Abstract
Shigella flexneri serotype 1b is among the most prominent serotypes in developing countries, followed by serotype 2a. However, only limited data is available on the global phenotypic and genotypic characteristics of S. flexneri 1b. In the present study, 40 S. flexneri 1b isolates from different regions of China were confirmed by serotyping and biochemical characterization. Antimicrobial susceptibility testing showed that 85% of these isolates were multidrug-resistant strains and antibiotic susceptibility profiles varied between geographical locations. Strains from Yunnan were far more resistant than those from Xinjiang, while only one strain from Shanghai was resistant to ceftazidime and aztreonam. Fifteen cephalosporin resistant isolates were identified in this study. ESBL genes (bla SHV, bla TEM, bla OXA, and bla CTX-M) and ampC genes (bla MOX, bla FOX, bla MIR(ACT-1), bla DHA, bla CIT and bla ACC) were subsequently detected among the 15 isolates. The results showed that these strains were positive only for bla TEM, bla OXA, bla CTX-M, intI1, and intI2. Furthermore, pulsed-field gel electrophoresis (PFGE) analysis showed that the 40 isolates formed different profiles, and the PFGE patterns of Xinjiang isolates were distinct from Yunnan and Shanghai isolates by one obvious, large, missing band. In summary, similarities in resistance patterns were observed in strains with the same PFGE pattern. Overall, the results supported the need for more prudent selection and use of antibiotics in China. We suggest that antibiotic susceptibility testing should be performed at the start of an outbreak, and antibiotic use should be restricted to severe Shigella cases, based on resistance pattern variations observed in different regions. The data obtained in the current study might help to develop a strategy for the treatment of infections caused by S. flexneri 1b in China.
Introduction
Shigella, the causative agent of human shigellosis, is a significant public health burden. Worldwide, 164.7 million Shigella episodes per year are estimated, of which 163.2 million are in developing countries. The majority of the infections and deaths caused by Shigella are in children <5 years old [1]. Moreover, a surveillance study in six Asian countries showed the global disease burden caused by shigellosis might be much higher than these estimates [2]. In China, shigellosis is one of the top four notifiable infectious diseases, with nearly half a million cases annually [3].
Shigella species are in fact clones of Escherichia coli [4]. Based on biochemical and serological properties, the genus Shigella is divided into four species or subgroups: S. dysenteriae, S. flexneri, S. boydii, and S. sonnei, among which, S. flexneri is the predominant species in developing countries [1]. A previous study showed that the annual shigellosis morbidity rate was 20.3 cases per 100,000 people in China using the national surveillance data from 2009, and S. flexneri (67.3%) and S. sonnei (32.7%) were two major causative species [5]. Because Shigella strains are all negative for H antigen, Shigella serotyping is based on the O antigen only. To date, 20 S. flexneri serotypes (1a, 1b, 1c (or 7a), 1d, 2a, 2b, 2v, 3a, 3b, 4a, 4av, 4b, 5a, 5b, X, Xv, Y, Yv, 6, and 7b) have been recognized [6,7]. Some S. flexneri serotypes are more prevalent than others, and S. flexneri 1b is among the most commonly encountered serotypes in developing countries, followed by serotype 2a [1].
Pickering LK showed that antimicrobial susceptibility patterns of Shigella species, were influenced by geographic location and other factors (year, classes of antimicrobial agents, pressure exerted by antimicrobial use, source of isolates) in endemic regions [8]. However, comparisons of associations between geographical diversity and antimicrobial resistance of serotype 1b strains are largely lacking. The objective of the present study was to evaluate the antimicrobial resistance profiles of S. flexneri 1b isolates from different regions of China, and guide the selection of the most effective antimicrobial agents for shigellosis treatment.
Materials and Methods
Bacterial isolates, serotyping, and biochemical characterization
During our routine surveillance of bacillary dysentery, fecal samples from individual outpatients with diarrhea or dysentery were collected and screened for Shigella species in sentinel hospitals based on a national pathogen monitoring system. Samples were streaked directly onto Salmonella-Shigella (SS) agar and incubated overnight at 37°C. Resultant colonies were picked and streaked directly onto SS agar and again incubated overnight at 37°C. The colonies were subcultured on Luria–Bertani agar plates and incubated overnight at 37°C, and were then sent to our laboratory for further confirmation. This study was approved by the ethics committee of the Academy of Military Medical Sciences (China). Written informed consent was obtained from the patients involved in this study. The isolates were confirmed using API 20E test strips (bioMerieux Vitek, Marcy-l’Etoile, France) following the manufacturer’s recommendations. Serotype identification was performed using two serotyping kits: a kit including antisera specific for all type and group-factor antigens (Denka Seiken, Tokyo, Japan), and a monoclonal antibody reagent kit (Reagensia AB, Stockholm, Sweden), specific for all S. flexneri type and group-factor antigens. Serological reactions were performed using the slide agglutination test, as described previously [9].
Antimicrobial susceptibility testing
MICs of 21 antimicrobial agents including ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), cefoperazone (CFP), cefazolin (CFZ), cefoxitin (FOX), imipenem (IPM), nitrofurantoin (NIT), piperacillin (PIP), ampicillin (AMP), ticarcillin (TIC), tetracycline (TE), tobramycin (TO), gentamicin (GEN), amikacin (AK), aztreonam (ATM), chloramphenicol (C), ticarcillin/clavulanic acid (TIM), levofloxacin (LEV), norfloxacin (NOR), and trimethoprim/sulfamethoxazole (SXT) were tested for each of the isolates. MICs were determined by broth microdilution using a 96-well microtiter plate (Sensititre, Thermo Fisher Scientific Ine, West Sussex, United Kingdom) according to the recommendations of the Clinical and Laboratory Standards Institute. E. coli ATCC 25922 was used as the control strain for susceptibility studies.
Pulsed-field gel electrophoresis (PFGE)
The isolates were analyzed by PFGE following digestion with NotI to generate DNA fingerprinting profiles according to the procedures developed by the CDC PulseNet program [10]. Salmonella enterica serotype Braenderup H9812 was used as the molecular size marker. PFGE patterns were interpreted using BioNumerics (Version 6.0) software (Applied Maths, Sint-Martens-Latem, Belgium). A tree indicating relative genetic similarity was constructed based on the unweighted pair group method of averages (UPGMA) and a position tolerance of 1.2%.
PCR amplification of the antibiotic-resistance determinants and integrons
PCR assays were carried out as described previously [11–16] to detect ESBL genes (bla SHV, bla TEM, bla OXA, and bla CTX-M) and ampC genes (bla MOX, bla FOX, bla MIR(ACT-1), bla DHA, bla CIT and bla ACC) which are responsible for resistance to cephalosporins. We also amplified the variable regions of class 1 and 2 integrons using primers listed in Table 1. The reverse primers aadA1, aadA2, aadA5, and cmlA1 were used with forward primer hep58 to amplify the gene cassettes of class 1 integron-positive strains. Forward primer hep74 and reverse primer hep51 were used to amplify the gene cassettes of class 2 integron-positive strains. Resultant PCR products were fully sequenced and analyzed by comparison with sequences in GenBank.
Table 1. Primers for detection of antibiotic-resistance determinants and integrons.
| Target | Primer sequence (5′ to 3′) | Reference |
|---|---|---|
| β-lactamases (bp) | ||
| bla CTX-M-1 group (873 bp) | F: GGTTAAAAAATCACTGCGTC | Matar et al. [15] |
| R: TTACAAACCGTCGGTGACGA | Matar et al. [15] | |
| bla CTX-M-9 group (868 bp) | F: AGAGTGCAACGGATGATG | Matar et al. [15] |
| R: CCAGTTACAGCCCTTCGG | Matar et al. [15] | |
| bla CTX-M-2/8/25 group (221 bp) | F: ACCGAGCCSACGCTCAA | This study |
| R: CCGCTGCCGGTTTTATC | This study | |
| bla TEM (1080 bp) | F: ATGAGTATTCAACATTTCCG | Tariq et al. [11] |
| R: CCAATGCTTAATCAGTGAGG | Tariq et al. [11] | |
| bla OXA (890 bp) | F: ATTAAGCCCTTTACCAAACCA | Ahmed et al. [12] |
| R: AAGGGTTGGGCGATTTTGCCA | Ahmed et al. [12] | |
| bla MOX (520 bp) | F: GCTGCTCAAGGAGCACAGGAT | F. Javier et al. [16] |
| R: CACATTGACATAGGTGTGGTGC | F. Javier et al. [16] | |
| bla FOX (190 bp) | F: AACATGGGGTATCAGGGAGATG | F. Javier et al. [16] |
| R: CAAAGCGCGTAACCGGATTGG | F. Javier et al. [16] | |
| bla MIR(ACT-1) (302 bp) | F: TCGGTAAAGCCGATGTTGCGG | F. Javier et al. [16] |
| R: CTTCCACTGCGGCTGCCAGTT | F. Javier et al. [16] | |
| bla DHA (405 bp) | F: AACTTTCACAGGTGTGCTGGGT | F. Javier et al. [16] |
| R: CCGTACGCATACTGGCTTTGC | F. Javier et al. [16] | |
| bla CIT (462 bp) | F: TGGCCAGAACTGACAGGCAAA | F. Javier et al. [16] |
| R: TTTCTCCTGAACGTGGCTGGC | F. Javier et al. [16] | |
| bla ACC (346 bp) | F: AACAGCCTCAGCAGCCGGTTA | F. Javier et al. [16] |
| R: TTCGCCGCAATCATCCCTAGC | F. Javier et al. [16] | |
| Integrons | ||
| IntI1 (569 bp) | F: ACATGTGATGGCGACGCACGA | Pan et al. [14] |
| R: ATTTCTGTCCTGGCTGGCGA | Pan et al. [14] | |
| Class 1 integron variable region | hep58: TCATGGCTTGTTATGACTGT | This study |
| hep59: GTAGGGCTTATTATGCACGC | This study | |
| Class 1 integron variable region | hep58: TCATGGCTTGTTATGACTGT | This study |
| aadA1: TGTCAGCAAGATAGCCAGAT | This study | |
| Class 1 integron variable region | hep58: TCATGGCTTGTTATGACTGT | This study |
| aadA2: TGATCTCGCCTTTCACAAA | This study | |
| Class 1 integron variable region | hep58: TCATGGCTTGTTATGACTGT | This study |
| aadA5: CATCTAACGCATAGTTGAGC | This study | |
| Class 1 integron variable region | hep58: TCATGGCTTGTTATGACTGT | This study |
| cmlA1: CAACGATTGGGATTTGACGTACTTT | This study | |
| IntI2 (789 bp) | F: CACGGATATGCGACAAAAAGGT | This study |
| R: GTAGCAAACGAGTGACGAAATG | This study | |
| Class 2 integron variable region | hep74: CGGGATCCCGGACGGCATGCACGATTTGTA | This study |
| hep51: GATGCCATCGCAAGTACGAG | This study |
Results
Bacterial isolation and biochemical characterization
We isolated and identified 40 S. flexneri 1b isolates from patients with either diarrhea or dysentery over the period from 2005 to 2013. The strain information is detailed in Table 2. Among them, 28 isolates were collected from Xinjiang from 2005–2012, 11 from Yunnan in 2009, and one from Shanghai in 2013. All S. flexneri serotype 1b strains examined possessed typical S. flexneri biochemical characteristics, and analysis of biochemical reactions indicated the presence of four biotypes among these serotype 1b strains (Table 3).
Table 2. Strain information of S. flexneri serotype 1b isolates from diarrheal patients.
| Original No | Species | Serotype | Source | Origin | Isolation time |
|---|---|---|---|---|---|
| SH13sh024 | S. flexneri | 1b | Human | Shanghai | 2013 |
| 2005126 | S. flexneri | 1b | Human | Xinjiang | 2005 |
| 2005173 | S. flexneri | 1b | Human | Xinjiang | 2005 |
| 2006230 | S. flexneri | 1b | Human | Xinjiang | 2006 |
| 2008026 | S. flexneri | 1b | Human | Xinjiang | 2008 |
| 2008046 | S. flexneri | 1b | Human | Xinjiang | 2008 |
| 2008076 | S. flexneri | 1b | Human | Xinjiang | 2008 |
| 2009158 | S. flexneri | 1b | Human | Xinjiang | 2009 |
| 2009378 | S. flexneri | 1b | Human | Xinjiang | 2009 |
| 2010026 | S. flexneri | 1b | Human | Xinjiang | 2010 |
| 2010144 | S. flexneri | 1b | Human | Xinjiang | 2010 |
| 2010230 | S. flexneri | 1b | Human | Xinjiang | 2010 |
| 2010236 | S. flexneri | 1b | Human | Xinjiang | 2010 |
| 2010294 | S. flexneri | 1b | Human | Xinjiang | 2010 |
| 2010297 | S. flexneri | 1b | Human | Xinjiang | 2010 |
| 2010315 | S. flexneri | 1b | Human | Xinjiang | 2010 |
| 2011032 | S. flexneri | 1b | Human | Xinjiang | 2011 |
| 2011033 | S. flexneri | 1b | Human | Xinjiang | 2011 |
| 2011052 | S. flexneri | 1b | Human | Xinjiang | 2011 |
| 2011065 | S. flexneri | 1b | Human | Xinjiang | 2011 |
| 20110163 | S. flexneri | 1b | Human | Xinjiang | 2011 |
| 20110176 | S. flexneri | 1b | Human | Xinjiang | 2011 |
| 20110187 | S. flexneri | 1b | Human | Xinjiang | 2011 |
| 2012059 | S. flexneri | 1b | Human | Xinjiang | 2012 |
| 2012061 | S. flexneri | 1b | Human | Xinjiang | 2012 |
| 2012063 | S. flexneri | 1b | Human | Xinjiang | 2012 |
| 2012073 | S. flexneri | 1b | Human | Xinjiang | 2012 |
| 2012133 | S. flexneri | 1b | Human | Xinjiang | 2012 |
| 2012147 | S. flexneri | 1b | Human | Xinjiang | 2012 |
| CDSF2 | S. flexneri | 1b | Human | Yunnan | 2009 |
| CDSF3 | S. flexneri | 1b | Human | Yunnan | 2009 |
| CDSF4 | S. flexneri | 1b | Human | Yunnan | 2009 |
| CDSF5 | S. flexneri | 1b | Human | Yunnan | 2009 |
| CDSF6 | S. flexneri | 1b | Human | Yunnan | 2009 |
| CDSF7 | S. flexneri | 1b | Human | Yunnan | 2009 |
| CDSF8 | S. flexneri | 1b | Human | Yunnan | 2009 |
| CDSF9 | S. flexneri | 1b | Human | Yunnan | 2009 |
| CDSF10 | S. flexneri | 1b | Human | Yunnan | 2009 |
| CDSF11 | S. flexneri | 1b | Human | Yunnan | 2009 |
| CDSF12 | S. flexneri | 1b | Human | Yunnan | 2009 |
Table 3. Biochemical patterns of S. flexneri serotype 1b strains.
| Biotype | Total No. % (n = 40) | Xinjiang No. % (n = 28) | Yunnan No. % (n = 11) | Shanghai No. % (n = 1) |
|---|---|---|---|---|
| B1 | 4 (10%) | 3 (10.7%) | 0 | 1 (100%) |
| B2 | 20 (50%) | 20 (71%) | 0 | 0 |
| B3 | 2 (5%) | 2 (7%) | 0 | 0 |
| B4 | 14 (35%) | 3 (10.7%) | 11 (100%) | 0 |
Biotype B1: mannose-, glucose-, and melibiose-positive; biotype B2: mannose-, glucose-, arabinose-, and melibiose-positive; biotype B3: mannose-, glucose-, and sucrose-positive; biotype B4: mannose-, glucose-, and arabinose-positive.
Antimicrobial susceptibility testing
The 40 S. flexneri 1b isolates were tested for susceptibility to 21 antimicrobials (Table 4). Resistance to TIC or AMP was the most common (36/40, 90%), followed by TE (35/40, 87.5%), C (33/40, 82.5%), SXT (20/40, 50%), PIP (16/40, 40%), TIM (12/40, 30%), and ATM (1/40, 2.5%). None of the isolates were resistant to NIT, IPM, FEP, FOX, AK, TO, or GEN. In addition, strains with intermediate resistance to TIM, ATM, and PIP were also observed.
Table 4. Comparison of susceptibility to 21 antibiotics among S. flexneri 1b isolates from different regions.
| Antimicrobial resistance rate No. (%) | ||||
|---|---|---|---|---|
| Antibiotic | Total (n = 40) | Xinjiang (n = 28) | Yunnan (n = 11) | Shanghai No. %(n = 1) |
| CAZ | 1 (2.5%) | 0 | 0 | 1 (100%) |
| CRO | 14 (35%) | 3 (10.7%) | 11 (100%) | 0 |
| PIP | 16 (40%) | 4 (14%) | 11 (100%) | 1 (100%) |
| TE | 36 (90%) | 24 (85.7%) | 11 (100%) | 1 (100%) |
| CFP | 14 (35%) | 3 (10.7%) | 11 (100%) | 0 |
| CFZ | 14 (35%) | 3 (10.7%) | 11 (100%) | 0 |
| TIC | 35 (87.5%) | 24 (85.7%) | 11 (100%) | 0 |
| TIM | 12 (30%) | 1 (3.6%) | 11 (100%) | 0 |
| ATM | 1 (2.5%) | 0 | 0 | 1 (100%) |
| AMP | 36 (90%) | 24 (85.7%) | 11 (100%) | 1 (100%) |
| C | 34 (85%) | 22 (78.5%) | 11 (100%) | 0 |
| SXT | 20 (50%) | 9(32.1%) | 11 (100%) | 0 |
| LEV | 0 | 0 | 0 | 0 |
| FEP | 0 | 0 | 0 | 0 |
| IPM | 0 | 0 | 0 | 0 |
| NIT | 0 | 0 | 0 | 0 |
| FOX | 0 | 0 | 0 | 0 |
| TO | 0 | 0 | 0 | 0 |
| GEN | 0 | 0 | 0 | 0 |
| NOR | 0 | 0 | 0 | 0 |
| AK | 0 | 0 | 0 | 0 |
Multidrug resistance (MDR; resistant to three or more classes of antimicrobials) was detected in 85% of the S. flexneri 1b isolates, and these isolates had diverse antibiotic-resistance profiles. About 78% of the isolates from Xinjiang were MDR isolates, dominated by resistance to AMP/TIC/TE/C (10/28, 35.7%), followed by AMP/TIC/TE/C/SXT (7/28, 25%), and CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM (3/28, 10.7%). All of the isolates from Yunnan presented the same resistance pattern, CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT, and the single strain from Shanghai exhibited a resistance profile of CAZ/CFP/CRO/CFZ/PIP/AMP/TIC/ATM (Table 5).
Table 5. Dominant antimicrobial resistance profiles of 40 S. flexneri 1b isolates from different regions of China.
| Antimicrobial resistance profiles | Total | Xinjiang | Sichuan |
|---|---|---|---|
| No. % (n = 40) | No. % (n = 28) | No. % (n = 11) | |
| Sensitive (n = 2) | 2 (5%) | 2 (7%) | 0 |
| TE (n = 2) | 2 (5%) | 2 (7%) | 0 |
| AMP/TIC (n = 1) | 1 (2.5%) | 1 (3.6%) | 0 |
| AMP/TIC/C (n = 1) | 1 (2.5%) | 1 (3.6%) | 0 |
| AMP/TIC/TE/C (n = 10) | 10 (25%) | 10 (35.7%) | 0 |
| AMP/TIC/TE/C/SXT (n = 7) | 7 (17.5%) | 7 (25%) | 0 |
| PIP/AMP/TIC/TE/SXT (n = 1) | 1 (2.5%) | 1 (3.6%) | 0 |
| AMP/TIC/TE/C/TIM/SXT (n = 1) | 1 (2.5%) | 1 (3.6%) | 0 |
| CAZ/CFP/CRO/CFZ/PIP/AMP/TIC/ATM (n = 1) | 1 (2.5%) | 0 | 0 |
| CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM (n = 3) | 3 (7.5%) | 3 (10.7%) | 0 |
| CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT (n = 11) | 11 (27.5%) | 0 | 11 (100%) |
Molecular analysis of antibiotic-resistance determinants and integrons
In this study, fifteen isolates were found to be resistant to cephalosporin antibiotics, and these strains were tested for antibiotic-resistance determinants and integrons, including the bla SHV, bla TEM, bla OXA, bla CTX-M, bla MOX, bla FOX, bla MIR(ACT-1), bla DHA, bla CIT, bla ACC, intI1, and intI2 gene regions (Table 6). PCR results showed that all 15 tested isolates were negative for bla SHV, bla MOX, bla FOX, bla MIR(ACT-1), bla DHA, bla CIT, bla ACC, but positive for bla TEM, bla OXA, bla CTX-M, intI1, and intI2 gene regions. Sequencing results of the bla TEM showed 100% identity with bla TEM-1. All isolates harbored bla OXA-1, except the only one isolate from Shanghai. Thirteen isolates harbored bla CTX-M-14; the only one strain from Shanghai contained bla CTX-M-79. Notably, one isolate, collected from Xinjiang, simultaneously harbored both bla CTX-M-28 and bla CTX-M-14. All isolates, except for the Shanghai isolate, contained class 1 integrons, among which, three strains from Xinjiang and two strains from Yunnan harbored both the bla OXA-30 and aadA1 gene cassettes. The bla OXA-30 and bla OXA-1 genes, which are contained on the Tn1409 and Tn2603 transposons respectively, differed by only a single nucleotide [17]. All 15 isolates harbored class 2 integrons with dfrA1, sat1, and aadA1 gene cassettes.
Table 6. Antibiogram and molecular analysis of the antibiotic-resistance determinants and integrons of 15 cephalosporin resistant isolates.
| Strain no. | Antibiogram | Antibiotic-resistant determinants and integrons present |
|---|---|---|
| SH13sh024 | CAZ/CFP/CRO/CFZ/PIP/AMP/TIC/ATM | IntI2 (dfrA1+sat1+aadA1), bla TEM-1, blactx-M-79 |
| 2010294 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM | IntI1 (bla OXA-30 +aadA1), IntI2 (dfrA1+sat1+aadA1), bla OXA-1, bla TEM-1, bla CTX-M-14 |
| 2010297 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM | IntI1 (bla OXA-30 +aadA1), IntI2 (dfrA1+sat1+aadA1), bla OXA-1, bla TEM-1, bla CTX-M-14 and 28 |
| 2010315 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM | IntI1 (bla OXA-30 +aadA1), IntI2 (dfrA1+sat1+aadA1), bla OXA-1, bla TEM-1, bla CTX-M-14 |
| CDSF2 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT | IntI1, IntI2 (dfrA1+sat1+aadA1), bla OXA-1, bla TEM-1, bla CTX-M-14 |
| CDSF3 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT | IntI1, IntI2 (dfrA1+sat1+aadA1), bla OXA-1, bla TEM-1, bla CTX-M-14 |
| CDSF4 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT | IntI1 (bla OXA-30 +aadA1), IntI2 (dfrA1+sat1+aadA1), bla OXA-1, bla TEM-1, bla CTX-M-14 |
| CDSF5 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT | IntI1, IntI2 (dfrA1+sat1+aadA1), bla TEM-1, bla CTX-M-14 |
| CDSF6 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT | IntI1, IntI2 (dfrA1+sat1+aadA1), bla TEM-1, bla CTX-M-14 |
| CDSF7 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT | IntI1, IntI2 (dfrA1+sat1+aadA1), bla OXA-1, bla TEM-1, bla CTX-M-14 |
| CDSF8 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT | IntI1, IntI2 (dfrA1+sat1+aadA1), bla OXA-1, bla TEM-1, bla CTX-M-14 |
| CDSF9 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT | IntI1, IntI2 (dfrA1+sat1+aadA1), bla OXA-1, bla TEM-1, bla CTX-M-14 |
| CDSF10 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT | IntI1 (bla OXA-30 +aadA1), IntI2 (dfrA1+sat1+aadA1), bla OXA-1, bla TEM-1, bla CTX-M-14 |
| CDSF11 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT | IntI1, IntI2 (dfrA1+sat1+aadA1), bla OXA-1, bla TEM-1, bla CTX-M-14 |
| CDSF12 | CFP/CRO/CFZ/PIP/AMP/TIC/TE/C/TIM/SXT | IntI1, IntI2 (dfrA1+sat1+aadA1), bla OXA-1, bla TEM-1, bla CTX-M-14 |
PFGE analysis
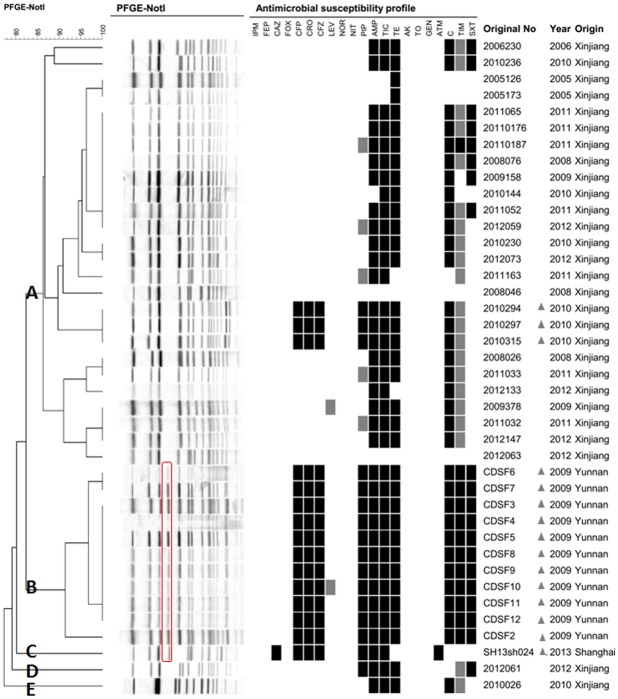
PFGE was performed to determine the genetic diversity of the S. flexneri 1b isolates. The 40 S. flexneri 1b isolates generated 18 PFGE patterns (Fig 1). All isolates could be divided into five main groups (A–E), with approximately 82% similarity. Notably, there was a high level of variation in the PFGE profiles of serotype 1b isolates between different regions of China, which contrasted with a previous study showing that 70 S. flexneri 1b strains isolated in Bangladesh shared the same PFGE profile [7]. In the present study, isolates from Xinjiang belonged to groups A, D, and E, whereas isolates from Yunnan and Shanghai belonged to groups B and C, respectively. Twenty-eight isolates from Xinjiang showed 14 different PFGE patterns; 26 of the 28 isolates formed a single cluster, with the two remaining isolates forming separate branches. Eleven isolates from Yunnan showed three different PFGE patterns, and the single isolate from Shanghai displayed a distinct PFGE pattern. Interestingly, the PFGE patterns of Xinjiang isolates were distinct from Yunnan and Shanghai isolates by one obvious, large, missing band indicated with a red border in Fig 1.
Fig 1. PFGE dendrogram and antibiotic resistance profile of 40 S. flexneri 1b isolates.
The original number, origin, and isolation year are indicated for each strain. The missing band of Xinjiang isolates was indicated with a red border and 15 cephalosporin resistant isolates were marked with triangles.
Among the isolates in different PFGE clusters, the prevalence of antibiotic resistance in clusters A, D, and E (Xinjiang) was much lower than that of isolates in cluster B (Yunnan). All 15 cephalosporin resistant isolates (marked with triangles in Fig 1) belonged to clusters B (Yunnan), C (Shanghai), and a small branch of cluster A (Xinjiang). All isolates in cluster B were resistant to cephalosporins, such as CFP, CRO, and CFZ, while the resistance rate of isolates in clusters A, D, and E (Xinjiang) to these antibiotics was 10.7%. Higher resistance rates to SXT, PIP, AMP, TIC, TE, C, and TIM were also observed in isolates from Xinjiang than in those from Yunnan. The isolate in cluster C (Shanghai) was the only isolate showing resistance to CAZ and ATM. In summary, similarities in resistance patterns were observed in strains with the same PFGE pattern.
Discussion
Approximately 85% of the strains in our study showed MDR profiles, and were highly resistant to the older-generation antimicrobials, such as AMP, C, TE, and TIC. So the treatment should be based on susceptibility patterns and antimicrobials with current resistance may turn to be effective in the future. This rapid accumulation of resistance in Shigella is probably the result of inappropriate prescription of, and easy access to, antimicrobials among outpatients in China [18]. Besides, the PFGE dendrogram showed that the S. flexneri type 1b isolates from Yunnan are closely related (95% similarity), indicating that they are possibly derived from a common parental strain. In contrast, the isolates from Xinjiang shared lower degrees of similarity, suggesting that they are likely derived from divergent sources. The antibiotic susceptibility profiles of the isolates also varied between the geographical locations; the antimicrobial resistance situation in Xinjiang does not seem to be as serious as that in Yunnan and Shanghai. Among the 40 isolates, only the isolate from Shanghai was resistant to CAZ and ATM, and isolates from Yunnan were more resistant to antibiotics CFP, CRO, CFZ, PIP, AMP, TIC, TE, C, TIM, and SXT than those from Xinjiang and Shanghai. The diverse antibiotic susceptibility may be attributed to the fact that mainland China covers a large territory and has many nationalities, with more than 1.3 billion people living in diverse areas, hence many varying factors such as customs, climates, economies, geographies may affect the antimicrobial susceptibility patterns.
In the present study, all 15 cephalosporin resistant isolates harbored bla CTX-M genes, most of which were bla CTX-M-14, followed by bla CTX-M-79, and bla CTX-M-28. A previous study indicated that plasmids carrying different bla CTX-M genes were introduced into isolates at different times [18]. The bla CTX-M-14 subtype was identified in 14 of these isolates, which indicated that these genes might have been circulating among Shigella isolates for a long time. In addition, bla TEM was detected in all 15 cephalosporin resistant isolates, and bla OXA was amplified in all such isolates, except the isolate from Shanghai. OXA-type β-lactamases are characterized by high hydrolytic activity against cloxacillin and oxacillin, which confer resistance to cephalothin and ampicillin [19]. Notably, all bla OXA isolates in this study carried bla OXA-1, suggesting a host specificity for this subtype in S. flexneri, which is consistent with a previous study by Siu et al. [17]. bla OXA-30 and aadA1 were also detected in the gene cassettes of class 1 integrons in five of the 15 cephalosporin resistant isolates in our study. These two gene cassettes appear to be coordinately excised or integrated, as described by a previous study [3]. Pan et al. reported that class 2 integrons were present in 87.9% of S. flexneri isolates [14]. In our study, all of the 15 strains examined harbored class 2 integrons, which followed the dfrA1+sat1+aadA1 gene cassette, conferring resistance to trimethoprim and streptomycin [20]. All of these various resistance genes facilitate the dissemination of resistance determinants and the survival of bacteria under the selective pressure of various antibiotics.
Fluoroquinolones and third-generation cephalosporins were two popular empirical options to treat severe gastrointestinal infections caused by pathogenic bacteria [21]. Ciprofloxacin is currently the first-line antibiotic recommended by the World Health Organization for treating shigellosis [22]. Fortunately, limited resistance to fluoroquinolones was observed in our study. The control of resistance to quinolones could be attributed to a relevant study on quinolone-induced cartilage toxicity [23]. Third-generation cephalosporins are considered alternative drugs for shigellosis treatment [22], but in our study, the isolates from Yunnan and Shanghai were all resistant to CFP, CRO, and CFZ, and the rate of resistance of isolates from Xinjiang to these cephalosporins was 10.7%. According to the literature, strains may acquire resistance to multiple antibiotics if exposed to a resistant gastrointestinal pathogen following antibiotic therapy [24]. Therefore, these ESBL-producing isolates, especially those from Yunnan and Shanghai, would put patients at high risk if third-generation cephalosporins were still used for empirical therapy. Moreover, the isolate from Shanghai also exhibited resistance to ATM and CAZ, which clearly raises the perennial issue of strict control of antimicrobial prescription in China.
In the present study, diverse antimicrobial resistance patterns were observed in different endemic areas, which support the need for prudent selection and use of antibiotics. Therefore, we suggest that antibiotic susceptibility testing should be performed at the start of an outbreak, and antibiotic use should be restricted to severe Shigella cases, based on variations in resistance patterns in different regions. Furthermore, resistance patterns should be recorded timely and locally, and empiric antibiotic therapy for Shigella cases should be changed accordingly. The data that we obtained here might provide a strategy for the treatment of infections caused by S. flexneri serotype 1b strains in China.
Acknowledgments
The authors thank XXY for participating in writing.
Data Availability
All relevant data are within the paper.
Funding Statement
This research was supported by the National Major Scientific and Technological Special Projects for Infectious Diseases during the Twelfth Five-year Plan Period (Nos. 2012ZX10004215 and 2013ZX10004607); the National Nature Science Foundation of China (Nos. 81371854, 81373053, 81202252 and 31200942); and the Beijing Science and Technology Nova program (No. xx2013061). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, et al. (1999) Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ 77: 651–666. [PMC free article] [PubMed] [Google Scholar]
- 2. von Seidlein L, Kim DR, Ali M, Lee H, Wang X, Thiem VD, et al. (2006) A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med 3: e353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ke X, Gu B, Pan S, Tong M (2011) Epidemiology and molecular mechanism of integron-mediated antibiotic resistance in Shigella . Arch Microbiol 193: 767–774. 10.1007/s00203-011-0744-3 [DOI] [PubMed] [Google Scholar]
- 4. Pupo GM, Lan R, Reeves PR (2000) Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A 97: 10567–10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sui J, Zhang J, Sun J, Chang Z, Zhang W, Wang Z (2010) Surveillance of bacillary dysentery in China, 2009. Disease Surveillance 25: 947–950. [Google Scholar]
- 6. Sun Q, Lan R, Wang J, Xia S, Wang Y, Wang Y, et al. (2013) Identification and characterization of a novel Shigella flexneri serotype Yv in China. PLoS One 8: e70238 10.1371/journal.pone.0070238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qiu S, Wang Y, Xu X, Li P, Hao R, Yang C, et al. (2013) Multidrug-resistant atypical variants of Shigella flexneri in China. Emerg Infect Dis 19: 1147–1150. 10.3201/eid1907.111221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pickering LK. Antimicrobial resistance among enteric pathogens; 2004. Elsevier; pp. 71–77. [DOI] [PubMed] [Google Scholar]
- 9. Talukder KA, Dutta DK, Safa A, Ansaruzzaman M, Hassan F, Alam K, et al. (2001) Altering trends in the dominance of Shigella flexneri serotypes and emergence of serologically atypical S. flexneri strains in Dhaka, Bangladesh. J Clin Microbiol 39: 3757–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. (2006) Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3: 59–67. [DOI] [PubMed] [Google Scholar]
- 11. Tariq A, Haque A, Ali A, Bashir S, Habeeb MA, Salman M, et al. (2012) Molecular profiling of antimicrobial resistance and integron association of multidrug-resistant clinical isolates of Shigella species from Faisalabad, Pakistan. Can J Microbiol 58: 1047–1054. 10.1139/w2012-085 [DOI] [PubMed] [Google Scholar]
- 12. Ahmed AM, Furuta K, Shimomura K, Kasama Y, Shimamoto T (2006) Genetic characterization of multidrug resistance in Shigella spp. from Japan. J Med Microbiol 55: 1685–1691. [DOI] [PubMed] [Google Scholar]
- 13. Galani I, Souli M, Mitchell N, Chryssouli Z, Giamarellou H (2010) Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae and Escherichia coli isolates possessing blaVIM-1 in Greece. Int J Antimicrob Agents 36: 252–254. 10.1016/j.ijantimicag.2010.05.004 [DOI] [PubMed] [Google Scholar]
- 14. Pan J- C, Ye R, Meng D- M, Zhang W, Wang H- Q, Liu KZ (2006) Molecular characteristics of class 1 and class 2 integrons and their relationships to antibiotic resistance in clinical isolates of Shigella sonnei and Shigella flexneri . Journal of Antimicrobial Chemotherapy 58: 288–296. [DOI] [PubMed] [Google Scholar]
- 15. Matar GM, Jaafar R, Sabra A, Hart CA, Corkill JE, Dbaibo GS, et al. (2007) First detection and sequence analysis of the bla-CTX-M-15 gene in Lebanese isolates of extended-spectrum-beta-lactamase-producing Shigella sonnei . Ann Trop Med Parasitol 101: 511–517. [DOI] [PubMed] [Google Scholar]
- 16. Perez-Perez FJ, Hanson ND (2002) Detection of Plasmid-Mediated AmpC-Lactamase Genes in Clinical Isolates by Using Multiplex PCR. Journal of Clinical Microbiology 40: 2153–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Siu LK, Lo JY, Yuen KY, Chau PY, Ng MH, Ho PL (2000) beta-lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like beta-lactamase, OXA-30. Antimicrob Agents Chemother 44: 2034–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang W, Luo Y, Li J, Lin L, Ma Y, Hu C, et al. (2011) Wide dissemination of multidrug-resistant Shigella isolates in China. J Antimicrob Chemother 66: 2527–2535. 10.1093/jac/dkr341 [DOI] [PubMed] [Google Scholar]
- 19. Bradford PA (2001) Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev 14: 933–951, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pan JC, Ye R, Meng DM, Zhang W, Wang HQ, Liu KZ (2006) Molecular characteristics of class 1 and class 2 integrons and their relationships to antibiotic resistance in clinical isolates of Shigella sonnei and Shigella flexneri . J Antimicrob Chemother 58: 288–296. [DOI] [PubMed] [Google Scholar]
- 21. Gendrel D, Cohen R, European Society for Pediatric Infectious D, European Society for Gastroenterology H, Nutrition (2008) [Bacterial diarrheas and antibiotics: European recommendations]. Arch Pediatr 15 Suppl 2: S93–96. 10.1016/S0929-693X(08)74223-4 [DOI] [PubMed] [Google Scholar]
- 22. Bhattacharya D, Purushottaman SA, Bhattacharjee H, Thamizhmani R, Sudharama SD, Manimunda SP, et al. (2011) Rapid emergence of third-generation cephalosporin resistance in Shigella sp. isolated in Andaman and Nicobar Islands, India. Microb Drug Resist 17: 329–332. 10.1089/mdr.2010.0084 [DOI] [PubMed] [Google Scholar]
- 23. Schaad UB (2005) Fluoroquinolone antibiotics in infants and children. Infect Dis Clin North Am 19: 617–628. [DOI] [PubMed] [Google Scholar]
- 24. Ahmed SF, Riddle MS, Wierzba TF, Messih IA, Monteville MR, Sanders JW, et al. (2006) Epidemiology and genetic characterization of Shigella flexneri strains isolated from three paediatric populations in Egypt (2000–2004). Epidemiol Infect 134: 1237–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.