Abstract
Structural and cellular attachment analysis identified overall bent helical regions of adhesive peptides identified within mussel adhesive protein (MAP) capable of also attaching cells. DOPA (L-DOPA, 3,4-dihydroxyphenylalanine) is frequently identified and credited for the attachment ability of several marine proteins [Olivieri, MP, et al. (2002), Biofouling 18, 149–159]. Newly designed cyclic peptides (DOPA-G-G-C-G-K-A-K-G-C [cyc-DOPA] & Y-G-G-C-G-K-A-K-G-C [cyc-Y]) derived from structurally conserved regions of several MAP peptides were examined to assist in the understanding of both surface and cellular attachment. Solution-state proton nuclear magnetic resonance (NMR) spectroscopy coupled with molecular modeling and dynamics revealed minimal differences in the structures of the proposed cellular attachment domain within these two peptides. Multiple attenuated internal reflection infrared (MAIR-IR) spectroscopy, ellipsometry and advancing contact angle analyses showed that formation of thin films by these peptides was L-DOPA and pH dependent. When compared to control surfaces, undifferentiated leukocyte cells (MOLT-4) significantly attached and spread onto films created from the cyc-DOPA. The culmination of these structural, biophysical and cellular attachment techniques reveal a conformation of cyc-DOPA that is capable of both adsorbing to surfaces and then attaching cells that spread. This work supports the sequence, K-A-K, as the cellular attachment domain, especially when held in a reliable structural conformation.,.
Keywords: peptide structure, protein adsorption, cell attachment, NMR, surface chemistry
The characterization of surface or cellular attachment to protein-like films is dependent on understanding significant binding interactions at the atomic level. The potential for controlling attachment is vast; often there is a need to enhance cellular attachment to surfaces, such as for implantable devices. Conversely, many seek the reduction of biofilm formation, perhaps in the oral cavity or in a marine environment. This work utilizes a molecule that was designed to examine both protein/surface as well as protein/cell interactions.
The common blue sea mussel, Mytilus edulis, produces mussel adhesive foot protein (mefp-1 or MAP) that is capable of attachment to various underwater surfaces [1,2] and various cell types [3]. This approximately 114 kDalton protein is generally comprised of a repeating decapeptide (alanine-lysine-proline-serine-tyrosine-hydroxyproline-hydroxyproline-threonine-L-DOPA–lysine; A-K-P-S-Y-Hyp-Hyp-T-DOPA-K; MAP-RD). Thin films of MAP have demonstrated the ability to attach various cell types to several substrata in vitro [3,4]. It was shown that several L-DOPA-containing molecules, including MAP, were dependent on alkaline conditions for adsorption and rinse resistant film formation [5,6]. Others have also reported the importance of maintaining pH levels to reduce the oxidation of the L-DOPA residues of peptides and proteins [7]. Since “conditioning” biofilm formation is prerequisite to cellular binding, the characterization of the protein adhesion process is essential to obtain a complete understanding of the cellular attachment process [8], especially in a wet environment [9].
Since MAP has already been examined as an adhesive in chondrocyte transplantation surgery [10] and tested as an attachment agent for corneal applications [11], understanding its abilities should allow direct application to applied research. Additionally, the three-dimensional structural elucidation of MAP with its remarkable surface as well as cellular binding properties would provide information on the general phenomenon of bioadhesion. This information could lead to the development of molecules designed to enhance tissue adhesive scaffolding for drug targeting. There is also interest in understand these proteins abilities as they are able to act as load-bearing junctions while joining functionally different biomacromolecules (i.e., mussel plaques to their collagen fibers [12]).
Although the structural analysis of MAP is incomplete, circular dichroism (CD), surface chemistry and molecular modeling determined that the sequential hydroxyprolines within MAP-derived peptides provided conformational rigidity likely imposing a poly-proline II conformation. These data further indicated that the polar side-chains of the remaining residues were presented to the solution, specifically the lysine side-chains [6]. The likelihood of the poly-proline II conformation was proposed [5] and was later affirmed [3,13,14].
Prior studies characterized the MAP-RD solution-state structure [13] and its uniform rinse resistant film properties [6]. Nuclear magnetic resonance (NMR) studies coupled with molecular modeling of this peptide revealed a bent helical conformation [13]. To continue the structural elucidation of MAP, a larger peptide structure was designed to sequentially internalize the terminal amino acids of MAP-RD (P-S-X-Hyp-Hyp-T-X-K-A-K-P-S-X-Hyp, (MAP-14) where X = (Y or L-DOPA)). Molecular modeling was used to examine similar structural overlapping regions of the MAP-RD [13] and MAP-14 to predict larger MAP fragments. It is evident from the accumulated information obtained through peptide design, NMR, molecular modeling, surface chemistry and cellular attachment data that the unique lysine-alanine-lysine (K-A-K) of the MAP-14 when able to maintain a reliable structure is potentially responsible for cellular attachment [3].
For this study, new peptide sequences were developed as biomimetics, materials designed to mimic compounds with biological origin [15], to incorporate the regions of MAP responsible for the substrate attachment {L-DOPA} and other regions responsible for cellular attachment {K-A-K}. From other studies, using peptides synthesized that contain L-DOPA for the adhesion process, and RGD for cellular attachment, DOPA-G-C-G-G-R-G-D-G-C we found that using peptides that were cyclic in design would limit flexibility and allow better monitoring of structure-function relationships [16].
MATERIALS AND METHODS
Materials
Two cyclic peptides were obtained from SynPep (Dublin, CA) after purification and characterization by high-pressure liquid chromatography and mass spectrometry (DOPA-G-G-C-G-K-A-K-G-C {cyc-DOPA} and Y-G-G-C-G-K-A-K-G-C {cyc-Y}). A-K-P-S-Y-Hyp-Hyp-T-DOPA-K was obtained in-kind from Christine Benidict and MAP was obtained from Collaborative Biomedical Products, Bedford, MA. Deuterium Oxide (99.99 %), needed for the NMR lock signal, was purchased from Cambridge Isotope Laboratories, Cambridge, MA. Cellular attachment assays were performed using six-well tissue culture polystyrene (TCPS) cell culture plates (Corning Glass Works, Corning, NY). Serum Free Hybridoma Medium (SFM), bovine serum albumin (BSA) and Dulbecco’s phosphate-buffered saline (PBS) were obtained from Life Technologies, Inc., Grand Island, NY. Toluidine blue, a protein-staining agent, was obtained from Carolina Biological (Burlington, NC). The lymphoid cell line MOLT - 4 was established in 1971 with cells from a 19 year old male donor with acute lymphoblastic leukemia (obtained from Dr. Ian McDonald, Roswell Park Cancer Institute, RPCI, Buffalo, NY). The cells are approximately 10 microns in diameter, of T-cell origin [17] and are often used as a negative control for cellular attachment due to their selective binding to VCAM-1 peptide films [18]. Cut and polished germanium (Ge) plates, 50 × 20 × 1 mm, used as substrates for surface chemistry experiments were obtained from Harrick Scientific Ossining, NY.
Peptide design
Two peptides were designed containing L-DOPA or tyrosine to monitor the peptide-surface adhesion process. The L-DOPA was created after subjecting tyrosine to mushroom tyrosinase in order to add a further OH group to carbon number four [19]. The peptides contained the structurally conserved region of MAP believed responsible for peptide-cellular adhesion (K-A-K) [3]. The peptides were designed as cyclic via disulfide bonding between residues 4 and 10 restricting conformational space in the K-A-K region. Several glycine residues were incorporated in the anticipation that some ring structural flexibility might be needed to expose the lysine side-chains to the solution side of a thin film.
Surface Chemistry Methods
Films of the cyc-Y and cyc-DOPA were created from solutions containing 1 mg/mL peptide/H2O. All samples were eventually diluted to a final concentration of 2 μg peptide/cm2 using H2O (pH 3) or 0.1 M sodium bicarbonate (pH 8), applied to germanium plates (Harrick Scientific, Ossining, NY), and allowed to dry. After drying they were leached, by gently applying 1.5 mL of distilled water on top of the film and then tilting the plate vertically for the loosely associate film components to be removed, and then dried. After which, the samples were strongly rinsed using a wash-bottle held 6 inches from the plates held in a vertical position for 10 s. All samples were analyzed nondestructively by both ellipsometry and MAIR-IR spectroscopy, before and after each physical challenge carried out on the coated germanium plates [20], thus, allowing for a qualitative and semi-quantitative assessment of the physical state of films in each circumstance. Ellipsometry was carried out on a Rudolf Research Thin Film Type 43702-200E Ellipsometer. Three spots per plate were measured, and the raw data processed to give film thickness (in Angstroms) with a National Bureau of Standards computer program [21]. This ellipsometer monitors films ranging between 2–2000 Å in thickness. MAIR-IR spectroscopy was carried out on a Perkin-Elmer Model 1420 Ratio Recording IR spectrophotometer with a dedicated computer capable of recording absorption spectra for films as thin as 10Å [22]. Finally, the advancing angle method of contact angle analysis was performed on a Rame-Hart contact angle goniometer at the conclusion of each MAIR-IR spectroscopic assessment [3]. Liquids used for contact angle assessment were the highest grade available. The liquids with corresponding surface tensions (in dynes/cm) were water (72.4), glycerol (64.8), formamide (58.9), thiodiglycol (53.5), methylene iodide (49.0), 1-bromo-naphthalene (45.0), 1-methyl-naphthalene (39.3), dicyclohexyl (32.7) and n- hexadecane (27.6), while other chemicals were standard reagent grade.
Cell Attachment Methods
Film Preparation. Cellular attachment assays were performed using 6-well cell culture plates. Volumes of 0.5 mL of cyc-Y and cyc-DOPA in a 0.1 M NaHCO3 (pH=8.1) buffered solution were applied to attain a surface concentration of 2 μg/cm2 into each of the six cell-wells per condition. These films were allowed to dry and were then rinsed three times with 1 mL of distilled water. Half of the treated plates were exposed to 0.5 mL of Bovine Serum Albumin (BSA;1 mg/mL) for 30 minutes followed by aspiration of the BSA solution. This resulted in 3 wells with sample films alone and 3 wells with an additional albumin exposure. The coating of surfaces with albumin before cellular introduction is used by many to distinguish between specific and non-specific cellular adhesion. It is generally believed that the albumin blocks the non-specific cellular attachment sites, while leaving areas available for subsequent “specific” cellular adhesion [23].
Cell Introduction
Approximately 120,000 cells in SFM were injected into each cell-well. Plates were incubated for one hour at 37°C. The SFM/cell solution was aspirated and each well was rinsed with 1 mL of PBS and fixed with 3% glutaraldehyde for one hour. The wells were stained with 0.025% toluidine blue and rinsed with 2 mL of distilled water. The cells were dehydrated using 1 mL each of 20, 50, 70 and 100% ethanol solutions for 10 minutes, and then aspirated. A total area of 4 cm2 per condition was counted using NIH image analysis on a Macintosh computer with the public domain NIH program (http://rsb.info.nih.gov/nih-image). Scanning electron microscopy (SEM) was then performed for each experimental condition. t-Tests (Microsoft Excel) were used to compare statistical significance (p < 0.05).
SEM Imaging Methods
The wells designated for SEM were fixed using the same procedures as above. The center 4 cm2 portion of each cell-well was cut out and affixed to a scanning electron microscope stub. Samples were then carbon coated to a few nanometers thickness, using an Enton Accum DV-502 Carbon Evaporator. The surfaces and attached cells were then examined using a Hitachi S-4000 SEM.
NMR and Molecular Modeling Methods
Nuclear magnetic resonance (NMR) spectra were collected for the pair of peptides. The L-DOPA-containing peptide sample was 5.4 mM at pH 2.74 in 90%H2O/10%D2O and the tyrosine-containing peptide samples were 5.4 mM at pH 3.18 in D2O and 5.4 mM at pH 2.78 90%H2O/10%D2O uncorrected for isotopic effects. These low pH conditions were used to maintain L-DOPA-peptide integrity over the lengthy time course of the 2D experiments. 1D and 2D correlated spectroscopy [COSY; 24] and rotating frame nuclear Overhauser effect spectroscopy (ROESY) experiments were collected on an AMX-600 MHz Brüker instrument at 295K. The 2D experiments were collected with a data matrix of 4096 × 512 complex data points in the timed proportional phase incrementation mode. The mixing time for the ROESY was 300 ms. The nuclear Overhauser effect (nOe) data was utilized in molecular modeling software [25] to develop structures of these peptides. Chemical shifts were referenced to H2O at 4.7421 ppm with respect to deuterated sodium 3-trimethylsilylpropionate-2,2,3,3-d4 (TSP).
NMR data and molecular modeling techniques were used to elucidate a family of possible conformations for the peptides. Molecular modeling was performed on a Silicon Graphics, Incorporated (Mountainview, CA) workstation with Sybyl software (St. Louis, MO). Amino acid starting conformations were extracted directly from the program’s protein dictionary. L-DOPA was created as a modification of tyrosine. The TRIAD component of Sybyl was used to process the NMR data. Specifically, ROESY data provided conformational constraints from nOe volumes that were divided into three categories with the corresponding distance ranges, strong (1.8–2.5 Å), medium (1.8–3.5 Å), and weak (1.8–5.0 Å). These constraints were then used to run DIANA, another component of Sybyl, which used distance geometry in dihedral space to generate a requested 100 structures. A family of the ten lowest energy structures underwent simulated annealing procedures. The molecules underwent 10 cycles of simulated annealing starting at 1000 K for 2 picoseconds (ps) and cooling to 300 K for 1 ps. X-PLOR [26] was used to calculate the root mean squared deviation (RMSD) of the family and to provide an overall averaged structure (Figure 6). X-PLOR’s averaging process created a strained structure that underwent molecular mechanics (MM) minimization procedures. This MM procedure consisted of a two-step optimization approach without constraints using 100 iterations of steepest descent minimization and then Powell minimization until convergence (gradient was less than 0.05 kcal/mol * Å) utilizing the Kollman forcefield contained within Sybyl.
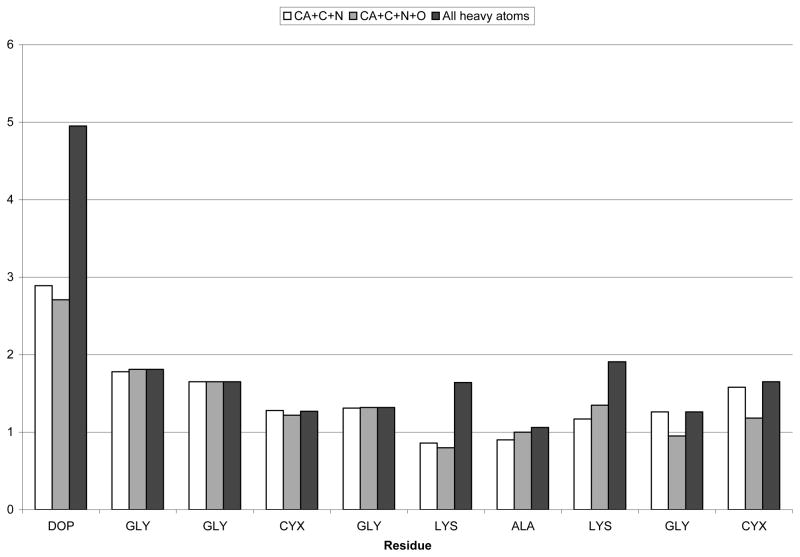
Figure 6.
RMSD per residue of the family of ten cyc-DOPA structures after simulated annealing. White is Cα-C-N, grey is Cα-C-O-N, black is all heavy atoms.
The minimized average structure was solvated using minimum periodic boundary conditions that placed water molecules around the peptide in a cube of approximately 32 Å per side. The solvated peptide was then subjected to molecular dynamics (MD) calculations. The volume and temperature (300 K) were held constant. The experiment was run for 200 ps and a snapshot was taken every ps. Gasteiger-Hückel charges were implemented with a constant dielectric function. Shake was implemented for all bonds and the nonbonded interactions were reset every 25 femptoseconds (fs). Initial velocities were generated using a random seed. An average structure was calculated using the 200 structures obtained from the snapshots. This structure underwent the MM procedure. The RMSD values of the backbone atoms of each snapshot structure were calculated using the average dynamics structure as the reference.
RESULTS
Surface Chemistry
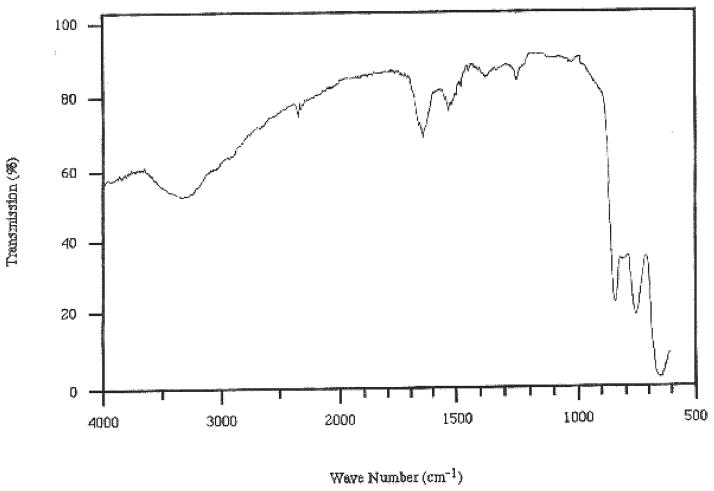
Ellipsometry, MAIR and contact angle analyses data provided evidence that rinse-resistant film formation was dependent on the presence of L-DOPA and the higher pH. The L-DOPA-containing films formed from a solution at pH 8 provided film thickness an order of magnitude greater than when formed from pH 3 solutions (119.0 ±63 Å vs. 11.6 ±6.4 Å; Table 1). MAIR absorbencies correlated with the pH 8 provided a greater mass at the prism surface; the Amide I band was (1650 cm−1; 0.093 ± 0.023 vs. 0.015 ± 0.01; Figure 1) and the Amide II band was (1550 cm−1; 0.061 ± 0.014 vs. .006± .005; Figure 1) [22]. These spectra are consistent with previous studies that indicate an absorption at 1260 cm−1 found for L-DOPA surface attachment to germanium oxide plates [5]. However, the cyc-Y did not result in this uniform rinse-resistant film formation regardless of pH. Contact angle analyses showed that samples even minimally capable of creating rinse-resistant films were polar, having high γ p values ranging from 30-40 dynes/cm (cyc-DOPA, γd: 21.6 ± 3.7, γp: 40.6 ± 5.4, γc:39.6 ± 3.1; cyc-Y γd: 26.0 ± 1.4, γp: 30.5 ± 1.2, γc: 42.3 ± 1.2).
Table 1.
Ellipsometry Thickness (Å) of Cyc-DOPA and Cyc-Y
| Cyclic-Dopa | Cyclic-Tyrosine | |
|---|---|---|
| pH: 3 | ||
| Dried | 356.6 ± 566.8 | 113.6 ± 219 |
| Leached | 13.43 ± 7.9 | 5.8 ± 6.4 |
| Rinsed | 11.67 ± 6.4 | |
| pH: 8 | ||
| Rinsed | 119.0 ± 63 | 11.6 ± 6.4 |
Figure 1.
MAIR-IR spectrum of a cyc-DOPA film created from 2μg/cm2 after rinsing.
Cell Attachment
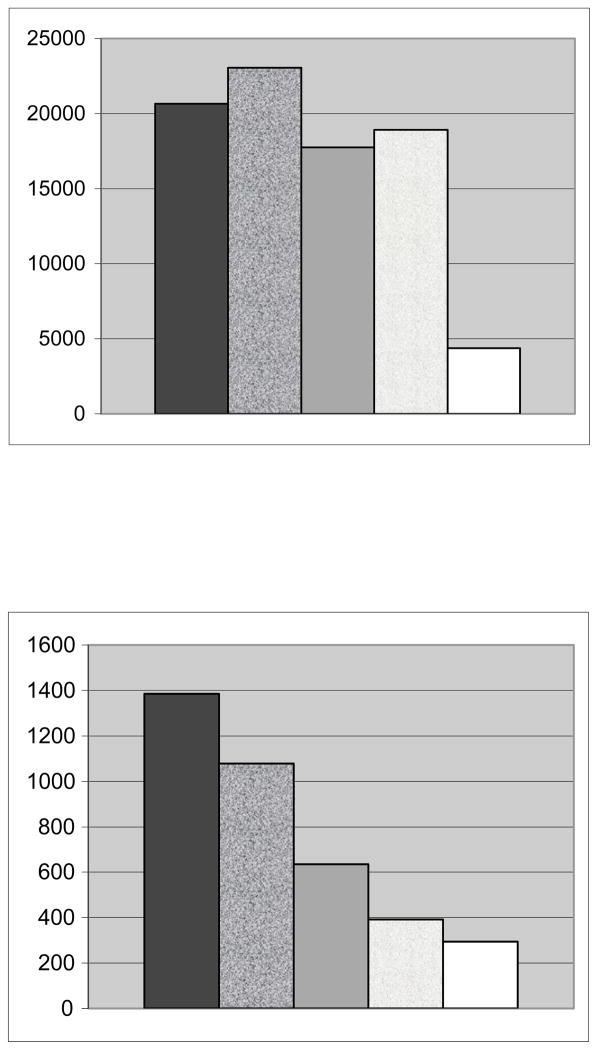
In the absence of BSA, cellular attachment for MAP-RD was 23,039, cyc-DOPA 20,670, TCPS 18,290, cyc-Y 17,767 and MAP 4,355 (Figure 2). For the MAP-RD films, only 50% of the cells which attached were spread compared to the cyc-DOPA where most, if not all, cells were seen through SEM imaging as uniformly distributed and spread (Figure 3). Although the TCPS attached more cells than cyc-Y and MAP, the cells for these three conditions were found to be rounded in shape and unevenly distributed. SEM showed that less than 25% of cells attached to the TCPS were spread. The decapeptide attached the most cells but not significantly more than the cyc-DOPA. In the absence of BSA, the only interface that showed consistent, uniform attachment and spreading of cells was the cyc-DOPA films.
Figure 2.
Number of MOLT-4 leukocytes attached over a total 8 cm2 region in the absence (top) and presence of albumin introduction (bottom). Going from left to right conditions included cyc-DOPA, MAP-RD, cyc-Y, TCPS, and MAP.
Figure 3.

SEM image of a MOLT-4 cell attached and spread onto a cyc-DOPA film.
In the presence of albumin, the cellular attachment was highest for cyc-DOPA (cyc-DOPA 1,384, MAP-RD 1,079, cyc-Y 634, TCPS 392, and MAP 294; Figure 2). However, only the cyc-DOPA films provided an environment that retained attached and spread cells throughout SEM sample preparation.
NMR
The data from HPLC, NMR and amino acid analysis performed on cyc-Y and cyc-DOPA validated amino acid composition and assured sample purity. One and two-dimensional NMR experiments allowed for the assignment of chemical shift values (supporting information). The extracyclic attachment residue (Tyr/DOPA) affected only minor differences in the chemical shift values in the N-H and C-H regions for like residues (+/−0.04 ppm). The COSY and ROESY (Figure 4), data coupled with semi-automatic algorithms, within TRIAD were sufficient to carefully assign the N-H protons in these overlapped spin systems. The nOe peaks having multiple assignments were manually edited and given the most conservative assignment (i.e., given the choice of neighboring vs. long range assignments, neighbors were assigned first). There were 39 (strong 19, medium 12, weak 8) and 40 (strong 25, medium 8, weak 7) inter-proton distance constraints for cyc-Y and cyc-DOPA respectively.
Figure 4.
Fingerprint region of a ROESY spectrum of cyc-DOPA at 600 MHz with a mixing time of 300ms.
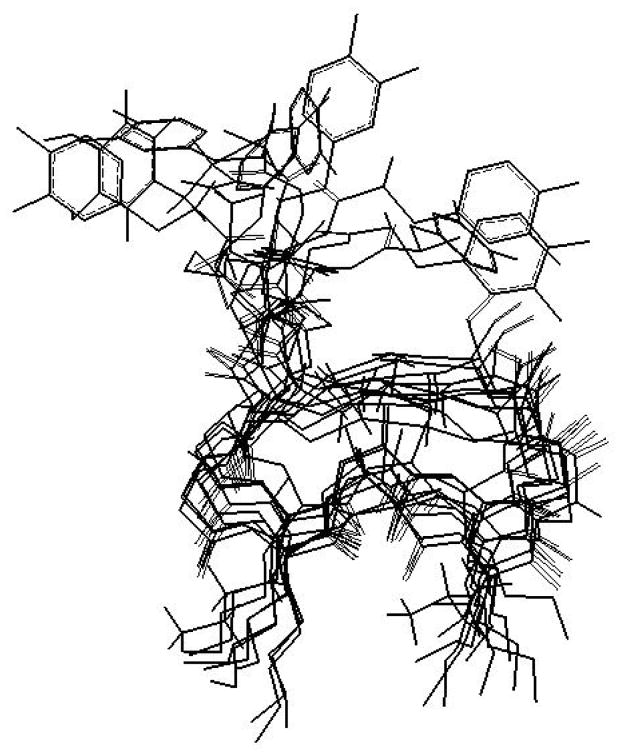
NMR data coupled with molecular modeling techniques have elucidated a family of ten structures for each peptide (Figure 5). After simulated annealing the 10 structures provided an average RMSD to their mean structure for non-H atoms of 2.85 Å and 1.46 Å for the backbone (NCC=O) atoms for cyc-Y and of 2.70 Å and 1.53 Å for cyc-DOPA (Figure 6). The structures show similar ring conformation throughout, with variability in the position of the extracyclic residues, permitting flexibility for the L-DOPA to position itself against the surface. The families were mathematically averaged to yield a single mean structure for each peptide that was subjected to energy minimization.
Figure 5.
Family of the 10 lowest energy structures resulting from nOe data, distance geometry calculations (DIANA) and molecular modeling of cyc-DOPA.
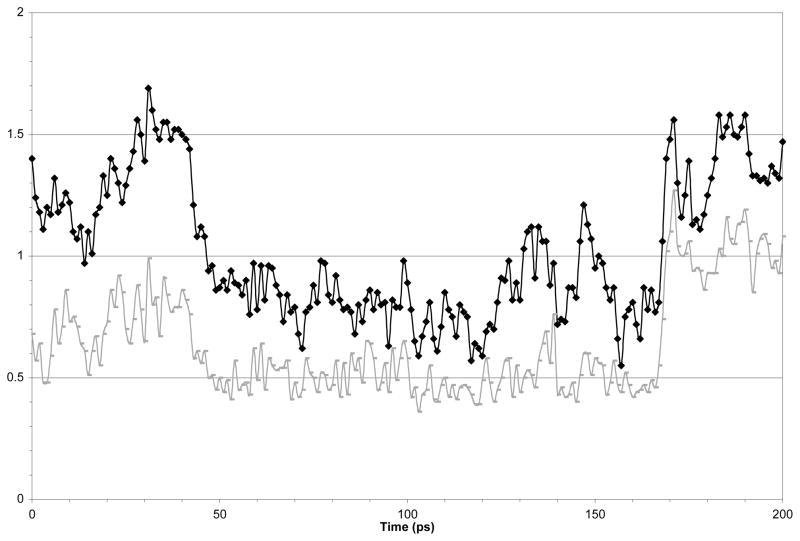
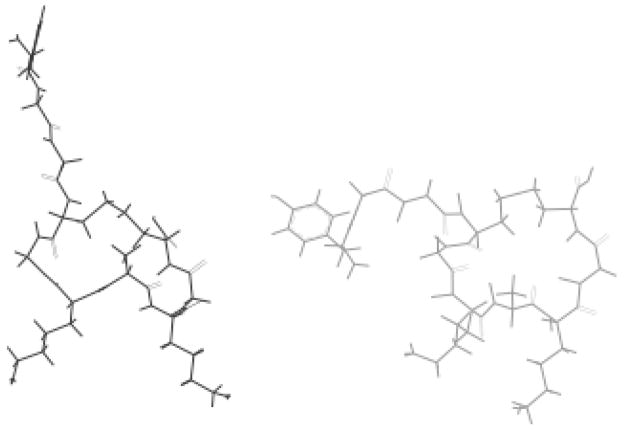
These structures were used as the starting conformations for molecular dynamics calculations. Figure 7 shows the RMSD for each dynamics time-point to the mean dynamics structure for the backbone and all heavy atoms. The data shows that after an initial equilibration period the peptides atoms have an RMSD value of 0.5 Å for the backbone atoms and 0.8 Å for heavy atoms. The RMSD values jump to about 1 Å after 170 ps, however inspection of the structures obtained from 100 and 190 ps clearly indicate that the variance is located in the side-chain region of the aromatic amino acids. Sybyl provided the average structure over the final 160 ps of the molecular dynamics experiment. The overall structures did not change significantly as a result of the dynamics calculations (Figure 8.). A list of the various energies associated with the minimized average structures before and after dynamics is given in Table 2, with the total energies providing 121.9 and 95.5 kcal/mol, respectively for cyc-Y and 240.2 and 58.8 kcal/mol for cyc-DOPA.
Figure 7.
RMSD for each dynamic time point to the mean dynamic structure for the backbone (black) and for all heavy atoms (grey) over the time course of a 200 ps dynamics experiment.
Figure 8.
The dynamics averaged structures of cyc-DOPA (left) and cyc-Y (right).
Table 2.
Energy Calculations for cyc-Y and cyc-DOPA before and After Molecular Dynamics Using a Kollman all-atom Force Field and Gasteiger-Hückel Charges
| Energy Type (kcals/mol) | cyc-Y | cyc-Y | cyc-DOPA | cyc-DOPA |
|---|---|---|---|---|
| Post MD | Post MD | |||
| Bond Stretching | 27.889 | 28.22 | 26.9 | 28.22 |
| Angle Bending | 100.532 | 99.359 | 228.81 | 98.359 |
| Torsional | 19.846 | 17.432 | 16.336 | 17.432 |
| Improper Torsional | 0.599 | 0.623 | 0.365 | 0.623 |
| 1-4 van der Waals | 17.107 | 16.77 | 39.327 | 16.77 |
| van der Waals | −3.098 | −11.743 | −8.142 | −11.743 |
| 1-4 Electrostatic | 25.452 | 81.551 | 94.233 | 81.551 |
| Electrostatic | −66.373 | −134.591 | −156.373 | −134.591 |
| H-Bond | −0.097 | −1.077 | −1.217 | −1.077 |
| Total | 121.857 | 95.543 | 240.24 | 95.543 |
The torsion angles of the final structures of each peptide after dynamics were compared to those of known secondary structure. Based on the torsion angles, bond angles and bond lengths, Recheck [27] found the mean conformations before and after dynamics is valid.
The final average minimized structure after dynamics was examined for possible H-bond associations. It was determined that 4 H-bonds were found for the cyc-Y (C4 O′ --> A7 HN, K6 O′ --> K8 H3N, A7 O′ --> G9 HN and G9 O′ --> C10 HOCAP) while two were found for cyc-DOPA (C4 O′ --> K6 HN and K6 O′ --> K8 HN).
DISCUSSION
Surface Chemistry
The cumulative surface chemistry data clearly indicate that the L-DOPA containing peptide delivered at an alkaline pH is necessary for surface adsorption. Both Cyc-Dopa and Cyc-Y films formed from solutions at a low pH provided films that were not uniform in thickness, ranging from almost no adsorption to over 1000 Å. Leaching of these films removed most loosely associated peptide, leaving virtually nothing behind on the Cyc-Y films. Films formed at a pH of 8 dried as too thick to measure, the rinsed films for both peptides masked the surfaces; however, the L-DOPA-containing films provided films that were thicker, and completely masked the surface (Table 1). Films created from these conditions were of similar characteristics to other proteinaceous cellular attachment films [1,3,5,6]. The particularly high γp values (30-40 dynes/cm) indicate a very polar surface and is resultant from the arrangement lysine R-groups outermost. Since poly-lysine is a well-known cellular attachment agent, the lysine residues are likely responsible for subsequent cell attachment [6]. A prior series of surface chemistry experiments identified the polar component of the surface free energy of films formed from MAP as linked to optimal cellular attachment (i.e., >12 dynes/cm) [4]. The 30-40 dynes/cm noted for cyc-DOPA films γp values are clearly above this range.
Cell Attachment
In the absence of BSA, there is little difference between the number of attached cells for TCPS and the cyc-Y. This is consistent with the surface chemistry data indicating that the cyclic peptides are dependent on L-DOPA for surface attachment and that cyc-Y is not capable of forming rinse resistant films. The decapeptide (MAP-RD) attached the most cells, but not significantly more than cyc-DOPA. Further, the MAP-RD attached cells that retained a rounded appearance that often could not withstand SEM sample preparation techniques as compared to the extended spread cells attached to the cyc-DOPA films. MAP attached few or no cells, rather it was a deterrent. This phenomenon has been associated with the proteins ability to aggregate at the surface, when allowed to completely dry down as in this study. These thicker films (sometimes greater than 1000Å) provide much-reduced polar component when compared to films created at the manufacturers specifications nearing monolayer in thickness [5].
In the presence of BSA, the K-A-K containing cyc-DOPA significantly enhanced cellular attachment when compare to each condition examined. The attachment of the cells in the presence of BSA is reported to be indicative of a specific interaction between the interface and the cell [23]. Although poly-lysine cellular interactions are not usually considered “specific” interactions, this data indicate that the arrangement of the K-A-K may play a roll in enhanced binding. It has been shown that the simultaneous addition of fetal bovine serum onto MAP films suggested that serum does reduce the adhesive nature of MAP [28] and RGD-containing peptides [4,29]. This reduction is also associated with a decrease in the polar components of resultant films, γp. [4,6]. It is believed that the addition of BSA to polar films aligns the BSA with its polar groups closest to the film surface leaving a resultant less polar interface for subsequent film/solution interaction [4]. Using this technology, layering of adhesive molecules could actually result in non-fouling surfaces [5].
Although MAP-RD and cyc-DOPA are similar in size, and contain equivalent amounts of lysines, BSA significantly reduced MAP-RD’s ability to attach cells and obliterates its ability to spread cells when compared to cyc-DOPA. The unique K-A-K sequence found in cyc-DOPA must be responsible for the enhanced cellular attachment and more noticeably cell spreading. It is the cellular attachment and spreading of cells on a surface which provides a stable foundation for subsequent surface masking, cellular growth and potential tissue integration.
NMR
Chemical shift assignments of both cyclic peptides have been made using a combination of solution-state NMR spectra. Based on the chemical shifts, the substitution of tyrosine for L-DOPA has little effect on the rest of the cyclic peptide. This suggests that the conformation of the cyclic portion is conserved regardless of the external attachment residue. The dynamics calculations of the cyc-DOPA provided a family of structures that indicated the L-DOPA residue had enough structural flexibility to encounter and adsorb to surfaces (Figure 5).
The molecular models of cyc-DOPA are consistent with our previous MAP-interface interaction model [6,13] in which the phenolic side chain functional groups are allowed to interact at a surface [13] and the lysine side chains remain outermost [6]. More recently, it reaffirmed that the (-OH) groups of the L-DOPA are likely responsible for surface adhesion [30].
Clearly, for the conditions examined, our results indicate that the presence of L-DOPA is required for peptide-surface adsorption. The NMR and amino acid analysis reveal only minor differences associated with the tyrosine and L-DOPA, therefore suggesting that the cyclic portion of each peptide is conserved allowing the polar side chains to remain outermost. It is further evident from the accumulated information obtained, that the unique lysine-alanine-lysine (K-A-K) of the cyc-DOPA is responsible for the specific cellular attachment noted here. Although others have suggested that the lysines are capable of charge associations between MAP and the surface [31], based on our observations, it seems more likely that the L-DOPA is responsible for surface attachment leaving the positively charged lysines outermost.
The significance and specification of the K-A-K binding domain has been further substantiated by its enhanced ability to attach cells when compared to lysine-alanine-ornathine (K-A-O) peptides [32]. Our continuing studies are examining K-A-K-containing film conditions that promote cellular attachment and growth. Promising results utilizing cyc-DOPA films to adhere rat retinal stem cells indicate that they clearly attach and spread forming multiple layers within some areas [33]. Ongoing studies are utilizing antibody-labeling techniques to identify possible cellular differentiation of these stem cells. Other studies examining surface modification have used L-DOPA films as a first layer for subsequent film absorption to create non-fouling resultant outerlayers [34, 35] while others have used dopamine [36] Some have had some success in using cyclic peptide coatings [37] or covalent grafting of cyclic RGD peptides to mediate specific cell adhesion [38], while others have used cyclic peptides to block binding sites on cells in solution to deter subsequent surface adhesion [39]. Structural analysis of these molecules often continue to rely on traditional solution-state NMR spectroscopy, however we are working toward an adsorbed-state protocol. Since these molecules are active while adsorbed to surfaces in wet environments, and very hygroscopic, using solid-state NMR techniques is difficult and perhaps lacking in significance. Solution state NMR techniques using peptides adsorbed to molecule size substrates, smaller nanoparticles, in solution may be the best way to monitor the adsorption process. Additionally, it may reveal the outermost structures of the wet adsorbed films that cells and other solute may encounter [40].
These biomimetic peptides, designed with the ability to adsorb to surfaces and then attach cells or to actually deter cellular attachment, could be utilized to mask surfaces of implantable devices reducing the inflammatory response, potentially design tissue adhesives, or assist in the regeneration process. These peptides could also be used in an in vitro environment where surface-fouling manipulation is desired.
CONCLUSIONS
These studies clearly demonstrate that peptides, which contain both L-DOPA and K-A-K moieties, can adhere to a surface and then attach cells. It has been reaffirmed that the L-DOPA residue is responsible for the peptides adsorption to the surface. However, it was only the cyc-DOPA peptide film that allowed cells to spread on the surface. The structural studies on this peptide show that the two moieties of interest are on opposite planes of the cyclized peptide. The L-DOPA portion shows a great deal of motion is possible. The freedom of movement along with the readily accessible (-OH) groups of the L-DOPA allow this peptide to adhere more strongly to the surface and its films more able to withstand SEM preparation methods. In the MAP-RD sequence, the L-DOPA residue is not at the end of the sequence and is therefore less accessible to the surface.
Also apparent from the structural studies is the arrangement of the K-A-K sequence. Not only is it on the opposite plane of the peptide, which allows presentation to the cells when the peptide is adhered to the surface, but also the side chains are presented like a pair of arms with the charges at the same relative distance from the ring. This is not the case with MAP-RD where the lysine side chains are at opposite ends of the sequence and not at the same distance from the surface. This is significant because the cyc-DOPA demonstrated the greatest cell specific attachment and was the only peptide capable of spreading of cells in the presence of BSA. This may be due to a cell specific attachment, at least in the case of the MOLT-4 cells used that recognize the specific spatial arrangement of the K-A-K peptide sequence. This cellular attachment and spreading of cells on a surface provides a stable foundation on which to build. These surfaces can subsequently be used for surface masking, cellular growth and potential tissue integration.
Acknowledgments
The research was supported by NSF Grant MCB 0132823 the NIH grant CA-16056 which supports in part the NMR Facility at Roswell Park Cancer Institute.
The authors thank Dr. Ian McDonald for MOLT-4 cells, Peter Bush and Edward Hurley for SEM imaging, Sabina Nagpal, Ryan Maine, Caroline Lopes, Margaret Miessner, Martin Lee, O’Dane Brady, Kathleen Anderson and Thomas Milano for their technical support. Dr. Robert Baier for helpful conversations regarding this work. We are especially grateful to the late Dr. James Alderfer that made his research facilities and excitement for science readily available to the students of our small undergraduate college.
Abbreviations
- MAP
Mussel Adhesive Protein
- L-DOPA
3,4-Dihydroxyphenylalanine
- cyc-DOPA
cyclic DOPA (DOPA-G-G-C-G-K-A-K-G-C)
- cyc-Y
cyclic Tyrosine (Y-G-G-C-G-K-A-K-G-C)
- NMR
Nuclear Magnetic Resonance
- MAIR-IR
Multiple attenuated internal reflection-infrared
- MAP-RD
decameric sequence of MAP (alanine-lysine-proline-serine-tyrosine-hydroxyproline-hydroxyproline-threonine-L-DOPA-lysine-COOH; A-K-PS-Y-Hyp-Hyp-T-DOPA-K)
- CD
Circular Dichroism
- MAP-14
14 amino acid peptide derived from MAP (P-S-X-Hyp-Hyp-T-X-K-A-K-P-S-X-Hyp)
- TCPS
Tissue Culture polystyrene
- SFM
Serum Free Hybridoma Media
- BSA
Bovine Serum Albumin
- PBS
Phosphate Buffered Saline
- SEM
Scanning Electron Microscopy
- COSY
Correlated Spectroscopy
- ROESY
Rotating frame nuclear Overhauser spectroscopy
- nOe
nuclear Overhauser effects
- TSP
Trimethylsilylpropionate
- RMSD
Root Mean Squared Deviation
- MM
Molecular Mechanics
- MD
Molecular dynamics
- HPLC
High Pressure Liquid Chromatography
References
- 1.Waite JH, Housley TJ, Tanzer ML. Biochem. 1985;24:5010–5014. doi: 10.1021/bi00340a008. [DOI] [PubMed] [Google Scholar]
- 2.Waite JH. Int J Adhesion and Adhesives. 1987;7:9–14. [Google Scholar]
- 3.Olivieri MP, Wollman RM, Hurley MI, Swartz MF. Biofouling. 2002;18:149–159. [Google Scholar]
- 4.Olivieri MP, Baier RE, Loomis RE. Quenching of surface polarity as a mechanism to reduce cell adhesion. Transactions of the Society For Biomaterials. 1990;XIII:78. [Google Scholar]
- 5.Olivieri MP, Loomis RE, Meyer AE, Baier RE. J Adhesion Science and Technology. 1990;4:197–204. [Google Scholar]
- 6.Olivieri MP, Baier RE, Loomis RE. Biomaterials. 1992;13:1000–1008. doi: 10.1016/0142-9612(92)90150-m. [DOI] [PubMed] [Google Scholar]
- 7.Lin Q, Gourdon D, Sun C, Holten-Anderson N, Anderson TH, Waite JH, Israelachvili JN. PNAS. 2007;104(10):3782–3786. doi: 10.1073/pnas.0607852104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baier RE. J Biomed Mater Res. 1982;16:173–175. doi: 10.1002/jbm.820160210. [DOI] [PubMed] [Google Scholar]
- 9.Zhao H, Robertson NB, Jewhurst SA, Waite JH. 2006;281(16):11090–11096. doi: 10.1074/jbc.M510792200. [DOI] [PubMed] [Google Scholar]
- 10.Grande DA, Pitman MI. Bull Hosp Joint Dis Orthop Inst. 1988;48:140–148. [PubMed] [Google Scholar]
- 11.Green K, Berdecia R, Cheeks L. Current Eye Research. 1987;6:835–838. doi: 10.3109/02713688709034851. [DOI] [PubMed] [Google Scholar]
- 12.Zhao H, Waite JH. Biochemistry. 2006;45:14223–14231. doi: 10.1021/bi061677n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olivieri MP, Wollman RM, Alderfer JL. J Peptide Res. 1997;50:436–442. doi: 10.1111/j.1399-3011.1997.tb01206.x. [DOI] [PubMed] [Google Scholar]
- 14.Kanyalkar M, Srivastava S, Coutinho E. Biomaterials. 2002;23:389–396. doi: 10.1016/s0142-9612(01)00117-x. [DOI] [PubMed] [Google Scholar]
- 15.Silverman HG, Roberto FF. Marine Biotechnology. 2007;9:661–681. doi: 10.1007/s10126-007-9053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagpal S, Wollman RM, Swartz MF, Maine RF, Rhodes TR, Olivieri MP. Using a RGD-Containing Peptide as an Adhesive. Surgical Applications of Tissue Sealants and Adhesives, (1st annual Gunther Schlag award recipient); New Orleans, LA. October 5–7, 2001. [Google Scholar]
- 17.Minowada J, Ohnuma H, Moore GE. J Natl Cancer Inst. 1972;49:891–895. [PubMed] [Google Scholar]
- 18.Makarem R, Newham P, Askari JA, Green LJ, Clements J, Edwards M, Humphries MJ, Mould AP. J Biol Chem. 1994;269:4005–4011. [PubMed] [Google Scholar]
- 19.Marumo K, Waite JH. Biochimica et Biophysica Acta. 1986;872:98–103. doi: 10.1016/0167-4838(86)90152-4. [DOI] [PubMed] [Google Scholar]
- 20.Christersson CE. PhD thesis, Swedish Dental Journal Supplement. Vol. 77. Lund University; Lund, Sweden: 1991. On salivary film formation and bacterial retention to solids. [PubMed] [Google Scholar]
- 21.McCrackin FL, Colson JP. National Bureau of Standards Misc Pub No 256. 1964. A FORTRAN program for analyses of ellipsometer measurements and calculation of reflection coefficients from thin films. [Google Scholar]
- 22.Harrick NJ. Internal Reflection Spectroscopy. Wiley Interscience; New York: 1967. [Google Scholar]
- 23.Pierschbacher M, Hayman EG, Ruoslahti E. Proc Natl Acad Sci. 1983;80:1224–1227. doi: 10.1073/pnas.80.5.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wüthrich K. NMR of Proteins and Nucleic Acids. John Wiley and Sons; New York: 1986. p. 17. [Google Scholar]
- 25.Sybyl Molecular Modeling System, Version 6.3. Tripos Associates, Inc; St. Louis Missouri: Jan, 1997. [Google Scholar]
- 26.Brünger AT. A System for X-Ray Crystallography and NMR. Yale University Press; New Haven and London: 1992. [Google Scholar]
- 27.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 28.Olivieri MP, Tweden KS, Rittle KH, Loomis RE. Biomaterials. 1992;13:201–208. doi: 10.1016/0142-9612(92)90185-q. [DOI] [PubMed] [Google Scholar]
- 29.Olivieri MP, Tweden KS. J of Biomaterials Res. 1999;43:355–359. doi: 10.1002/(sici)1097-4636(19990905)46:3<355::aid-jbm7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 30.Zhao H, Waite HW. J Biol Chem. 2006;281(36):26150–26158. doi: 10.1074/jbc.M604357200. [DOI] [PubMed] [Google Scholar]
- 31.Suci PA, Geesey J Colloid Interf Sci. 2000;230:340. doi: 10.1006/jcis.2000.7120. [DOI] [PubMed] [Google Scholar]
- 32.Olivieri MP, Swartz MF, Hurley MI, Wollman RM, Maine RF, Link PA. Cellular Attachment to MAP-binding domain and derivatives. Transactions of the Society for Biomaterials, 7th World Congress; Sydney, Australia. May 17–21, 2004. [Google Scholar]
- 33.Olivieri MP, Hurley MI, Swartz MF, Seigel GM, Strom MJ, Maine RF, Gurita C. Attachment of retinal stem cells to MAP cellular binding domain. Society for Biomaterials, Biomaterials in Regenerative Medicine: The Advent of Combination Products; October 16–18, 2004; Philadelphia, P A. [Google Scholar]
- 34.Strom M, Olivieri MP. 3,4 Dihydroxyphenylalanine (L-DOPA) Film as Potential Non-fouling Surface. 19th Annual Meeting of the Protein Society; Boston, MA. July 31– August 3, 2005. [Google Scholar]
- 35.Trawally L, Hurley MI, Gridley CL, Shapiro CD, Gurita C, Olivieri MP. Peptide Films as Non-Attachment Coatings. Society for Biomaterials World Congress; Amsterdam, The Netherlands. May 2008. [Google Scholar]
- 36.Lee H, Dellatore SM, Miller WM, Messersmith PB. Science. 2007;381:426–430. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeschke B, Meyer J, Jonczyk A, Kessler H, Adamietz P, Meenen NM, Kantlehner M, Goepfert C, Nies B. Biomaterials. 2002;23:3455–3463. doi: 10.1016/s0142-9612(02)00052-2. [DOI] [PubMed] [Google Scholar]
- 38.Lieb E, Hacker M, Tessmar J, Kunz-Schughart LA, Fiedler J, Dahmen C, Hersel U, Kessler H, Schulz MB, Göpferich A. Biomaterials. 2005;26:2333–2341. doi: 10.1016/j.biomaterials.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 39.Huang J, Gräter SV, Corbellini F, Rinck S, Bock E, Kemkemer R, Kessler H, Ding J, Spatz JP. Nano Letters. 2009;9:1111–1116. doi: 10.1021/nl803548b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olivieri MP, Wollman RM, Swartz MF. Nanoparticles Enable NMR Spectroscopy to Monitor Molecular Interactions at an Interface. Society For Biomaterials Annual Meeting; St. Paul, Minnesota. April 24–29, 2001. [Google Scholar]