Abstract
The tandem PHD (plant homeodomain) fingers of the CHD4 (chromodomain helicase DNA-binding protein 4) ATPase are epigenetic readers that bind either unmodified histone H3 tails or H3K9me3 (histone H3 trimethylated at Lys9). This dual function is necessary for the transcriptional and chromatin remodelling activities of the NuRD (nucleosome remodelling and deacetylase) complex. In the present paper, we show that calixarene-based supramolecular hosts disrupt binding of the CHD4 PHD2 finger to H3K9me3, but do not affect the interaction of this protein with the H3K9me0 (unmodified histone H3) tail. A similar inhibitory effect, observed for the association of chromodomain of HP1γ (heterochromatin protein 1γ) with H3K9me3, points to a general mechanism of methyl-lysine caging by calixarenes and suggests a high potential for these compounds in biochemical applications. Immunofluorescence analysis reveals that the supramolecular agents induce changes in chromatin organization that are consistent with their binding to and disruption of H3K9me3 sites in living cells. The results of the present study suggest that the aromatic macrocyclic hosts can be used as a powerful new tool for characterizing methylation-driven epigenetic mechanisms.
Keywords: calixarene, chromodomain helicase DNA-binding protein 4 (CHD4), histone, inhibitor, methylation, plant homeodomain (PHD)
INTRODUCTION
Epigenetic marks, including PTMs (post-translational modifications) of histones, regulate diverse processes in the eukaryotic cell [1,2]. PTMs provide unique binding sites for protein effectors and alter interactions between histones and DNA. PTMs are written or removed by enzymes named writers and erasers respectively, whereas protein domains that recognize these PTMs are referred to as readers. Binding to PTMs recruits reader-containing proteins and their host complexes to particular genomic sites, which is required for propagating signals essential in chromatin remodelling, gene transcription, and DNA repair, recombination and replication [3]. The four core histones (H3, H4, H2A and H2B) and a linker histone H1 contain multiple lysine residues that undergo methylation at the ε-amino group. This reaction is catalysed by HKMTs (histone lysine methyltransferases), whereas KDMs (lysine demethylases) reverse this process by removing the methylation PTMs. Currently, eight lysine residues in the tails of histones H3, H4 and H1 are known to be mono-, di- or tri-methylated. Among these PTMs, H3K4me3 (histone H3 trimethylated at Lys4), H3K9me3 (histone H3 trimethylated at Lys9) and H3K27me3 (histone H3 trimethylated at Lys27) are the most thoroughly characterized methyl-lysine marks. H3K4me3 is typically found at promoters of actively transcribed genes, whereas H3K9me3 is associated with the condensed heterochromatic regions and H3K27me3 indicates transcriptionally repressive chromatin.
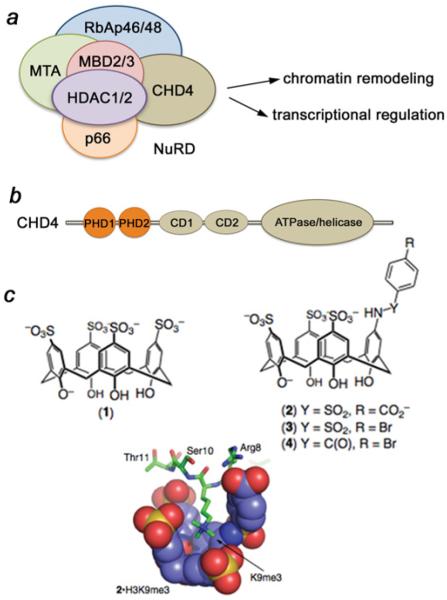
The NuRD (nucleosome remodelling and deacetylase) complex is involved in a wide array of nuclear signalling events. It regulates chromatin organization, gene transcription, genomic stability and developmental signalling. Aberrant activity of NuRD has been linked to human diseases, such as immunological abnormalities, aging and cancer, and thus NuRD components are emerging as potential therapeutic targets [4]. NuRD contains six core subunits, including an ATPase and a HDAC (histone deacetylase), and remains the only chromatin regulatory complex imparting two enzymatic functions (Figure 1a) [5–8]. One of the catalytic subunits of the complex, HDAC1 (or HDAC2), deacetylates acetylated lysine residues of histones and non-histone proteins. The second catalytic subunit, CHD4 [CD (chromodomain) helicase DNA-binding protein 4)], hydrolyses the ATP that is necessary for DNA sliding and repositioning of nucleosomes. CHD4 is a large multidomain protein, which consists of the catalytic ATPase/helicase module, two atypical DNA-binding CDs and two histone readers, i.e. tandem PHD (plant homeodomain) fingers (Figure 1b). The structurally and functionally similar PHD fingers recognize the H3K9me0 (unmodified histone H3) or H3K9me3 tails [9,10]. Binding to H3K9me0 and H3K9me3 has been shown to play a critical role in the repressive transcriptional and chromatin remodelling functions of CHD4 [11].
Figure 1. The CHD4–NuRD complex and supramolecular hosts used in the present study.
(a) Core components and functions of NuRD. (b) CHD4 domain architecture. (c) The trimethyl-lysine-binding supramolecular hosts include the parent compound 1 and compounds 2–4. The molecular model shows the predicted binding mode of 2 in complex with an H3K9me3-derived peptide sequence as determined by NMR (complexation-induced chemical shifts) and molecular modelling (MMFFaq as implemented in Spartan). Also see Supplementary Figure S1 (http://www.biochemj.org/bj/459/bj4590505add.htm).
In the last few years much effort has been put forth to generate small-molecule inhibitors that disrupt epigenetic interactions through occupying PTM-binding sites of readers [12–16]. However, a reverse approach to design chemicals that bind to a certain PTM has not been as well developed. This approach has a high potential and can be applied for targeting of a wide range of biomolecules which lack defined binding pockets and thus are impossible to probe with conventional small-molecule inhibitors. Supramolecular hosts, i.e. macrocycles bearing concave scaffolds that bind guest molecules non-covalently, have long been used to co-ordinate organic cations and ammonium-mimicking biomolecules, including cholines and catecholamines, and synthetic calixarenes have shown promising results in the recognition of methyl-lysine [17–23]. However, the supramolecular hosts are inherently less selective in comparison with small-molecule agents. We have previously demonstrated that the aromatic macrocycle p-sulfonatocalix[4]arene (1), which mimics the natural aromatic cage of methyl-lysine-binding readers, does not discriminate between peptides containing trimethylated lysine residues and binds H3K4me3, H3K9me3 and H3K27me3 equally well [24]. Furthermore, calix[4]arenes have been known to associate promiscuously with various cationic protein surface sites, which has limited their potential in biological applications up to now [17,25]. In the present study, we report a new set of calixarene-based hosts and show their applicability in characterizing the functions of methyl-lysine-specific epigenetic readers. We demonstrate that calixarenes readily disrupt binding of the CHD4 PHD2 finger to H3K9me3 without affecting binding of this protein to H3K9me0. This ability to separate multiple biological functions of the CHD4 ATPase could be imperative in understanding the role of CHD4/NuRD in regulation of chromatin structure and gene expression patterns.
MATERIALS AND METHODS
HPLC purifications
Compounds 2–4 were purified by RP-HPLC (reverse-phase HPLC) on a preparative Apollo C18 column (5 μm; 22 mm × 250 mm; Alltech) using a Shimadzu HPLC or a Thermo-Dionex HPLC–mass spectrometer with a preparative Luna C18 column (5 μm; 21.2 mm × 250 mm; Phenomenex), detecting at 280 nm. Compounds were purified by running a gradient from 90:10 0.1 % TFA (trifluoroacetic acid) in water/0.1 % TFA in MeCN (acetonitrile) to 10:90 0.1 % TFA in water/0.1 % TFA in MeCN over 35 min.
General synthesis
Compound 1 was purchased from TCI America. ESI–MS was performed on a Finnigan LCQ MS. Compounds 2 and 5 (for structure of 5, see Supplementary Figure S1 at http://www.biochemj. org/bj/459/bj4590505add.htm) were made according to a published procedure [19].
Compound 3
Compound 5 (20 mg; 2.9 × 10 −5 M) and p-bromobenzene sulfonyl chloride (1.1 eq.; 8 mg; 3.13 × 10 −5 M; TCI America) were dissolved in 2.5 ml of 0.1 M phosphate buffer (pH 8) and stirred at room temperature (22 °C) overnight. The aqueous solution was extracted with dichloromethane (2 × 10 ml) and ethyl acetate (2 × 10 ml). The aqueous layer was evaporated to dryness under reduced pressure and the crude solid subjected to RP-HPLC purification as outlined above. Fractions containing product were pooled and freeze-dried to afford 5 mg (19 % yield) as an off-white solid. NMR (300 MHz, 2H2O, δ p.p.m.): 7.76 (s, 4H), 7.13 (s, 2H), 7.10 (d, 8.1 Hz, 2H), 6.92 (s, 2H), 6.69 (d, 9.0 Hz, 2H), 4.11 and 3.94 (br, 8H). ESI–MS [M–H]− calculated for C34H27BrNO15S4− 895.95, 897.94 and found, 896.27, 898.20.
Compound 4
Compound 5 (20 mg; 2.9 × 10 −5 M) and p-bromobenzoyl chloride (1.1 eq.; 7.1 mg; 3.24 × 10 −5 M) were dissolved in 2.5 ml of 0.1 M phosphate buffer (pH 8) and stirred at room temperature overnight. The aqueous solution was extracted with dichloromethane (2 × 10 ml) and ethyl acetate (2 × 10 ml). The aqueous layer was evaporated to dryness under reduced pressure and the crude solid was subjected to RP-HPLC purification as outlined above. Fractions containing product were pooled and freeze-dried to afford 12 mg (45 % yield) as an off-white solid. NMR (300 MHz, 2H2O, δ p.p.m.): 7.75 (s, 4H), 7.65 (s, 2H), 7.32 (s, 2H), 7.26 (d, 8.1 Hz, 2H), 6.48 (d, 8.9 Hz, 2H), 4.11 (br, 8H). ESI–MS [M–H]− calculated for C35H27BrNO14 S3− 859.98, 861.98, and found, 860.27, 862.27.
Peptide synthesis
For fluorescence assays, peptides were made using a CEM Liberty1 microwave peptide synthesizer, using standard Fmoc-solid phase synthesis employing HBTU [2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate] as a coupling agent and DIEA (di-isopropylethyl amine) as an activator base. Fmoc-Lys(me3)-OH was purchased from GL Biochem and all other reagents were purchased from Sigma–Aldrich or ChemImpex. Peptides were made as H3 1–12 sequences [H-ARTKQTARK(me3)STGY-NH2 or H-ARTKQTARKSTGY-NH2], with an N-terminal free amine, C-terminal amide and tyrosine added to the C-terminus to aid in UV detection for purification and concentration determination. Peptides were purified by HPLC as described above, and identity was confirmed by ESI–MS. The concentration of peptide was determined by dissolving the freeze-dried solid into water and using the molar absorption coefficient of tyrosine at 280 nm (Spectramax M5; Molecular Devices). For NMR experiments, peptides (amino acids 1–12 of H3) were synthesized by the UCD Biophysics Core Facility.
Fluorescence assay and determination of Kassoc values between calixarene and LCG and between calixarene and peptides
This assay was adapted from an indicator displacement assay [22,23]. LCG (lucigenin) dye was purchased from Invitrogen. Calixarene hosts were made in stock solutions from weighed solid and dissolved in distilled water. All further dilutions were done using this stock. LCG was stored as a 5 mM stock protected from light, and diluted to a 5 μM stock solution in distilled water on the day of the experiment. Phosphate buffer (0.2 M) at pH 7.4 was made from the corresponding salts and used as is; 10 μl amounts were used in every well to furnish a final buffer concentration of 10 mM. Peptide solutions were freshly prepared and concentrations were confirmed as outlined above. All titrations were performed in 96-well Nunc™ optically clear-bottomed black-walled plates and read using a Spectramax M5 (Molecular Devices) plate reader.
Using a calixarene stock solution, various concentrations of calixarenes were made (0.05–50 mM). The first well was a blank solution that contained 10 μl of 0.2 M phosphate buffer, 20 μl (2.5 or 5 μM) of LCG and 170 μl of distilled water. Subsequent wells (2–12) contained the same amounts except for 150 μl of distilled water and 20 μl of calixarene (in increasing concentrations). Titration of peptides was performed using various peptide concentrations (20 μl; 1–100 μM) and 130 μl of distilled water. The plate emission was read (100 reads/well) between 445 and 600 nm and excitation was set to 369 nm. The λmax of the fluorescence readout was selected (485 nm) and these data was fitted with the program Equilibria (http://www.sseau.unsw.edu.au) [26]. See Figure 2(a) and Supplementary Table S1 for results and Supplementary Figure S2 for all titration data and fits (Supplementary Online data available at http://www.biochemj.org/bj/459/bj4590505add.htm).
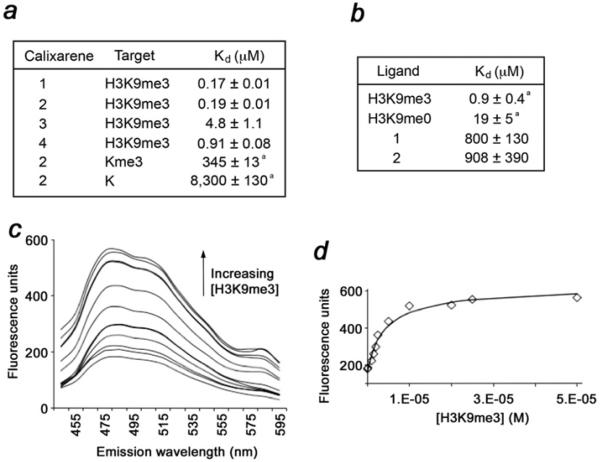
Figure 2. Compounds 1–4 bind robustly to H3K9me3 peptide.
(a) Binding affinities±S.D. of 1–4 for the indicated targets as determined by fluorescence assays. a Taken from [19]. (b) Binding affinities±S.D. of CHD4 PHD2 measured by NMR and fluorescence. a Taken from [9]. (c) Fluorescence data arising from titration of H3K9me3 into a solution of 2 (0.5 μM) and LCG (250 nM). Emission spectra (Iexc 369 nm) show displacement of LCG from the calixarene by the added H3K9me3 (0.1–50 μM). (d) Emission intensities at 485 nm (points) are fitted (line) to give a value for Kassoc. See also Supplementary Figure S2 and Table S1 (http://www.biochemj.org/bj/459/bj4590505add.htm).
Protein purification
The CHD4 PHD2 finger and CD of HP1γ (heterochromatin protein 1γ) were expressed in Escherichia coli BL21(DE3) pLysS cells grown in 15NH4Cl-supplemented minimal medium. Bacteria were harvested by centrifugation after induction with IPTG (0.5 mM) and lysed by sonication. The 15N-labelled GST (glutathione transferase)-fusion proteins were purified on glutathione–Sepharose 4B beads (Amersham Biosciences). The GST tag was cleaved with PreScission protease. The proteins were concentrated into 20 mM Tris/HCl (pH 6.8) in the presence of 150 mM NaCl and 3 mM DTT.
NMR spectroscopy
NMR experiments were carried out at 298 K on a 500 MHz Varian INOVA spectrometer. Chemical shift perturbation analyses were performed on uniformly 15N-labelled CHD4 PHD2 (0.15 mM) and HP1γ CD (0.1 mM). 1H,15N HSQC spectra were recorded in the presence of increasing concentrations of unmodified histone H3 or H3K9me3 peptides (synthesized by the UCD Biophysics Core Facility; amino acids 1–12 of histone H3), followed by the addition of 1–4. Kd values were calculated by a non-linear least-squares analysis in Kaleidagraph using the equation:
where [L] is the concentration of the peptide, [P] is the concentration of the protein, Δδ is the observed chemical shift change and Δδmax is the normalized chemical shift change at saturation. The normalized chemical shift change was calculated using equation [(ΔδH)2 + (ΔδN/5)2]0.5, where δ is chemical shift in p.p.m.
Western blot analysis
GST-fusion CHD4 PHD2 was incubated with C-terminal biotinylated peptides (Upstate Biotechnology) corresponding to the unmodified histone H3 (residues 1–20) and singly modified H3K4me3 (residues 1–20), H3K9me3 (residues 1–21) and H3K27me3 (residues 21–44) histone tails in the presence of streptavidin–Sepharose beads (GE Healthcare) and with and without 5 μl of 50 mM 2 in BB [binding buffer; 50 mM Tris/HCl (pH 7.5), 150 mM NaCl, 3 mM DTT and 0.05% Nonidet P40] at room temperature. The beads were collected via centrifugation and washed. The bound protein was detected by Western blotting using HRP (horseradish peroxidase)-conjugated anti-GST monoclonal antibody (GE Healthcare). Negative controls, GST–CHD4 PHD2 and GST–CHD4 PHD2 with 2 but without peptides, were run in parallel to ensure that the protein does not bind to the beads.
Pull-down
His-tagged HP1γ CD was incubated with the C-terminal biotinylated H3K9me3 (residues 1–21) peptide (Upstate Biotechnology) in the presence of streptavidin–Sepharose beads (GE Healthcare) and with and without 5 μl of 50 mM 2 in BB at room temperature. The beads were collected via centrifugation and washed. The bound protein was detected by Coomassie Blue staining of an SDS/PAGE gel. His–HP1γ CD with and without 2 but without peptide was used as a negative control.
Inhibitor treatment in cells and immunofluorescence
HEK (human embryonic kidney)-293T cells were purchased from the A.T.C.C. (Manassas, VA, U.S.A.) and maintained in Dulbecco’s modified Eagle’s medium/nutrient mixture F-12 (Gibco) supplemented with 10 % FBS. Cells (5 × 105) were seeded in each well of a six-well plate in 2 ml of medium. Inhibitors were added to each well at a final concentration of 0.2, 1, 5, 10 or 15 μM in the presence of 1 % DMSO. For the control sample only 1 % DMSO was added to the well. Cells were trypsinized after 48 h and deposited on to microscope slides by cytospin (200 rev./min for 10 min). For GFP visualization, cells were transfected using Lipofectamine™ 2000 according to the manufacturer’s protocol (Invitrogen). Transfected HEK-293T cells were FACS-sorted on the basis of GFP intensity (BD FACSAria II). Cytospinning, fixation and immunofluorescence were performed as described previously [27]. The primary antibodies used for immunofluorescence include anti-H3K9me3 (catalogue number 07-523; Millipore), anti-H4K20me3 (histone H4 trimethylated at Lys20; catalogue number 39180; Active Motif) and anti-HP1γ (catalogue number MAB3450; Millipore). Images were collected on a Zeiss Axiovert 200 imaging system equipped with an Axiocam MR digital camera controlled by AxioVision software or on a Zeiss LSM Pascal confocal microscope.
RESULTS AND DISCUSSION
Synthesis of trisulfonated calix[4]arenes
To improve the binding potential of p-sulfonatocalix[4]arene (1), we synthesized a set of analogues (2–4) carrying a single amide- or sulfonamide-linked functional group at the upper rim of the calixarene skeleton (Figure 1c). The three remaining sulfonate groups were left unmodified in order to maintain the favourable co-ordinating capability of the macrocycle, as well as to promote solubility in water. The bulkiness and electrostatic properties of the substituent varied, with the overall charge of the resulting compounds being −4 (3 and 4) or −5 (1 and 2) at neutral pH. The amide group in 4 provides a rigid linker to appended functionality, whereas the sulfonamides in 2 and 3 are more flexible and have different conformational preferences. We rationalized that the alterations in functionality, size and charge of the substituents would modify the host’s ability to disrupt the protein–guest complexes.
The synthesis of the supramolecular hosts relied on a single calixarene precursor bearing three sulfonates and a single amine group [19] (see the Materials and methods section and Supplementary Figure S1). This strategy allowed for the introduction of the key aromatic moiety with various substituents that can form additional contacts with the guest residues and alter the host’s affinity and selectivity.
To assess whether 2–4 retain methyl-lysine-binding capability, we examined their interactions with the H3K9me3 peptide (residues 1–12) using fluorescence assays (Figure 2 and Supplementary Figure S2). The measured equilibrium dissociation constants (Kd values) were found to be in the range of 0.2–0.9 μM for the interactions of 1, 2 and 4, and slightly higher for the interaction of 3 (Figure 2a). The relatively similar Kd values indicate that modified calixarenes co-ordinate the methyllysine peptide essentially as well as unmodified 1 does, and the substituents appear to have no significant effect on this interaction. Surprisingly, we found that binding to H3K9me3 is ~103-fold stronger than binding to trimethyl-lysine as a free amino acid (Figure 2a). Although we cannot exclude possible electrostatic repulsion between calixarenes and the carboxylate of the free lysine, this can also imply additional energetically favourable interactions between the host and the peptide residues surrounding trimethylated Lys9. The negatively charged sulfonate groups at the upper rim of 1–4 can make contact with the positively charged residues of H3K9me3, including Arg8 and Lys4, thus providing an opportunity for the use of these compounds to modulate the binding activities of methyl-lysine-specific histone readers.
p-Sulfonatocalix[4]arenes inhibit binding of CHD4 to H3K9me3
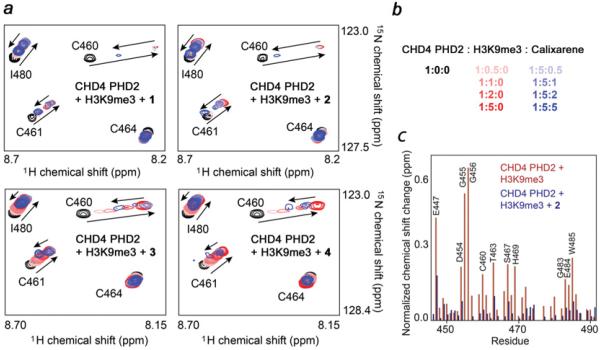
We examined the ability of synthesized calixarenes to disrupt the function of the CHD4 PHD2 finger, a known histone reader, by NMR (Figure 3). 1H,15N HSQC spectra of the uniformly 15N-labelled protein were collected while histone H3K9me3 peptide was gradually added to the NMR sample (Figure 3a). Large perturbations in the PHD2 finger amide resonances induced by the peptide indicated direct interaction with H3K9me3 (Figure 3a, red gradient colours). The binding was in intermediate exchange regime on the NMR time scale, which is characterized by progressive shifting and broadening of cross-peaks and consistent with a binding affinity of ~1 μM [9]. Subsequent addition of calixarenes caused the amide resonances to shift back to their positions in the ligand-free state of the protein, indicating complex dissociation (Figure 3a, blue gradient colours). Among the four calixarenes tested, 2 induced the largest changes, almost completely restoring the spectrum of the ligand-free PHD2 finger at a protein/H3K9me3/2 ratio of 1:5:5 (Figure 3b). Compound 1 showed a slightly weaker inhibitory activity, followed by 3 and 4. These results demonstrate that calixarenes are capable of disrupting the interaction between CHD4 PHD2 and histone H3K9me3 in vitro, and the bulky negatively charged substituent enhances this ability, whereas hydrophobic substituents reduce it.
Figure 3. Compounds 1–4 disrupt binding of the CHD4 PHD2 finger to H3K9me3.
(a) Superimposed 1H,15N HSQC spectra of 0.15 mM CHD4 PHD2 collected while first H3K9me3 peptide and then 1–4 were titrated in. Spectra are colour-coded according to the protein/peptide/calixarene molar ratio. (b) Numerical values of the protein/peptide/calixarene molar ratio. (c) The normalized chemical shift changes observed in the CHD4 PHD2 finger upon binding to H3K9me3 (red) and inhibition by 2 (blue) as a function of residue. See also Supplementary Figure S3 (http://www.biochemj.org/bj/459/bj4590505add.htm).
We noticed that the majority of chemical shift changes induced in PHD2 by calixarenes reversed the changes caused by H3K9me3; however, several amide cross-peaks of PHD2 moved to different positions (Figure 3c). In particular, Lys473, Asn474, Arg489 and Thr491 showed significant differences in either the direction or magnitude of the chemical shift changes, suggesting that these residues are involved in off-target interaction with calixarenes (Supplementary Figure S3 at http://www.biochemj.org/bj/459/bj4590505add.htm). Titrating calixarenes into the unbound PHD2 finger in the absence of H3K9me3 resulted in a similar pattern of chemical shift changes for Lys473, Asn474, Arg489 and Thr491, confirming that these residues are involved in non-specific interactions with calixarenes (Supplementary Figure S3b). Plotting chemical shift changes for these residues against the ligand concentration yielded Kd values of 800–900 μM (Figure 2b), which is consistent with affinities previously measured for non-specific associations of calixarenes with proteins [17], but ~103-fold weaker than the interaction with the targeted guest H3K9me3. Interestingly, the only lysine residue of the PHD finger, Lys453, was essentially unperturbed, supporting the idea that calixarenes do not indiscriminately bind surface residues that are positively charged, but instead recognize regions that have appropriate complementarity in charge, hydrophobicity and shape.
p-Sulfonatocalix[4]arenes do not disrupt the CHD4–H3K9me0 complex
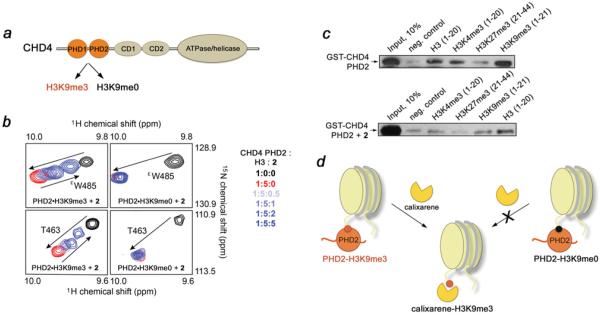
In addition to recognizing H3K9me3, the CHD4 PHD2 finger binds to H3K9me0, and this interaction has been shown to be essential for the transcriptional activity of the CHD4/NuRD complex (Figure 4a) [11]. We therefore tested whether calixarenes can disrupt the association of CHD4 with H3K9me0 using 1H,15N HSQC titration and pull-down experiments (Figures 4b and 4c). Gradual addition of the H3K9me0 peptide to the CHD4 PHD2 finger resulted in large chemical shift perturbations that were comparable with those seen upon binding of H3K9me3, indicating that formation of the CHD4–H3K9me0 complex (Kd = 18 μM) does not rely on Lys9 methylation (Figure 4b, compare black and red cross-peaks). However, titration of 2 to this complex caused no reverse chemical shift changes, indicating that calixarene does not abrogate binding of the CHD4 PHD2 finger to unmodified H3 (Figure 4b, right-hand panels, blue gradient colours). The CHD4–H3K9me0 complex was unperturbed even upon addition of a 5-fold excess of 2.
Figure 4. Compound 2 abrogates interaction of the CHD4 PHD2 finger with H3K9me3, but does not perturb the CHD4–H3K9me0 complex.
(a) The functions of CHD4. (b) Overlays of 1H,15N HSQC spectra of the H3K9me3-bound (left-hand panels) and H3K9me0-bound (right-hand panels) CHD4 PHD2 recorded during the gradual addition of 2. A lack of chemical shift changes in the CHD4 PHD2–H3K9me0 complex indicates no inhibitory effect. The spectra are colour-coded as shown on the right-hand side. (c) Binding of the GST-fusion CHD4 PHD2 finger to the indicated biotinylated histone peptides in the absence of 2 (upper panel) and the presence of 2 (lower panel). (d) Strong selectivity of 2 provides a unique tool to separately characterize multiple biological functions of CHD4. The nucleosomes are shown in light yellow.
The discrimination between the trimethylated and unmodified histone tails was supported by pull-down assays. We incubated GST-fusion CHD4 PHD2 with biotinylated histone peptides corresponding to unmodified H3 (residues 1–20) and singly modified H3K4me3 (residues 1–20), H3K9me3 (residues 1–21) and H3K27me3 (residues 21–44), and with streptavidin– Sepharose beads in the absence and presence of 2. After collecting the beads through centrifugation, the histone peptide-bound protein was detected by Western blot analysis. As shown in Figure 4(c), in the absence of 2 the PHD2 finger recognizes unmodified H3 and binds to H3K9me3 slightly stronger, corroborating previously measured binding affinities of this module (Kd = 19 μM and 0.9 μM for H3K9me0 and H3K9me3 respectively) [9]. As expected, binding to H3K4me3 was significantly weaker and no interaction with H3K27me3 was observed. However, in the presence of 2, GST–CHD4 PHD2 was able to bind only unmodified H3K9me0, whereas its interaction with H3K9me3 was abolished. The weaker complex between H3K4me3 and CHD4 PHD2 (Kd = ~2 mM [9]) was also disrupted, supporting the earlier observations that calixarenes lack of specificity for a particular trimethyl-lysine site [24].
The CHD4 PHD2 finger is a non-canonical H3K9me3 reader because it has only one aromatic residue, Phe451, involved in cation-π and hydrophobic contacts with trimethylated Lys9. Nevertheless 2 readily eliminates this interaction, most probably by forming a supramolecular complex between the calixarene and the H3K9me3 peptide, in which an aromatic cage of the calixarene host surrounds the trimethyl-lysine guest. Together, the NMR and pull-down data demonstrate that calixarene inhibits interaction of the CHD4 PHD2 finger with H3K9me3 without affecting its interaction with another physiologically relevant binding partner, H3K9me0. This ability offers a unique tool for separate characterization of multiple biological functions of CHD4, which would be impossible to reproduce with more conventional agents that target the binding surface of PHD2 itself.
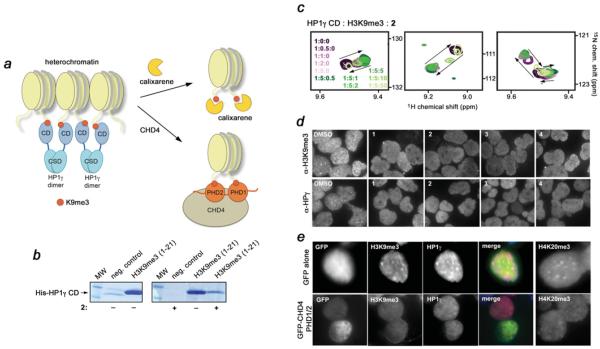
Inhibition of the CD of HP1γ
H3K9me3 is a well-established mark of pericentric heterochromatin. We have shown previously that the CHD4 PHD fingers can bind H3K9me3, displacing heterochromatic protein HP1γ from H3K9me3-enriched regions [11]. HP1γ contains a CD, whose H3K9me3-binding activity is required for the assembly and maintenance of the condensed transcriptionally inactive chromatin [28–32], whereas the displacement of HP1γ by CHD4 induces changes in the heterochromatin structure and leads to the dispersion of H3K9me3 [11] (Figure 5a). The HP1γ CD module utilizes a typical aromatic cage, consisting of three aromatic residues and a glutamate, to recognize di/trimethylated Lys9 [29,30]. We tested whether calixarenes can inhibit binding of HP1γ CD to H3K9me3 in vitro using pull-down assays and NMR (Figures 5b and 5c). As shown in Figure 5(b), His–HP1γ CD interacts strongly with the biotinylated H3K9me3 peptide in the pull-down assays; however, the addition of 2 abrogated this interaction. To confirm the displacement of HP1γ, we generated an 15N-labelled CD and examined its activity by 1H,15N HSQC titration experiments. The H3K9me3 peptide induced substantial changes in the 1H,15N HSQC spectra of the HP1γ CD, indicating robust and direct interaction (Figure 5c, purple gradient colours). Titration of 2 into the NMR sample, however, reversed these changes, driving the CD back to its apo-state and pointing to inhibition (Figure 5c, green gradient colours). These results demonstrate that calixarenes are capable of extracting methyllysine from a well-defined aromatic cage of the reader and suggest a general mode for the 2–H3K9me3 supramolecular assembly.
Figure 5. Compounds 1–4 disrupt pericentric heterochromatin.
(a) A model for the displacement of HP1γ (blue) from the nucleosomes (light yellow), enriched in H3K9me3 (red), by calixarenes (yellow) or by the tandem PHD1/2 fingers (orange) of CHD4. HP1γ associates with two spatially proximal nucleosomes via binding of its CD to H3K9me3 and dimerization through its chromoshadow domain (CSD) [37], providing a mechanism for the formation and spreading of heterochromatin. 1–4 or PHD1/2 compete with the CD of HP1γ for H3K9me3. The antagonistic action of 1–4 or CHD4 PHD1/2 leads to the disruption of pericentric heterochromatin assembly and the subsequent dispersion of H3K9me3 and HP1γ observed by light microscopy (d and e). (b) Binding of the His–HP1γ CD to the biotinylated H3K9me3 peptide in the absence or presence of 2, as indicated by +/− below the gels, was examined by pull-down assays. (c) Superimposed 1H,15N HSQC spectra of 0.1 mM CD of HP1γ collected while first H3K9me3 peptide and then 2 were titrated in. Spectra are colour-coded according to the HP1γ CD/H3K9me3/2 molar ratio. (d) Representative images of immunofluorescence performed in HEK-293T cells upon the treatment with DMSO or 1–4. Cells were spotted on to slides and stained using anti-H3K9me3 and anti-HP1γ antibodies followed by Alexa Fluor™ 488-conjugated anti-mouse or rabbit IgG. (e) Representative images of immunofluorescence performed in sorted GFP only and GFP–CHD4 PHD1/2 cells. The pericentric heterochromatin marks were visualized using anti-H3K9me3, anti-H4K20me3 and anti-HP1γ antibodies. The merged image displays H3K9me3 (blue), HP1γ (red) and GFP/GFP–CHD4 PHD1/2 (green). See also Supplementary Figure S4 (http://www.biochemj.org/bj/459/bj4590505add.htm).
p-Sulfonatocalix[4]arenes disrupt H3K9me3 binding in vivo
To determine whether the calixarene inhibitors can displace HP1γ and cause the redistribution of H3K9me3 in cells, we incubated HEK-293T cells with 1–4 or DMSO as a control and assessed the localization pattern of H3K9me3 and HP1γ (Figure 5d and Supplementary Figure S4 at http://www.biochemj.org/bj/459/bj4590505add.htm). The cell permeability of calixarenes, owing to their compact structure and low number of rotatable bonds compared with typical small-molecule agents, has been demonstrated previously [33]. As expected, in cells incubated with DMSO, we observed the normal localized focal accumulation of H3K9me3 and HP1γ. However, in cells incubated with 1–4, we observed more diffuse nuclear localization of HP1γ and H3K9me3. The response of HEK-293T cells to added calixarenes was as acute as the effect of the overexpressed GFP-fused tandem CHD4 PHD fingers (PHD1/2) on the global localization of the major heterochromatin markers, i.e. H3K9me3, HP1γ and H4K20me3 (Figure 5e). Notably, the ability of these calixarenes to disrupt heterochromatin markers in the cells correlated well with their ability to abrogate the interaction of the CHD4 PHD finger with H3K9me3; calixarenes with the strongest and weakest inhibitory effects, 2 and 4, perturbed pericentric heterochromatin to the greatest and least degrees respectively (Figure 5d). A dose–response study of 2 confirmed the concentration dependence of these effects (Supplementary Figure S4).
Together, these results demonstrate that calixarenes can disturb the H3K9me3-binding activities of the PHD2 finger of CHD4 and CD of HP1γ and suggest a common inhibitory mechanism. We concluded that the second-generation sulfonatocalixarenes could be used to probe and alter biological interactions in biochemical assays and in living cells. Since these host agents target methylated, but not unmodified, lysine in the context of the intact histone tails, they can be invaluable in investigating separate functions of chromatin-targeting protein complexes that rely on interactions with unmodified and methylated histone tails.
Concluding remarks
Supramolecular agents that target artificially introduced bio-orthogonal structural motifs have made a substantial impact in chemical biology [34,35]; however, examples that work on biomolecules in their native cell environment remain exceedingly rare [36]. In the present study we show that the calixarene-based macrocycle hosts selectively disrupt binding of the CHD4 PHD2 finger to H3K9me3 while not affecting the interaction of this protein with H3K9me0. A similar inhibitory effect, observed for another methyl-lysine reader, the CD of HP1γ, points to a common mechanism for the host–guest supramolecular assembly and suggests high potential of these compounds in biochemical applications. The present study also demonstrates that supramolecular hosts can target trimethyl-lysine in vivo and alternate native biological processes within a living cell. We anticipate that this approach, applied broadly to methylated histones and other nuclear proteins, will yield a new generation of tools to help unravel the intricate network of PTM-driven signalling in molecular epigenetics.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by the NIH (National Institutes for Health) [grant number GM096863 and GM101664 (to T.G.K)], WestCoast Ride to Live (to K.D.D. and F.H.), the Prostate Cancer Foundation of BC (to K.D.D.), a Prostate Cancer Canada Pilot Grant (to F.H.), and by the Intramural Research Program of the NIEHS (National Institute of Environmental Health Sciences) [grant number Z01ES101965 (to P.A.W.)]. H.F.A. is supported by a Neuroscience Graduate Training Grant [grant number T32HD041697]. F.H. is a Canada Research Chair.
Abbreviations
- BB
binding buffer
- CD
chromodomain
- CHD4
CD helicase DNA-binding protein 4
- GST
glutathione transferase
- HDAC
histone deacetylase
- HEK
human embryonic kidney
- H3K4me3
histone H3 trimethylated at Lys4
- H3K9me0
unmodified histone H3
- H3K9me3
histone H3 trimethylated at Lys9
- H3K27me3
histone H3 trimethylated at Lys27
- H4K20me3
histone H4 trimethylated at Lys20
- HP1γ
heterochromatin protein 1γ
- LCG
lucigenin
- MeCN
acetonitrile
- NuRD
nucleosome remodelling and deacetylase
- PHD
plant homeodomain
- PTM
post-translational modification
- RP-HPLC
reverse-phase HPLC
- TFA
trifluoroacetic acid
REFERENCES
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen HF, Wade PA, Kutateladze TG. The NuRD architecture. Cell. Mol. Life Sci. 2013;70:3513–3524. doi: 10.1007/s00018-012-1256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 6.Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 7.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 9.Musselman CA, Mansfield RE, Garske AL, Davrazou F, Kwan AH, Oliver SS, O’Leary H, Denu JM, Mackay JP, Kutateladze TG. Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem. J. 2009;423:179–187. doi: 10.1042/BJ20090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansfield RE, Musselman CA, Kwan AH, Oliver SS, Garske AL, Davrazou F, Denu JM, Kutateladze TG, Mackay JP. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J. Biol. Chem. 2011;286:11779–11791. doi: 10.1074/jbc.M110.208207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musselman CA, Ramirez J, Sims JK, Mansfield RE, Oliver SS, Denu JM, Mackay JP, Wade PA, Hagman J, Kutateladze TG. Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression. Proc. Natl. Acad. Sci. U.S.A. 2012;109:787–792. doi: 10.1073/pnas.1113655109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao C, Herold JM, Kireev D, Wigle T, Norris JL, Frye S. Biophysical probes reveal a “compromise” nature of the methyl-lysine binding pocket in L3MBTL1. J. Am. Chem. Soc. 2011;133:5357–5362. doi: 10.1021/ja110432e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herold JM, Wigle TJ, Norris JL, Lam R, Korboukh VK, Gao C, Ingerman LA, Kireev DB, Senisterra G, Vedadi M, et al. Small-molecule ligands of methyl-lysine binding proteins. J. Med. Chem. 2011;54:2504–2511. doi: 10.1021/jm200045v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 15.James LI, Barsyte-Lovejoy D, Zhong N, Krichevsky L, Korboukh VK, Herold JM, MacNevin CJ, Norris JL, Sagum CA, Tempel W, et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat. Chem. Biol. 2013;9:184–191. doi: 10.1038/nchembio.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner EK, Nath N, Flemming R, Feltenberger JB, Denu JM. Identification and characterization of small molecule inhibitors of a plant homeodomain finger. Biochemistry. 2012;51:8293–8306. doi: 10.1021/bi3009278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGovern RE, Fernandes H, Khan AR, Power NP, Crowley PB. Protein camouflage in cytochrome c-calixarene complexes. Nat. Chem. 2012;4:527–533. doi: 10.1038/nchem.1342. [DOI] [PubMed] [Google Scholar]
- 18.Perret F, Coleman AW. Biochemistry of anionic calix[n]arenes. Chem. Commun. 2011;47:7303–7319. doi: 10.1039/c1cc11541c. [DOI] [PubMed] [Google Scholar]
- 19.Daze KD, Ma MC, Pineux F, Hof F. Synthesis of new trisulfonated calix[4]arenes functionalized at the upper rim, and their complexation with the trimethyllysine epigenetic mark. Org. Lett. 2012;14:1512–1515. doi: 10.1021/ol300243b. [DOI] [PubMed] [Google Scholar]
- 20.Martos V, Bell SC, Santos E, Isacoff EY, Trauner D, de Mendoza J. Molecular recognition and self-assembly special feature: calix[4]arene-based conical-shaped ligands for voltage-dependent potassium channels. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10482–10486. doi: 10.1073/pnas.0813396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beshara CS, Jones CE, Daze KD, Lilgert BJ, Hof F. A simple calixarene recognizes post-translationally methylated lysine. ChemBioChem. 2010;11:63–66. doi: 10.1002/cbic.200900633. [DOI] [PubMed] [Google Scholar]
- 22.Florea M, Kudithipudi S, Rei A, Gonzalez-Alvarez MJ, Jeltsch A, Nau WM. A fluorescence-based supramolecular tandem assay for monitoring lysine methyltransferase activity in homogeneous solution. Chemistry. 2012;18:3521–3528. doi: 10.1002/chem.201103397. [DOI] [PubMed] [Google Scholar]
- 23.Guo D-S, Uzunova VD, Su X, Liu Y, Nau WM. Operational calixarene-based fluorescent sensing systems for choline and acetylcholine and their application to enzymatic reactions. Chem. Sci. 2011;2:1722–1734. [Google Scholar]
- 24.Daze KD, Pinter T, Beshara CS, Ibraheem A, Minaker SA, Ma MC, Courtemanche RJ, Campbell RE, Hof F. Supramolecular hosts that recognize methyllysines and disrupt the interaction between a modified histone tail and its epigenetic reader protein. Chem. Sci. 2012;3:2695–2699. [Google Scholar]
- 25.Daze KD, Hof F. The cation-π interaction at protein–protein interaction interfaces: developing and learning from synthetic mimics of proteins that bind methylated lysines. Acc. Chem. Res. 2013;46:937–945. doi: 10.1021/ar300072g. [DOI] [PubMed] [Google Scholar]
- 26.Young PG, Jolliffe KA. Selective recognition of sulfate ions by tripodal cyclic peptides functionalised with (thio)urea binding sites. Org. Biomol. Chem. 2012;10:2664–2672. doi: 10.1039/c2ob06964d. [DOI] [PubMed] [Google Scholar]
- 27.Helbling Chadwick L, Chadwick BP, Jaye DL, Wade PA. The Mi-2/NuRD complex associates with pericentromeric heterochromatin during S phase in rapidly proliferating lymphoid cells. Chromosoma. 2009;118:445–457. doi: 10.1007/s00412-009-0207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 31.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 32.Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lalor R, Baillie-Johnson H, Redshaw C, Matthews SE, Mueller A. Cellular uptake of a fluorescent calix[4]arene derivative. J. Am. Chem. Soc. 2008;130:2892–2893. doi: 10.1021/ja0782596. [DOI] [PubMed] [Google Scholar]
- 34.Logsdon LA, Schardon CL, Ramalingam V, Kwee SK, Urbach AR. Nanomolar binding of peptides containing noncanonical amino acids by a synthetic receptor. J. Am. Chem. Soc. 2011;133:17087–17092. doi: 10.1021/ja207825y. [DOI] [PubMed] [Google Scholar]
- 35.Lee DW, Park KM, Banerjee M, Ha SH, Lee T, Suh K, Paul S, Jung H, Kim J, Selvapalam N, et al. Supramolecular fishing for plasma membrane proteins using an ultrastable synthetic host-guest binding pair. Nat. Chem. 2011;3:154–159. doi: 10.1038/nchem.928. [DOI] [PubMed] [Google Scholar]
- 36.Ghang Y, Schramm M, Zhang F, Acey R, David C, Wilson E, Wang Y, Cheng Q, Hooley R. Selective cavitand-mediated endocytosis of targeted imaging agents into live cells. J. Am. Chem. Soc. 2013;135:7090–7093. doi: 10.1021/ja401273g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.