Abstract
Scedosporium boydii is a pathogenic filamentous fungus that causes a wide range of human infections, notably respiratory infections in patients with cystic fibrosis. The development of new therapeutic strategies targeting S. boydii necessitates a better understanding of the physiology of this fungus and the identification of new molecular targets. In this work, we studied the conidium-to-germ tube transition using a variety of techniques including scanning and transmission electron microscopy, atomic force microscopy, two-phase partitioning, microelectrophoresis and cationized ferritin labeling, chemical force spectroscopy, lectin labeling, and nanoLC-MS/MS for cell wall GPI-anchored protein analysis. We demonstrated that the cell wall undergoes structural changes with germination accompanied with a lower hydrophobicity, electrostatic charge and binding capacity to cationized ferritin. Changes during germination also included a higher accessibility of some cell wall polysaccharides to lectins and less CH3/CH3 interactions (hydrophobic adhesion forces mainly due to glycoproteins). We also extracted and identified 20 GPI-anchored proteins from the cell wall of S. boydii, among which one was detected only in the conidial wall extract and 12 only in the mycelial wall extract. The identified sequences belonged to protein families involved in virulence in other fungi like Gelp/Gasp, Crhp, Bglp/Bgtp families and a superoxide dismutase. These results highlighted the cell wall remodeling during germination in S. boydii with the identification of a substantial number of cell wall GPI-anchored conidial or hyphal specific proteins, which provides a basis to investigate the role of these molecules in the host-pathogen interaction and fungal virulence.
Introduction
Scedosporium species are filamentous fungi commonly isolated from polluted soils and water, but paradoxically infrequent in the air and indoor environment [1–5]. Recent taxonomic studies revealed that Pseudallescheria boydii, now called Scedosporium boydii [6], and Scedosporium apiospermum which was initially considered its asexual state, are two distinct species. These two along with three other closely related species, S. dehoogii, S. aurantiacum and S. minutisporum, constitute the Scedosporium apiospermum species complex [7–9].
Depending on the portal of entry and the patient’s immune status, these usually saprophytic fungi may be at the origin of a wide variety of human infections, ranging from localized infections subsequent to traumatic inoculation of fungal elements as in subcutaneous mycetomas, to disseminated infections in immunocompromised individuals [1, 10].
In the past two decades, these fungi gained worldwide recognition as the second most frequently isolated filamentous fungi in the airways of patients with cystic fibrosis, the most common autosomal recessive disease in Caucasian populations [11–14]. In CF patients, these fungi usually colonize the respiratory tract and may contribute to the progressive deterioration of the lung function as suggested by recent works on other fungal species like Candida albicans and Aspergillus fumigatus [15–17]. In addition, this chronic colonization of the airways constitutes a risk factor for severe and often fatal disseminated infections in patients undergoing lung transplantation, which remains the ultimate treatment in CF [18, 19]. Until now, the diagnosis of Scedosporium infections remains challenging mainly because of the similarities of clinical features and histopathology with other relatively common hyaline hyphomycetes like Aspergillus or Fusarium species. Add to this, Scedosporium species exhibit low susceptibility to amphotericin B and current triazole drugs as well as primary resistance to echinocandins [20, 21]. There is, therefore, an urgent need for a better understanding of the fungal biology in order to define new therapeutic strategies.
One of the most attractive targets for the development of new antifungal agents is the cell wall, mainly because of the uniqueness of many of its components with respect to mammalian cells [22]. The cell wall plays a critical role during morphogenesis and fungal growth since it changes accordingly to fit survival needs [23]. It protects the fungus from a wide range of environmental stresses such as desiccation, osmotic stresses and temperature variations. In pathogenic fungi it also provides the means to sustain fungal presence inside the human host by allowing adherence to the host tissues and evasion from the host immune response.
In S. boydii, which is the most prevalent species within the S. apiospermum species complex in CF [24], the conidial and mycelial cell walls were shown to contain N- and O-linked peptidorhamnomannans (PRM) having a branched structure of α-Rhap-(1→3)-α-Rhap- side chain epitope linked (1→3) to a (1→6)-linked α-Manp core [25]. Unlike α-glucans, isolated from both conidial and hyphal cell walls of S. boydii, glucosylceramides could only be obtained from mycelial samples [26, 27]. However a more recent study again showed that glucosylceramides were also detectable on the conidial surface of the fungus [28].
We previously demonstrated that the conidial cell wall of S. boydii contains dihydroxynaphtalene (DHN)-melanin and that the cell wall content in melanin and mannose-containing glycoconjugates increases during maturation of conidia along with the cell surface physical properties [29]. Here, we tracked the cell wall modifications during the germination process using various approaches, including investigation, at the molecular level, of glycosylphosphatidylinositol (GPI)-anchored proteins in conidial and hyphal walls as these integral cell wall proteins (CWPs) play a major role in normal morphology and virulence in other fungal models [30–32].
Materials and Methods
Strain and culture conditions
The fungal strain S. boydii IHEM 15155 (formerly Pseudallescheria boydii) was used throughout this study [29]. It was maintained on yeast extract-peptone-dextrose (YPD; 0.5% w/v yeast extract, 2% w/v glucose, 1% w/v peptone, 0.05% w/v chloramphenicol) agar plates.
Cultures were incubated for 7 days at 37°C, then conidia were harvested by flooding the agar surface with sterile Milli-Q water and filtered through a 20-μm pore size nylon filter. Conidia were washed twice, pelleted at 5000 X g for 5 min at 4°C, resuspended in 10 ml sterile water and finally counted with a hemocytometer.
Kinetics of germination
To study the kinetics of germination, three conditions were tested: the effect of age of cultures, culture medium and incubation temperature. First, cultures on YPD agar medium were incubated at 37°C for 5, 9 and 14 days, afterwards conidia were isolated and resuspended in YPD liquid medium (20 ml per Petri dish) at a concentration of 2 x 106 conidia/ml and kept at 37°C. To study the effects of culture medium, the same settings were applied except that the fungus was cultivated on Malt (1.5% w/v malt extract, 0.05% w/v chloramphenicol) or YPD agar and then conidia were resuspended in Malt or YPD liquid media which were incubated at 37°C. Finally, for the incubation temperature, conidia taken from cultures on YPD agar at 37°C were resuspended in YPD liquid medium which was incubated at 20°C, 25°C, or 37°C. In all cases, germination in liquid media was monitored over 8 h and five pictures were taken every 2 h, the presence of mycelia was also checked after 16 h. The percentage of germination was determined after counting at least 100 cells from each picture.
Scanning and transmission electron microscopy
Resting conidia or germ tubes cultured in YPD medium for 6, 8, 10 or 24 hours were washed twice in Milli-Q water and once in 0.1 M cacodylate buffer, and then incubated in the fixative solution (2.5% (w/v) glutaraldehyde, 2% (w/v) paraformaldehyde, 0.1 M cacodylate buffer) for 24 h at room temperature under vacuum. After washing with cacodylate buffer, samples were incubated for 24 h in 2% KMnO4 in cacodylate buffer at 4°C, washed and post-fixed for 2 h at room temperature in 2% osmium tetroxide. Then samples were washed in Milli-Q water and finally dehydrated through a series of ethanol-water solutions (50, 70, 95% ethanol, 2 x 30 min each) and then 100% ethanol (3 x 20 min).
For scanning electron microscopy (SEM), samples underwent two baths of graded ethanol-hexamethyldisilazane (HDMS) solutions (50/50, then 25/70 proportions, 45 min each) followed by immersion in pure HMDS baths (3 x 45 min). Processed samples were mounted on aluminium stubs, coated with carbon, and stored in a desiccator until studied. Observations were made on a JSM 6301F scanning electron microscope (Jeol, Paris, France) operating at 3 kV and equipped with digital imaging.
For transmission electron microscopy (TEM), ethanol was replaced by propylene oxide (3 x 20 min) and samples were impregnated overnight in a propylene oxide-Epon mixture (1:1 v/v) and then in pure Epon for 16 h and 8 h. After polymerization (24 h at 37°C, 24 h at 45°C and then 48 h at 60°C), thin sections were directly examined on a JEM-1400 transmission electron microscope (Jeol, Paris, France; 120 kV) except for life cycle studies of S. boydii where thin sections were contrasted with uranyl acetate and lead citrate prior examination.
Ferritin labeling
Cationized ferritin is a positively charged ligand that allows visualization of anionic sites at the cell surface under physiological pH and ionic strength [33]. Scedosporium boydii germ tubes were examined by TEM after labeling with cationized ferritin, and controls consisted in incubation of fungal elements with native ferritin (lacking a positive charge) and in pretreatment of germ tubes with neuraminidase (type X) in order to remove sialic acids (all products purchased from Sigma-Aldrich, St Quentin Fallaviers, France). To do this, S. boydii germ tubes were washed 3 times with Milli-Q water, centrifuged and then incubated with cationized or native ferritin (1 mg/ml in phosphate buffered saline 150 mM) for 1 h at room temperature with agitation. To remove sialic acids, cells were first incubated with neuraminidase (1 U/ml in 0.1 M acetate buffer pH 5, supplemented with 40 mM CaCl2) for 30 min at room temperature with shaking, washed twice and then incubated with cationized ferritin as described above. Cells in the three conditions were finally washed twice in Milli-Q water and treated as described earlier for transmission electron microscopy.
Cell surface charge and hydrophobicity measurement
Cell surface charge and hydrophobicity were evaluated by microelectrophoresis and two-phase partitioning as described previously [29]. For evaluation of the cell surface charge, resting or germinating conidia (106 cells) were washed and resuspended in Milli-Q water containing 1 mM NaCl, then their electrophoretic mobility was measured at 25°C using Zetasizer Nano ZS (Malvern Instruments Ltd, Malvern, UK). The cell surface hydrophobicity (CSH) was determined using the water/hexadecane system. Briefly, germ tube and conidial suspensions in PBS were topped (except for control samples) with hexadecane, vortexed and then allowed to stand at room temperature for 3 min; then 1 ml of the aqueous phase (bottom) was transferred into a new tube and vortexed and finally the aqueous phase was transferred to a microplate and read at an optical density (OD) of 405 nm. The percentage difference in optical density readings between test samples and controls was considered as the hydrophobic index. All experiments were performed in triplicate.
Lectin labeling
Germ tubes were washed with Tris buffer (0.5 mM Tris, 100 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, pH 7.0) and then incubated for 30 min at 37°C under continuous rotatory mixing with gold-conjugated concanavalin A (Con A-gold 5 nm from Biovalley, Marne la Vallée, France; 1:50 dilution in Tris buffer) or with FITC-conjugated Con A, peanut agglutinin (PNA) or wheat germ agglutinin (WGA) at a final concentration of 100 μg/ml (all fluorescent lectins from Sigma-Aldrich) [29]. Control samples consisted in incubation of fungal elements together with the lectins and a large excess (0.2 M) of the lectin-specific carbohydrates (α-methyl D mannopyranoside for Con A, N-acetyl glucosamine for WGA, and galactose for PNA) added immediately before lectins. Finally, samples were washed three times in Tris buffer, and observed under fluorescence microscope (Leica DMR, Leipzig, Germany) for FITC-conjugated lectins or processed as described earlier for TEM without contrasting with uranyl acetate and lead citrate.
Chemical force spectroscopy (CFS) measurements
The surface of S. boydii resting or germinated conidia was imaged using a NanoWizard atomic force microscope (JPK Instruments AG, Berlin, Germany) operating in intermittent contact mode under ambient conditions. A standard rectangular cantilever (Nanosensors NCL-W) was used for imaging, with a free resonance frequency of 165 kHz and a typical spring constant of about 40 N/m. The radius curvature of the tip was ~10 nm. The detailed analysis of chemical force spectroscopy images was performed using JPK Data Processing software (JPK Instruments AG). Hydrophilic and hydrophobic adhesions were obtained in ultrapure water from force-distance curves measured on the surface of both conidia and germ tubes using functionalized cantilevers. Gold-coated cantilevers (Olympus, Hambourg, Germany) with spring constants of 0.01 N/m were immersed either in 1 mM solutions of 1-dodecanethiol or in 11-mercapto-1-undecanol (Sigma-Aldrich) in ethanol for 14 h and then rinsed with ethanol prior their use. From force-curve measurements (2048 measurements), the mean hydrophilic and hydrophobic adhesions were extracted from gaussian fits performed on the histograms. Before probing the conidial or germ tube surface, the cantilevers functionality was tested by measuring their adhesion to hydrophobic or hydrophilic flat surfaces.
Protein extraction, identification and analysis
Protein extraction
Extraction was performed according to Damveld et al. [34] with modifications. Frozen conidia or germ tubes were ground with a mortar and pestle in liquid nitrogen, then crushed in a cell homogenizer (Braun Melsungen model MSK, Melsungen, Germany) for 1.5 min for conidia or 1 min for germ tubes (150 mg dry material) under a current of CO2 cooling in the presence of a protease inhibitor cocktail (15 ml; 1X in Milli-Q water, cOmplete, EDTA-free, Roche, Meylan, France) and a mix of 1 mm and 0.25 mm diameter glass beads. Glass beads were removed by filtration through 41-μm-pore size sterile nylon filters and cell breakage was confirmed by phase-contrast microscopy (> 95%). Suspensions were centrifuged at 13500 X g for 10 min at 4°C. After lyophilization, cell debris (500–900 mg) were washed 5 times with 50 mM Tris-HCl buffer pH 7.8 (25 μl per mg dry weight) and pelleted at 18000 X g for 10 min at 4°C. Cytosolic contaminants, membrane proteins and disulfide-linked cell wall proteins were removed by boiling five times (2 min each) with SDS-extraction buffer (50 mM Tris-HCl pH 7.8, 2% w/v SDS, 0.1 M Na-EDTA, and 1.6 μl β-mercaptoethanol; 25 μl per mg dry weight). Then cell wall debris were washed six times with Milli-Q water, lyophilized and weighed. To extract cell wall GPI-anchored proteins, freeze-dried cell wall debris were incubated with hydrofluoric acid (HF)-pyridine (10 μl per mg dry weight) for 3 h at 0°C [35]. Then the suspension was centrifuged at 18000 X g for 10 min at 4°C and proteins were precipitated from the supernatant by the addition of 9 volumes of 100% methanol-Tris buffer (100% v/v methanol, 50 mM Tris-HCl pH 7.8) followed by incubation for 2 h at 0°C. After centrifugation, the pellet was washed twice with 90% methanol-Tris buffer (90% v/v methanol, 50 mM Tris-HCl pH 7.8) and lyophilized. The resulting extracts represented 10.0% and 10.2% of the initial dry weight of cell wall debris after removal of cytosolic contaminants in conidia and germ tubes, respectively. Finally, proteins were deglycosylated with peptide-N-glycosidase F (PNGase F glycerol free, New England Biolabs, Evry, France) according to the manufacturer’s recommendation. Deglycosylation was performed in glass vials by the addition of 5000 units PNGase F to 50–100 mg protein extract, followed by incubation for 3 h at 37°C. Proteins were precipitated and washed as described earlier using the methanol-Tris buffers and finally lyophilized.
Trypsin digestion
Lyophilized samples were suspended in 50 mM ammonium bicarbonate and incubated with 7.2 mM dithiothreitol (DTT) during 15 min at 37°C. They were next incubated with 13.5 mM iodoacetamide during 15 min at room temperature in the dark. Samples were then digested overnight with 4 ng/μl of sequencing grade modified trypsin (Promega, Madison, WI, USA) at 37°C.
Nano liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS)
Tryptic peptides were separated on a nanoflow high-performance liquid chromatography (nano-HPLC) system (Dionex, Villebon sur Yvette, France; LC Packings Ultimate 3000) connected to a hybrid LTQ-OrbiTrap XL (Thermo Fisher Scientific, Villebon sur Yvette, France) equipped with a nanoelectrospray ion source (New Objective, Wil, Switzerland). They were first concentrated onto a trapping precolumn (5 mm x 300 μm i.d., 300 Å pore size, Pepmap C18, 5 μm) and then separated and eluted by reverse-phase using an analytical column (15 cm x 300 μm i.d., 300 Å pore size, Pepmap C18, 5 μm; Dionex, LC Packings) with a 140 min, 2–90% acetonitrile gradient in 0.05% formic acid at a flow rate of 0.25 μl/min. The mass spectrometer was operated in its data-dependent mode by automatically switching between full survey scan MS and consecutive MS/MS acquisition. Survey full scan MS spectra (mass range 400–2000) were acquired in the OrbiTrap section of the instrument with a resolution of R = 60000 at m/z 400. The ten most intense peptide ions in each survey scan with an intensity above 2000 counts and a charge state = 2 were sequentially isolated and fragmented in the linear ion trap by collision induced dissociation (CID). For OrbiTrap measurements, an external calibration was used before each injection series ensuring an overall error mass accuracy below 5 ppm for the detected peptides. MS data were saved in RAW file format (Thermo Fisher Scientific) using XCalibur 2.0.7 with tune 2.5.5 SP1.
Protein identification
The Proteome Discoverer 1.2 software was used to submit MS/MS data to the translated genome of S. apiospermum IHEM 14462 [36] completed with Sus scrofa trypsin and Elizabethkingia miricola PNGase F (10829 sequences) using the Mascot search engine (Mascot server v2.2; http://www.matrixscience.com). Parameters were set as follows: trypsin as enzyme with one allowed miscleavage, carbamidomethylation of cysteins as fixed modification and methionine oxidation as variable modifications. Mass tolerance for MS and MS/MS was set at 10 ppm and 0.5 Dalton, respectively. Identified rank 1 peptides were filtered based on the Mascot score to obtain a false discovery rate of 1%.
Protein analysis
Identified protein sequences were analyzed for the presence of GPI anchor using Big PI fungal predictor (http://mendel.imp.ac.at/gpi/fungi_server.html), signal peptide using Signal P (http://www.cbs.dtu.dk/services/SignalP/), transmembrane helices using TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/), N- and O-glycosylation sites using NetNGlyc and NetOGlyc services (http://www.cbs.dtu.dk/services/), pI using ProtParam (http://web.expasy.org/protparam/) and grand average of hydropathy values (GRAVY) to evaluate the hydrophilic or hydrophobic character of a protein along its amino acid sequence (http://www.bioinformatics.org/sms2/protein_gravy.html). For pipeline filtering of proteins the new web tool proFasta was also used (http://www.bioinformatics.nl/tools/profasta/). Functional domain analysis was performed using Interpro (http://www.ebi.ac.uk/interpro/). Protein sequence similarities were searched by using Blastp in NCBI website (http://blast.ncbi.nlm.nih.gov) against all the non-redundant protein sequences and then against species-specific databases Aspergillus fumigatus, Neurospora crassa, Magnaporthe oryzae and Saccharomyces cerevisiae. Specific websites were also used to perform Blastp analysis against Candida albicans (http://www.candidagenome.org), Colletotrichum graminicola and Colletotrichum higginsianum genomes (http://www.broadinstitute.org/annotation/genome/colletotrichum_group/Blast.html).
Statistical analysis
For studies of the kinetics of germination, two-way analysis of variance (ANOVA) was used with the Bonferroni post-hoc test. For cell surface charge and hydrophobicity studies, results were analyzed using the Student t-test. P-values less than 0.05 were considered significant.
Results
Germination of S. boydii
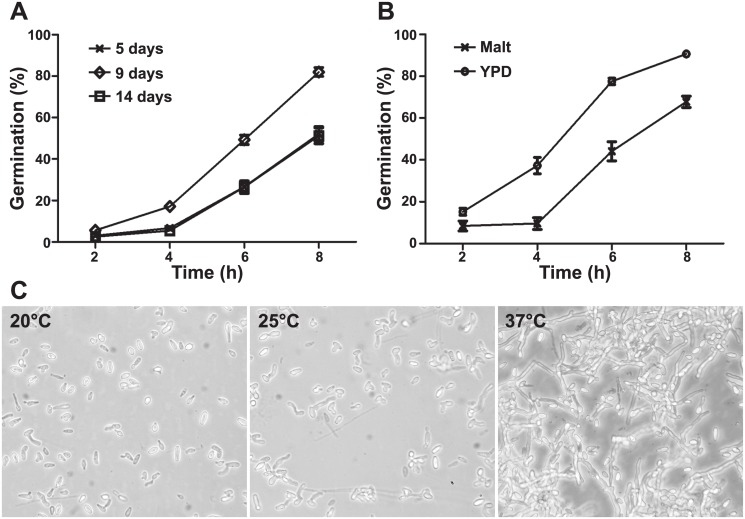
In order to define the best conditions for germination of S. boydii, three different parameters were first investigated: the age of culture, the culture medium and incubation temperature. Tracking germination during 8 h showed significant differences (P < 0.01 at 4 h and P < 0.001 at 6 and 8 h) in the percentages of germination between conidia recovered from 5-, 9-, and 14-day old cultures (Fig 1A). Germination started after 4 h of incubation and conidia recovered from 9-day-old cultures showed highest rates of germination as well as more homogeneity in terms of length of germ tubes. Prolonging the duration of cultivation to 14 days resulted in a decrease in the germination rate possibly because conidia started entering a state of long dormancy or because they were dying. After 16 h of incubation all cultures showed mycelial agglomerations and the percentage of germination could no more be determined.
Fig 1. Kinetics of germination of S. boydii in various conditions.
(A) Conidia isolated from 5-, 9- and 14-day-old cultures on yeast peptone dextrose (YPD) agar were incubated in YPD liquid medium over 8 h at 37°C. (B) Conidia isolated from 9-day-old cultures on Malt or YPD agar were incubated in Malt or YPD liquid media over 8 h at 37°C. (C) Conidia isolated from 9-day-old cultures on YPD agar were incubated in YPD liquid medium for 16 h at 20°C, 25°C or 37°C (200X).
Germination was also affected by the culture medium. Higher germination rates were observed for conidia recovered from 9-day-old cultures and incubated in YPD liquid medium compared to malt liquid medium (Fig 1B; P < 0.001). Finally, incubation temperature greatly affected the kinetics of germination since the number and length of germ tubes progressively increased with the increase in incubation temperature from 20°C to 25°C and 37°C, with the presence of long branched intermixed hyphae after 16 h of incubation at 37°C (Fig 1C). Therefore germination was performed by incubation of conidia from 9-day old cultures in YPD liquid medium at 37°C for all subsequent experiments.
The process of germination in S. boydii starts by the protrusion of a germ tube from the mother cell without significant differences in the conidial size before and after initiation of the germination process. As illustrated in Fig 2A, resting conidia measured 5.31 ± 0.92 x 2.74 ± 0.57 μm (length x width; 7 conidia studied), a size which remains essentially the same after germination (Fig 2B; 4.79 ± 0.36 x 2.13 ± 0.34 μm; length x width; 7 conidia measured). Most germination events occurred laterally rather than along the axis of the mother cell: Among 111 germinated cells only 29 cells (26.1%) germinated along the axis whereas the rest of conidia (73.9%) germinated laterally as shown in Fig 2B and 2C. With the progression of germination, branching occurred and the first branching site was seen very close to the mother cell (Fig 2D and 2E, arrow). Finally, more branching appeared along the filaments, at the subapical region of the articles (Fig 2E and 2F, arrowheads), and filaments elongated until the mother cell was no more distinguished.
Fig 2. The life cycle of S. boydii under scanning electron microscopy.
After release, conidia (A) germinate (B) and the hyphal part of germ tubes elongates (C) until a first branch emerges near the mother cell (D). Both hyphae grow and more branching sites appear on filaments at the subapical region of the articles (E) until the mother cell can no more be distinguished (F). Arrows indicate sites of first branching, and later branching are indicated by arrowheads. All cultures were performed in YPD broth with incubation at 37°C for 6h (B), 8h (C), 10 h (D and E) or 24 h (F). Bars: 1 μm in A, B and C; 0.5 μm in D; and 5 μm in E and F.
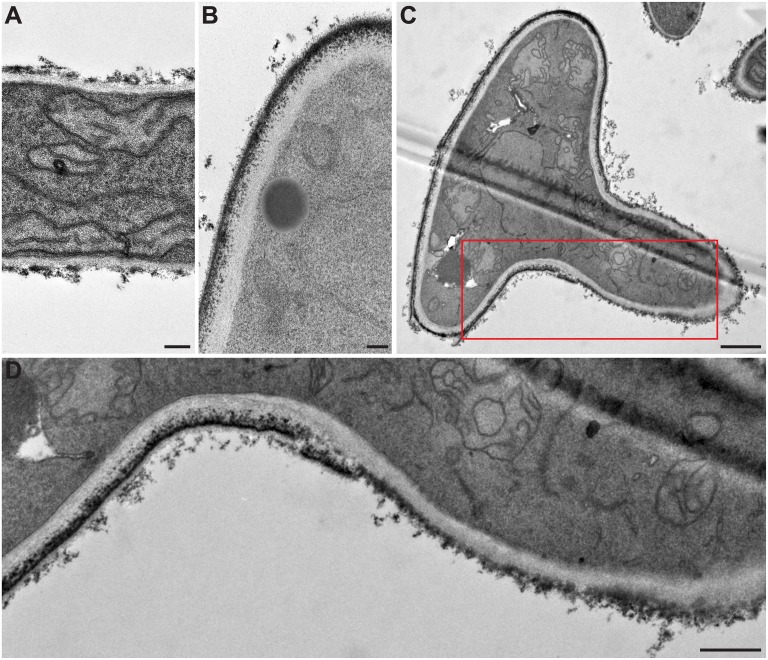
TEM examination of the different morphological stages of the fungus showed important ultrastructural changes in the cell wall with germination (Fig 3). The fungal cell wall appeared to be composed of two layers, a large electron transparent inner layer and a thinner electron dense outer layer. Comparing the cell wall of hyphae (Fig 3A) to that of conidia (Fig 3B) showed a homogeneous thickness of the outer cell wall layer in resting conidia and mother cells of germ tubes, whereas its thickness greatly varied in hyphae. The outer cell wall layer of the filament was thin or even absent in some restricted areas. Of note, the cell wall surface of the mother cell seemed to be covered by a thin electron dense layer which was less continuous at the surface of the hyphal part of germ tubes, possibly in relation with the necessary plasticity of the cell wall for hyphal elongation (Fig 3C and 3D).
Fig 3. Cell wall modifications during germination of S. boydii under transmission electron microscopy.
(A) hyphal cell wall; (B) conidial cell wall; and (C) cell wall of a germinating conidium. (D) Enlarged part of (C) highlighting the cell wall structural modifications during germination, particularly the electron dense outer layer being less continuous at the surface of the hyphal part of germ tubes. Bars: 0.2 μm in A and B; 1 μm in C; and 0.5 μm in D.
Changes in cell surface physical properties during germination
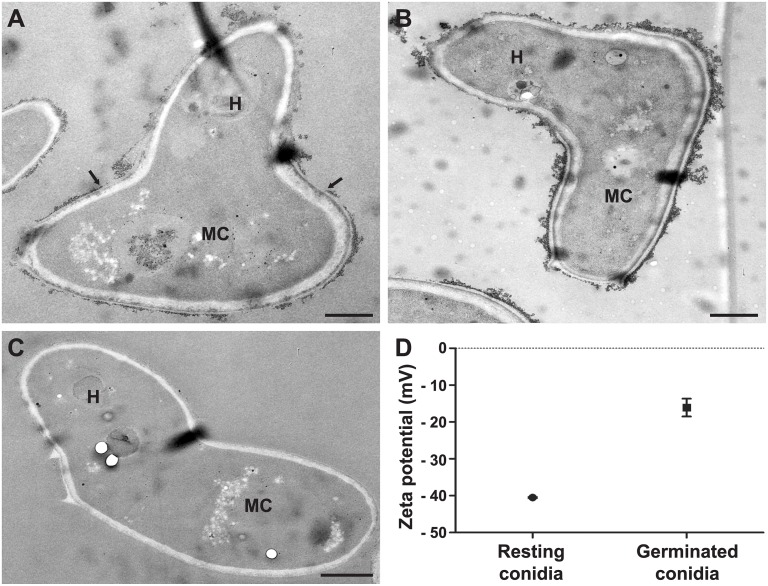
The differences in the biochemical composition of the cell wall between conidia and hyphae reflected by the ultrastructural changes were also revealed by modifications of the surface physical properties as can be seen in Fig 4A. TEM after cationized ferritin labeling showed a greater density of negatively charged areas at the surface of mother cells of germ tubes compared to their hyphal part. Neuraminidase treatment before incubation with cationized ferritin did not reduce the binding of cationized ferritin, suggesting that the surface electronegative charge was not connected to sialic acids (Fig 4B). The use of native ferritin confirmed that binding of cationized ferritin was related to its electrostatic charge and not to the ferritin molecule itself (Fig 4C). Zeta potential measurements also provided evidence for the difference in the surface electronegative charge between resting and germinated conidia as seen in Fig 4D (- 40.50 mV vs.—16.10 mV; P = 0.0005).
Fig 4. Surface charge modifications during germination of S. boydii by ferritin labeling and zeta potential measurements.
TEM images of germ tubes labeled with cationized ferritin (A), germ tubes treated with neuraminidase prior cationized ferritin labeling (B) or germ tubes incubated with native ferritin (C). (D) Comparison of the surface electrostatic charge of resting and germinated conidia calculated from the electrophoretic mobility of 10 000 cells using Zetasizer Nano ZS (P = 0.0005). H: hyphal part of germ tube; MC: mother cell of germ tube. Bars: 1 μm.
Similarly, a significant difference in the cellular hydrophobicity between resting and germinated conidia was revealed by two-phase partitioning using the water/hexadecane system. The mean percentage of resting conidia excluded from the aqueous medium, which reflects the number of hydrophobic cells within the whole population, was 37.8 ± 0.7 compared to 2.9 ± 1.3 for germinated conidia (P < 0.0001).
Changes in the cell surface composition during germination
Lectin binding to cell wall carbohydates
The accessibility of cell wall carbohydrates to mannose-binding, chitin-binding [37] and galactose-binding [38] lectins (Con A, WGA and PNA, respectively) was investigated during germination of S. boydii. While most of the mother cells were not labeled with Con A, as previously shown for resting conidia [29], the hyphal part of almost all germ tubes was intensely labeled (Figs 5A, 5C and 6), suggesting unmasking of the mannose-containing glycoconjugates by the loss of the surface electron dense film seen on the mother cells. A similar binding pattern was also seen for WGA (Fig 5B and 5D), whereas no fluorescence was observed after incubation with PNA either on the mother cell or the hyphal part of germ tubes.
Fig 5. Fluorescence labeling of S. boydii surface carbohydrates with FITC-conjugated lectins.
Germ tubes after labeling with concanavalin A (A and C) or wheat germ agglutinin (B and D) lectins. The same fields are presented under fluorescence (A and B) and phase contrast microscopy (C and D) respectively. Arrows indicate mother cells.
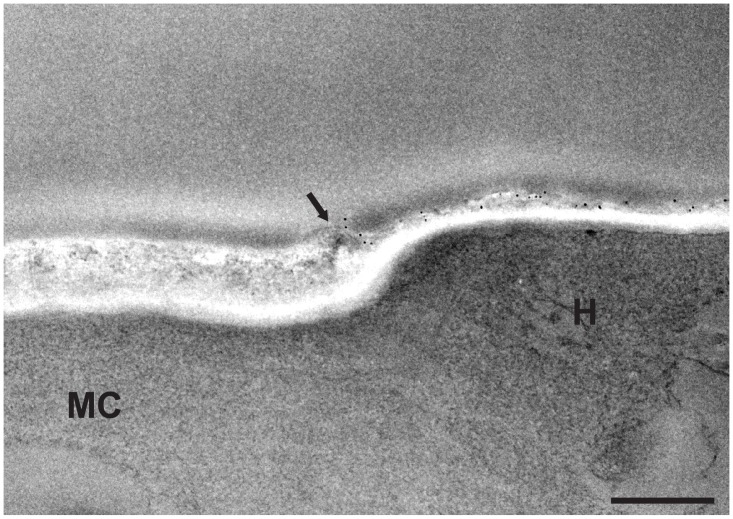
Fig 6. Gold labeling of cell wall mannan groups in S. boydii germ tubes.
Germ tubes labeled with gold-conjugated concanavalin A (Con A; 5-nm gold particles) showing higher affinity of gold particles to the hyphal part (H) of germ tubes compared to the mother cell (MC) under transmission electron microscopy. Arrow indicates the limit of the outer cell wall layer of the mother cell. Bar: 0.5 μm.
Detection of hydrophobic/hydrophilic adhesions at high spatial resolution in conidia and hyphae
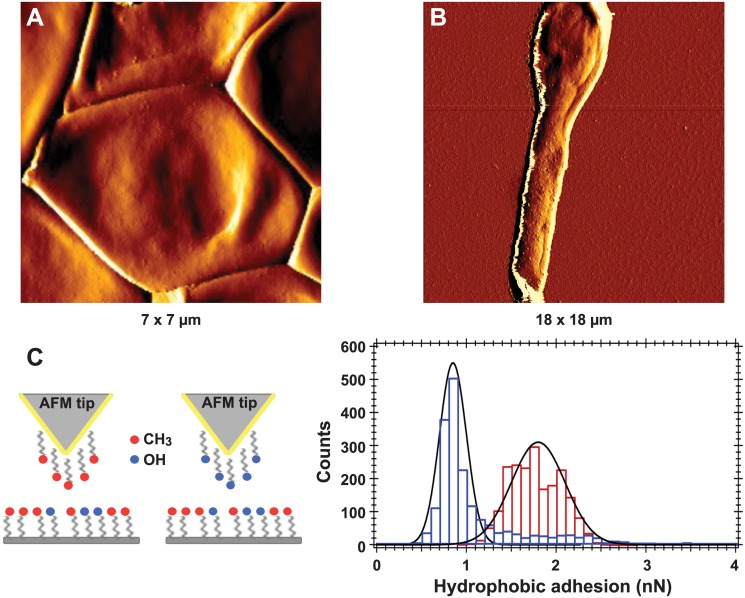
AFM images presented on Fig 7A and 7B revealed a smooth conidial cell wall surface, devoid of any peculiar organization, conversely to that of A. fumigatus conidia which is totally covered by rodlets [39]. To investigate the chemical nature of S. boydii cell surface, non-specific force-curves were measured with OH- or CH3-modified tips (Fig 7C) to determine the distribution of hydrophobic or hydrophilic components at the surface of individual resting conidia and its evolution with germination.
Fig 7. High resolution imaging and chemical force spectroscopy analysis of S. boydii resting conidia and germ tubes.
AFM amplitude images of a resting (A) or germinated (B) S. boydii conidium. (C) Left, scheme for chemical functionalization of AFM tips. Gold-coated tips were modified with CH3-terminated alkanethiols or OH-terminated alkanethiols. (C) Right, histograms of hydrophobic adhesion forces measured on the surface of a resting conidium (1.8 ± 0.3 nN, in red) and the hyphal part of a germinated conidium (0.85 ± 0.15 nN, in blue).
The use of CH3-modified probes (to measure hydrophobic adhesion forces) revealed the presence of hydrophobic components, such as glycoproteins. Force-curves recorded on the surface of resting conidia with CH3 tips showed large adhesion forces of 1.8 ± 0.3 nN, whereas a lower value of 0.85 ± 0.15 nN was obtained on the hyphal surface (Fig 7C), which was in agreement with the diminished cell surface hydrophobicity of germ tubes compared to resting conidia measured by two-phase partitioning. The values for CH3/CH3 interactions (hydrophobic adhesions) measured on the trunk and the apex of the germ tube were similar (0.8 to 0.9 nN) suggesting that the same hydrophobic components (glycoprotein) covered the whole hyphal surface.
The use of OH-modified probes revealed hydrophilic adhesion forces mainly due to the presence of polysaccharides. On the contrary to results of CH3/CH3 interaction measurements, no significant difference in OH/OH interactions (hydrophilic adhesions) was observed between resting (1.2 ± 0.4 nN) and germinated conidia (1.0 ± 0.2 nN). Also, the same OH/OH adhesion values (1.0 nN) were calculated for the trunk and apex of the hyphal part of germ tubes.
Characteristics of the identified cell wall proteins
GPI-anchored proteins selectively released with HF-pyridine from SDS buffer washed cell wall fragments, were first analysed on SDS-PAGE. Nevertheless, extracted proteins were revealed only by silver staining. Due to the small amount of proteins, analysis of glycosylation was not performed and attention was focused on the identification of the extracted proteins by nanoLC-MS/MS. Consequently deglycosylated HF-extracts were analysed with nanoLC-MS/MS which detected two hundred and fifty four proteins from both the conidial and germ tube extracts taken together. In silico analysis of these proteins identified 32 proteins with a signal peptide among which 20 proteins had a GPI-anchor following analysis with BigPI software (Table 1 and S1 Table). Seven out of the 20 GPI-anchored proteins were detected in both conidial and germ tube extracts, 12 only in the germ tube extract and one only in the conidial extract. All identified GPI-anchored proteins were predicted to be glycosylated (Table 2), and all but one (glycine-rich protein, accession number KEZ42341.1) had no transmembrane helices after excluding 45 N-terminal and 35 C-terminal amino acids. One transmembrane helix was found for the glycine-rich protein (accession number KEZ42341.1). All the twenty proteins had a pI < 5 except for the glycine-rich protein whose pI was 6.89. To analyze the serine (S)—threonine (T) content of our proteins, we first considered the overall S/T content of proteins (results indicated in Table 2). A further analysis was performed with proFasta using additional filters (S/T content > 10%, Start position of scan 26 and End position of scan -26) to exclude the S/T content of N- and C-terminal signal sequences. This analysis showed that all identified GPI-anchored proteins had an S/T content higher than 10%.
Table 1. GPI-anchored proteins identified in conidial and/or germ tube extracts.
| Protein (accession number) | Protein family | Function | Ref. | Fungal extract a | Sequence b |
|---|---|---|---|---|---|
| Glucan endo-1,3-β-D-glucosidase (KEZ41172.1) | Bglp/Bgtp (Bgt2p) GH 17 | Cell wall remodeling: branching β-(1,3) glucan through the formation of β-(1,6) linkage | [73, 74] | RC | AAQGLDGTNGAFNSAR |
| GT | ISPTGIANKEFAGANPDTLVGYIK | ||||
| EFAGANPDTLVGYIK | |||||
| AAQGLDGTNGAFNSAR | |||||
| SQSDFEAEFKAAQGLDGTNGAFNSAR | |||||
| CFEM protein (KEZ46909.1) | CFEM domain | Fungus-host interaction (plant infection, heme- uptake, biofilm structure) | [77–80] | RC | AGEFGcQSTDVAcLcR |
| SRDFVYGIR | |||||
| GT | AGEFGcQSTDVAcLcR | ||||
| SRDFVYGIR | |||||
| DFVYGIR | |||||
| CFEM protein (KEZ46627.1) | CFEM domain | Fungus-host interaction (plant infection, heme- uptake, biofilm structure) | [77–80] | RC | QGDWYcGcQPDNmSK |
| GT | QGDWYcGcQPDNMSK | ||||
| IQGAATNcVIEAcGGAAGALAVITEVQGIcEEALK | |||||
| CFEM protein (KEZ44163.1) | CFEM domain | Fungus-host interaction (plant infection, heme- uptake, biofilm structure) | [77–80] | RC | IPEcANScVTQATSGNK |
| GT | IPEcANScVTQATSGNK | ||||
| IAGcNQGDIK | |||||
| GDSL_like lipase (KEZ43142.1) c | Unknown function in fungi | RC | SQKVVLVDFR | ||
| GT | MAALLFDGINNAASR | ||||
| Glycine-rich protein (KEZ42341.1) | Unknown function in fungi | RC | GGSSSSSSSSSRPGSPGFAGSGAPR | ||
| GT | GGSSSSSSSSSRPGSPGFAGSGAPR | ||||
| Unknown (KEZ44256.1) | Unknown function | RC | NTcEALcPGAAK | ||
| GT | YYSASLYSFVcQEAFK | ||||
| NTcEALcPGAAK | |||||
| CRH1_transglycosylase (KEZ42985.1) | Crhp (Crh1p) GH16 | Cell wall remodeling: cross link chitin to β-(1,3) and β-(1,6) glucan | [68–71] | GT | GAVFSIANEK |
| LGSWVAGR | |||||
| GGKTYPQTPMQVK | |||||
| TYPQTPMQVK | |||||
| DcPADPAIGGDFTVDFTK | |||||
| GH17 family protein (SAPIO_CDS10506) | Bglp/Bgtp (Bgt2p), GH17 | Cell wall remodeling: branching β-(1,3) glucan through the formation of β-(1,6) linkage | [73, 74] | GT | DSNPDNKMQFAITK |
| NAPGKFNAVR | |||||
| AGIGADPSVLVGFIGDYR | |||||
| Unknown (KEZ45212.1) | Unknown function | GT | AcGATDYDcQcAAQQAISTcYNNcPGDSRK | ||
| AcGATDYDcQcAAQQAISTcYNNcPGDSR | |||||
| 1,3-β-glucanosyltransferase gel4 (KEZ46619.1) | Gelp/Gasp, (AfGel5p) GH72 | Cell wall remodeling: elongation of β(1–3)glucan | [30], [71–73] | GT | GIAYQQNTGAAGAGVQDAK |
| FFYENGTQFYIK | |||||
| Unknown (KEZ45428.1) | Unknown function | GT | QGLLGVVANAEDGVLYAcSQVK | ||
| CFEM protein (KEZ43031.1) | CFEM domain | Fungus-host interaction (plant infection, heme- uptake, biofilm structure) | [77–80] | GT | cVVDGITAIGcTVEDTAcAcTTENLAK |
| Unknown (KEZ44206.1) | Unknown function | GT | TPTKDELVPAGK | ||
| Glucanosyltransferase; Glyco_hydro_72 (KEZ46098.1) | Gelp/Gasp (AfGel2p) GH 72 | Cell wall remodeling: elongation of β(1–3) glucan | [30], [71–73] | GT | ILSAGVKPAPSGK |
| Unknown (KEZ43170.1) | Unknown function | GT | cDQGDGSESATLAYSNcLQK | ||
| cINScPATDVNcLAHcTPVPSPNEDNLNKLHDcAAK | |||||
| Unknown (SAPIO_CDS2081) | Unknown function | GT | GLTSMQTSIQQNcANVR | ||
| Unknown (SAPIO_CDS8694) | Unknown function | GT | SGIcGGEGVVSLYKK | ||
| Cerato platanin (SAPIO_CDS5955) | Fungus-host interaction (carbohydrate binding) | [82] | GT | VLTDAPVYNVQYGSGK | |
| Cu/Zn superoxide dismutase (KEZ44265.1) | Cu/Zn SOD | Fungus-host interaction (antioxidant, degradation of superoxide anions) | [65, 66] | RC | TLAHLDPFIR |
a RC: resting conidia; GT: germ tube.
b For more details on the identification of each protein refer to S1 Table and for the MS/MS spectra of proteins identified with a single peptide refer to S2 Fig.
c GDSL: is the consensus sequence of Glycine (G), Aspartate (D), Serine (S), and Leucine (L) amino acids around the active site serine
Table 2. Sequence analysis of the identified GPI-anchored proteins.
| Protein (accession) | Fungal extract a | S/T b (%) | pI | Omega site (ω-5 to ω+5) c | GRAVY value d | N- and O- glycosylation |
|---|---|---|---|---|---|---|
| Glucan endo-1,3-β-D-glucosidase (KEZ41172.1) | RC/GT | 22.0 | 4.41 | QQAAPS AGSSNN | -0.413 | 7 N + 68 O |
| CFEM protein (KEZ46909.1) | RC/GT | 26.2 | 4.01 | TQGPGNTGGAQQS | -0.062 | 0 N + 40 O |
| CFEM protein (KEZ46627.1) | RC/GT | 17.6 | 4.37 | SSFPTA GAGSIA | -0.242 | 2 N + 24 O |
| CFEM protein (KEZ44163.1) | RC/GT | 22.5 | 4.24 | SAPTSS GAAGVV | 0.453 | 1 N + 25 O |
| GDSL_like lipase (KEZ43142.1) | RC/GT | 19 | 4.45 | KAGDQG SGAVRV | -0.031 | 2 N +18 O |
| Glycine-rich protein (KEZ42341.1) | RC/GT | 20.1 | 6.89 | VPADESGARSV | -0.160 | 5 N + 4 O |
| Unknown (KEZ44256.1) | RC/GT | 21.6 | 4.25 | GSEDSSSGDNKEGAAASV | -0.205 | 2 N + 9 O |
| CRH1_transglycosylase (KEZ42985.1) | GT | 23.3 | 4.69 | TGSQDS GASLVQ | -0.270 | 2 N + 31 O |
| GH17 family protein (SAPIO_CDS10506) | GT | 20.3 | 4.78 | ATGADS SASGYT | -0.222 | 1 N + 32 O |
| Unknown (KEZ45212.1) | GT | 16.1 | 4.40 | SETSK G GAAELA | -0.035 | 1 N + 14 O |
| 1,3-β-glucanosyl transferase; gel4 (KEZ46619.1) | GT | 13.2 | 4.70 | NKKEDD SSAVRF | -0.275 | 6 N + 13 O |
| Unknown (KEZ45428.1) | GT | 19.8 | 4.19 | GSGDEG GAAALA | 0.086 | 3 N + 20 O |
| CFEM protein (KEZ43031.1) | GT | 23.5 | 3.90 | GDDNGNGSGTSGAVVN | 0.144 | 3 N + 30 O |
| Unknown (KEZ44206.1) | GT | 19.9 | 4.69 | SPVPTN GAARSA | 0.090 | 2 N + 27 O |
| Glucanosyl transferase Glyco-hydro 72 (KEZ46098.1) | GT | 11.6 | 4.81 | STSKED AGAFLR | -0.222 | 3 N + 7 O |
| Unknown (KEZ43170.1) | GT | 25.2 | 4.20 | ATGTGS SASATE | -0.112 | 0 N + 30 O |
| Unknown (SAPIO_CDS2081) | GT | 20.5 | 4.72 | FDIVASPSAHL | -0.243 | 0 N + 18 O |
| Unknown (SAPIO_CDS8694) | GT | 21 | 3.40 | GPVEVS AAGRNT | -1.021 | 0 N + 120 O |
| Cerato platanin (SAPIO_CDS5955) | GT | 30.2 | 4.35 | VTAAQSAGRRQ | -0.213 | 0 N + 94 O |
| Cu/Zn superoxide dismutase (KEZ44265.1) | RC | 14.1 | 4.47 | TNLPEG SAAVSS | -0.314 | 4 N + 10 O |
a RC: resting conidia; GT: germ tube.
b S: serine; T: threonine.
c Bold: best predicted ω site. Underlined: alternative ω site (second best). Italic: basic amino acids.
d GRAVY index > 0: hydrophobic; GRAVY index < 0: hydrophilic.
Analysis of S. apiospermum proteome also revealed the presence of 100 gene sequences coding for GPI-anchored proteins (S1 Fig), which meant that 20% of GPI-anchored proteins were extracted in our conditions.
Discussion
Germination and invasiveness are tightly interwound in fungal infections since the presence of hyphae in tissue sections represents a major indicator of fungal burden [40–42]. In filamentous fungi, the conidia-to-hyphae transition is accompanied by cell wall modifications to allow adaptation to new environments. As a matter of fact, in A. fumigatus, the cell wall of resting conidia is composed of three layers; during germination, the outer hydrophobin/melanin-rich cell wall layer is shed, leading to major changes in the cell wall ultrastructure and physical properties [39, 43–45]. Other major changes during germination of A. fumigatus include the emergence of β-1,3-glucans to the cell surface leading to the selective recognition of hyphae by Dectin-1 and the decrease in laminin receptors that mediate the adherence to basement membranes [42, 46]. In C. albicans, Castillo et al. [31] also showed that hyphae produced additional GPI-anchored CWPs, Als3 and Rbt1, that are not detected in the cell wall of blastospores. In another study, hyphal induction in C. albicans was shown to modulate a larger number of GPI-anchored CWPs such as the Als3, Hwp2, Hyr1, Plb5, Sod5, Rhd3, Sod4 and Ywp1 proteins [47].
In this study, we first showed that the cell wall of S. boydii, which is composed of two layers, undergoes ultrastructural modifications during germination, demonstrated by the transition from a compact electron dense outer cell wall layer in resting conidia and mother cells of germ tubes to a more diffuse and irregular outer layer in hyphae. However, unlike A. fumigatus that passes by the swelling step during germination, no major changes in the cell size were observed nor any cytoplasm vacuolization, and the outer cell wall layer of the mother cell remained attached to the electron transparent inner layer after germination. Physical properties of the cell surface were also affected by the germination process: conidia and mother cells of germ tubes were more electronegatively charged than hyphae as attested by electrophoretic mobility measurements and cationized ferritin binding. Consequently, the presence of the negatively charged sialic acids was investigated since it could affect the surface charge and was previously correlated to fungal pathogenesis [48] and adhesion of A. fumigatus conidia to the host basal lamina [49]. In S. boydii, the neuraminidase treatment did not reduce the binding of cationized ferritin to mother cells suggesting that the surface electronegative charge was not connected to sialic acids. On the contrary, the inhibition of DHN-melanin synthesis in S. boydii [29] significantly reduced the surface electronegative charge of conidia suggesting an important effect of melanin which is consistent with the reduction of the electronegative charge upon germination and production of hyaline hyphae. Glucuronic acid has also been reported in the cell wall of different fungal species. Therefore involvement of uronic acids in the electronegative charge of S. boydii cell wall should be investigated. However, to our knowledge, in all studies that have been performed on Aspergillus fumigatus [50] or Fusarium species [51–53], glucuronic acid was detected from hyphae and not from resting conidia. Therefore it is likely that uronic acids do not contribute in S. boydii to the higher electronegative charge of conidia compared to hyphae.
In a previous study we demonstrated that the amount of mannose-containing glycoconjugates in the cell wall increased with the maturation of conidia of S. boydii, but the accessibility of these molecules to Con A was hampered by the accumulation of melanin [29]. As conidia germinate, these mannose-containing glycoconjugates as well as the chitin molecules became unmasked in the hyphal part of S. boydii germ tubes, thus markedly increasing the binding of Con A and WGA.
The hydrophobic adhesion forces recorded on the conidial surface of S. boydii (1.8 ± 0.3 nN) were significantly lower than those recorded by Dague et al. [39] for A. fumigatus conidia (3 ± 0.4 nN). Scedosporium boydii conidia lacked any peculiar structures formed by rodlet-forming hydrophobins that render the aspergillar surface homogeneously hydrophobic [54]. However, the measurement of CH3/CH3 (hydrophobic) and OH/OH (hydrophilic) interactions at the surface of conidia and germ tubes showed non-zero values indicating that the whole surface of the conidial and hyphal structures are composed of a mixture of hydrophobic and hydrophilic components. After germination, CH3/CH3 interactions diminished suggesting a decrease in the content of some hydrophobic glycoproteins. The inhibition of DHN-melanin synthesis in conidia did not affect these hydrophobic adhesion forces, which meant that these interactions were not linked to melanin (data not shown). On the other hand, the OH/OH interactions remained the same before and after germination reflecting no change in some polysaccharide components.
Changes in the cell wall during germination were also illustrated by the analysis of GPI-anchored CWPs. Among the 254 proteins detected from cell wall extracts, only 20 had a GPI-anchor. The presence of non-GPI anchored proteins or atypical proteins was in agreement with some studies [31, 55] where such proteins were consistently found in cell wall extracts even after varying the methodologies of extraction. However in their studies, de Groot et al. [35, 56], did not detect atypical proteins.
In yeasts and filamentous fungi, GPI-anchored proteins may be classified into two groups according to their subcellular localization: the plasma membrane proteins (GPI-PMP) and the cell wall proteins (GPI-CWP). Cell wall proteins have abundant N- and/or O-linked glycosylation sites, a signal peptide [57], and no transmembrane helixes, which was the case for 19 out of the 20 extracted GPI-anchored proteins. Moreover, Pittet and Conzelmann [58] reported that GPI-CWPs had pIs of 4.87 ± 0.22 whereas PMPs had significantly higher pIs of 6.67 ± 0.95. However, a recent study on Pichia pastoris showed that proteins in the cell wall or in the plasma membrane did not have different pIs [59]. In our case, all the 19 GPI-anchored proteins had a pI value less than 5.
There is growing evidence that the proximal and distal sequences to the GPI-attachment site (called the ω site), found close to the C-terminus of the protein, exert a major effect on the subcellular localization of a GPI-anchored protein. The presence of a high S/T content (≥ 10%) or stretches of ST upstream the ω-proximal region has been recognized as a marking feature for CWPs [60, 61] since it can override ω-proximal signals like dibasic amino acid signals (arginine (R), histidine (H) and lysine (K)) at ω-1 and ω-2 sites that direct GPI-containing proteins to the plasma membrane [62]. Analysis of the S/T content of GPI-anchored proteins of A. fumigatus Af293 using proFasta yielded an overall S/T content ≥ 10% for most identified proteins [61]. For the 20 extracted proteins and after the exclusion of N- and C- terminal sequences, the S/T content remained higher than 10%. Moreover, no dibasic amino acids were found at the ω-1 and ω-2 sites (Table 2). Ouyang et al. [63] mentioned that a monobasic amino acid can be sufficient for retaining a GPI-anchored protein in the plasma membrane, as was the case for 3 of our extracted proteins, but again the S/T rich regions can override such signal. Add to this, the presence of valine (V), isoleucine (I) or leucine (L) at ω-4 and ω-5 as well as tyrosine (Y) or asparagine (N) at ω-2 was also suggested to act positively for the cell wall localization of proteins according to Hamada et al. [64], but these conditions did not apply to our protein sequences neither did they apply to some cell wall proteins in A. fumigatus like the AfMp1p for example [63]. The ω-proximal signals and cell wall or membrane localization of proteins remain a matter of debate especially that the same GPI-anchored protein might be present in both the cell wall and the membrane compartments [60, 63].
The different families to which these proteins belonged to were also analyzed. Among the 20 identified GPI-anchored proteins identified, one was found only in conidial extracts, whereas 12 were found only in the germ tube extracts and 7 in both extracts. The protein identified only in conidial extracts (KEZ44265.1) carried a Cu/Zn-superoxide dismutase domain. Superoxide dismutases (SOD) are antioxidant enzymes involved in the degradation of superoxide anions. The identified protein in S. boydii belonged to a particular class of extracellular SODs containing a signal peptide and predicted to have a GPI anchor. These SODs were first described in the opportunistic fungal pathogen C. albicans that expressed three GPI-anchored SOD1 related proteins: CaSod4p, CaSod5p and CaSod6p [65]. Strains lacking CaSod5p were more susceptible to killing by macrophages and neutrophils and exhibited a decreased virulence in mouse models [65]. Sod5p was suggested to be involved in the degradation of superoxide anions produced by the host cells [66]. In S. apiospermum genome, seven sequences were predicted to have a superoxide dismutase domain, three of them are Cu/Zn-SODs. Two of the Cu/Zn-SODs have a predicted signal peptide but only one has a predicted GPI-anchor. To be noted, the extracted protein in this study was different from the one previously identified by Lima et al. [67] in S. apiospermum (KEZ41328.1) that did not contain a signal peptide, a transmembrane helix, or a GPI anchor.
Twelve GPI-anchored proteins were only found in germ tube extracts and among these, four proteins belonged to known GPI-anchored protein families implicated in cell wall biosynthesis activities, one to CFEM (Common in Fungal Extracellular Membrane)-domain containing proteins, one to cerato-platanin proteins and the rest had no known functions. The first group of proteins contained a protein similar to Crhp proteins (KEZ42985.1) that are suggested to be involved in the linkage of cell wall polysaccharides β(1–6)glucan and β(1–3)glucan to chitin [68–70]. Crhp proteins are classified in the glycoside hydrolases 16 family (GH16) of the CAZy Database (http://www.cazy.org/). They were originally studied in the yeast S. cerevisiae but their biochemical function in the fungus remains unknown and single deletions of these genes in A. fumigatus were not associated to any phenotype changes [71]. Two proteins similar to proteins of the Gel/Gas family were also detected in this study in the germ tube cell wall extract (KEZ466191.1, KEZ46098.1). Proteins of this family were particularly well studied in A. fumigatus, Schizosaccharomyces pombe, S. cerevisiae and C. albicans [71]. Gel/Gas proteins belong to the glycoside hydrolase family 72 (GH72) and were shown to perform elongation of β(1–3)glucan chains. To date, only Gel1p, Gel2p and Gel4p in A. fumigatus have been studied and AfGel2p was shown to be important for cell wall morphogenesis and virulence in a mouse model of invasive aspergillosis while AfGEL4 was shown to be an essential gene for cell wall remodeling [30, 72, 73]. In S. boydii germ tube extract, a protein (SAPIO_CDS10506) similar to proteins of the Bgtp/Bglp family was also identified. These proteins belong to the glycoside hydrolase 17 family (GH17) and were studied in S. cerevisiae and A. fumigatus. AfBgt2p displays a branching activity in the cell wall by cleaving two residues of a β(1–3)glucan chain and transferring them to another chain of β(1–3)glucan with a β(1–6) linkage in vitro [74, 75]. However the single Afbgt2 mutant strain did not display a differential phenotype with respect to the wild-type strain, thus suggesting that there were other proteins with β(1–3)glucan branching activity in the cell wall [74].
A protein with a predicted CFEM domain was also detected in the hyphal extract. The CFEM domain contains around 60 amino acids, predominantly hydrophobic, and eight cysteine residues with a conserved spacing [76]. These domains are found mainly in GPI-CWPs and most CFEM-containing proteins studied to date are involved in host-pathogen interaction and virulence. In Magnaporthe grisea, Pth11p, which contains a CFEM domain, is required for plant infection [77]. Likewise, three proteins with CFEM domain were shown to be involved in cell wall stabilization in A. fumigatus, but they did not play a role in cell wall morphogenesis or as virulence factors [78]. In C. albicans proteins containing CFEM domains were suggested to play a role in heme uptake and biofilm structure [79, 80].
In agreement with a previous proteomic study on S. boydii secreted proteins [81], a cerato-platanin protein was also detected here, from the germ tube extract exclusively. Cerato-platanins are poorly studied proteins, they are thought to be carbohydrate binding virulence factors involved in fungal-plant interactions [82]. They were shown to self-assemble at hydrophobic/hydrophilic interfaces and form protein layers. Under AFM, they were shown to form branched structures or large agglomerates characterized by a disordered assembly of protruding segments [83]. In contrast to the resting conidia, examining the surface of hyphal parts of the germ tubes in S. boydii under AFM showed the presence of structures of no specific orientation assembled in an isotropic manner on the surface (results not shown). The identification of a cerato-platanin protein in our hyphal extract presents a possible explanation for such structures, but further investigations are needed to confirm such link.
We identified seven proteins present in both conidial and germ tube extracts of which three had no known function or domain and three had CFEM domains. The remaining protein was similar to AfBgt2p (KEZ41172.1) and shared 31.4% homology with its S. boydii paralogue detected in germ tube extracts mentioned earlier (SAPIO_CDS10506). Both proteins (KEZ41172.1 and SAPIO_CDS10506) contained the two conserved glutamic acid residues of the catalytic site described for proteins of GH17 family [84].
Finally, no hydrophobins could be identified in our extracts using the hydrophobin conserved eight-cystein pattern in proFasta although two genes encoding putative hydrophobins were found in the fungal genome. In A. fumigatus, Dague et al [54] showed that the presence of RodAp hydrophobin accounted for the high hydrophobic adhesions (CH3/CH3 interactions) measured on the conidial surface. RodAp is a moderately hydrophobic protein with a GRAVY value of 0.245 (A. fumigatus Af293, protein accession P41746.2) and a suggested GPI-anchor [85]. After analyzing the GRAVY values of the extracted GPI-anchored proteins, only one protein in the conidial extract presented a hydrophobic character (GRAVY > 0, Table 2). Interestingly, this protein was twice more hydrophobic than RodAp (GRAVY = 0.453) and had a CFEM domain (KEZ44163.1). CFEM domains, as previously mentioned, have a conserved eight-cysteine pattern that is distinct from that of hydrophobins; they are commonly identified in GPI-anchored cell wall proteins extracts and have predominant hydrophobic amino acid residues in their sequences (32%- 45% of the total amino acids) [76]. The identified CFEM (KEZ44163.1) was present in both conidial and germ tube extracts, but its relative amount with respect to the extracted conidial or hyphal GPI-anchored proteins (calculated after analyzing the average intensity of the strongest peptides) was twice higher in the conidial extract (2.054%) than in the germ tube extract (1.055%). Even though this remains speculative, this may account for the higher cell surface hydrophobicity and CH3/CH3 interactions observed on the conidial surface and suggests a hypothesis for future investigations on fungi exhibiting high hydrophobic adhesion forces at the conidial surface and no rodlet layer as S. boydii.
The analysis of the genome of S. apiospermum also revealed the presence of 100 gene sequences coding for GPI-anchored proteins which is similar to the number identified in other fungal genomes like A. fumigatus strains Af293 and A1163 which contain 91 and 85 sequences, respectively [61]. Twenty percent of the proteins identified in silico were extracted which is similar to results in other studies. If we take C. albicans for example, in one study [35], 12 GPI-anchored proteins were extracted out of 104 predicted GPI-anchored proteins (11.5%) while in another study [31], 19 GPI-anchored proteins were extracted after using multiple techniques. In S. pombe and S. cerevisiae, four and twelve GPI-anchored proteins could be extracted, respectively [86, 87]. The low number of identified GPI-anchored proteins may be related to some technical limitations due to the low abundance of certain proteins, the difficulty of ionizing some peptides, the high O-glycosylation that renders trypsin digestion and peptide identification more difficult or to the need of some particular culture conditions for the expression of these proteins as suggested by the comparison of the results from conidial and hyphal extracts [31].
All these results demonstrate that the cell wall in S. boydii is a highly dynamic structure whose components, when taken together, give the physical, chemical and molecular fungal cell wall imprint. Today, mapping interactions at the surface of microbial cells at high spatial resolution and correlating such information to molecular data is highly valuable to our understanding of the pathogenesis of fungi.
Supporting Information
(PDF)
Accession numbers of analyzed sequences are available on GenBank database (Scedosporium apiospermum genome accession number JOWA00000000).
(PDF)
The major fragmentation series (y-carboxy and b-amino) are annotated; the deduced sequence, m/z value and charge state from the precursor ion are indicated.
(PPTX)
Acknowledgments
The authors would like to thank Dr. Emmanuelle Com and the Proteomics Core Facility Biogenouest, IRSET-Inserm U1085 (Rennes, France) for the assistance to identify proteins in our extracts and the thoughtful discussions for the protein analysis. The authors also acknowledge Dr. Patrick Saulnier (L'UNAM Université, Université d'Angers, INSERM U646, France) for the use of the Zetasizer Nano ZS.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Région Pays de la Loire as part of the Myco-AFM research program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Guarro J, Kantarcioglu AS, Horré R, Luis Rodriguez-Tudela J, Cuenca Estrella M, Berenguer J, et al. 2006. Scedosporium apiospermum: changing clinical spectrum of a therapy-refractory opportunist. Med Mycol 44:295–327. [DOI] [PubMed] [Google Scholar]
- 2. Kaltseis J, Rainer J, De Hoog GS. 2009. Ecology of Pseudallescheria and Scedosporium species in human-dominated and natural environments and their distribution in clinical samples. Med Mycol 47:398–405. 10.1080/13693780802585317 [DOI] [PubMed] [Google Scholar]
- 3. Harun A, Gilgado F, Chen SC, Meyer W. 2010. Abundance of Pseudallescheria/Scedosporium species in the Australian urban environment suggests a possible source for scedosporiosis including the colonization of airways in cystic fibrosis. Med Mycol 48 (Suppl 1):S70–S76. 10.3109/13693786.2010.515254 [DOI] [PubMed] [Google Scholar]
- 4. Rougeron A, Schuliar G, Leto J, Sitterlé E, Landry D, Bougnoux M-E, et al. 2015. Human-impacted areas of France are environmental reservoirs of the Pseudallescheria boydii/Scedosporium apiospermum species complex. Environ Microbiol 17:1039–1048. 10.1111/1462-2920.12472 [DOI] [PubMed] [Google Scholar]
- 5. Beguin H, Nolard N. 1994. Mould biodiversity in homes I. Air and surface analysis of 130 dwellings. Aerobiologia 10: 157–166. [Google Scholar]
- 6. Lackner M, de Hoog SG, Yang L, Ferreira Moreno L, Ahmed SA, Andreas F, et al. Proposed nomenclature for Pseudallescheria, Scedosporium and related genera. Fungal Diversity, 25 July 2014. 25722662 [Google Scholar]
- 7. Gilgado F, Cano J, Gené J, Guarro J. 2005. Molecular phylogeny of the Pseudallescheria boydii species complex: proposal of two new species. J Clin Microbiol 43:4930–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gilgado F, Cano J, Gené J, Sutton DA, Guarro J. 2008. Molecular and phenotypic data supporting distinct species statuses for Scedosporium apiospermum and Pseudallescheria boydii and the proposed new species Scedosporium dehoogii . J Clin Microbiol 46:766–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gilgado F, Gené J, Cano J, Guarro J. 2010. Heterothallism in Scedosporium apiospermum and description of its teleomorph Pseudallescheria apiosperma sp. nov. Med Mycol 48:122–128. 10.3109/13693780902939695 [DOI] [PubMed] [Google Scholar]
- 10. Cortez KJ, Roilides E, Quiroz-Telles F, Meletiadis J, Antachopoulos C, Knudsen T, et al. 2008. Infections caused by Scedosporium spp. Clin Microbiol Rev 21:157–197. 10.1128/CMR.00039-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cimon B, Carrère J, Vinatier JF, Chazalette JP, Chabasse D, Bouchara JP. 2000. Clinical significance of Scedosporium apiospermum in patients with cystic fibrosis. Eur J Clin Microbiol Infect Dis 19: 53–56. [DOI] [PubMed] [Google Scholar]
- 12. Pihet M, Carrère J, Cimon B, Chabasse D, Delhaes L, Symoens F, et al. 2009. Occurrence and relevance of filamentous fungi in respiratory secretions of patients with cystic fibrosis—a review. Med Mycol 47:387–397. 10.1080/13693780802609604 [DOI] [PubMed] [Google Scholar]
- 13. Müller FM, Seidler M. 2010. Characteristics of pathogenic fungi and antifungal therapy in cystic fibrosis. Expert Rev Anti Infect Ther 8:957–964. 10.1586/eri.10.72 [DOI] [PubMed] [Google Scholar]
- 14. Middleton PG, Chen SC, Meyer W. 2013. Fungal infections and treatment in cystic fibrosis. Curr Opin Pulm Med 19:670–675. 10.1097/MCP.0b013e328365ab74 [DOI] [PubMed] [Google Scholar]
- 15. Amin R, Dupuis A, Aaron SD, Ratjen F. 2010. The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in patients with cystic fibrosis. Chest 137:171–176. 10.1378/chest.09-1103 [DOI] [PubMed] [Google Scholar]
- 16. Chotirmall SH, O'Donoghue E, Bennett K, Gunaratnam C, O'Neill SJ, McElvaney NG. 2010. Sputum Candida albicans presages FEV₁ decline and hospital-treated exacerbations in cystic fibrosis. Chest 138:1186–1195. 10.1378/chest.09-2996 [DOI] [PubMed] [Google Scholar]
- 17. Fillaux J, Brémont F, Murris M, Cassaing S, Rittié JL, Tétu L, et al. 2012. Assessment of Aspergillus sensitization or persistent carriage as a factor in lung function impairment in cystic fibrosis patients. Scand J Infect Dis 44:842–847. 10.3109/00365548.2012.695454 [DOI] [PubMed] [Google Scholar]
- 18. Symoens F, Knoop C, Schrooyen M, Denis O, Estenne M, Nolard N, et al. 2006. Disseminated Scedosporium apiospermum infection in a cystic fibrosis patient after double-lung transplantation. J Heart Lung Transplant 25:603–607. [DOI] [PubMed] [Google Scholar]
- 19. Hosseini-Moghaddam SM, Husain S. 2010. Fungi and molds following lung transplantation. Semin Respir Crit Care Med 31:222–233. 10.1055/s-0030-1249118 [DOI] [PubMed] [Google Scholar]
- 20. Alastruey-Izquierdo A, Cuenca-Estrella M, Monzón A, Rodriguez-Tudela JL. 2007. Prevalence and susceptibility testing of new species of Pseudallescheria and Scedosporium in a collection of clinical mold isolates. Antimicrob Agents Chemother 51:748–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lackner M, de Hoog GS, Verweij PE, Najafzadeh MJ, Curfs-Breuker I, Klaassen CH, et al. 2012. Species-specific antifungal susceptibility patterns of Scedosporium and Pseudallescheria species. Antimicrob Agents Chemother 56:2635–2642. 10.1128/AAC.05910-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Latgé J-P. 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol 66:279–290. [DOI] [PubMed] [Google Scholar]
- 23. Latgé J-P, Beauvais A. 2014. Functional duality of the cell wall. Curr Opin Microbiol 20:111–117. 10.1016/j.mib.2014.05.009 [DOI] [PubMed] [Google Scholar]
- 24. Sedlacek L, Graf B, Schwarz C, Albert F, Peter S, Würstl B, et al. 2015. Prevalence of Scedosporium species and Lomentospora prolificans in patients with cystic fibrosis in a multicenter trial by use of a selective medium. J Cyst Fibros 14:237–241. 10.1016/j.jcf.2014.12.014 [DOI] [PubMed] [Google Scholar]
- 25. Lopes LCL, da Silva MID, Bittencourt VCB, Figueiredo RT, Rollin-Pinheiro R, Sassaki GL, et al. 2011. Glycoconjugates and polysaccharides from the Scedosporium/Pseudallescheria boydii complex: structural characterisation, involvement in cell differentiation, cell recognition and virulence. Mycoses 54:28–36. 10.1111/j.1439-0507.2011.02141.x [DOI] [PubMed] [Google Scholar]
- 26. Bittencourt VCB, Figueiredo RT, Silva RB da, Mourão-Sá DS, Fernandez PL, Sassaki GL, et al. 2006. An α-glucan of Pseudallescheria boydii is involved in fungal phagocytosis and toll-like receptor activation. J. Biol. Chem. 281:22614–22623. [DOI] [PubMed] [Google Scholar]
- 27. Pinto MR, Rodrigues ML, Travassos LR, Haido RMT, Wait R, Barreto-Bergter E. 2002. Characterization of glucosylceramides in Pseudallescheria boydii and their involvement in fungal differentiation. Glycobiology 12:251–260. [DOI] [PubMed] [Google Scholar]
- 28. Rollin-Pinheiro R, Liporagi-Lopes LC, de Meirelles JV, Souza LM de, Barreto-Bergter E. 2014. Characterization of Scedosporium apiospermum glucosylceramides and their involvement in fungal development and macrophage functions. PLoS ONE 9:e98149 10.1371/journal.pone.0098149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ghamrawi S, Renier G, Saulnier P, Cuenot S, Zykwinska A, Dutilh BE, et al. 2014. Cell wall modifications during conidial maturation of the human pathogenic fungus Pseudallescheria boydii . PLoS ONE 9:e100290 10.1371/journal.pone.0100290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mouyna I, Fontaine T, Vai M, Monod M, Fonzi WA, Diaquin M, et al. 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J Biol Chem 275:14882–14889. [DOI] [PubMed] [Google Scholar]
- 31. Castillo L, Calvo E, Martínez AI, Ruiz-Herrera J, Valentín E, Lopez JA, Sentandreu R. 2008. A study of the Candida albicans cell wall proteome. Proteomics 8:3871–3881. 10.1002/pmic.200800110 [DOI] [PubMed] [Google Scholar]
- 32. Free SJ. 2013. Fungal cell wall organization and biosynthesis, p. 33–82. In Friedmann T, Dunalp JC and Goodwin SF (eds.), Advances in Genetics, Vol. 81 Academic Press; 10.1016/B978-0-12-407677-8.00002-6 [DOI] [PubMed] [Google Scholar]
- 33. Bouchara JP, Sanchez M, Esnault K, Tronchin G. 1999. Interactions between Aspergillus fumigatus and host matrix proteins. Contrib Microbiol 2:167–181. [DOI] [PubMed] [Google Scholar]
- 34. Damveld RA, Arentshorst M, VanKuyk PA, Klis FM, van den Hondel CAMJJ, Ram AFJ. 2005. Characterisation of CwpA, a putative glycosylphosphatidylinositol-anchored cell wall mannoprotein in the filamentous fungus Aspergillus niger . Fungal Genet Biol 42:873–885. [DOI] [PubMed] [Google Scholar]
- 35. De Groot PWJ, de Boer AD, Cunningham J, Dekker HL, de Jong L, Hellingwerf KJ, et al. 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot Cell 3:955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vandeputte P, Ghamrawi S, Rechenmann M, Iltis A, Giraud S, Fleury M, et al. 2014. Draft genome sequence of the pathogenic fungus Scedosporium apiospermum . Genome Announc 2:e00988–14. 10.1128/genomeA.00988-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bloch R, Burger MM. 1974. Purification of wheat germ agglutinin using affinity chromatography on chitin. Biochem Biophys Res Commun 58:13–19. [DOI] [PubMed] [Google Scholar]
- 38. Lotan R, Skutelsky E, Danon D, Sharon N. 1975. The purification, composition, and specificity of the anti-T lectin from peanut (Arachis hypogaea). J Biol Chem 250:8518–8523. [PubMed] [Google Scholar]
- 39. Dague E, Alsteens D, Latgé J-P, Dufrene YF. 2008. High-resolution cell surface dynamics of germinating Aspergillus fumigatus conidia. Biophys J 94:656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jabado N, Casanova J-L, Haddad E, Dulieu F, Fournet J-C, Dupont B, et al. 1998. Invasive pulmonary infection due to Scedosporium apiospermum in two children with chronic granulomatous disease. Clin Infect Dis 27:1437–1441. [DOI] [PubMed] [Google Scholar]
- 41. Kepez Yildiz B, Hasanreisoglu M, Aktas Z, Aksu G, Kocak BC, Akata F. 2014. Fungal keratitis secondary to Scedosporium apiospermum infection and successful treatment with surgical and medical intervention. Int Ophthalmol 34:305–308. 10.1007/s10792-013-9777-8 [DOI] [PubMed] [Google Scholar]
- 42. Sundaram C, Shantveer Gu, Umabala P, Lakshmi V. 2014. Diagnostic utility of melanin production by fungi: Study on tissue sections and culture smears with Masson-Fontana stain. Ind J Pathol Microbiol 57:217. [DOI] [PubMed] [Google Scholar]
- 43. Tronchin G, Bouchara J-P, Ferron M, Larcher G, Chabasse D. 1995. Cell surface properties of Aspergillus fumigatus conidia: correlation between adherence, agglutination, and rearrangements of the cell wall. Can J Microbiol 41:714–721. [DOI] [PubMed] [Google Scholar]
- 44. Pihet M, Vandeputte P, Tronchin G, Renier G, Saulnier P, Georgeault S, et al. 2009. Melanin is an essential component for the integrity of the cell wall of Aspergillus fumigatus conidia. BMC Microbiol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tronchin G, Esnault K, Renier G, Filmon R, Chabasse D, Bouchara JP. 1997. Expression and identification of a laminin-binding protein in Aspergillus fumigatus conidia. Infect Immun 65:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gersuk GM, Underhill DM, Zhu L, Marr KA. 2006. Dectin-1 and TLRs permit macrophages to distinguish between different Aspergillus fumigatus cellular states. J Immunol 176:3717–3724. [DOI] [PubMed] [Google Scholar]
- 47. Heilmann CJ, Sorgo AG, Siliakus AR, Dekker HL, Brul S, Koster CG de, et al. 2011. Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology 157:2297–2307. 10.1099/mic.0.049395-0 [DOI] [PubMed] [Google Scholar]
- 48. Wasylnka JA, Simmer MI, Moore MM. 2001. Differences in sialic acid density in pathogenic and non-pathogenic Aspergillus species. Microbiology 147:869–877. [DOI] [PubMed] [Google Scholar]
- 49. Wasylnka JA, Moore MM. 2000. Adhesion of Aspergillus species to extracellular matrix proteins: evidence for involvement of negatively charged carbohydrates on the conidial surface. Infect Immun 68:3377–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fontaine T, Lamarre C, Simenel C, Lambou K, Coddeville B, Delepierre M, et al. 2009. Characterization of glucuronic acid containing glycolipid in Aspergillus fumigatus mycelium. Carbohydr Res 344:1960–1967. 10.1016/j.carres.2009.07.012 [DOI] [PubMed] [Google Scholar]
- 51. Barran LR, Schneider EF, Wood PJ, Madhosingh C, Miller RW. 1975. Cell wall of Fusarium sulphureum; I. Chemical composition of the hyphal wall. Biochim Biophys Acta 392:148–158. [DOI] [PubMed] [Google Scholar]
- 52. Barbosa IP, Kemmelmeier C. 1993. Chemical composition of the hyphal wall from Fusarium graminearum . Experiment Mycol 17:274–283. [Google Scholar]
- 53. Schoffelmeer EA, Klis FM, Sietsma JH, Cornelissen BJ. 1999. The cell wall of Fusarium oxysporum . Fungal Genet Biol 27:275–282. [DOI] [PubMed] [Google Scholar]
- 54. Dague E, Alsteens D, Latgé J-P, Verbelen C, Raze D, Baulard AR, et al. 2007. Chemical force microscopy of single live cells. Nano Lett 7:3026–3030. [DOI] [PubMed] [Google Scholar]
- 55. Nombela C, Gil C, Chaffin WL. 2006. Non-conventional protein secretionin yeast. Trends Microbiol 14:15–21. [DOI] [PubMed] [Google Scholar]
- 56. De Groot PWJ, Brandt BW, Horiuchi H, Ram AFJ, de Koster CG, Klis FM. 2009. Comprehensive genomic analysis of cell wall genes in Aspergillus nidulans . Fungal Genet Biol 46 Suppl 1:S72–81. [DOI] [PubMed] [Google Scholar]
- 57. Bowman SM, Free SJ (2006) The structure and synthesis of the fungal cell wall. BioEssays 28: 799–808. [DOI] [PubMed] [Google Scholar]
- 58. Pittet M, Conzelmann A. 2007. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae . Biochim Biophys Acta 1771:405–420. [DOI] [PubMed] [Google Scholar]
- 59. Zhang L, Liang S, Zhou X, Jin Z, Jiang F, Han S, et al. 2013. Screening for glycosylphosphatidylinositol-modified cell wall proteins in Pichia pastoris and their recombinant expression on the cell surface. Appl Environ Microbiol 79:5519–5526. 10.1128/AEM.00824-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Frieman MB, Cormack BP. 2004. Multiple sequence signals determine the distribution of glycosylphosphatidylinositol proteins between the plasma membrane and cell wall in Saccharomyces cerevisiae . Microbiology 150:3105–3114. [DOI] [PubMed] [Google Scholar]
- 61. De Groot PWJ, Brandt BW. 2012. ProFASTA: A pipeline web server for fungal protein scanning with integration of cell surface prediction software. Fungal Genet Biol 49:173–179. 10.1016/j.fgb.2011.12.009 [DOI] [PubMed] [Google Scholar]
- 62. Eisenhaber B, Schneider G, Wildpaner M, Eisenhaber F. 2004. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe . J Mol Biol 337:243–253. [DOI] [PubMed] [Google Scholar]
- 63. Ouyang H, Chen X, Lu Y, Wilson IBH, Tang G, Wang A, et al. 2013. One single basic amino acid at the ω-1 or ω-2 site is a signal that retains glycosylphosphatidylinositol-anchored protein in the plasma membrane of Aspergillus fumigatus . Eukaryot Cell 12:889–899. 10.1128/EC.00351-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hamada K, Terashima H, Arisawa M, Yabuki N, Kitada K. 1999. Amino acid residues in the ω-minus region participate in cellular localization of yeast glycosylphosphatidylinositol-attached proteins. J Bacteriol 181:3886–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Frohner IE, Bourgeois C, Yatsyk K, Majer O, Kuchler K. 2009. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol Microbiol 71:240–252. 10.1111/j.1365-2958.2008.06528.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gleason JE, Galaleldeen A, Peterson RL, Taylor AB, Holloway SP, Waninger-Saroni J, et al. 2014. Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. Proc Natl Acad Sci USA 111:5866–5871. 10.1073/pnas.1400137111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lima OC, Larcher G, Vandeputte P, Lebouil A, Chabasse D, Simoneau P, et al. 2007. Molecular cloning and biochemical characterization of a Cu,Zn-superoxide dismutase from Scedosporium apiospermum . Microbes Infect 9:558–565. [DOI] [PubMed] [Google Scholar]
- 68. Cabib E, Blanco N, Grau C, Rodriguez-Pena JM, Arroyo J. 2007. Crh1p and Crh2p are required for the cross-linking of chitin to beta(1–6)glucan in the Saccharomyces cerevisiae cell wall. Mol Microbiol 63:921–935. [DOI] [PubMed] [Google Scholar]
- 69. Cabib E, Farkas V, Kosik O, Blanco N, Arroyo J, McPhie P. 2008. Assembly of the yeast cell wall. Crh1p and Crh2p act as transglycosylases in vivo and in vitro . J Biol Chem 283:29859–29872. 10.1074/jbc.M804274200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cabib E. 2009. Two novel techniques for determination of polysaccharide cross-links show that Crh1p and Crh2p attach chitin to both beta(1–6)- and beta(1–3)glucan in the Saccharomyces cerevisiae cell wall. Eukaryot Cell 8:1626–1636. 10.1128/EC.00228-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mouyna I, Hartl L, Latgé J-P. 2013. β-1,3-glucan modifying enzymes in Aspergillus fumigatus . Front Microbiol 4:81 10.3389/fmicb.2013.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mouyna I, Morelle W, Vai M, Monod M, Lechenne B, Fontaine T, et al. 2005. Deletion of GEL2 encoding for a beta(1–3)glucanosyltransferase affects morphogenesis and virulence in Aspergillus fumigatus . Mol Microbiol 56:1675–1688. [DOI] [PubMed] [Google Scholar]
- 73. Gastebois A, Fontaine T, Latgé J-P, Mouyna I. 2010. beta(1–3)Glucanosyltransferase Gel4p is essential for Aspergillus fumigatus . Eukaryot Cell 9:1294–1298. 10.1128/EC.00107-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gastebois A, Mouyna I, Simenel C, Clavaud C, Coddeville B, Delepierre M, et al. 2010. Characterization of a new beta(1–3)-glucan branching activity of Aspergillus fumigatus . J Biol Chem 285:2386–2396. 10.1074/jbc.M109.077545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mouyna I, Hartland RP, Fontaine T, Diaquin M, Simenel C, Delepierre M, et al. 1998. A 1,3-beta-glucanosyltransferase isolated from the cell wall of Aspergillus fumigatus is a homologue of the yeast Bgl2p. Microbiology 144:3171–3180. [DOI] [PubMed] [Google Scholar]
- 76. Kulkarni RD, Kelkar HS, Dean RA. 2003. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem Sci 28:118–121. [DOI] [PubMed] [Google Scholar]
- 77. DeZwaan TM, Carroll AM, Valent B, Sweigard JA. 1999. Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell 11:2013–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vaknin Y, Shadkchan Y, Levdansky E, Morozov M, Romano J, Osherov N. 2014. The three Aspergillus fumigatus CFEM-domain GPI-anchored proteins (CfmA-C) affect cell-wall stability but do not play a role in fungal virulence. Fungal Genet Biol 63:55–64. 10.1016/j.fgb.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 79. Kuznets G, Vigonsky E, Weissman Z, Lalli D, Gildor T, Kauffman SJ, et al. 2014. A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PLoS Pathog 10:e1004407 10.1371/journal.ppat.1004407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Perez A, Pedros B, Murgui A, Casanova M, Lopez-Ribot JL, Martinez JP. 2006. Biofilm formation by Candida albicans mutants for genes coding fungal proteins exhibiting the eight-cysteine-containing CFEM domain. FEMS Yeast Res 6:1074–1084. [DOI] [PubMed] [Google Scholar]
- 81. Da Silva BA, Sodré CL, Souza-Gonçalves AL, Aor AC, Kneipp LF, Fonseca BB, et al. 2012. Proteomic analysis of the secretions of Pseudallescheria boydii, a human fungal pathogen with unknown genome. J Proteome Res 11:172–188. 10.1021/pr200875x [DOI] [PubMed] [Google Scholar]
- 82. Gaderer R, Bonazza K, Seidl-Seiboth V. 2014. Cerato-platanins: a fungal protein family with intriguing properties and application potential. Appl Microbiol Biotechnol 98: 4795–4803. 10.1007/s00253-014-5690-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sbrana F, Bongini L, Cappugi G, Fanelli D, Guarino A, Pazzagli L, et al. 2007. Atomic force microscopy images suggest aggregation mechanism in cerato-platanin. Eur Biophys J 36:727–732. [DOI] [PubMed] [Google Scholar]
- 84. Mouyna I, Monod M, Fontaine T, Henrissat B, Lechenne B, Latgé JP. 2000. Identification of the catalytic residues of the first family of beta(1–3)glucanosyltransferases identified in fungi. Biochem J 347:741–747. [PMC free article] [PubMed] [Google Scholar]
- 85. Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, et al. 2009. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117–1121. 10.1038/nature08264 [DOI] [PubMed] [Google Scholar]
- 86. De Groot PWJ, Yin QY, Weig M, Sosinska GJ, Klis FM, et al. 2007. Mass spectrometric identification of covalently bound cell wall proteins from the fission yeast Schizosaccharomyces pombe . Yeast 24:267–278. [DOI] [PubMed] [Google Scholar]
- 87. Yin QY, Groot PWJ de, Dekker HL, Jong L de, Klis FM, et al. 2005. Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls identification of proteins covalently attached via glycosylphosphatidylinositol remnants or mild alkali-sensitive linkages. J Biol Chem 280:20894–20901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Accession numbers of analyzed sequences are available on GenBank database (Scedosporium apiospermum genome accession number JOWA00000000).
(PDF)
The major fragmentation series (y-carboxy and b-amino) are annotated; the deduced sequence, m/z value and charge state from the precursor ion are indicated.
(PPTX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.