Abstract
Purpose
We initiated a personalized medicine program in the context of early clinical trials, using targeted agents matched with tumor molecular aberrations. Herein, we report our observations.
Patient and Methods
Patients with advanced cancer were treated in the Clinical Center for Targeted Therapy. Molecular analysis was conducted in the MD Anderson Clinical Laboratory Improvement Amendments (CLIA) -certified laboratory. Patients whose tumors had an aberration were treated with matched targeted therapy, when available. Treatment assignment was not randomized. The clinical outcomes of patients with molecular aberrations treated with matched targeted therapy were compared with those of consecutive patients who were not treated with matched targeted therapy.
Results
Of 1,144 patients analyzed, 460 (40.2%) had 1 or more aberration. In patients with 1 molecular aberration, matched therapy (n = 175) compared with treatment without matching (n = 116) was associated with a higher overall response rate (27% vs. 5%; P < 0.0001), longer time-to-treatment failure (TTF; median, 5.2 vs. 2.2 months; P< 0.0001), and longer survival (median, 13.4 vs. 9.0 months; P= 0.017). Matched targeted therapy was associated with longer TTF compared with their prior systemic therapy in patients with 1 mutation (5.2 vs. 3.1 months, respectively; P < 0.0001). In multivariate analysis in patients with 1 molecular aberration, matched therapy was an independent factor predicting response (P = 0.001) and TTF (P = 0.0001).
Conclusion
Keeping in mind that the study was not randomized and patients had diverse tumor types and a median of 5 prior therapies, our results suggest that identifying specific molecular abnormalities and choosing therapy based on these abnormalities is relevant in phase I clinical trials.
Introduction
The identification of pathways involved in the pathophysiology of carcinogenesis, metastasis, and drug resistance, as well as the emergence of technologies enabling tumor molecular analysis and the discovery of targeted therapies, have stimulated research focusing on the optimal use of targeted agents. The discovery of imatinib for the successful treatment of Philadelphia chromosome–positive chronic myeloid leukemia (1) was one factor that encouraged researchers to identify molecular aberrations in solid tumors (2–7).
In 2007, we initiated a personalized medicine program for patients referred to the Phase I Clinic at The University of Texas MD Anderson Cancer Center (Houston, TX). Our goal was to observe whether molecular analysis of advanced cancer and use of targeted therapy to counteract the effects of specific aberrations would be associated with improved clinical outcomes. Indeed, the allocation of patients with solid tumors to treatment with specifically targeted therapies has proven to be efficacious, with examples including the use of BRAF and ALK inhibitors based on the presence of a BRAF mutation or ALK rearrangement, respectively (8–10). However, this approach involves molecular screening for a single aberration and matching with a single targeted drug. Because most aberrations of cancer-related genes are rare, sequential single-aberration screening is unlikely to be practical in clinical practice. Rather, profiling for multiple aberrations and assigning an appropriately targeted drug or drugs, from a portfolio of agents, will likely be needed. We undertook such a strategy. This article is a report of the outcomes of our approach. We evaluated, in a nonrandomized study, the outcomes of patients tested for tumor molecular aberrations and treated accordingly in comparison with the outcomes of patients who were treated without regard to their tumors' molecular profiles. All patients were participants in early-phase clinical trials, as described in the Patients and Methods section.
Patients and Methods
Patients
Patients referred to the Phase I Clinic were of various ages, had advanced or metastatic cancer that was refractory to standard therapy, had relapsed after standard therapy, or had a tumor for which there was no standard therapy available.
All protocols required that participants have evidence of evaluable or measurable disease according to Response Evaluation Criteria in Solid Tumors (RECIST) guidelines (11, 12) and an Eastern Cooperative Oncology Group performance status of 0 to 2. Additional eligibility criteria varied according to the protocol on which the patient was enrolled. All patients provided written informed consent before enrollment onto a trial. All trials, as well as this analysis, were conducted with the approval of and in accordance with the guidelines of the MD Anderson Cancer Center Institutional Review Board.
Assignment to a clinical trial was determined after clinical, laboratory, and pathologic data from all available patient records were reviewed. Consecutive patients who had tumor tissue that could be tested or had been tested for molecular aberrations were included in the analyses. Patients whose tumors had a molecular aberration were preferably treated on a clinical trial with a matched targeted agent, when available.
If 2 or more molecular aberrations were present, patients were preferably treated with a matched trial that targeted both aberrations. If such a trial was unavailable, physician choice of a matched trial targeting 1 aberration was permitted.
Therapy
Patients treated on protocol over a 4-year period were included (∼2,350 patients were seen in our clinic during this time period and enrolled on a protocol).
The treatment regimens included 1 or more drugs. The allocation of patients to investigational treatment varied over time according to protocol availability, eligibility criteria, histologic diagnosis, the patient's prior response to therapy, potential toxicity, insurance coverage, and patient preference or physician choice. In addition, molecular profiling was added as a screening procedure. Physicians prioritized matched therapy (vs. nonmatched therapy) on the basis of the following criteria: (i) patients had an “actionable” molecular aberration; (ii) matched targeted therapy was available; (iii) patients met the eligibility criteria; (iv) insurance coverage was obtained; and (v) patients agreed to comply with study requirements. Because of the “3+3” design in most phase I clinical trials (requiring monitoring of 3 patients for a month before dose escalation and, therefore, sometimes resulting in a lack of immediate protocol availability), the multi-institutional study design in several sponsored studies (further limiting the number of patients enrolled per institution), and restrictions associated with eligibility criteria, not all patients with an “actionable” aberration could be treated on a protocol with matched therapy.
The clinical trials that patients were treated on are listed in Supplementary Table S1. Patients were treated with a variety of regimens that included, but were not limited to, agents targeting PIK3CA, mTOR, BRAF, MEK, multikinases, KIT, EGFR, and RET. Many of the targeted agents had multi-kinase inhibitory activity. A patient's tumor was considered matched with a targeted therapy if a drug in the trial was known to inhibit the aberration at low nmol/L concentrations. PIK3CA mutations and PTEN loss could be targeted by inhibitors of AKT and mTOR, as well as PI3K, as AKT and mTOR are downstream of activated PIK3CA and as both PIK3CA mutations and PTEN loss (which usually reflects PTEN mutation) activate PI3K. GNAQ, RAS, and BRAF mutations could be targeted by inhibitors of MEK. BRAF mutations could also be targeted by BRAF inhibitors. Other aberrations, such as RET, EGFR, KIT, and MET mutations, were targeted by drugs inhibiting the respective activated kinase with IC50 in the low nmol/L range. EGF receptor (EGFR) could also be targeted by anti-EGFR antibodies. TP53 mutations were not considered actionable by drugs available in our trials.
The best phase I therapy [based on longest time-to-treatment failure (TTF)] was considered for analysis. Patients treated with regional therapy were excluded from the analysis.
Endpoints and statistical methods
All statistical analyses were conducted by S. Wen and D. Berry (biostatisticians) using SAS 9.1 (SAS Institute) and S-Plus, version 7.0 (Insightful Corp.) software.
The objectives of this study were the following: (i) to establish a program for the molecular profiling of patients with advanced cancer; (ii) to provide a comprehensive characterization of the molecular profiles of individual patients; (iii) to correlate molecular profile with clinical outcomes of patients treated in the Phase I Clinical Trials Program; and (iv) to assign patients to matched targeted therapy based on their “actionable” molecular aberrations (www.clinicaltrials.gov, NCT00851032). To assess the antitumor effects of treatments, tumor responses were evaluated using the RECIST criteria (11, 12). TTF and overall survival were also analyzed. Molecular profiles of patients with evidence of antitumor activity [complete response (CR), partial response (PR), orprolonged stable disease (SD] were assessed to define subsets of patients who responded to specific therapies in clinical trials of novel agents.
The clinical outcomes of patients with molecular aberrations treated with matched therapy were compared with those of consecutive patients seen during the same time period who were not treated with matched therapy.
Best response was assessed using imaging studies conducted every 2 cycles (1 cycle = 3–4 weeks, depending on the protocol) by an MD Anderson radiologist. Tumor measurements were confirmed independently by a physician in the response assessment clinic within our department using RECIST guidelines applicable at the time of the patient's response assessment (11, 12). Finally, objective responses (CR/PR) were assessed by an MD Anderson radiologist either in our multidisciplinary conference or during imaging review rounds. Stable disease lasting 6 or more months was not considered as an initial endpoint, but it was felt that its inclusion adds to the description of the results. Waterfall plot analysis was used to illustrate response, if any, as previously described (13). Responses shown in the waterfall plot were also grouped according to RECIST guidelines. Survival was measured from the date of treatment on the first phase I clinical trial until death from any cause or last follow-up. TTF was measured from the first day of treatment on a clinical trial until the patient came off study (for toxicity, disease progression, or death). The decision to discontinue treatment on protocol was made by the treating physician and based on the patient's history, clinical presentation, and imaging studies (response assessment using RECIST criteria). These assessments could not be blinded. The criterion for taking patients off study (at least 20% disease progression) was applied uniformly and strictly across all the studies.
Patients' characteristics were analyzed using descriptive statistics. Categorical data were described using contingency tables, including counts and percentages. Continuously scaled measures were summarized by median and range. Bar-plots, waterfall plots, and a CONSORT diagram were used to show the data. The association between 2 categorical variables was examined using the χ2 test. Survival and hazard functions were estimated using the Kaplan-Meier method, and survival between groups was compared using the 2-sided log-rank test.
A paired time-to-event analysis was used for comparing TTF between phase I studies and patients' prior systemic therapy (14).
The multivariate Cox proportional hazards regression model was used to adjust for other risk factors related to survival and TTF in addressing the role of matched therapy. All P values presented are 2-sided and statistical significance means P ≤ 0.05.
Analysis of molecular aberrations
Molecular profiling was conducted in the Clinical Laboratory Improvement Amendments (CLIA)-certified Molecular Diagnostics Laboratory at MD Anderson using standard operating procedures and PCR-based sequencing technology for all tests, except for RET testing, which was conducted in a non-CLIA setting for patients with medullary thyroid cancer (15–17). DNA was extracted from microdissected paraffin-embedded tumor samples, and analysis was conducted on the coding regions for specific exons, depending on the test ordered, for the following genes: PIK3CA (exon 9: codons 532–554; exon 20: codons 1011–1062); BRAF (exon 15: codons 595–600); KRAS and NRAS (exon 2: codons 12, 13, and 61); EGFR (exons 18–21 of the kinase domain); KIT (exons 9, 11, 13, and 17); GNAQ (exon 5); TP53 (exons 4–9); MET (exon 2: codon 375; exon 11: codon 848; exon 14: codons 988 and 1010; exon 16: codons 1112 and 1124; exon 19: codons 1248,1253, and 1268); and RET (exon 10: codons 609, 611, 618, and 620; exon 11: codon 634; exon 16: codon 918).
The sensitivity of the mutation assays for detection of PIK3CA, BRAF, KRAS, NRAS, GNAQ, and MET mutations was approximately 1 in 10 mutation-bearing cells in the microdissected area. For EGFR, KIT, and TP53 mutations, the lower limit of detection was approximately 1 in 5 mutation-bearing cells. For RET analysis (Sanger sequencing; ref. 18), the sensitivity was 1 in 5 for a homozygous mutation or 2 in 5 for a heterozygous mutation. The loss of expression of the tumor suppressor nuclear protein PTEN was determined using immunohistochemical staining with the monoclonal mouse anti-human PTEN clone 6H2 (code M3627; Dako). Anaplastic lymphoma kinase (ALK) translocation was assessed using FISH (commercial probe, Abbott Molecular).
The CLIA pathology laboratory prioritized the panel of molecular aberrations for development on the basis of their known frequency in cancer and/or whether they were perceived as actionable or as having other clinical relevance to patients. The treating physicians requested all available molecular tests that were CLIA-certified at MD Anderson at the time a patient who was interested in receiving treatment in the Phase I Clinical Trials Program presented to the Phase I Clinic. If tissue available for analysis was limited, the treating physician prioritized molecular testing on the basis of tumor type and the availability of clinical trials that could impact specific targets.
Results
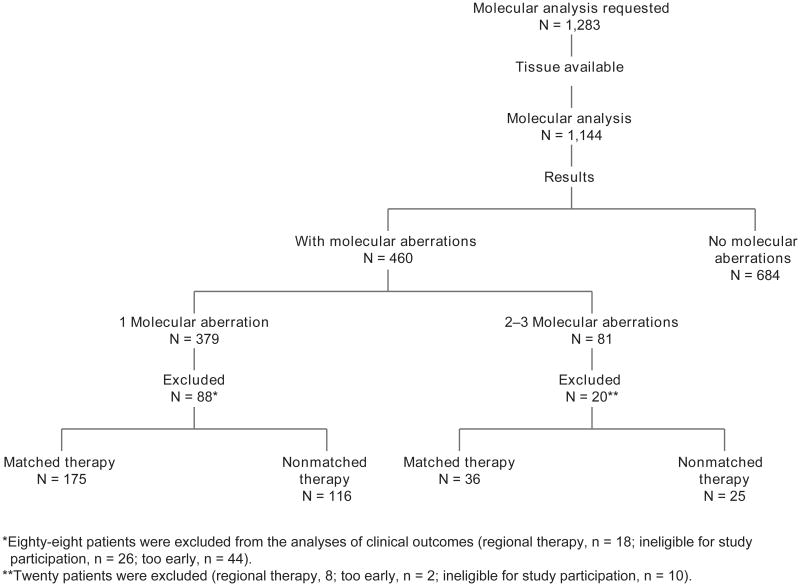
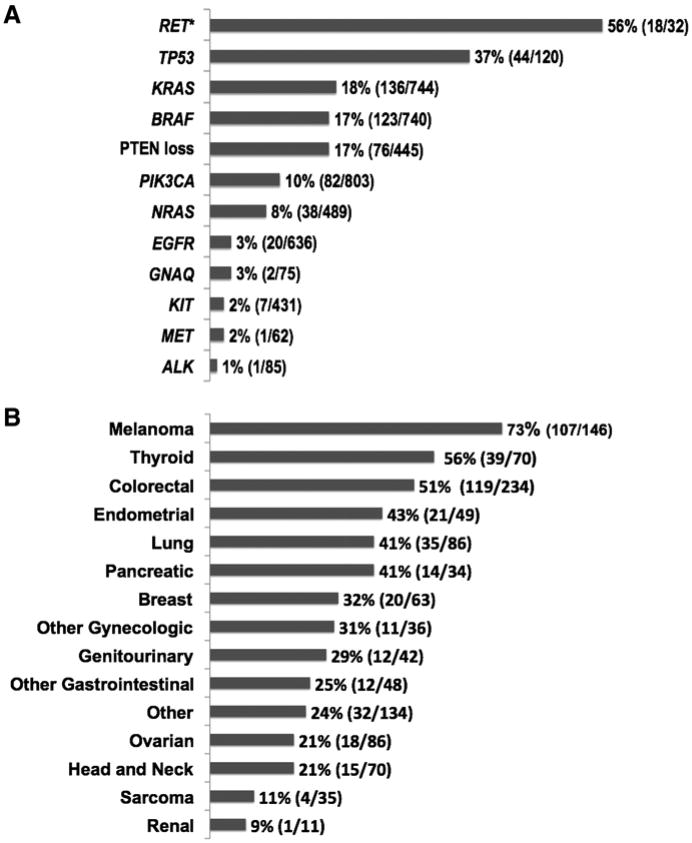
Molecular analysis testing was requested for 1283 consecutive patients who were seen in the Phase I Clinic and were interested in participating in clinical trials. Of 1,144 (89.2%) patients who had adequate tissue available for molecular analysis, 460 (40.2%) patients had 1 or more aberrations (Table 1; Fig. 1). The proportions of the different molecular aberrations in 1,144 patients and the distribution of molecular analysis by diagnosis are shown in Fig. 2A and B, respectively. It was not possible to test all patients for all aberrations because of the large amount of tissue needed to test for multiple abnormalities. Of 460 patients with 1 or more aberrations, 379 patients had a single aberration, 73 had 2 aberrations, and 8 had 3 aberrations.
Table 1. Tissue molecular aberrations.
| No. of patients (%)a | |
|---|---|
| Molecular analysis ordered | 1,283 |
| Adequate tissue available | 1,144(89.2) |
| No. of aberrations | |
| 0 | 684 (59.8) |
| 1 | 379 (33.1) |
| 2 | 73 (6.4) |
| ≥3 | 8(0.7) |
| No. of patients with aberration | 460 (40.2) |
pProportion was calculated for patients whose tissue was analyzed for 1 or more molecular aberration
Figure 1.
CONSORT diagram.
Figure 2.
A, proportions of molecular aberrations (N = 1,144). Bars indicate proportions of patients whose tumors had a molecular aberration (number of patients with aberration/number of patients tested). Asterisk indicates patients with medullary thyroid cancer analyzed for RET mutation. B, molecular aberrations by tumor type (N = 1,144). Bars indicate percentages of patients whose tumors had genetic aberrations by type of cancer (number of patients with molecular aberrations/number of patients analyzed for the specific aberration).
Among the patients tested, the most common aberrations were as follows: TP53 mutation (44/120; 36.7%); KRAS mutation (136/744; 18.3%); PTEN loss (76/445; 17.0%); BRAF mutation (123/740; 16.6%); and PIK3CA mutation (82/803; 10.2%). RET mutations were found in 56.3% (18/32) of patients, but unlike the other aberrations, which were assessed in diverse tumors, RET was tested only in thyroid cancer (Fig. 2A). The distribution of molecular aberrations by tumor type is shown in Supplementary Tables S2 and S3.
The cancers in which aberrations were most commonly found were melanoma (73% of patients tested), thyroid cancer (56%), and colorectal cancer (51%; Fig. 2B). More than 30% of patients with endometrial, lung, pancreatic, and breast cancers also had discernible aberrations.
Patients with one molecular aberration
Characteristics
Because the majority of patients (n = 379) had 1 aberration, we primarily focused our analyses on those patients, who were enrolled on a total of 51 trials (Supplementary Table SI). One hundred and seventy-five patients were treated with matched therapy; 116 were treated with nonmatched therapy. Eighty-eight patients were excluded from the analyses of clinical outcomes (regional therapy, n = 18; ineligible for study participation, n = 26; too early, n = 44).
Of 175 patients treated in the matched therapy group, 80 (46%) patients were treated in the escalation phase. Of 116 patients treated in the nonmatched therapy group, 56 (48%) patients were treated in the escalation phase.
The matched and nonmatched therapy groups had similar pretreatment characteristics, with the exception of lactate dehydrogenase (LDH) levels (Table 2). Tumor types are listed in Supplementary Table S4. The median number of prior therapies in both groups was 5 (P = 0.59). Of 175 patients treated with matched therapy, 24 (14%) also received a cytotoxic agent as part of their regimens; of 116 patients treated with nonmatched therapy, 29 (25%) also had a cytotoxic agent in their regimens.
Table 2. Baseline characteristics of 291 patients with one mutation by the type of therapy.
| Covariate | Matched (%) (n = 175) |
Nonmatched (%) (n = 116) |
P | |
|---|---|---|---|---|
| Age, y | <60 | 96 (55) | 61 (53) | 0.80 |
| ≥60 | 79 (45) | 55 (47) | ||
| Sex | Male | 86 (49) | 47 (41) | 0.19 |
| Female | 89 (51) | 69 (59) | ||
| Number of prior therapies | ≤3 | 55 (31) | 33 (28) | 0.68 |
| >3 | 120 (69) | 83 (72) | ||
| Performance status | 0 | 52 (30) | 38 (33) | 0.77 |
| 1 | 113 (65) | 73 (63) | ||
| 2+ | 10 (6) | 5 (4) | ||
| Platelet count, × 109/L | <140 | 25 (14) | 20 (17) | 0.78 |
| ≥140–<440 | 143 (82) | 92 (79) | ||
| ≥440 | 7 (4) | 4 (3) | ||
| Number of metastatic sites | ≤2 | 101 (58) | 71 (61) | 0.64 |
| >2 | 74 (42) | 45 (39) | ||
| Liver metastases | Yes | 97 (55) | 74 (64) | 0.20 |
| No | 78 (45) | 42 (36) | ||
| Lactate dehydrogenase ≥ 618 IU/L | Yes | 50 (29) | 47 (41) | 0.05 |
| No | 125 (71) | 69 (59) | ||
| Albumin < 3.5 g/dL | Yes | 17 (10) | 8 (7) | 0.53 |
| No | 158 (90) | 108 (93) | ||
| Royal Marsden Hospital score | 0 or 1 | 133 (76) | 92 (79) | 0.61 |
| 2 or 3 | 42 (24) | 24 (21) |
Responses
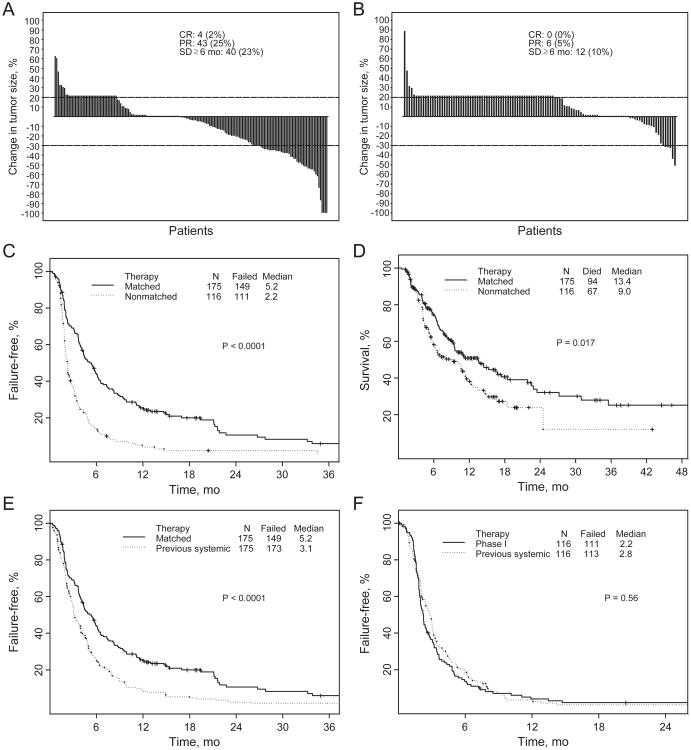
The overall response rate in the 175 patients treated with matched therapy was 27% (CR, 2%; PR, 25%), and it was 5% (all PRs) in the 116 patients treated with nonmatched therapy (P < 0.0001). SD lasting 6 or more months was noted in 23% and 10% of patients in each group, respectively (Fig. 3A and B).
Figure 3.
A, best response by RECIST of 175 patients with one molecular aberration treated with matched therapy: changes from baseline in tumor measurements (waterfall plot). Patients with new lesions and/or clinical progression were illustrated as 20% progression (≥20% increase indicates progression; >30% decrease indicates partial response). B, best response by RECIST of 116 patients with 1 molecular aberration treated without molecular matching: changes from baseline in tumor measurements (waterfall plot; ≥20% increase indicates progression; ≥30% decrease indicates partial response). C, TTF of patients with 1 molecular aberration [matched targeted therapy: n = 175, median TTF = 5.2 months (95% CI, 4.3–6.2); therapy without matching: n = 116, median TTF = 2.2 months (95% CI, 2.0–2.7; P < 0.0001)]. D, survival of patients with 1 molecular aberration [matched targeted therapy: n = 175, median survival = 13.4 months (95% CI, 9.5–18.5); therapy without matching: n = 116, median survival = 9.0 months (95% CI, 5.9–11.7; P = 0.017)]. E, TTF of 175 patients with 1 molecular aberration treated with matched therapy (median, 5.2 months; 95% CI, 4.3–6.2) versus prior therapy (median, 3.1 months; 95% CI, 2.8–4.0; P < 0.0001). F, TTF of 116 patients with 1 molecular aberration treated with nonmatched therapy (median, 2.2 months; 95% CI, 2.0–2.7) versus prior therapy (median, 2.8 months; 95% CI, 2.2–3.2; P = 0.35).
There was no correlation between response and number of prior therapies in 175 patients treated with matched therapy (P = 0.73) or in 116 patients treated with non-matched therapy (P = 0.99).
In patients with a BRAF mutation, the response rate with matched therapy was 37%, compared with 0% with non-matched therapy (P = 0.004). However, when these patients were subtracted from the analysis, matched therapy still resulted in a significantly higher response rate than nonmatched therapy (20% vs. 6%; P = 0.003; Table 3).
Table 3. Response in patients with a BRAF mutation, a mutation other than BRAF, and all patients by type of therapy (nonrandomized).
| 1 aberration | Type of therapy | No. of treated patients | CR/PR (%) | P |
|---|---|---|---|---|
| BRAF | Matched | 70 | 26 (37) | 0.004 |
| Nonmatched | 14 | 0 (0) | ||
| Non-BRAF | Matched | 105 | 21 (20) | 0.003 |
| Nonmatched | 102 | 6 (6) | ||
| Total | Matched | 175 | 47 (27) | <0.0001 |
| Nonmatched | 116 | 6 (5) |
Subset analyses showed that when RET mutant patients were excluded from the analyses, response rates were still higher in the matched non-RET-mutated patients with one mutation than in the nonmatched patients [40/158 (25%) vs. 6/116 (5%), respectively, P < 0.0001]. Similarly, when patients with PTEN loss, PI3K aberrations, or KRAS aberrations were excluded from the analyses, patients with one mutation treated with matched therapy had higher response rates than those treated with nonmatched therapy (P < 0.0001, P < 0.0001, and P = 0.002, respectively; Supplementary Table S5). The small number of patients with individual molecular aberrations precluded a robust analysis of response to each individual therapy and a comparative analysis.
TTF
The median TTF in patients treated with matched targeted therapy was 5.2 months [95% confidence interval (CI), 4.3–6.2], versus 2.2 months (95% CI, 2.0–2.7) in patients treated without matching (P < 0.0001; Fig. 3C).
There were 17 patients in the matched therapy group (n = 175) and 2 patients in the nonmatched therapy group (n = 116) who failed treatment and were taken off study owing to toxic effects. When the TTF analysis was conducted excluding these patients, the median TTF was 5.2 months (95% CI, 4.1–6.1) in the matched therapy group and 2.3 months (95% CI, 2.0–2.8) in the nonmatched therapy group (P < 0.0001).
Survival
The median follow-up was 15 months. The median survival duration of 175 patients treated with matched therapy was 13.4 months (95% CI, 9.5–18.5), compared with 9.0 months (95% CI, 5.9–11.7) for 116 patients treated without matching (P = 0.017; Fig. 3D)
TTF compared with previous therapy
In the 175 patients with one molecular aberration treated with matched targeted therapy, median TTF (5.2 months, 95% CI, 4.3–6.2) was longer than that associated with the patients' previous systemic antitumor therapies (3.1 months; 95% CI, 2.8–4.0; P < 0.0001; Fig. 3E), where 120 patients (69%) had longer TTF with matched targeted therapy. In patients treated with nonmatched therapy, there was no statistical difference in TTF between patients' nonmatched therapy and their prior systemic therapy (2.2 vs. 2.8 months, respectively; 95% CI, 2.2–3.2; P= 0.35; Fig. 3F), where 55 patients (47%) had longer TTF with nonmatched therapy. The P value is descriptive only and has no inferential interpretation because this was not a randomized study.
Multivariate analyses
To address whether the effect of matched therapy was due to other covariates, we conducted multivariate analyses. Ten covariates were included in the model: age, sex, number of prior therapies, performance status, number of metastatic sites, platelet count, levels of lactate dehydrogenase (LDH) and albumin, type of therapy (matched vs. nonmatched therapy), and inclusion of cytotoxic therapy in the regimen.
Independent factors predicting response were matched therapy (P < 0.001) and normal LDH levels (P = 0.045; Table 4). Independent factors predicting longer TTF were matched therapy (P < 0.0001), normal levels of albumin (P < 0.0001), normal platelet counts (P = 0.001), and 2 or more metastatic sites (P = 0.003). Normal LDH levels were associated with a trend towards longer TTF (P = 0.08). Independent factors predicting longer survival were normal platelet counts (P < 0.0001), normal LDH (P < 0.0001) and albumin (P < 0.001) levels, and 2 or fewer metastatic sites (P = 0.008). Matched therapy was associated with a trend towards longer survival (P = 0.06; Table 4).
Table 4. Multivariate analyses for treated patients with 1 molecular aberration (N = 291).
| ORa | 95% CI | P | |
|---|---|---|---|
| Response | |||
| Matched therapy (vs. nonmatched) | 6.33b | 2.65–15.13 | <0.001 |
| Lactate dehydrogenase ≤ 618 IU/L (vs. > 618 IU/L) | 2.16 | 1.02–.60 | 0.045 |
| TTF | HRc | ||
| Matched therapy (vs. nonmatched) | 0.42d | 0.32–0.55 | <0.0001 |
| Albumin ≥ 3.5 g/dL (vs. < 3.5) | 0.45 | 0.29–0.68 | <0.0001 |
| Platelet count ≤ 440 × 109/L (vs. > 440) | 0.34 | 0.18–0.65 | 0.001 |
| Metastatic sites ≤ 2 (vs. >2) | 0.68 | 0.53–0.88 | 0.003 |
| Lactate dehydrogenase < 618 IU/L (vs. > 618) | 0.79 | 0.60–1.03 | 0.076 |
| Survival | HRc | ||
| Platelet count ≤ 440 × 109/L (vs. > 440 × 109/L) | 0.21 | 0.11–0.42 | <0.0001 |
| Lactate dehydrogenase ≤ 618 IU/L (vs. > 618 IU/L) | 0.56 | 0.40–0.77 | <0.0001 |
| Number of metastatic sites ≤ 2 | 0.65 | 0.47–0.89 | 0.008 |
| Albumin ≥ 3.5 g/dL (vs. < 3.5) | 0.40 | 0.24–0.65 | 0.009 |
| Matched therapy (vs. nonmatched) | 0.73e | 0.53–1.01 | 0.06 |
0R (>1 is associated with higher response)
OR in univariate logistic model for response = 6.72 (P < 0.0001)
HR (<1 is associated with longer TTF or survival)
HR in univariate Cox model for TTF = 0.43 (P < 0.0001)
HR in univariate Cox model for survival = 0.68 (P = 0.019)
The remaining factors that were included in the model were not significant for response, TTF, or survival.
Patients with 2 or 3 molecular aberrations
Of 81 patients with 2 or 3 molecular aberrations, 20 patients were excluded (regional therapy, n = 8; too early, n = 2; ineligible for study participation, n = 10). Of the 61 evaluable patients, the CR plus PR rate was 14% in the matched therapy group (n = 36) compared with 0% in the nonmatched therapy group (n = 25; P = 0.14). The median TTFs were 3.02 months and 2.7 months for the matched and nonmatched groups, respectively (P = 0.79). The median survival durations were 10.6 months and 17.0 months, respectively (P = 0.28).
Patients with ≥1 molecular aberrations
When patients with 1 or ≥2 molecular aberrations were analyzed together, the CR plus PR rate was 25% in the matched therapy group compared with 4% in the non-matched therapy group (P< 0.0001).The median TTFs were 4.36 months and 2.26 months for the matched and non-matched groups, respectively (P < 0.0001). The median survival durations were 11.4 months and 10.2 months for the matched and nonmatched groups, respectively (P = 0.04).
Patients with no molecular aberrations
In the 684 patients without a known aberration, the rates of CR and PR were 0% and 6%, respectively. Their median survival was 9.0 months (95% CI: 8.1–10.4 months) and their median TTF was 3.0 months (95% CI: 2.7–3.4 months).
Discussion
The optimal therapy for patients with advanced cancer who have experienced treatment failure with conventional agents has not been defined. Numerous therapeutic strategies are available. Despite some responses noted, the outcomes of those patients have been disappointing. The median survival in our historical data was 9 months (19). Keeping in mind that the study was not randomized, and patients had diverse tumor types and a median of 5 prior therapies, our results are encouraging. In patients with 1 molecular aberration, matched therapy compared with treatment without matching was associated with a higher overall response rate (27% vs. 5%; P < 0.0001), longer TTF (median, 5.2 vs. 2.2 months; P < 0.0001), and longer survival (median, 13.4 vs. 9.0 months; P= 0.017). Matched targeted therapy was also associated with longer TTF compared with prior systemic therapy (5.2 vs. 3.1 months; P < 0.0001). In multivariate analyses, matched therapy was an independent factor predicting response (P= 0.001) and TTF (P= 0.0001).
The proportion of patients with elevated (>618 IU/L) LDH levels was lower in the matched therapy group compared with the nonmatched therapy group (29% vs. 41%, P = 0.05). However, in multivariate analyses, matched targeted therapy was an independent factor predicting response (P < 0.001) and TTF (P < 0.0001), whereas LDH levels were less significant (response, P = 0.045; TTF, P = 0.08; Table 4). Multivariate analyses for survival showed that lower LDH level was one of the independent factors predicting longer survival (P < 0.0001). Other independent factors predicting longer survival were normal platelet counts (P < 0.0001), normal albumin levels (P < 0.001), and 2 or fewer metastatic sites (P = 0.008; Table 4).
Of 1,144 patients, the analysis was focused on 460 (40.2%) patients with 1 or more molecular aberrations because the majority of the patients did not have an aberration in their tumors. In contrast, Von Hoff and colleagues reported that 98% of patients in their study had a defined tumor aberration (20). The difference may be attributable to the fact that our targeted molecular studies were limited to mutations, with the exception of PTEN loss (which reflects mutation or epigenetic changes leading to loss of PTEN function), whereas the Von Hoff study included overexpression of genes in tumor compared with control organ tissue (20). As strong correlations between mutations and response have been shown for several cancers, the difference in study design may also account for the fact that the median TTF for patients with a molecular aberration treated with a matched phase I agent in our study was 1.68 times that of their TTF on previous systemic therapy, whereas in the Von Hoff study, only 27% of patients (18/66) had a progression-free survival duration 1.3 times or more on their prior treatment.
Tumor types most often linked to mutational abnormalities in our population were melanoma, thyroid and colorectal cancers, and gynecologic malignancies. In some tumor types, such as colorectal cancer, there are fewer “targetable” aberrations. Patients with thyroid cancer that were referred to us frequently were known to have mutations. The most common actionable molecular aberrations were BRAF (3), KRAS and PIK3CA mutations, and PTEN loss (21, 22). Taking into consideration that tumors were tested for only 1 to 12 molecular aberrations (each test was run separately and tumor tissue was generally insufficient for all tests), and the lack of complete characterization of other molecular abnormalities that are likely involved in tumor progression, it seems likely that, as more sophisticated molecular technology emerges (23, 24), the proportion of patients who test positive for driver mutations will increase. However, the complexity of the genomic landscape of aberrations is also likely to make analysis more challenging (25, 26). Indeed, recent publications suggest that advanced molecular technology reveals significant tumor heterogeneity, even in individual patients (27).
The significant difference in survival between patients treated with matched targeted therapy and those treated without matching, along with the higher rates of response and TTF associated with matched therapy, even as compared to the patients' previous conventional treatment, confirm the need to further investigate study designs that incorporate novel technologies for molecular profiling into the process of making treatment decisions. Indeed, other investigators have shown that specific molecular testing for these aberrations and use of agents to target these aberrations is associated with improved clinical outcomes (8, 10, 28–34). For instance, the use of BRAF inhibitors in patients with BRAF-mutated melanoma and ALK inhibitors in patients with ALK-rearranged lung cancer (6) showed remarkably high response rates, even in the phase I setting. Specifically, vemurafenib induced an overall response rate of 81% (26/32) in patients with melanoma bearing the V600E BRAF mutation (8). In addition, the ALK inhibitor crizotinib induced an overall response rate of 57% (47/82) in patients with ALK-rearranged non-small cell lung cancer (10). A key difference between these studies and the current one is that, at the time of referral, we screened patients for multiple, rather than single, aberrations and matched them with one of several targeted drugs, as appropriate. Such a comprehensive approach is likely to be more efficient and informative than sequential screening for individual aberrations.
There are several limitations to our study. The inclusion of a variety of molecular aberrations, and targeted agents, while suggesting that the concept of individualized cancer therapy maybe generalized, does not provide the controlled setting of a single drug and target. Furthermore, as this trial was not randomized or blinded, unknown confounding factors may have contributed to higher rates of response, TIT, and survival in patients with molecular aberrations treated with matched targeted therapy compared with those treated without matching. Because some protocols included more than 1 agent, such as a targeted agent combined with a cytotoxic agent, some responses might have been due to the cytotoxic drugs or to synergy. Another possible confounding effect on TTF is the possibility that the patients treated with matched targeted therapy had a more favorable prognosis than the non-matched patients by virtue of the biomarker selection. For example, it is well known that EGFR mutations confer a more favorable prognosis (35). These factors would not, however, account for the observation that patients treated on our trials without molecular matching had significantly lower response rates, nor for the fact that, in a paired analysis, patients treated with molecular matching had a higher TTF with their matched targeted treatment than with their prior systemic therapy, while patients treated without matching did not show this longer TTF. Finally, multivariate analysis identified matched targeted therapy as the major independent factor predicting higher rates of response (P = 0.001) and TTF (P = 0.0001), as well as showing a trend towards predicting longer survival (P = 0.06).
There are also several features of this study that may have attenuated the benefits of matched targeted therapy. For instance, because the patients were enrolled on phase I trials and the dose levels varied, some individuals may have received low doses and/or targeted agents that were ultimately proven to perform poorly in the human setting. The response rates might also have been diminished by the fact that patients were heavily pretreated (median of 5 prior treatments). Finally, even though participants had at least one detectable aberration, it is plausible that other driver mutations coexisted and were not discerned. Indeed, it was not possible to test many patients for all aberrations because of limited tissue availability.
In conclusion, our observational study suggests that identifying specific molecular abnormalities and choosing therapy based on these abnormalities is associated with longer TTF in the phase I setting compared with that of previous systemic therapy. Furthermore, in the nonrandomized setting, rates of response, TTF, and survival were higher with matched targeted therapy than those observed without matching. Our results are sufficiently supportive of the benefit of matching therapy to molecular aberrations that we plan a randomized trial to test this hypothesis.
Supplementary Material
Translational Relevance.
Our results support the approach of matching drugs to molecular aberrations and suggest that it is relevant in phase I clinical trials. Patients with refractory, advanced cancer treated with molecularly matched targeted therapy had higher rates of response and longer time-to-treatment failure (TTF) and survival than patients treated without matching. In addition, our results suggest that identifying specific molecular abnormalities and choosing therapy on the basis of those abnormalities is associated with longer TTF in the phase I setting compared with that of previous systemic therapy.
Acknowledgments
The authors thank Dr. Waun Ki Hong for his vision, guidance, and support in his capacity as the Division Head of Cancer Medicine at The University of Texas MD Anderson Cancer Center.
Grant Support: The study was in part supported by seed funding from the Institute for Personalized Cancer Therapy (IPCT), The University of Texas MD Anderson Cancer Center; CTSA Grant Award 5ULlRR024148-04(PP-2), Center for Clinical and Translational Sciences; and CTSA Supplemental Grant Award 3UL1RR024148-04SI.
Disclosure of Potential Conflicts of Interest: A.M. Tsimberidou received MD Anderson seed funding from Institute for Personalized Cancer Therapy and is a consultant/advisory board member of Caris Life Sciences, Baxter Healthcare. F. Janku has a commercial research grant from Novartis. D. Berry is a paid consultant and co-owner of Berry Consultants, whose clients include some of the sponsors of clinical trials with results included in this article. R. Kurzrock has commercial research grants from Amgen, Amptimen, Angiochem, Aronex, AstraZeneca, Callisto, Centocor, Concordia, Enzon, Exelixis, Genentech, GSK, Globomax, BPC Biotech, Hoffman LaRoche, Merck & Co., MGI Pharmaceuticals, Myriad, NCCN, NCI, Nereus, Novartis, Otsuka, Pfizer, Ziopharm, and Wyeth, is a consultant/advisory board member of AACR, Enzon, Health Advances, LLC, Johnson & Johnson, Merck, and SAIC-Frederick, Inc., and has a Honoraria from Speakers' Bureau from AACR-MCT, Alfred Mann Foundation, Amgen, ASCO Program Committee, CTCA, CTRT-EAB, Exelixis, Georgetown University, MK7A, Novartis Oncology, OptumHealth, Pfizer, Pharmacogenomic Medicine, SWOG, UT Health Science Center (San Antonio, TX), Yokohama City University, and Yonsei Cancer Center.
Footnotes
Prior presentations: This article was presented orally at the American Society of Clinical Oncology (ASCO) 2011 Annual Meeting (Session Title: Clinical Science Symposium: Personalized Medicine) and was highlighted in ASCO's inaugural official Press Program. It was also presented at the 2011 American Society of Hematology/ASCO joint symposium as an invited paper.
Authors' Contributions: Conception and design: A.M. Tsimberidou, R. Kurzrock
Development of methodology: A.M. Tsimberidou, R. Luthra, D. Berry
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): A.M. Tsimberidou, D.S. Hong, J.J. Wheler, G.S. Falchook, S. Fu, S.A. Piha-Paul, A. Naing, F. Janku, D. Berry
Analysis and interpretation of data (e.g., statistical analysis, biosta-tistics, computational analysis): A.M. Tsimberidou, S. Wen, D. Berry
Writing, review, and/or revision of the manuscript: A.M. Tsimberidou, N.G. Iskander, D.S. Hong, J.J. Wheler, G.S. Falchook, S. Fu, S.A. Piha-Paul, A. Naing, F. Janku, Y. Ye, S. Wen, D. Berry, R. Kurzrock
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): A.M. Tsimberidou, N.G. Iskander, F. Janku, R. Luthra, Y. Ye
Study supervision: A.M. Tsimberidou
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
References
- 1.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–7. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 2.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CAgene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 3.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 4.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 6.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu K, Bimbaum D, Ruley MA, Fasano O, Suard Y, Edlund L, et al. Structure of the Ki-ras gene of the human lung carcinoma cell line Calu-1. Nature. 1983;304:497–500. doi: 10.1038/304497a0. [DOI] [PubMed] [Google Scholar]
- 8.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 12.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:22847. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Ratain MJ, Eisen T, Stadler WM, Flaherty KT, Kaye SB, Rosner GL, et al. Phase II placebo-controlled randomized discontinuation trial of sorafenib in patients with metastatic renal cell carcinoma. J Clin Oncol. 2006;24:2505–12. doi: 10.1200/JCO.2005.03.6723. [DOI] [PubMed] [Google Scholar]
- 14.Wei U, Lin DY, Weissfeld L. Regression analysis of multivariate incomplete failure time data by modeling marginal distributions. J Am Stat Assoc. 1989;84:1065–73. [Google Scholar]
- 15.Zuo Z, Chen SS, Chandra PK, Galbincea JM, Soape M, Doan S, et al. Application of COLD-PCR for improved detection of KRAS mutations in clinical samples. Mod Pathol. 2009;22:1023–31. doi: 10.1038/modpathol.2009.59. [DOI] [PubMed] [Google Scholar]
- 16.Janku F, Tsimberidou AM, Garrido-Laguna I, Wang X, Luthra R, Hong DS, et al. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther. 2011;10:558–65. doi: 10.1158/1535-7163.MCT-10-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janku F, Wheler JJ, Westin SN, Moulder SL, Naing A, Tsimberidou AM, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012;30:777–82. doi: 10.1200/JCO.2011.36.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margraf RL, Durtschi JD, Dames S, Pattison DC, Stephens JE, Mao R, et al. Multi-sample pooling and illumina genome analyzer sequencing methods to determine gene sequence variation for database development. J Biomol Tech. 2010;21:126–40. [PMC free article] [PubMed] [Google Scholar]
- 19.Wheler J, Tsimberidou AM, Hong D, Naing A, Jackson T, Liu S, et al. Survival of patients in a Phase 1 Clinic: the M. D. Anderson Cancer Center experience. Cancer. 2009;115:1091–9. doi: 10.1002/cncr.24018. [DOI] [PubMed] [Google Scholar]
- 20.Von Hoff DD, Stephenson JJ, Jr, Rosen P, Loesch DM, Borad MJ, Anthony S, et al. Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010;28:4877–83. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- 21.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105:2652–7. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28:1075–83. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coriess CL. Medicine. Personalized cancer diagnostics. Science. 2011;334:1217–8. doi: 10.1126/science.1216427. [DOI] [PubMed] [Google Scholar]
- 24.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 25.Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bemabe RR, et al. International network of cancer genome projects. Nature. 2010;464:993–8. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juric D, Baselga J. Tumor genetic testing for patient selection in Phase I clinical trials: The case of PI3K inhibitors. J Clin Oncol. 2012;30:765–6. doi: 10.1200/JCO.2011.39.6390. [DOI] [PubMed] [Google Scholar]
- 27.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baselga J, Bradbury I, Eidtmann H, Di Cosimo S, de Azambuja E, Aura C, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379:633–40. doi: 10.1016/S0140-6736(11)61847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baselga J. Targeting the phosphoinositide-3 (PI3) kinase pathway in breast cancer. Oncologist. 2011;16(Suppl 1):12–9. doi: 10.1634/theoncologist.2011-S1-12. [DOI] [PubMed] [Google Scholar]
- 30.Arteaga CL, Baselga J. Impact of genomics on personalized cancer medicine. Clin Cancer Res. 2012;18:612–8. doi: 10.1158/1078-0432.CCR-11-2019. [DOI] [PubMed] [Google Scholar]
- 31.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–30. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 32.Lennerz JK, Kwak EL, Ackerman A, Michael M, Fox SB, Bergethon K, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol. 2011;29:4803–10. doi: 10.1200/JCO.2011.35.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallberg B, Palmer RH. Crizotinib latest champion in the cancer wars? N Engl J Med. 2010;363:1760–2. doi: 10.1056/NEJMe1010404. [DOI] [PubMed] [Google Scholar]
- 34.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 35.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–9. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.