Janiszewski M, Pontieri V, et al. Platelet-derived exosomes from septic shock patients induce myocardial dysfunction. Crit Care. 2007;11:R120. doi: 10.1186/cc6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andre F, Schartz NE, Chaput N, et al. Tumor-derived exosomes: a new source of tumor rejection antigens. Vaccine. 2002;20(Suppl 4):A28–31. doi: 10.1016/s0264-410x(02)00384-5. [DOI] [PubMed] [Google Scholar]
- 48.Hulsmans M, Holvoet P. MicroRNA-containing microvesicles regulating inflammation in association with atherosclerotic disease. Cardiovasc Res. 2013;100:7–18. doi: 10.1093/cvr/cvt161. [DOI] [PubMed] [Google Scholar]
- 49.Bakhti M, Winter C, Simons M. Inhibition of myelin membrane sheath formation by oligodendrocyte-derived exosome-like vesicles. J Biol Chem. 2011;286:787–96. doi: 10.1074/jbc.M110.190009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng YH, Rome S, Jalabert A, et al. Endometrial exosomes/microvesicles in the uterine microenvironment: a new paradigm for embryo-endometrial cross talk at implantation. PLoS ONE. 2013;8:e58502. doi: 10.1371/journal.pone.0058502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 2013;91:431–7. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bobrie A, Krumeich S, Reyal F, et al. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72:4920–30. doi: 10.1158/0008-5472.CAN-12-0925. [DOI] [PubMed] [Google Scholar]
- 53.Principe S, Jones EE, Kim Y, et al. In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics. 2013;13:1667–71. doi: 10.1002/pmic.201200561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simona F, Laura S, Simona T, Riccardo A. Contribution of proteomics to understanding the role of tumor-derived exosomes in cancer progression: state of the art and new perspectives. Proteomics. 2013;13:1581–94. doi: 10.1002/pmic.201200398. [DOI] [PubMed] [Google Scholar]
- 55.Choi DS, Park JO, Jang SC, et al. Proteomic analysis of microvesicles derived from human colorectal cancer ascites. Proteomics. 2011;11:2745–51. doi: 10.1002/pmic.201100022. [DOI] [PubMed] [Google Scholar]
- 56.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–20. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 57.Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28:1551–8. doi: 10.3892/or.2012.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 59.Bobrie A, Colombo M, Raposo G, Thery C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–68. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 60.Ratajczak J, Miekus K, Kucia M, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–56. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 61.Leibovici J, Itzhaki O, Huszar M, Sinai J. The tumor microenvironment: part 1. Immunotherapy. 2011;3:1367–84. doi: 10.2217/imt.11.111. [DOI] [PubMed] [Google Scholar]
- 62.Kato Y, Ozawa S, Miyamoto C, et al. Acidic extracellular microenvironment and cancer. Cancer Cell Int. 2013;13:89. doi: 10.1186/1475-2867-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–91. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou W, Fong MY, Min Y, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–15. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ono M, Kosaka N, Tominaga N, et al. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci Signal. 2014;7:ra63. doi: 10.1126/scisignal.2005231. [DOI] [PubMed] [Google Scholar]
- 66.Khan S, Aspe JR, Asumen MG, et al. Extracellular, cell-permeable survivin inhibits apoptosis while promoting proliferative and metastatic potential. Br J Cancer. 2009;100:1073–86. doi: 10.1038/sj.bjc.6604978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Higginbotham JN, Demory Beckler M, Gephart JD, et al. Amphiregulin exosomes increase cancer cell invasion. Curr Biol. 2011;21:779–86. doi: 10.1016/j.cub.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deng Z, Cheng Z, Xiang X, et al. Tumor cell cross talk with tumor-associated leukocytes leads to induction of tumor exosomal fibronectin and promotes tumor progression. Am J Pathol. 2012;180:390–8. doi: 10.1016/j.ajpath.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 69.Sheldon H, Heikamp E, Turley H, et al. New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. Blood. 2010;116:2385–94. doi: 10.1182/blood-2009-08-239228. [DOI] [PubMed] [Google Scholar]
- 70.Dolo V, Ginestra A, Cassara D, Ghersi G, Nagase H, Vittorelli ML. Shed membrane vesicles and selective localization of gelatinases and MMP-9/TIMP-1 complexes. Ann N Y Acad Sci. 1999;878:497–9. doi: 10.1111/j.1749-6632.1999.tb07707.x. [DOI] [PubMed] [Google Scholar]
- 71.Graves LE, Ariztia EV, Navari JR, Matzel HJ, Stack MS, Fishman DA. Proinvasive properties of ovarian cancer ascites-derived membrane vesicles. Cancer Res. 2004;64:7045–9. doi: 10.1158/0008-5472.CAN-04-1800. [DOI] [PubMed] [Google Scholar]
- 72.Whiteside TL. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes) Biochem Soc Trans. 2013;41:245–51. doi: 10.1042/BST20120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Epple LM, Griffiths SG, Dechkovskaia AM, et al. Medulloblastoma exosome proteomics yield functional roles for extracellular vesicles. PLoS ONE. 2012;7:e42064. doi: 10.1371/journal.pone.0042064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu C, Yu S, Kappes JR, et al. Expansion of spleen myeloid suppressor cells represses NK cell cytotoxicity in tumor-bearing host. Blood. 2007;109:4336–42. doi: 10.1182/blood-2006-09-046201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Corcoran C, Friel AM, Duffy MJ, Crown J, O'Driscoll L. Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 2011;57:18–32. doi: 10.1373/clinchem.2010.150730. [DOI] [PubMed] [Google Scholar]
- 76.Lotvall J, Valadi H. Cell to cell signalling via exosomes through esRNA. Cell Adh Migr. 2007;1:156–8. doi: 10.4161/cam.1.3.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koles K, Budnik V. Exosomes go with the Wnt. Cell Logist. 2012;2:169–73. doi: 10.4161/cl.21981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ekstrom K, Omar O, Graneli C, Wang X, Vazirisani F, Thomsen P. Monocyte exosomes stimulate the osteogenic gene expression of mesenchymal stem cells. PLoS ONE. 2013;8:e75227. doi: 10.1371/journal.pone.0075227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou H, Cheruvanky A, Hu X, et al. Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int. 2008;74:613–21. doi: 10.1038/ki.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simpson RJ, Kalra H, Mathivanan S. ExoCarta as a resource for exosomal research. J Extracell Vesicles. 2012;1:18374. doi: 10.3402/jev.v1i0.18374. http://dx.doi.org/10.3402/jev.v1i0.18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Graner MW, Alzate O, Dechkovskaia AM, et al. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009;23:1541–57. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stasevich TJ, McNally JG. Assembly of the transcription machinery: ordered and stable, random and dynamic, or both? Chromosoma. 2011;120:533–45. doi: 10.1007/s00412-011-0340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahmed KA, Xiang J. Mechanisms of cellular communication through intercellular protein transfer. J Cell Mod Med. 2011;15:1458–73. doi: 10.1111/j.1582-4934.2010.01008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Unique proteins identified in the Vesiclepedia database.
Table S2. Transcriptional regulators identified in the Vesiclepedia composite database.


