Abstract
The vast majority of microbial species remain uncultivated and, until recently, about half of all known bacterial phyla were identified only from their 16S ribosomal RNA gene sequence. With the advent of single-cell sequencing, genomes of uncultivated species are rapidly filling in unsequenced branches of the microbial phylogenetic tree. The wealth of new insights gained from these previously inaccessible groups is providing a deeper understanding of their basic biology, taxonomy and evolution, as well as their diverse roles in environmental ecosystems and human health.
Historically, microbial research was limited by the need to grow bacteria in culture. Even in the modern era of genomics, DNA sequencing requires large amounts of DNA template obtained from homogeneous cell cultures. However, as it is estimated that >99% of all bacterial species remain uncultivated owing to unknown growth requirements, most of these species could not be sequenced. The development of 16S ribosomal RNA gene PCR analysis revolutionized the field of microbial genomics, as it enabled the amplification and sequencing of a highly informative gene from most bacterial species (FIG. 1a). However, although the 16S rRNA gene sequence allowed construction of phylogenetic trees, analyses were limited to this single gene. Another major advancement was enabled by metagenomics, a method in which total DNA from an environmental sample is sequenced. Metagenomics has the advantage of providing sequences for the entire gene content of environmental communities (FIG. 1b). However, given the high diversity of many microbial communities, the assembly of genes and individual genomes remains a challenge. Metagenomics predominately uses next-generation sequencing with short reads. As the diversity of the microbial community increases, the technique becomes limiting in its ability to accurately identify variations between bacterial strains, such as the presence of novel genes.
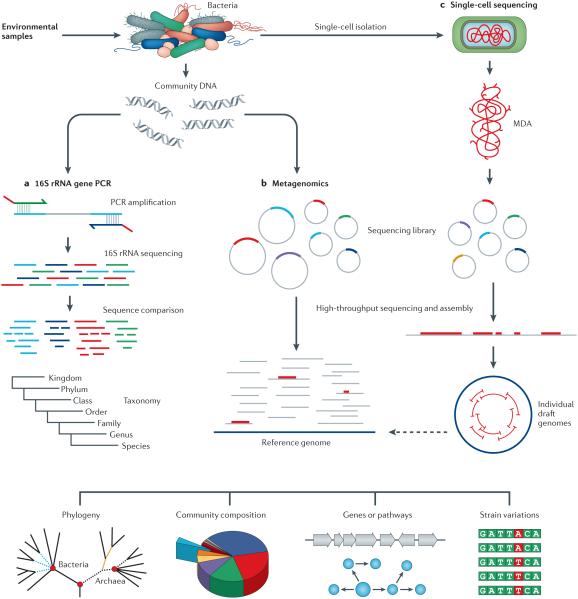
Figure 1. Complementary methods used to investigate the genomics of uncultivated bacteria.
a | PCR amplification of the 16S ribosomal RNA gene can be carried out from most novel bacteria using primers that anneal to sequences which are highly conserved across bacterial species. Variable regions of the 16S rRNA gene can then be used to derive a phylogenetic tree. b | Metagenomics is based on sequencing of total DNA extracted from the environment. Gene content and frequencies are obtained for the entire ecosystem; however, assembly of the sequencing reads from the sequencing library inserts (which are indicated by coloured sequences) into the genomes of individual species is complicated by the large number of organisms that contribute to the DNA in the sample. c | Single-cell sequencing typically does not recover the entire genome; however, the reads obtained in the sequencing library are all genetically linked, which facilitates genome assembly. These genomes represent the integrated genetics and biochemistry of individual microbial species. Single-cell genomes can serve as a reference genome to aid the assembly of sequencing data for uncultivated species that are closely related. DNA or cDNA sequencing reads obtained through shotgun metagenomic or metatranscriptomic approaches can be mapped to the reference genomes (dashed arrow). Single-cell reference genomes can also provide phylogenetic placement for a substantial portion of the metagenomic reads that have not been previously assigned a taxonomy. 16S rRNA gene PCR is a rapid and inexpensive method to assign phylogeny and community composition. Metagenomics also provides community composition and gene content. Single-cell sequencing provides assembled contigs to determine genes and pathways within an individual cell and is best suited for the evaluation of strain variations. MDA, multiple displacement amplification.
The development of the multiple displacement amplification (MDA)1,2 reaction constituted a ‘quantum leap’ forward for microbial research. This method enables amplification of a single bacterial genome by >1 billion-fold3, which avoids the need to develop cultivation methods to elucidate the genome. High-throughput automated processes are often used to carry out MDA from bacteria that are sorted into microtitre plates by fluorescence-activated cell sorting (FACS). Cell lysis and MDA are followed by screening of the amplified DNA using cycle sequencing of PCR products for the 16S rRNA gene to identify the taxonomy of the cells, thus allowing whole-genome sequencing efforts to be focused only on the bacteria of interest. When MDA is combined with next-generation sequencing and bioinformatic assembly methods, this approach can yield partial to near-complete bacterial genomes of high quality (FIG. 1c). Single-cell genomic sequencing is thus rapidly transforming our understanding of the vast numbers of microorganisms in the environment. In an acknowledgement of its recent impact on many scientific fields, single-cell sequencing was named method of the year in 2013 (REF. 4).
The methodology for sequencing uncultivated microbial genomes from single cells and its relevance for a broad range of applications have been reviewed in detail elsewhere5. Here, we provide an update on recent progress in capturing novel genomes, large-scale environmental studies, research relating to human health and recent improvements in the methods used for sequencing DNA from uncultivated bacterial cells.
Sequencing uncultivated bacteria
As single-cell sequencing does not require prior cultivation, this method has the potential to immediately contribute many new genomes of uncultivated strains and to reveal intra-species variations by refining the identified ‘core genes’ that are essential to a species and by expanding the total identified gene diversity within species, which is referred to as the ‘pan genome’. Genetic variations between bacterial strains provide crucial insights into biological function and adaptations, and are particularly important for the study of pathogen infectivity, transmission and development of antibiotic resistance.
New phylum-level genomes
The first reference genomes for many candidate phyla, which were previously known only from their 16S rRNA gene sequences, have been obtained directly from single cells. These genome assemblies are rapidly filling in many branches of the bacterial and archaeal tree of life that did not have representative genomes (FIG. 2) and are also revealing many previously unknown genes and functions. The first elusive candidate phyla sequenced using this approach was TM7, and the representative cells were obtained from the human oral cavity6 and soil7. Over the past several years, partial genomes have been reconstructed from other candidate phyla, including SR-1 from the oral cavity8, TM6 from a biofilm in a hospital sink9, OP11 from an anoxic spring10 and ‘Candidatus phylum Atribacteria’ (OP9) from hot spring sediments11. A proposal was recently put forth to designate a new bacterial candidate phylum — Tectomicrobia — on the basis of genome assemblies from sponge symbionts with highly distinct gene clusters that are involved in biosynthesis of secondary metabolites (which are often referred to as natural products)12. These are an important source of new drugs, including antibiotics and antitumour agents, as well as of other commercially useful chemicals. The gene clusters are deeply branched from other bacterial taxa, which supports the proposal to designate this group as a new phylum.
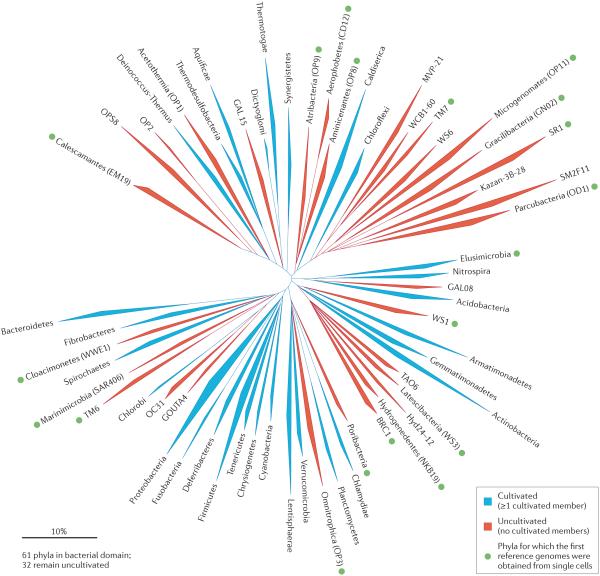
Figure 2. Filling in the bacterial tree of life.
The 16S ribosomal RNA gene maximum likelihood phylogenetic inference for the domain Bacteria13 highlights the uncultivated phyla and the impact of single-cell genomics. Of 61 known phyla in the bacterial domain, 32 still have no cultivated representatives (red branches). The majority of genomes from the uncultivated candidate phyla were captured using multiple displacement amplification (MDA)-based single-cell approaches (green dot). Sequences are grouped on the phylum level, except for the phylum Proteobacteria, which is polyphyletic in this analysis. Groups containing 16S rRNA genes from cultured isolates are in blue. The scale bar represents 10% estimated sequence divergence.
At the US Department of Energy Joint Genome Institute, the largest study so far has been carried out to sequence genomes from major uncultivated bacterial and archaeal lineages13 using optimized high-throughput procedures14. Nine environmental samples — including marine, brackish, freshwater and hydrothermal samples — were used13. Thousands of single-cell MDA reactions were typed by the 16S rRNA gene, and ~200 of these cells were deeply sequenced. The partially assembled genomes, which range from 148 kb to 2.4 Mb, represent 29 major uncharted branches of the evolutionary tree (FIG. 2). These are the first substantive genomic data for candidate bacterial phyla SAR406 (Marine Group A), OP3, OP8, WS1, WS3, BRC1, CD12, EM19, EM3, NKB19 and Oct-Spa1-106, as well as for several highly divergent archaeal groups related to Nanoarchaeota. This study revealed unexpected metabolic features, including a complete sigma factor in archaea that is similar to those in bacteria, a novel amino acid use for the opal stop codon and an archaeal-type purine synthesis in bacteria13.
Partial genome assemblies
Even when single-cell assemblies are highly fragmented, the new information gained on metabolic pathways and adaptations to the environment is useful and can assist in the development of cultivation approaches, which is still a crucial goal for many biological investigations. For the rare and yet uncultivated candidate phylum TM6, the near completion of an assembled genome from single cells9 revealed homology to sequences such as ankyrin repeat domains, which are enriched in bacterial genomes of known intracellular symbionts of amoeba. This finding suggests that TM6 is an endosymbiont that might resist in vitro growth until its host is identified and used to assist cultivation.
Development of cultivation methods for novel bacteria is also aided by insights into their metabolic capabilities, which can be obtained from only partial reconstruction of biochemical pathways. For example, the sequencing of 70% of the genome from MDA-amplified Beggiatoa spp. cells, which could not be cultivated previously, revealed crucial enzymes for sulphur oxidation, nitrate and oxygen respiration, and carbon dioxide fixation, and confirmed a putative chemolithoautotrophic physiology15. Uncultivated bacteria and archaea may also be highly adapted to particular biological niches in which the interdependence between members of the microbial community prevents cultivation of pure populations. Determination of missing biochemical substrates or special adaptations to the environment should be useful in developing cultivation methods.
Given these insights, partial single-cell genome drafts tend to be published and shared in spite of missing sequences and the higher likelihood of assembly errors that result from amplification bias, chimaera formation or reconstruction of consensus genomes from multiple single-cell amplifications. Efforts are currently underway, for example, in the Human Microbiome Project funded by the US National Institutes of Health (BOX 1), to formalize the criteria for naming and assessing the quality of single-cell genome assemblies.
Box 1. The Human Microbiome Project.
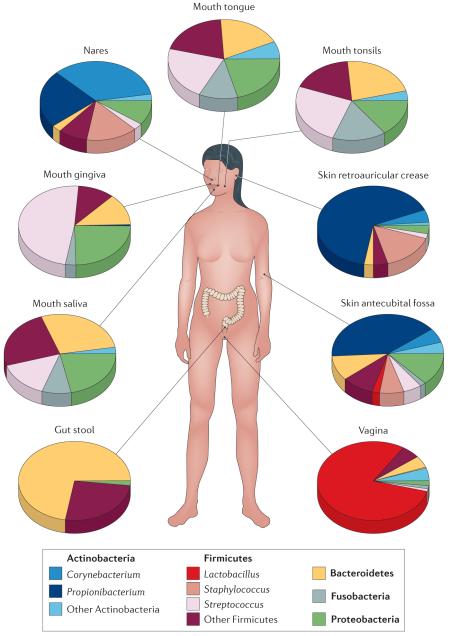
The Human Microbiome Project (HMP) funded by the US National Institutes of Health identified the sequencing of uncultivated bacteria as a crucial unmet goal. To address this need, single-cell multiple displacement amplification (MDA) reactions and 16S ribosomal RNA gene screening were carried out on cells from human stool and oral swabs using a high-throughput automated platform25. The 16S rRNA gene sequences from these cells (see HMMDA16S — HMP single cell MDA 16S rRNA Sanger sequencing) were used to identify the MDA-amplified DNA from species of greatest interest. As a result, >50 initial genome drafts were made publicly available to the research community, and ~350 more drafts are currently being deposited as part of HMP Reference Genomes. Many of the amplified genomes were on a prioritized list of human-related taxa that lack reference genomes, which are referred to as the ‘100 most wanted’ (REF. 45). The uncultivated bacterial genomes derived from the human body will be a powerful tool to investigate the community structure of the microbiome and can act as key references to aid the analysis of metagenomic and metatranscriptomic data sets. A large majority of human-associated microorganisms remain uncultivated (see the figure; pie charts represent relative abundance at the phylum level for each major body site sampled in the HMP). Single-cell sequencing allows genome recovery from species that are distantly related to current isolates and provides new key reference genomes to aid the characterization of healthy and disease states.
New microbial environments
A growing range of environments are being studied with single-cell sequencing strategies in order to investigate uncultivated microbial species.
Marine environments
A partial genome was assembled for ‘Candidatus Poribacteria’, which are symbiotically associated with marine sponges16. More recent single-cell sequencing of Poribacteria yielded several near-complete genomes, which revealed a specialized metabolism and supports their role as efficient scavengers and recyclers of a particular suite of carbon compounds that are unique to sponges17. Single-cell sequencing has also been used to investigate changes through time that occur in microbial communities. The Deepwater Horizon oil spill in the Gulf of Mexico resulted in a bloom of uncultured and uncharacterized members of the Oceanospirillales18. Single-cell genomes revealed genes that encode n-alkane and cycloalkane degradation enzymes, which are thought to be involved in a rapid response to the aliphatic hydrocarbons that would be introduced by an oil spill. Although metagenomic surveys might detect the presence of these genes in the DNA of the total community, single-cell sequencing associates the gene with individual strains and reveals its context within gene networks, which may aid function prediction.
Human indoor environments
Hospital-acquired infections and the emergence of antibiotic-resistant strains present a serious threat. Despite their importance for human health, bacteria that reside in human indoor environments have so far been understudied. Genomic analyses in the hospital environment have been severely limited by technical obstacles. Moreover, a critical segment of the pathogen life cycle has been nearly invisible to us owing to the low abundance of these pathogens on hospital surfaces, on medical devices and within reservoirs such as biofilms. Importantly, biofilms are thought to be reservoirs of disease-causing organisms at barely detectable levels in both outdoor and indoor environments, including those within water distribution systems, such as Legionella pneumophila19, Escherichia coli, Vibrio cholerae20 and Helicobacter pylori21. New tools such as single-cell sequencing are needed to allow genomic analyses of strain variants, as culturing will yield only a fraction of the species present. Obtaining genomes without prior cultivation provides a direct unbiased sampling of cells in a given environment, as culturing on only one or a few media types could favour known and fast-growing species. Moreover, PCR of virulence genes or marker genes is mainly focused on a small number of known species, and shotgun metagenomics is limited in its ability to detect strain variations. Even when culturing is possible, growth biases can result in selection for genome alterations such as gene loss22, and single-cell sequencing of the source organism is therefore desirable.
Many critical pathogens (for example, Legionella spp., Francisella spp. and Burkholderia spp.) are difficult to culture because they reside at low abundance within vectors such as amoeba, which are a key mode of transmission for these pathogens23. This endosymbiotic lifestyle is thought to contribute to the evolutionary changes that are necessary for human intracellular pathogenesis24. The problem is compounded in hospital water distribution systems by the presence of pathogen-laden amoeba that may contain levels of organisms well within the infective dose. Low-abundance novel bacteria that reside only within amoeba may be missed from conventional culturing and other identification assays but could represent emerging pathogens. Direct sequencing of pathogens without the requirement of cultivation will give insights into life outside the human host and to factors that influence infection, virulence and transmission.
Recently, pathogen genomes and the first genome assemblies from the candidate phylum TM6 were obtained from a biofilm in a hospital sink drain9. An automated system was used to generate thousands of single-cell MDA reactions from bacteria in a hospital sink biofilm, which yielded ~400 amplified genomes of interest from 25 different genera9,25. These represent environmental species, human commensals and opportunistic pathogens. Three amplified genomes25 were obtained of the human pathogen Porphyromonas gingivalis, which had previously been sequenced only from cultured isolates from patients with periodontitis. The genomes from the environmental biofilm were the first for P. gingivalis obtained outside a human host. Increased confidence in the genome data was gained by sequencing multiple single cells. For example, the three independent single-cell genomes of P. gingivalis were confirmed to be highly clonal, and the largest de novo assembly was able to generate a near-complete genome25. Several complementary strategies were used to analyse the deeply sequenced cells (FIG. 3). Genetic diversity between the cells captured from the environment, including single-nucleotide polymorphisms in virulence factors, was found through read mapping to the known reference P. gingivalis genomes (FIG. 3A). MDA-optimized de novo genome assembly tools26 were used to reveal variant genes in the genome (FIG. 3B). For example, the apparent loss of genes involved in capsule formation and the lack of clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) genes were consistent with life outside the host, as a previous study showed that loss of these factors increases biofilm formation in other bacteria27.
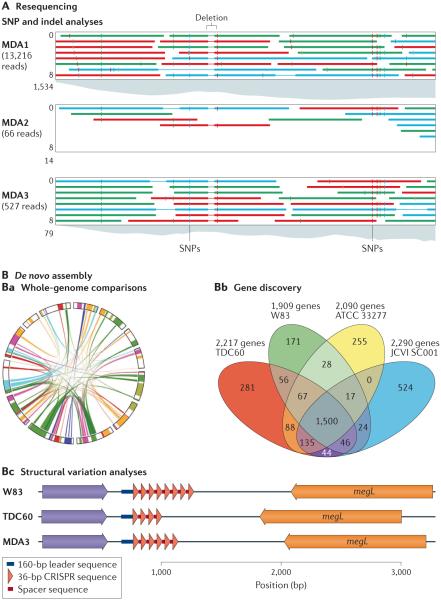
Figure 3. Comparative genomics using single-cell DNA amplification.
Amplification of DNA by multiple displacement amplification (MDA) can achieve single-nucleotide resolution in resequencing studies (that is, studies in which a known reference genome is available) for use in analyses of single-nucleotide polymorphisms (SNPs), insertions and deletions (indels), and structural variations, as well as in whole-genome comparisons from de novo assembled sequences. A | SNP and indel analyses have been carried out on a virulence-related gene (the reference sequence is shown at the top of each panel) of Porphyromonas gingivalis for three independent amplified single cells (MDA1, MDA2 and MDA3) that were captured from a complex biofilm in a hospital25, which revealed shared variants (that is, SNPs) within this gene. B | With advances in MDA-optimized assembly tools such as SPAdes26, the technique is approaching the level expected for sequencing cultured strains, which is improving whole-genome comparisons of synteny (that is, the order of genes in a genome) (part Ba; syntenic blocks that are shared between genomic regions are connected with coloured ribbons). Similarly, this technique can improve gene discovery (part Bb). In this example, an assembled draft genome (JCVI SC001), which consists of the SPAdes assembly of MDA3, is compared against reference genomes from the P. gingivalis strains TDC60, W83 and ATCC 33277 (REF. 25). De novo assembly of regions that contain multiple repeats and that are difficult to assemble, such as the clustered regularly interspaced short palindromic repeat (CRISPR) region, can be resolved (part Bc). For example, de novo assembly of the repeats in CRISPR region 36–30 showed that the repeat regions in MDA3 were identical to sequenced genomes from cultured pathogen isolates (W83 and TDC60) but contained variable spacer sequences, which is indicative of phage predation in a different environment25. megL, methionine gamma-lyase. Parts A and Bc are adapted with permission from REF. 25, Cold Spring Harbor Laboratory Press.
Overall, these near-complete genomes captured from difficult sample types (such as biofilms) highlight the usefulness of single-cell sequencing for the investigation of low-abundance pathogens and their transmission between the environment and the host. Biofilms within a human host are crucial to many disease processes, such as infection of the mucosa, and clinical samples should now be amenable to single-cell genomic studies to reveal genetic polymorphisms in a pathogen population.
Technical advances
The methods used for single-cell genome amplification, sequencing and assembly are still under rapid improvement.
Combining single-cell genomic data and metagenomic data
Filling in the branches of the bacterial and archaeal tree of life with genomes of uncultivated species, as well as understanding their adaptations and dependence on the community, can be accelerated by combining single-cell genomic data with metagenomic data or metatranscriptomic data from bulk environmental samples (FIG. 1a). For example, single-cell contigs were used to recruit up to an additional 20% of metagenomic reads obtained from previously unsequenced organisms13. Although most metagenomic data are used for population-level analyses of gene diversity and metabolic potential, several complete individual genomes of candidate phyla and other uncultivated species have also been recently assembled from deep metagenomic sequencing28,29. It will be increasingly powerful to use genome assemblies derived from metagenomic and single-cell sources to validate each other.
‘Mini-metagenomes’
A novel promising approach is ‘mini-metagenomics’ (REF. 9), which is intermediate between the use of single cells and the use of the thousands of microbial species that can contribute to metagenomic data. Limited pools of FACS-sorted cells from the environment are amplified by MDA for sequencing. Large numbers of cells can be processed; however, the reduced diversity of the pools, compared with whole-community metagenomics, makes it simpler to deconvolute individual genomes. For this approach to be successful, specialized MDA-optimized assembly methods30, combined with recent advances in contig classification and binning9,29,31, are required to improve genome recoveries.
Immunomagnetic separation
A new culture-free strategy — immunomagnetic separation (IMS) coupled with MDA32 — was able to capture low numbers of specific pathogens from clinical specimens that are highly contaminated with human DNA. Chlamydia trachomatis is an obligate intracellular pathogen and is problematic to culture. A small pool of C. trachomatis cells from patients was enriched by antibody capture using magnetic beads, and the DNA from this pool was amplified by MDA to allow sequencing of novel strains32. Although the method would need to be optimized to capture each pathogen of interest, it could have the advantage of ease of use for multiple patients. It can yield complete genome assemblies, as MDA is carried out on DNA from multiple cells, which reduces amplification bias compared with sequencing from a single cell. However, if there are multiple genetic polymorphisms within the cell pool that has been isolated, then it can be difficult to determine which of these polymorphisms occur together in individual cells. By sequencing one cell, the genetic linkage can be obtained, whereas sequencing multiple cells gives the total range of polymorphisms in the population.
Remaining technical challenges
Amplification bias remains a complicating factor for single-cell sequencing. Several strategies are currently being pursued to address this issue.
Amplification bias and de novo assembly
Although MDA has fairly low amplification bias for a whole-genome amplification method33, there is still highly uneven coverage from single cells partly as a result of random variation in the rate of exponential amplification across the single DNA template. It was recognized early in the development of de novo genome assembly from single cells34 that this high variation in coverage creates a challenge for available assembly programs, which were designed for use with the fairly uniform sequence coverage obtained from unamplified genomic DNA templates. Both laboratory methods for reducing amplification bias and computational methods for managing variable read depth are helping to overcome this problem.
Laboratory methods
Limited clonal growth within agarose beads35 of cells that are difficult to culture reduces MDA bias by providing multiple DNA template copies. It is likely that many species that are difficult to culture can still progress through several cell divisions within the beads, and this could therefore be a promising strategy for sequencing novel bacteria. Another approach is to inhibit cell division to generate multiple genome copies within single cells36. This was demonstrated in a test case by blocking the bacterial cytoskeleton protein FtsZ in Bacillus subtilis using an inhibitory compound. In principle, it may be possible to block cell division in various bacterial species found within complex natural communities and then isolate them by flow cytometry on the basis that larger cells contain more genome copies. Another laboratory strategy reduces amplification bias by carrying out the MDA reaction in microfluidic chambers, which is thought to favour more complete coverage37. One new microfluidic system produced 98–99% of reads that correctly mapped to a reference genome and >90% assembly from single E. coli cells38.
Bioinformatic methods
Recent advances in computational methods have been focused on improving genome assemblies from biased sequencing data. Fragment assembly tools typically assume the near-uniform coverage obtained with unamplified DNA templates. For example, these tools may assume that reads are erroneous for genome regions with coverage that is below average for the whole data set. However, a large portion of valid reads are filtered out when a single coverage cutoff is used. Recently, a method was introduced for varying the cutoff in order to use the low-coverage regions created by MDA39. A newer version of this method called SPA des26,40 improves upon the use of non-uniform coverage and addresses chimerism that results from MDA-generated DNA rearrangements41, as well as from read pairs sampled from distant regions of the genome. SPAdes also improves assemblies40 from mini-metagenomes9. Another new assembly tool called IDBA-UD also reports improved results for highly uneven coverage of single-cell sequencing42. A period of improvements and testing of new computational methods is likely to drive progress in genome assembly from amplified DNA.
Conclusions
In the past few years, there has been a large increase in the number of single-cell studies, and many taxonomic groups have received their first reference genome. Large-scale studies have been carried out in many environments, including the human microbiome (BOX 1). Recent studies of human pathogens and the human microbiome establish single-cell sequencing as a powerful method to compare genomic polymorphism between strains through both read mapping to a close reference and de novo assembly, which enables whole-genome comparisons. Sequencing of single eukaryotic cells is also improving, which has exciting prospects for research into human cell development and disease. DNA amplification methods that were originally developed for bacteria are becoming reliable enough for use with diploid cells43,44, which require sufficient coverage of both parental chromosomes for analysis of heterozygous alleles. There is also great potential for single-cell studies of microbial eukaryotes, some of which are important pathogens, as well as of unicellular and multicellular plants.
Looking to the future, new whole-genome amplification methods may reduce amplification bias and chimeric DNA rearrangements. Research designs will also continue to improve for de novo assembly of DNA sequences of uncultured bacteria. Considering the potential of single-cell, mini-metagenomic, metagenomic and metatranscriptomic data to address important questions in microbial physiology, ecology and evolution, there is an enormous opportunity to advance the strategies and computational tools that we use. These methods each have strengths and limitations, and several examples demonstrate their use in combination5. We look forward to an exciting era of single-cell biology.
Acknowledgements
The authors acknowledge discussions with G. Tesler, S. Yooseph and J. Badger. They also acknowledge assistance with the phylogenetic tree from C. Rinke and T. Woyke. This work was supported by grants to R.S.L. from the Alfred P. Sloan Foundation (Sloan Foundation-2007-10-19) and the US National Institutes of Health (NIH 2R01 HG003647 and NIH-HHSN272200900007C), and by grants to J.S.M. from the US National Institute of General Medical Sciences (NIH 1R01GM095373).
Glossary
- 16S ribosomal RNA gene PCR analysis
A method in which primers designed for highly conserved regions of the 16S rRNA gene enable PCR from most bacteria, and variable regions of the sequence can be used for taxonomic identification
- Amplification bias
Uneven representation of regions of the DNA template in amplified DNA
- Bacterial and archaeal tree of life
The phylogenetic tree of all known bacteria and archaea based on the 16S ribosomal RNA gene
- Biofilm
A layered aggregate of microorganisms. These adherent cells are frequently embedded within a self-produced extracellular matrix that is generally composed of DNA, proteins and polysaccharides
- Candidate phyla
Uncultivated microbial groups that branch independently from known sequences near the base of the bacterial clade
- Chimaera
A recombinant molecule of DNA composed of segments from more than one source; multiple displacement amplification (MDA) can generate chimaeras that are predominantly inversions through its branching mechanism of DNA replication
- Endosymbiont
An organism that lives within the body or cells of another organism; it can include facultative or obligate symbionts
- Metagenomics
The study of the collective genomes contained in environmental samples using shotgun sequencing of DNA extracted from such samples
- Metatranscriptomic data
The set of all mRNA molecules or transcripts produced in a population of cells; they are typically obtained by shotgun sequencing of cDNA from a mixed microbial community
- Multiple displacement amplification
(MDA). A whole-genome DNA amplification method in which a DNA polymerase (usually the highly processive, strand-displacing Φ29 DNA polymerase) extends random primers while concurrently displacing the older products of downstream priming, which results in an exponential branching mechanism of DNA replication
Footnotes
Competing interesss statement
The authors declare no competing interests.
References
- 1.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raghunathan A, et al. Genomic DNA amplification from a single bacterium. Appl. Environ. Microbiol. 2005;71:3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chi KR. Singled out for sequencing. Nature Methods. 2014;11:13–17. doi: 10.1038/nmeth.2768. [DOI] [PubMed] [Google Scholar]
- 5.Lasken RS. Genomic sequencing of uncultured microorganisms from single cells. Nature Rev. Microbiol. 2012;10:631–640. doi: 10.1038/nrmicro2857. [DOI] [PubMed] [Google Scholar]
- 6.Marcy Y, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl Acad. Sci. USA. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Podar M, et al. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl. Environ. Microbiol. 2007;73:3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell JH, et al. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc. Natl Acad. Sci. USA. 2013;110:5540–5545. doi: 10.1073/pnas.1303090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLean JS, et al. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc. Natl Acad. Sci. USA. 2013;110:E2390–E2399. doi: 10.1073/pnas.1219809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Youssef NH, Blainey PC, Quake SR, Elshahed MS. Partial genome assembly for a candidate division OP11 single cell from an anoxic spring (Zodletone Spring, Oklahoma) Appl. Environ. Microbiol. 2011;77:7804–7814. doi: 10.1128/AEM.06059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dodsworth JA, et al. Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nature Commun. 2013;4:1854. doi: 10.1038/ncomms2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson MC, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 13.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 14.Rinke C, et al. Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nature Protoc. 2014;9:1038–1048. doi: 10.1038/nprot.2014.067. [DOI] [PubMed] [Google Scholar]
- 15.Mussmann M, et al. Insights into the genome of large sulfur bacteria revealed by analysis of single filaments. PLoS Biol. 2007;5:e230. doi: 10.1371/journal.pbio.0050230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegl A, et al. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 2011;5:61–70. doi: 10.1038/ismej.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamke J, et al. Single-cell genomics reveals complex carbohydrate degradation patterns in poribacterial symbionts of marine sponges. ISME J. 2013;7:2287–2300. doi: 10.1038/ismej.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mason OU, et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 2012;6:1715–1727. doi: 10.1038/ismej.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Declerck P. Biofilms: the environmental playground of Legionella pneumophila. Environ. Microbiol. 2010;12:557–566. doi: 10.1111/j.1462-2920.2009.02025.x. [DOI] [PubMed] [Google Scholar]
- 20.Shikuma NJ, Hadfield MG. Marine biofilms on submerged surfaces are a reservoir for Escherichia coli and Vibrio cholerae. Biofouling. 2010;26:39–46. doi: 10.1080/08927010903282814. [DOI] [PubMed] [Google Scholar]
- 21.Percival SL, Thomas JG. Transmission of Helicobacter pylori and the role of water and biofilms. J. Water Health. 2009;7:469–477. doi: 10.2166/wh.2009.070. [DOI] [PubMed] [Google Scholar]
- 22.Karch H, Meyer T, Russmann H, Heesemann J. Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect. Immun. 1992;60:3464–3467. doi: 10.1128/iai.60.8.3464-3467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown MR, Barker J. Unexplored reservoirs of pathogenic bacteria: protozoa and biofilms. Trends Microbiol. 1999;7:46–50. doi: 10.1016/s0966-842x(98)01425-5. [DOI] [PubMed] [Google Scholar]
- 24.Horwitz MA. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLean JS, et al. Genome of the pathogen Porphyromonas gingivalis recovered from a biofilm in a hospital sink using a high-throughput single-cell genomics platform. Genome Res. 2013;23:867–877. doi: 10.1101/gr.150433.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zegans ME, et al. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 2009;191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrighton KC, et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science. 2012;337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 29.Kantor RS, et al. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio. 2013;4:e00708–e00713. doi: 10.1128/mBio.00708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nurk S, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013;20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albertsen M, et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nature Biotech. 2013;31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 32.Seth-Smith HM, et al. Whole-genome sequences of Chlamydia trachomatis directly from clinical samples without culture. Genome Res. 2013;23:855–866. doi: 10.1101/gr.150037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosono S, et al. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang K, et al. Sequencing genomes from single cells by polymerase cloning. Nature Biotech. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 35.Fitzsimons MS, et al. Nearly finished genomes produced using gel microdroplet culturing reveal substantial intraspecies genomic diversity within the human microbiome. Genome Res. 2013;23:878–888. doi: 10.1101/gr.142208.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dichosa AE, et al. Artificial polyploidy improves bacterial single cell genome recovery. PLoS ONE. 2012;7:e37387. doi: 10.1371/journal.pone.0037387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcy Y, et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3:1702–1708. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gole J, et al. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nature Biotech. 2013;31:1126–1132. doi: 10.1038/nbt.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chitsaz H, et al. Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nature Biotech. 2011;29:915–921. doi: 10.1038/nbt.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nurk S, et al. Research in Computational Molecular Biology. Springer; 2013. pp. 158–170. [Google Scholar]
- 41.Lasken RS, Stockwell TB. Mechanism of chimera formation during the Multiple Displacement Amplification reaction. BMC Biotechnol. 2007;7:19. doi: 10.1186/1472-6750-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng Y, Leung HC, Yiu SM, Chin FY. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 43.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nature Rev. Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 44.McConnell MJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fodor AA, et al. The “most wanted” taxa from the human microbiome for whole genome sequencing. PLoS ONE. 2012;7:e41294. doi: 10.1371/journal.pone.0041294. [DOI] [PMC free article] [PubMed] [Google Scholar]