Abstract
A stochastic model of nuclear receptor-mediated transcription was developed based on activation of the aryl hydrocarbon receptor (AHR) by 2,3,7,8-tetrachlorodibenzodioxin (TCDD) and subsequent binding the activated AHR to xenobiotic response elements (XREs) on DNA. The model was based on effects observed in cells lines commonly used as in vitro experimental systems. Following ligand binding, the AHR moves into the cell nucleus and forms a heterodimer with the aryl hydrocarbon nuclear translocator (ARNT). In the model, a requirement for binding to DNA is that a generic coregulatory protein is subsequently bound to the AHR-ARNT dimer. Varying the amount of coregulator available within the nucleus altered both the potency and efficacy of TCDD for inducing for transcription of CYP1A1 mRNA, a commonly used marker for activation of the AHR. Lowering the amount of available cofactor slightly increased the EC50 for the transcriptional response without changing the efficacy or maximal response. Further reduction in the amount of cofactor reduced the efficacy and produced non-monotonic dose-response curves (NMDRCs) at higher ligand concentrations. The shapes of these NMDRCs were reminiscent of the phenomenon of squelching. Resource limitations for transcriptional machinery are becoming apparent in eukaryotic cells. Within single cells, nuclear receptor-mediated gene expression appears to be a stochastic process; however, intercellular communication and other aspects of tissue coordination may represent a compensatory process to maintain an organism’s ability to respond on a phenotypic level to various stimuli within an inconstant environment.
Introduction
As science begins to comprehend the workings of transcription through new techniques such as microarrays, chromatin immuno-precipitation (ChIP), and fluorescence visualization techniques, the overall and inescapable conclusion is that the inside of the nucleus is a very busy place. Transcription factors appear to interact with DNA via a number of interdependent mechanisms that occur on multiple time scales, and these complex dynamics may be important for producing appropriate and coordinated gene expression programs;[1] these mechanisms include chromatin and nucleosome remodeling, modulation of RNA polymerase activity, and alterations in epigenetic features including acetylation and methylation. ChIP methods provide a population-level approach to the interaction of proteins with DNA and show fluctuations of DNA binding on a time scale of minutes or hours. Single cell methods typically use fluorescence microscopy and, in contrast with the ChIP results, suggest that interactions of regulatory proteins with DNA are short-lived with dwell times on the order of milliseconds.[2–6] Multiple and differing time scales and the number of unknown features of transcriptional regulation present a challenge to the development of gene expression models.
The basic helix-loop-helix Per-ARNT-Sim (bHLH-PAS) family of transcription factors occurs ubiquitously in eukaryotes. [7–9]. Cells rely on these transcription factors that dimerize and then bind to response elements on DNA. The AHR is a member of the PAS Superfamily of proteins that plays a role in the detection of and adaptation to environmental change. The name “PAS” derives from the three founding members of the family, PER, ARNT and SIM.[7] Unlike a number of the nuclear receptor transcription factors with identified endogenous ligands, e.g, hormones like corticosteroids, estrogen, testosterone, the AHR PAS protein is a transcription factor without known endogenous or natural ligands or a well-defined physiological role. [8–10] Also, the AHR has distinct evolutionary origins and protein folding patterns. Up to now, the transcriptional response to AHR activation has been investigated largely by focusing on toxicity associated with chlorinated polyhalogenated hydrocarbons and not the modulatory role of natural or endogenous ligands in various tissues for either greater understanding of normal physiology or pharmaceutical development.[11–13]
The molecular machinery of transcription also includes a number of cofactors.[14, 15] The various functions of these cofactors include chromatin remodeling, histone modification, scaffolding to enable binding of yet other cofactors, and other necessary functions[16]. For example, the Mediator cofactor is a large multi-subunit complex that produced transcription in yeast and mammalian cell extracts reconstituted with RNA polymerase and other initiation factors. Mediator appears to facilitate chromatin looping. [17, 18] Recently, the idea of “transcription factories” has been advanced; these transcription factories are assemblies of nuclear receptors, cofactors, RNA polymerases, and other co-regulatory proteins.[19–23] This organization would maximize the shared utility of transcriptional resources.
Despite these mechanisms for resource allocation, competition for transcriptional resources among independent pathways of gene expression has been observed for a number of years.[24–28] In the case of ligand-activated nuclear receptors such as the aryl hydrocarbon receptor (AHR), a plethora of molecular interactions occur in simply moving the ligand-bound receptor to the nucleus where it can dimerize with the aryl hydrocarbon nuclear translocator (ARNT) and then the liganded dimer functions as a transcription factor.[29–33]
The aryl hydrocarbon receptor (AHR) is one of the most intensively studied transcription factors. [13] After binding to a ligand, the AHR moves into the nucleus. Once there, the liganded AHR sheds the chaperone proteins including a heat shock protein (HSP90), the phosphoprotein p23, and the hepatitis-B X-associated protein 2 (XAP2).[34] In the nucleus, after losing the chaperones, the AHR binds to its heterodimer partner, ARNT.[35] The liganded AHR-ARNT heterodimer recruits a number of transcription cofactors; these include steroid receptor coactivator 1 (SRC1), the nuclear receptor coactivator protein (NCoA), the histone acetyl transferase p300/CBP, and the thyroid hormone-associated protein (TRAP220) subunit of the Mediator complex.[36–38]
Recently, there has been much discussion about nonmonotonic dose-response curves (NMDRCs), especially with regard to endocrine disrupting chemicals (EDCs).[39–45] A highly precautionary non-threshold framework approach to the regulation of presumed EDCs by the European Union prompted a letter criticizing this approach from editors of 18 prominent toxicology journals.[46] Evolutionarily, the ability to distinguish biologically relevant signals produced by endogenous hormones from signals produced by exogenous chemicals that may have weak hormonal effects—or no effect at all—would be an adaptive trait. The ability to distinguish endogenous from exogenous signals is inherent in the differences in affinity of a receptor for various ligands and the principles of chemical mass action.[47] Modeling chemical kinetics with a macroscopic approach that uses time-dynamic differential equations cannot address the stochastic nature of gene expression that has motivated concern about the lack of knowledge of potential gene expression effects in the low dose region [36, 37].
Here, we chose to examine the binding of a ligand to a nuclear receptor and subsequent transcription of mRNA to determine the likelihood of low dose effects. To do so, a model of AHR-mediated transcription of mRNA from cytochrome p450 1A1 (CYP1A1) gene was developed. The model uses the Gillespie stochastic simulation algorithm (SSA) [48, 49] to keep track of the changes in the number of molecules of the various reactants and products within a single cell. The stochastic nature of the model is an attempt to capture the transcriptional “noise” that may underlie some NMDRCs and provides another line of evidence to determine whether the concerns about NMDRCs and potential low-dose effects are indeed warranted.[41, 46, 50–53]
While this model can faithfully reproduce some actual measurements of AHR activation and resulting transcriptional activity, the real benefit of this exercise is the increase in understanding of the process of ligand-activated transcription. While this model may not be generalizable to all nuclear receptors and co-regulatory proteins, competition for cellular transcription resources demonstrated by the model results has been observed for other nuclear receptors. In fact, competition for cellular transcription resources is one proposed mechanism that may account for thresholds and NMDRCs in ligand-induced transcriptional responses. Regarding this competition, the relative scarcity of transcription cofactors, RNA polymerase and other transcription-associated molecules within the nucleus or “transcription factory” is increasingly recognized as a mechanism for altering gene expression.[24–28, 54–56] Competition may be the reason that these scarce resources are organized into “transcription factories” to be shared amongst a number of genes.[19, 20, 57, 58]
The model is used to demonstrate that the efficacy and potency of transcriptional responses may vary based on the availability of transcriptional resources. In some circumstances, the modeled transcriptional response shows a biphasic response with decreasing transcription at higher doses. This phenomenon, known as “squelching” has been observed for a number of transcriptional responses.[59–63] Squelching is repression of transcription at high concentrations of ligand by sequestering limiting components (e.g. coactivators) required for transcriptional activation away from the promoter in the affected gene.[63]
Methods
The model simulates the effect of 2,3,7,8-tetrachlorodibenzodioxin (TCDD) on transcription in T47D cells.[64] The model includes binding of ligand to AHR, its movement into the nucleus and binding to the aryl hydrocarbon nuclear translocator (ARNT), binding of a generic cofactor to the heterodimer, binding of the activated heterodimer to xenobiotic response elements (XREs) associated with the CYP1A1 gene and those associated with other AHR-induced genes, binding of RNA polymerase, transcription initiation and termination.
Details of the Model
The model was developed from details of the well-known response of many cells to TCDD application. The model was based on ChIP experiments using T47D cells.[37, 64]
TCDD enters the cell by diffusion and once inside binds to the AHR. The ligand-bound receptor can either undergo degradation or move to the nucleus where it binds to ARNT to form a heterodimer. The ligand-bound AHR can also undergo degradation in the cytoplasm. AHR is newly synthesized at a constant rate.
Ligand-bound AHR can also move to the nucleus where it can bind to ARNT or undergo degradation. The ligand-bound dimer bind a generic cofactor to form a transcription complex. The complex can then bind to xenobiotic response elements on DNA (XREs). Two of these XREs are the TATA box and enhancer region associated with CYP1A1. [37] These response elements are referred to as “XREs” throughout this paper as many different xenobiotic chemicals in addition to dioxins bind to the AHR. [65, 66]
The transcription complex, consisting of AHR, ARNT and the cofactor, is also capable of binding to other AREs regulating other genes. The model assumed 470 AREs for other genes from estimates ranging between 400 and 900.[67–70]
The exact number of available RNA polymerase molecules available for AHR-activated gene expression is not known. Some RNA polymerase molecules will be bound to chromatin and transcribing genes associated with normal maintenance of the cell. Kimura et al. (1999) used saponin lysis to release soluble RNA polymerase molecules from HeLa cells and observed between 2000 and 4000 soluble molecules of RNA Pol II that were the size expected of the holoenzyme.[71] Hence, a value of 4000 molecules of RNA polymerase was assumed to be the upper limit for that available for AHR-mediated transcription.
Once the transcription complex is bound to an XRE, RNA polymerase II can also bind. Then the XRE-bound transcription complex can undergo transcription initiation. Once initiated, the complex is capable of transcription. ARNT may dissociate and transcription may still continue.[37] Transcription is terminated in a separate reaction. Within the transcription complex, another co-regulator protein could reversibly bind to the initiated transcription complex and slightly increase the rate of transcription.
We considered inclusion of negative feedback by the aryl hydrocarbon receptor repressor (AHRR) in the model.[72] However, the role of the AHRR is not sufficiently well understood that details of its action could be reproduced.[73] Further, AHRR may not play a role in the modeled response: in MCF-7 cells, similar in origin to T47D cells modeled here, AHRR protein expression in response to TCDD is not measurable before 24 hours and the simulation time here was six hours.[64, 73]Also, AHRR expression does not correlate well with TCDD-induced expression of CYP1A1 mRNA in a variety of mouse tissues and in human dermal fibroblasts.[74, 75]
Transcription itself was modeled with irreversible mass action kinetics. Any transcribed mRNA was subject to either degradation or export from the transcription complex. The kinetics of transcription are complex and include both early termination, pauses and varying rates of nucleotide addition to the end of the message.[76–84] The model includes four states of the transcription complex capable of producing mRNA, each state having a slightly different rate of transcription. These four states are included in the model in an attempt to reproduce, at some level, the complexity of transcription.
The model consists of 4 compartments, 29 molecular species, and 32 reactions. Full details of the model are provided in the Supporting Information.
Simulation Methods
The model was developed and exercised with the Simbiology module of MATLAB. Simbiology automates the use of compiled executable files and the model runs were much faster than without compilation. Model code is available from the corresponding author and in the SI.
The simulation method used here is direct method (DM) of Gillespie’s stochastic simulation algorithm (SSA).[49] This method relies on the fact that for any biochemical reaction (e.g., ligand binding), the time to the next transition will be an exponentially distributed random variable with a mean value equal to the reciprocal of the rate constant. The Gillespie SSA is fully implemented in Simbiology.[85] As noted, the use of ordinary differential equations (ODEs) does not capture the stochastic nature of responses. Knowledge of the complexity of events occurring in the crowded space of the nucleus remains incomplete.[1] However, the use of propensities as stochastic rate constants likely comes closer to reality than macroscopic kinetics.
For simple ligand binding, two mass action reactions are involved can be shown as reversible as follows:
The mass action expression for the forward reaction and the forward rate constant is in units of (conc.-time)-1. Because there is only a single species participating in the backward reaction the rate constant is in units of time-1. To use Gillespie’s method, the macroscopic rate constants must be converted to propensities. The propensity of a reaction is defined as the probability of the reaction occurring in the next time interval. [48, 49, 86] The time interval to the next transition is an exponentially distributed random variable; the mean of this distribution is the reciprocal of the product of the rate constant and the concentration of one or more reactants. [48]
100 runs of the model were conducted for each TCDD concentration. The simulation time was 6 hours and time unit used was one second. Because the time vectors associated with each run were different because of the probabilistic calculation for the propensity, linear interpolation was used to obtain the mean and variance of the numbers of molecules for each species throughout the course of the run from the 100 runs that represent a population of 100 cells.
Results
Comparison of Measured vs. Modeled Transcription
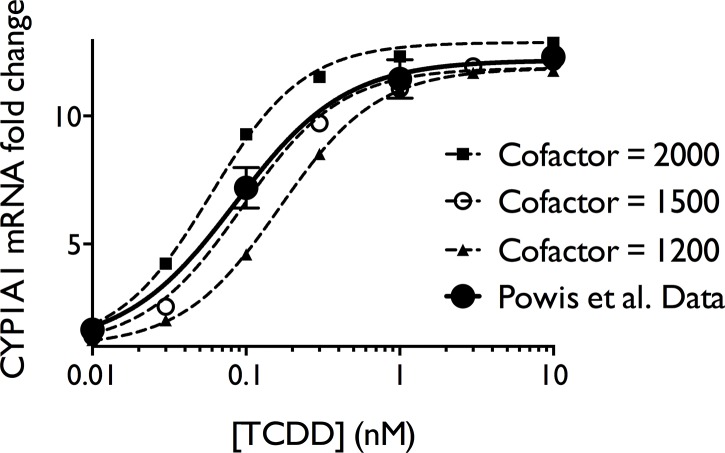
The model was exercised with a range of TCDD concentrations and a range of cofactor amounts. The model faithfully reproduces the transcriptional dose-response for CYP1A1 mRNA at 6 hours from Powis et al. (2011)[64] when 1500 molecules of cofactor, 1535 molecules of competing non-AHR binding proteins (Other) and 4000 molecules of RNA polymerase are present in the nucleus (Fig 1). Also shown are the modeled transcription dose-response curves for 2000, 1500 and 1200 molecules of cofactor. Fitting the Powis et al. (2011) data to a Hill dose-response model yielded an EC50 value of 0.086 nM and a Hill coefficient of 1.12. Fitting the model results with 1500 molecules of cofactor yielded an EC50 value of 0.094 nM and a Hill coefficient of 1.35 (Table 1) and the two plots are almost indistinguishable (Fig 1). Estimates of transitional dose values[87, 88] were determined by baseline projection from the ligand concentration needed to produce 21% of the maximal response (EC21) using S4 Equation in S1 File (Table 1).
Fig 1. Measured and modeled transcriptional dose-response to TCDD.
The larger filled black circles show the transcriptional response of CYP1A1 at 6 hours in T47-D cells. These data were digitally extracted from Fig 1 in Powis et al. (2011). [64] The smaller symbols and dotted lines show the modeled transcriptional dose response at three different amounts of cofactor present. When 1500 molecules of cofactor were present, the modeled response is very similar to the observed response in T47-D cells by Powis et al. (2011). [64]
Table 1. Hill equation fits of modeled data at a range of cofactor amounts along with the fit to the transcriptional response in Fig 1 in Powis et al. (2011) [61].
| Altered Amount (molecules) | Bmax (fold change) (mean ± SE) 1 | Hill Coefficient (mean ± SE) 1 | EC50 (nM) (mean ± SE) 1 | Transitional Dose Value from EC21 2 (nM) |
|---|---|---|---|---|
| Powis et al. data | 11.18 ± 0.1880 | 1.212 ± 0.1202 | 0.08508 ± 0.00592 | 0.02580 |
| Modeled results from altering cofactor amounts (Fig 1 and Fig 3A) | ||||
| 200 | 1.421 ± 0.0566 | 1.826 ± 0.2768 | 0.09873 ± 0.0091 | 0.04275 |
| 400 | 2.547 ± 0.1550 | 1.637 ± 0.3258 | 0.1050 ± 0.0153 | 0.04139 |
| 800 | 10.92 ± 0.2650 | 1.252 ± 0.0431 | 0.3134 ± 0.0164 | 0.07515 |
| 1000 | 10.33 ± 0.1531 | 1.418 ± 0.0684 | 0.2101 ± 0.0088 | 0.06443 |
| 1200 | 10.87 ± 0.0583 | 1.360 ± 0.0312 | 0.1659 ± 0.00327 | 0.05148 |
| 1300 | 10.94 ± 0.1599 | 1.310 ± 0.0826 | 0.1319 ± 0.0074 | 0.04105 |
| 1500 | 10.87 ± 0.2173 | 1.350 ± 0.1243 | 0.09446 ± 0.00756 | 0.03166 |
| 2000 | 11.87 ± 0.1820 | 1.431 ± 0.1089 | 0.05903 ± 0.00384 | 0.02182 |
| 3000 | 11.37 ± 0.2045 | 1.437 ± 0.1316 | 0.04963 ± 0.00386 | 0.01862 |
| 6000 | 11.62 ± 0.2525 | 1.519 ± 0.1722 | 0.05223 ± 0.00481 | 0.02054 |
| Modeled results from altering the number of Other cofactor binding sites (Fig 3B) | ||||
| 50000 | 9.335 ± 0.3997 | 1.784 ± 0.2106 | 0.1746 ± 0.0156 | 0.06815 |
| 20000 | 8.889 ± 0.2663 | 1.757 ± 0.1422 | 0.1782 ± 0.01112 | 0.07346 |
| 10000 | 10.07 ± 0.3268 | 1.565 ± 0.1191 | 0.1978 ± 0.0135 | 0.06736 |
| 7500 | 10.78 ± 0.3106 | 1.358 ± 0.07192 | 0.2395 ± 0.0149 | 0.06803 |
| 2500 | 10.60 ± 0.03626 | 1.276 ± 0.01765 | 0.1702 ± 0.00217 | 0.04929 |
| 1535 | 10.87 ± 0.2173 | 1.350 ± 0.1243 | 0.09446 ± 0.00756 | 0.03166 |
| 750 | 11.60 ± 0.1565 | 1.351 ± 0.08818 | 0.05439 ± 0.003197 | 0.01913 |
| 250 | 11.79 ± 0.1223 | 1.285 ± 0.0618 | 0.05867 ± 0.002662 | 0.01952 |
Fitted parameters are shown as the best-fit value ± standard error. The upper part of the table shows fits for a series of varying cofactor amounts. The lower part of the table shows fits for a series of varying competing non-AHR cofactor binding sites (Other). Fitting was conducted with Graphpad Prism. The rising portion of the curve was fit. It was not possible to obtain a Hill equation fit to the modeled results at 60 molecules of cofactor. The lower part of the table shows the effect of changing the number of competing binding sites for the cofactor.
1 For 1000 cofactor molecules, points below 3 nM TCDD were fit, and for 800 molecules and lesser amounts, points below 1 nM TCDD were fit. It was not possible to obtain a Hill equation fit to the modeled results at 60 molecules of cofactor.
2 Transitional dose values (TDVs) as a measure of threshold were estimated by projecting to the background response using the methods for the Hill model described in Simon et al., (2014). [78] The equations for estimating TDVs using background projection from Simon et al., 2014 are shown in Equation D in S1 File.
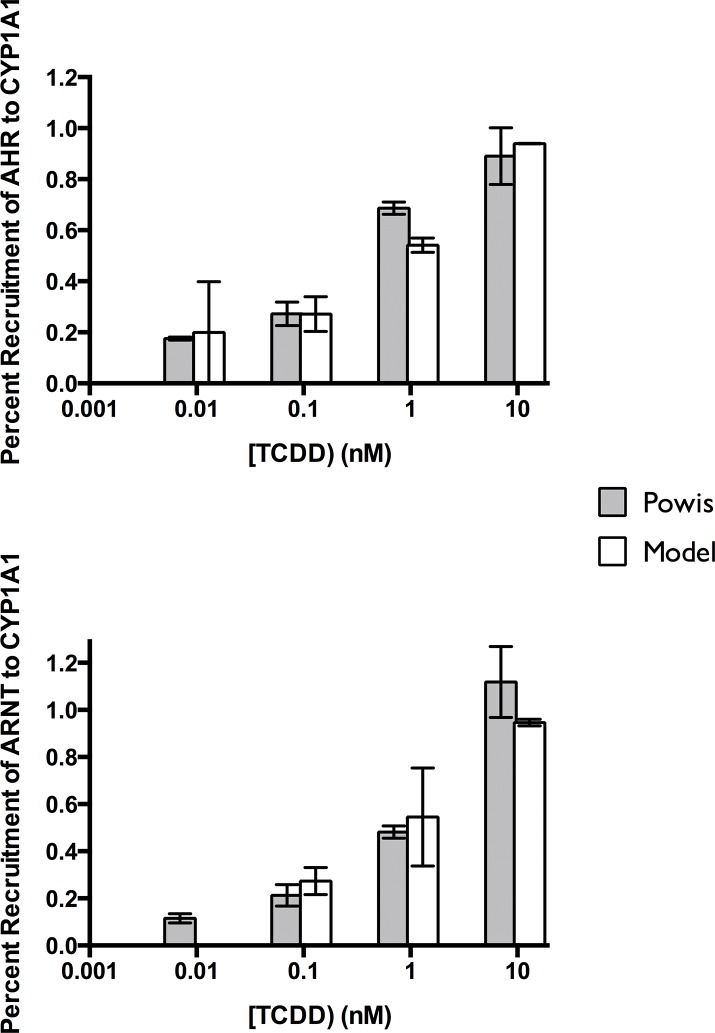
Comparison of Measured and Modeled Binding to DNA
A comparison of measurements of binding of both AHR and ARNT to XREs associated with CYP1A1 measured by ChIP experiments and corresponding model results is shown in Fig 2. The modeled result was very close to the measured ChIP results.
Fig 2. Comparison of ChIP results for AHR and ARNT from Fig 2A of Powis et al. (2011) [64] with those of the model.
ChIP results were estimated with Equation A and Equation B in S1 File. (A) Percent recruitment of AHR to CYP1A1; (B) Percent recruitment of ARNT to CYP1A1.
The value of percent recruitment represents the fraction of a particular transcription factor bound to DNA.[64] The calculation of percent recruitment from the model results is shown in the Equation A in S1 File.
The Effect of Altering Cofactor Concentrations or the Number of Binding Partners for the Cofactor other than AHR-ARNT
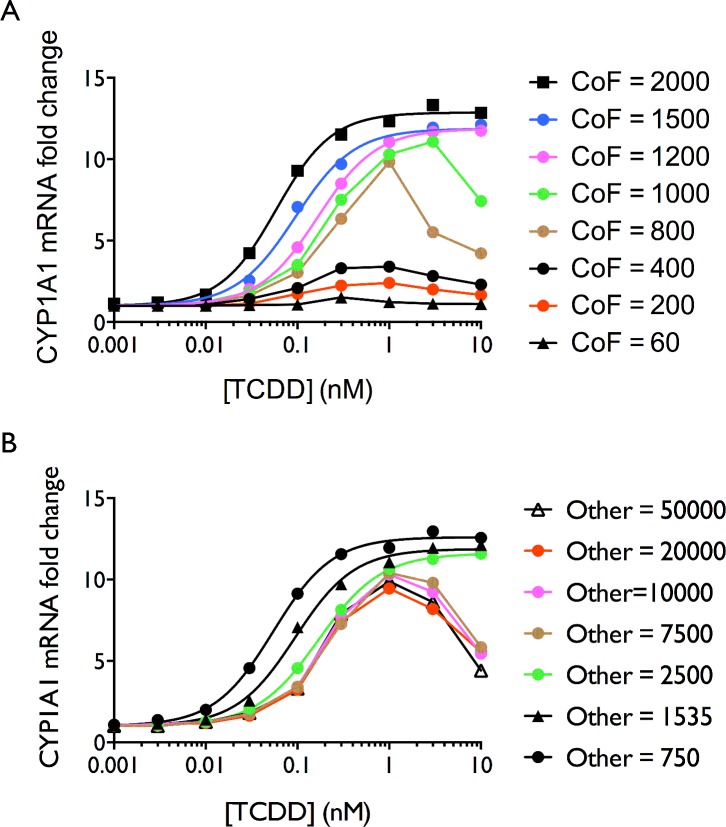
Squelching is apparent at cofactor amounts of 1000 molecules or less and appears as a reduction in the response at TCDD concentrations of 1 nM or greater (Fig 3A). The EC50 can change approximately four fold between cofactor amounts of 1000 and 3000 molecules without a change in the efficacy of the response (Table 1). When squelching occurs, the trend of EC50 values reverses and a reduction in EC50 values, i.e. higher potency, occurs with decreasing cofactor amounts when cofactor amounts are less than 1000 molecules.
Fig 3. Modeled transcriptional dose-response plots at varying amounts of cofactor and non-AHR-ARNT cofactor binding sites.
(A) Reduction in the amount of cofactor (CoF) at a constant concentration/amount of competing non-AHR binding proteins (1535 molecules). The modeled response and Hill equation fits are shown for cofactor amounts of 2000, 1500 and 1200 molecules. At 1000 molecules of cofactor and less, squelching was apparent, shown by a reduction in the responses at higher TCDD concentrations and the biphasic appearance of the DR curves. (B) Increasing the amount of competing non-AHR binding proteins (Other) also produced a squelching-like response at high ligand concentrations with squelching occurring at 7500 or more molecules of non-AHR cofactor binding proteins. The amount of cofactor was kept constant at 1500 molecules. The Hill equation fits are shown for competing non-AHR binding site (Other) amounts of 2500 or less.
Squelching is a reduction in the response at higher concentrations and results in the appearance of NMDRCs. Squelching was also observed to occur in the model in response to increasing the cofactor binding species other than AHR-ARNT (Table 1; Fig 3B) (Species #8 in Table A in S1 File). Squelching is a well-documented observation for the estrogen receptor and has been observed in many ToxCast results.[89]
Increasing the number of competing non-AHR cofactor binding sites (Other), induced squelching and NMDRCs at amounts of competing non-AHR binding sites at 7500 or more (Fig 3B).
The Roles of Cofactor and Competition in Squelching
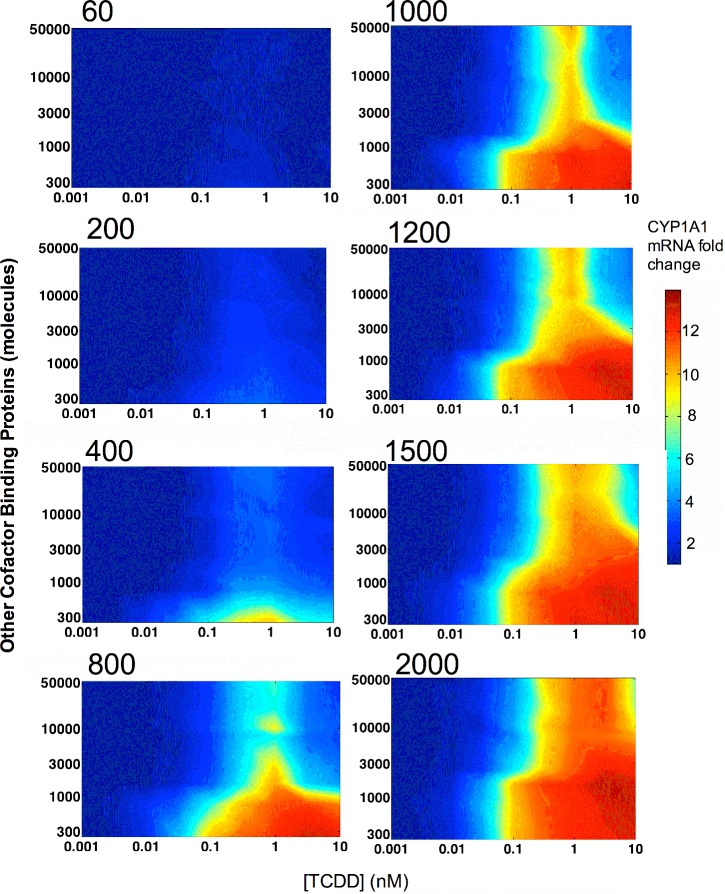
To demonstrate the interaction of cofactor availability, the model was exercised at a range of TCDD concentrations and various combinations of the amounts of cofactor and non-AHR cofactor binding proteins that would compete with the AHR-ARNT complex for available cofactor. With these model results, contour maps of the transcriptional response were developed for varying amounts of both cofactor and non-AHR cofactor binding proteins (Fig 4) (Species #7 and #8 in Table A in S1 File). Squelching is apparent at all concentrations of cofactor. Hence, squelching occurs as an interaction between the amount of cofactor and the amount of competing non-AHR binding proteins.
Fig 4. Contour plots of the modeled transcriptional responses showing the relationship between number of cofactor molecules and the number of competing non-AHR (Other) binding proteins.
The x-axes show the applied concentration of TCDD. The y-axes show the amount of other binding proteins available in the cell. The fold change in CYP1A1 mRNA is represented by the colors on the plots and the color bar to the right. The number at the upper left of each plot shows the number of molecules of cofactor.
The Effect of Altering Ligand Activated AHR on Dose-Response Curves and Squelching
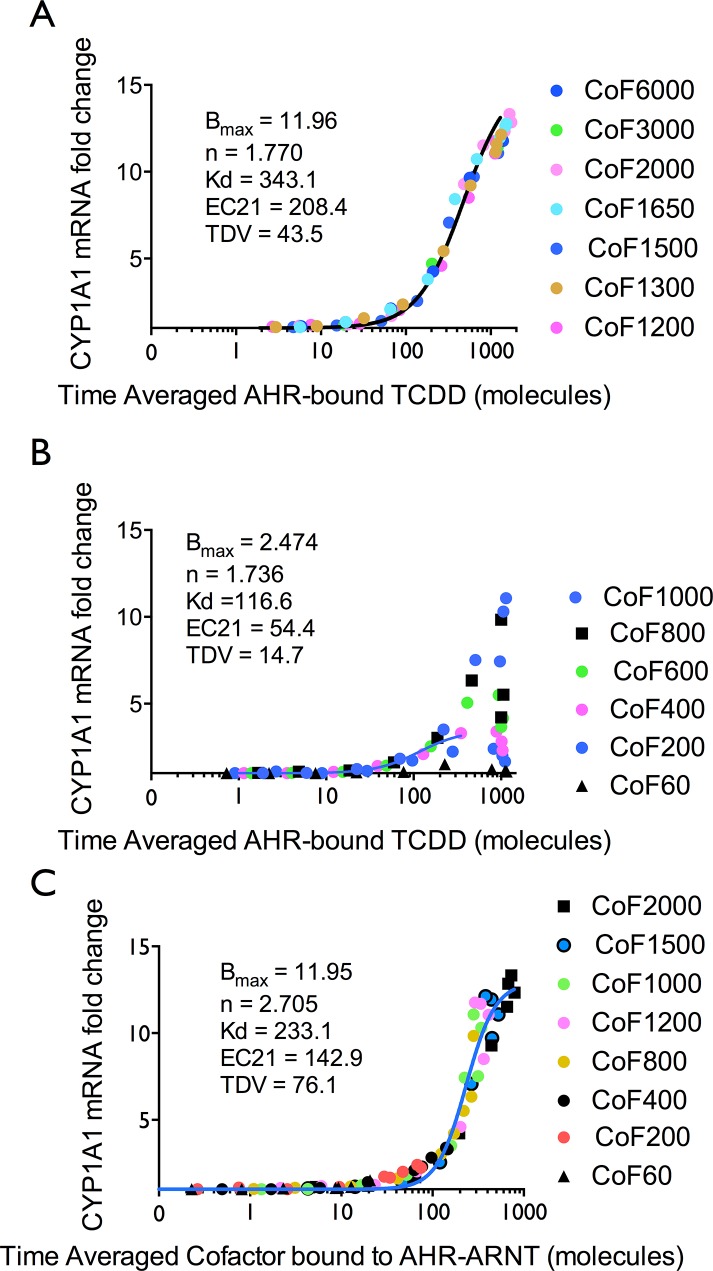
To explore how cofactor availability acts to produce shifts in the dose-response curves and squelching, the transcriptional response of the model was plotted against the time-averaged number of ligand-activated AHR molecules. Hence, the total number of ligand-activated AHR molecules as a time average was used as the dose term and the mRNA fold change was used as the response term. A first order Hill model was fit to the dose-response of the aggregated data for responses when no squelching was apparent (Fig 5A) and for responses showing squelching (Fig 5B). When no squelching occurred, the aggregate response was well fit by a Hill equation and the transcriptional responses were consistent at all doses of TCDD (Fig 5A). When squelching did occur (1000 cofactor molecules and less), considerable divergence in the responses to higher TCDD concentrations was observed (Fig 5B). A Hill model was fit to the data with 400 ligand-bound AHR molecules or less and the fit indicated an approximately fivefold reduction in efficacy in the squelched responses.
Fig 5. Transcriptional dose-response using time-averaged species from the model results to demonstrate that squelching occurs at the cofactor-binding step.
Each plot was fit to a Hill function (details in text) and the EC21 and transitional dose values are shown. [87, 88] (A) Plot of CYP1A1 mRNA fold change vs. time-averaged ligand-bound AHR for responses without squelching. (B) Plot of time-averaged ligand-bound AHR for responses with squelching. (C) Plot of time-averaged cofactor bound to AHR-ARNT and thus contributing to CYP1A1 transcription.
When the time-averaged amount of cofactor bound to AHR-ARNT anywhere in the cell (Equation C in S1 File) is used as the dose term, the responses become very different, and the aggregate dose-response, both for those squelching and non-squelching responses remain consistent and were fit to a Hill function (Fig 5C). Thus, these plots demonstrate that the locus at which squelching occurs is cofactor binding to AHR-ARNT.
Discussion and Conclusions
Modulation and Variability of the Dose-Response for Receptor-mediated Effects
When one observes the dose-response for the formation of all ligand-bound AHR averaged over the duration of simulation (Fig 5A; Equation B in S1 File), no squelching is observed, and the dose-response curves have sufficiently similar values for EC50 and the Hill coefficient that these parameters can be fit in common. (Fig A in S1 File). Hence, squelching occurs at a later step than ligand-receptor binding.
Lately there has been much discussion about the lack of knowledge of events in the low dose region—especially regarding binding of nuclear receptors and subsequent gene expression.[39] This model demonstrates that because of the limited availability of transcription cofactors and the stochastic nature of molecular events involved in gene expression, transitional dose values that represent response thresholds need to be considered as variable or stochastic values rather than a fixed value.[87, 88] Stochastic variation in a population of T-cells has been observed and limited numbers of regulator molecules associated with transcription has been observed to introduce noise into cellular processes.[90–92]
Histograms of transcriptional responses of all 100 cells indicate that only a single cell shows an increase in mRNA copy number at the lowest dose modeled (Fig B in S1 File). Some concern has been expressed about endocrine effects in the low dose region.[39, 41] In the low dose region, greater variability in mRNA copy number can be observed in the model results than predicted by Poisson statistics (Table E in S1 File, 4th and 5th columns). However, when considered the overall response of the modeled tissue or cell population, consisting of 100 modeled cells, the total number of mRNA copies is well within the expected range (Table E in S1 File, last two columns). Hence, the model results indicate that on a tissue or cell population basis, gene expression responses will likely be very small or non-existent in the low dose region and that these concerns may be misplaced.
As well as TCDD concentration, the transcriptional response depends to a fairly large extent on the availability of cofactor and other cellular resources. However, at the lowest TCDD concentration at which the model was executed, the overall response of the tissue is absent (Fig 4). The availability of cellular resources can affect both the efficacy and potency of the response and this fact should be considered for interpretation of in vitro high throughput assays.[89, 93, 94] Recently, assays have been developed for transcription cofactors and these may have the potential for judging the performance of in vitro assays that are increasingly being used for regulatory purposes.[95–97]
Because of the changing needs of organisms, the context for appropriate gene expression is inconstant, and this context, reflected by the panoply of genes expressed given the limitations on resources, will result in ever changing levels of available cofactor and thus ever changing “thresholds.” Stochastic variation in these “thresholds” combined with the difficulty in knowing what level of response is biological significant in what particular context is one of the factors that accounts for the focus on discovering meaningful methods for filtering gene expression data. [98–101]
A method for estimating transitional dose values (TDVs) for dose-response data that follows a Hill equation as potential estimates of thresholds has been developed and was used here to estimate transitional dose values for TCDD concentration thresholds for the transcriptional response (Table 1).[87, 88] In addition, TDVs were estimated for the time-averaged number of ligand-bound AHR molecules and the number of AHR-ARNT complexes with bound cofactor. The method and equations are provided in Equation D in S1 File. The calculated TDVs for TCDD concentration are shown in Table 1.
Identity of the Cofactor and Other Binding Sites
This model was developed to explore the role of competition in ligand-activated gene transcription. Hankinson (2005) provides a list of possible candidates for the cofactor including histone acetylases, histone methyl transferases or receptor-interacting protein 140 (RIP140).[36] Recently, four cellular factors were identified in Hepa1c1c7 cells as crucial for CYP1A1 mRNA induction independent of effects on AHR expression. These factors are SIN3 homolog A (SIN3A), phosphoducin (PDC), transmembrane protein 5 (TMEM5), and the CD9 cell surface glycoprotein (CD9). [102] Although the model could be altered to include cofactor involvement in AHR induction, some cofactors such as transmembrane factor 20 (TCF20) and crystalline gamma D (CRYGD) are necessary for induction of both CYP1A1 and AHR mRNA and protein. [102]
Nuclear factor erythroid 2-realted factor 2 (Nrf2) interacts with the AHR in its role in the expression of both CYP1A1 and NQO1.[103–105] Nrf2 is a transcription factor involved in responses to oxidative and electrophilic stress, and inflammation.[106] Nrf2 interacts with both the AHR and the estrogen receptor. [107] In Nrf2-null mice, the transcriptional response to TCDD for CYP1A1 was reduced compared to that in wild-type mice. [103, 105, 107] Hence, Nrf2 is another potential candidate for the identity of the cofactor in the model.
Yet another candidate for the identity of the cofactor in the model is nuclear factor-κB (NF-κB). Inhibition of NF-κB reduced the expression of mRNA for both AHR and CYP1A1 in U-937 derived dendritic cells.[108]
2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) co-localizes with AHR in the nucleus. TiPARP enhances the production of CYP1A1 mRNA but also increases the proteolytic destruction of ligand-bound AHR. [109] This negative feedback mechanism could also be included in the model.
Oscillations in ChIP Data
At present, data on binding of transcription factors to DNA is obtained by two primary methods—chromatin immuno-precipitation and fluorescence techniques—with the former conducted on populations of cells and the latter on single cells. These two methods give very different kinetic results. Hager (2009) has proposed that several explanations for the oscillations of transcription factor binding observed in ChIP experiments.[4] ChIP experiments are often expressed as fractional or percent recruitment to a single XRE or limited set of XREs with total chromatin input as the denominator.
Oscillations in the binding of either AHR, ARNT, cofactor or RNA polymerase to CYP1A1 were not observed in any of the model runs.
Modeling Transcription
Current knowledge of the process of transcription suggests it is complex and not yet well understood[78, 82–84, 110, 111] In the model, transcription of CYP1A1 was represented by four different promoter states, each with a slightly different rate constant. This simplified model was an attempt to introduce variable transcription rates into the model. For the sake of parsimony, more complex models of transcription were not used.[83, 110, 111] Rates of mRNA export from the transcription complex and degradation within the complex were determined empirically to match mRNA fold change data at 6 hours in T47-D cells.[64] Distributions of CYP1A1 mRNA copy number per cell at various TCDD concentrations were consistent with Poisson distributions similar to that measured in other eukaryotic cells and observed by other modelers (Fig B in S1 File; Table E in S1 File).[112–115]
Examples of Possible Competition for Cofactors
Whether ARNT is considered a transcription factor or a cofactor is somewhat moot. The basic helix-loop-helix-PAS proteins appear to have been evolutionarily conserved and serve multiple roles. ARNT dimerizes with the bHLH-PAS protein hypoxia inducible factor 1 (HIF-1) that acts to produce cellular responses to hypoxia.[36, 116] ARNT also acts as a coactivator for both ERα- and ERβ-mediated transcription.[117, 118] AHR-mediated transcription can be modulated by recruitment of a variety of cofactors including CBP/p300, p160/SRC-1, NCoA2, p-CIP and RIP140.[16, 34, 37]
Nonmonotonic U-shaped dose response curves have been shown to occur for steroid hormone-mediated gene expression. Such shapes in the dose-response occur when there exist receptors unoccupied by endogenous hormones and recruitment of cofactors by receptors bound to xenobiotic ligands is weaker than to hormone-bound receptor.[44] For the androgen receptor, the presence of mixed ligand heterodimers that are transcriptionally inactive along with homodimers with bound hormone and xenobiotic ligand lead to J-shaped dose-response curves reminiscent of hormesis.[45] Receptor dimerization appears to be an inherently non-linear process and the normal milieu of endogenous hormones along with a xenobiotic ligand produce a variety of shapes of dose-response curves.[43] The situation modeled here, competition for one or more coregulatory proteins, only occurs at high concentrations of ligand and high receptor occupancy. At low concentrations of ligand, coregulatory proteins are sufficiently available to ligand-bound receptors for normal transcriptional/signaling responses.[119, 120]
ARNT serves as a coactivator for ERα and ERβ, similar to the generic cofactor modeled here.[117] Hence, one could imagine a situation where the physiological need for estrogen-mediated gene expression might result in a competition for ARNT such that the AHR response to endogenous or exogenous ligands might be muted.
Other Possible Uses of the Model
Transforming growth factor-β1 (TGF-β1) appears to act as an inhibitor or repressor of AHR-mediated effects in a cell line derived from human prostate cells.[121] The cross-talk between TGF-β1 and AHR is complex, but this could be explored initially with this model as a second cofactor with repressive effects.
2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) co-localizes with AHR in the nucleus. TiPARP enhances the production of CYP1A1 mRNA but also increases the proteolytic destruction of ligand-bound AHR.[109] This negative feedback mechanism could also be included in the model.
Other processes that could be included in the model are:
Induction of AHR by including AHR synthesis dependent on AHR activation level;
Repression of AHR by TIPARP[73] or other possible mechanisms; and,
Transcription of other genes in the AHR core battery at different transcription rates.
While the model is somewhat complex, the actual inter-relationships between receptors involved in gene induction are even more extensive and inter-related. Extensive crosstalk and overlap exists between the AHR, ERα and Nrf2.[107] A specific model of this crosstalk could potentially be developed and could be used to explore the effects of ligands for each of these receptors presented alone or in combination.
Stochastic gene expression in evolution
Stochastic effects resulting from transcription factor binding likely produce heterogeneity in gene expression across cells within organs or tissues in metazoans. This heterogeneity could result from stochasticity in gene activation and inactivation processes and could be an overall compensatory mechanism for the limitations on resources within a single cell. Hence, at the organ tissue levels, the ability to respond to stimuli or environmental changes by gene expression would be maintained.[113, 122–124]
Within a single cell, a small number of mRNA molecules can potentially be amplified into a sufficient number of protein molecules to produce a phenotypic change in that cell. Cell-to-cell signaling may occur either by extracellular chemical signals such as cytokines or through gap junctions between adjacent cells. One possible function of such signaling could be to regulate gene expression on a tissue-wide basis. The fact that gene expression in metazoans occurs within nuclear structures that maintain locally high concentrations of transcription components is testament to these resource limitations.[19, 20, 125] The model results suggest that these resource limitations would limit any tissue-level phenotypic responses.
If the oscillations observed in ChIP data represent different levels of nuclear receptor binding in cell populations within tissues, this could be another aspect of this mechanism of tissue-wide coordination.[4] What these model results also suggest is that generalizations about mechanisms or even responses across different levels of biological organization (e.g., molecule, cell, tissue, organism) may lead to faulty conclusions and begs the question: What else is going that leads to an observed phenotypic change?
Supporting Information
This file contains: Code A, MATLAB Script for Model Runs. Code B, SBML Code for the Model. Equation A, Calculation of Percent Recruitment from the model results. Equation B, Calculation of Time-Averaged Numbers of ligand-bound AHR. Equation C, Calculation of Time-Averaged Numbers of cofactor bound to AHR-ARNT. Equation D, Equations used for Baseline Projection to determine Transitional Dose Values from Simon et al., (2014). Fig A, Dose-response to the number of ligand-bound AHR molecules as a function of TCDD concentration and fitted Hill functions. Fig B, Histograms of CYP1A1 mRNA copy number induced by TCDD. Table A, Compartments and Species. Table B, Reactions and Rate Constants. Table C, Comparison of classical rate constants, molecular rate constants and propensities. Table D, Hill Model fits of the Percentage of Cells with one or more CYP XRE bound. Table E, Goodness of fit of Poisson distribution for modeled CYP1A1 mRNA copy number.
(DOCX)
Acknowledgments
We wish to thank Dr. Stoney S. Simons, Jr. for his helpful comments on an earlier draft of this paper. This work was funded by a grant to TWS from the Dow Chemical Company. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
The MATLAB script used to run the model is included in the Supporting Information.
Funding Statement
This work was funded by a grant to TWS from the Dow Chemical Company through PO93437034. Ted Simon LLC & The Dow Chemical Company provided support in the form of salaries for authors TWS, RAB and JCR retrospectively, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the author contributions section.
References
- 1. Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15(2):69–81. 10.1038/nrg3623 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolitz SE, Lauffenburger DA. Measurement and Modeling of Signaling at the Single-Cell Level. Biochemistry. 2012. 10.1021/bi300846p . [DOI] [PMC free article] [PubMed]
- 3. Coulon A, Chow CC, Singer RH, Larson DR. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet. 2013;14(8):572–84. 10.1038/nrg3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35(6):741–53. 10.1016/j.molcel.2009.09.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Selimkhanov J, Hasty J, Tsimring LS. Recent advances in single-cell studies of gene regulation. Curr Opin Biotechnol. 2012;23(1):34–40. 10.1016/j.copbio.2011.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bakstad D, Adamson A, Spiller DG, White MRH. Quantitative measurement of single cell dynamics. Curr Opin Biotechnol. 2012;23(1):103–9. 10.1016/j.copbio.2011.11.007 . [DOI] [PubMed] [Google Scholar]
- 7. Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: sensors of environmental and developmental signals. Annual review of pharmacology and toxicology. 2000;40:519–61. 10.1146/annurev.pharmtox.40.1.519 . [DOI] [PubMed] [Google Scholar]
- 8. Hahn ME. Aryl hydrocarbon receptors: diversity and evolution. Chem Biol Interact. 2002;141(1–2):131–60. . [DOI] [PubMed] [Google Scholar]
- 9. Wincent E, Amini N, Luecke S, Glatt H, Bergman J, Crescenzi C, et al. The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-formylindolo[3,2-b]carbazole is present in humans. J Biol Chem. 2009;284(5):2690–6. Epub 2008/12/05. doi: M808321200 [pii] 10.1074/jbc.M808321200 . [DOI] [PubMed] [Google Scholar]
- 10. Wincent E, Bengtsson J, Mohammadi Bardbori A, Alsberg T, Luecke S, Rannug U, et al. Inhibition of cytochrome P4501-dependent clearance of the endogenous agonist FICZ as a mechanism for activation of the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A. 2012;109(12):4479–84. 10.1073/pnas.1118467109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sorg O. AhR signalling and dioxin toxicity. Toxicol Lett. 2014;230(2):225–33. 10.1016/j.toxlet.2013.10.039 . [DOI] [PubMed] [Google Scholar]
- 12. Bock KW. The human Ah receptor: hints from dioxin toxicities to deregulated target genes and physiological functions. Biol Chem. 2013;394(6):729–39. 10.1515/hsz-2012-0340 . [DOI] [PubMed] [Google Scholar]
- 13. Budinsky RA, Schrenk D, Simon T, Van den Berg M, Reichard JF, Silkworth JB, et al. Mode of action and dose-response framework analysis for receptor-mediated toxicity: The aryl hydrocarbon receptor as a case study. Crit Rev Toxicol. 2014;44(1):83–119. 10.3109/10408444.2013.835787 . [DOI] [PubMed] [Google Scholar]
- 14. Aoyagi S, Archer TK. Dynamics of coactivator recruitment and chromatin modifications during nuclear receptor mediated transcription. Mol Cell Endocrinol. 2008;280(1–2):1–5. 10.1016/j.mce.2007.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J, Kinyamu HK, Archer TK. Changes in attitude, changes in latitude: nuclear receptors remodeling chromatin to regulate transcription. Mol Endocrinol. 2006;20(1):1–13. 10.1210/me.2005-0192 . [DOI] [PubMed] [Google Scholar]
- 16. Beischlag TV, Wang S, Rose DW, Torchia J, Reisz-Porszasz S, Muhammad K, et al. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol Cell Biol. 2002;22(12):4319–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen W, Roeder RG. Mediator-dependent nuclear receptor function. Semin Cell Dev Biol. 2011;22(7):749–58. 10.1016/j.semcdb.2011.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19(5):631–42. 10.1016/j.molcel.2005.07.018 . [DOI] [PubMed] [Google Scholar]
- 19. Edelman LB, Fraser P. Transcription factories: genetic programming in three dimensions. Curr Opin Genet Dev. 2012;22(2):110–4. 10.1016/j.gde.2012.01.010 . [DOI] [PubMed] [Google Scholar]
- 20. Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145(5):787–99. 10.1016/j.cell.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reddy KL, Singh H. Using molecular tethering to analyze the role of nuclear compartmentalization in the regulation of mammalian gene activity. Methods. 2008;45(3):242–51. 10.1016/j.ymeth.2008.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21(23):3027–43. 10.1101/gad.1604607 . [DOI] [PubMed] [Google Scholar]
- 23. Stein GS, van Wijnen AJ, Imbalzano AN, Montecino M, Zaidi SK, Lian JB, et al. Architectural genetic and epigenetic control of regulatory networks: compartmentalizing machinery for transcription and chromatin remodeling in nuclear microenvironments. Crit Rev Eukaryot Gene Expr. 2010;20(2):149–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Assenza S, Gutiérrez R, Gómez-Gardeñes J, Latora V, Boccaletti S. Emergence of structural patterns out of synchronization in networks with competitive interactions. Sci Rep. 2011;1:99 10.1038/srep00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De Vos D, Bruggeman FJ, Westerhoff HV, Bakker BM. How molecular competition influences fluxes in gene expression networks. PLoS One. 2011;6(12):e28494 10.1371/journal.pone.0028494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Genot AJ, Fujii T, Rondelez Y. Computing with competition in biochemical networks. Phys Rev Lett. 2012;109(20):208102 . [DOI] [PubMed] [Google Scholar]
- 27. Rowland MA, Fontana W, Deeds EJ. Crosstalk and competition in signaling networks. Biophys J. 2012;103(11):2389–98. 10.1016/j.bpj.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Treuter E, Johansson L, Thomsen JS, Wärnmark A, Leers J, Pelto-Huikko M, et al. Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of TRAP and p160 coactivator complexes. J Biol Chem. 1999;274(10):6667–77. . [DOI] [PubMed] [Google Scholar]
- 29. Simons SS Jr. The importance of being varied in steroid receptor transactivation. TIPS. 2003;24(5):253–9. 10.1016/S0165-6147(03)00101-9 [DOI] [PubMed] [Google Scholar]
- 30. Simons SS Jr. How much is enough? Modulation of dose-response curve for steroid receptor-regulated gene expression by changing concentrations of transcription factor. Curr Top Med Chem. 2006;6(3):271–85. Epub 2006/03/07. . [DOI] [PubMed] [Google Scholar]
- 31. Simons SS Jr. What goes on behind closed doors: physiological versus pharmacological steroid hormone actions. Bioessays. 2008;30(8):744–56. Epub 2008/07/16. 10.1002/bies.20792 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ong KM, Blackford JA Jr, Kagan BL, Simons SS Jr, Chow CC. A theoretical framework for gene induction and experimental comparisons. Proc Natl Acad Sci U S A. 2010;107(15):7107–12. Epub 2010/03/31. doi: 0911095107 [pii] 10.1073/pnas.0911095107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lebedeva G, Yamaguchi A, Langdon SP, Macleod K, Harrison DJ. A model of estrogen-related gene expression reveals non-linear effects in transcriptional response to tamoxifen. BMC Syst Biol. 2012;6(1):138 10.1186/1752-0509-6-138 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit Rev Eukaryot Gene Expr. 2008;18(3):207–50. Epub 2008/06/11. doi: 6f28b0540a5e6e63,5ec7b3e06964879d [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Puga A, Ma C, Marlowe JL. The aryl hydrocarbon receptor cross-talks with multiple signal transduction pathways. Biochem Pharmacol. 2009;77(4):713–22. Epub 2008/09/27. doi: S0006-2952(08)00590-X [pii] 10.1016/j.bcp.2008.08.031 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hankinson O. Role of coactivators in transcriptional activation by the aryl hydrocarbon receptor. Arch Biochem Biophys. 2005;433(2):379–86. Epub 2004/12/08. 10.1016/j.abb.2004.09.031 . [DOI] [PubMed] [Google Scholar]
- 37. Matthews J, Wihlen B, Thomsen J, Gustafsson JA. Aryl hydrocarbon receptor-mediated transcription: ligand-dependent recruitment of estrogen receptor alpha to 2,3,7,8-tetrachlorodibenzo-p-dioxin-responsive promoters. Mol Cell Biol. 2005;25(13):5317–28. Epub 2005/06/21. doi: 25/13/5317 [pii] 10.1128/MCB.25.13.5317-5328.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pandey PK, Udayakumar TS, Lin X, Sharma D, Shapiro PS, Fondell JD. Activation of TRAP/mediator subunit TRAP220/Med1 is regulated by mitogen-activated protein kinase-dependent phosphorylation. Mol Cell Biol. 2005;25(24):10695–710. 10.1128/MCB.25.24.10695-10710.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vandenberg LN, Colborn T, Hayes TB, Heindel JJ, Jacobs DR, Lee D-H, et al. Hormones and endocrine-disrupting chemicals: low-dose effects and nonmonotonic dose responses. Endocr Rev. 2012;33(3):378–455. 10.1210/er.2011-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vandenberg LN. Low-dose effects of hormones and endocrine disruptors. Vitam Horm. 2014;94:129–65. 10.1016/B978-0-12-800095-3.00005-5 . [DOI] [PubMed] [Google Scholar]
- 41. Birnbaum LS. Environmental chemicals: evaluating low-dose effects. Environ Health Perspect. 2012;120(4):A143–4. 10.1289/ehp.1205179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rhomberg LR, Goodman JE. Low-dose effects and nonmonotonic dose-responses of endocrine disrupting chemicals: has the case been made? Regul Toxicol Pharmacol. 2012;64(1):130–3. 10.1016/j.yrtph.2012.06.015 . [DOI] [PubMed] [Google Scholar]
- 43. Li L, Andersen ME, Heber S, Zhang Q. Non-monotonic dose-response relationship in steroid hormone receptor-mediated gene expression. J Mol Endocrinol. 2007;38(5):569–85. 10.1677/JME-07-0003 . [DOI] [PubMed] [Google Scholar]
- 44. Kohn MC, Melnick RL. Biochemical origins of the non-monotonic receptor-mediated dose-response. J Mol Endocrinol. 2002;29(1):113–23. . [DOI] [PubMed] [Google Scholar]
- 45. Conolly RB, Lutz WK. Nonmonotonic dose-response relationships: mechanistic basis, kinetic modeling, and implications for risk assessment. Toxicol Sci. 2004;77(1):151–7. 10.1093/toxsci/kfh007 . [DOI] [PubMed] [Google Scholar]
- 46. Dietrich DR, von Aulock S, Marquardt H, Blaauboer B, Dekant W, Kehrer J, et al. Scientifically unfounded precaution drives European Commission's recommendations on EDC regulation, while defying common sense, well-established science and risk assessment principles. Toxicol In Vitro. 2013;27(7):2110–4. 10.1016/j.tiv.2013.07.001 . [DOI] [PubMed] [Google Scholar]
- 47. Borgert CJ, Baker SP, Matthews JC. Potency matters: thresholds govern endocrine activity. Regul Toxicol Pharmacol. 2013;67(1):83–8. 10.1016/j.yrtph.2013.06.007 . [DOI] [PubMed] [Google Scholar]
- 48. Ullah M, Schmidt H, Cho K- H, Wolkenhauer O. Deterministic modelling and stochastic simulation of biochemical pathways using MATLAB. IEE Proceedings—Systems Biology. 2006;153(2):53 10.1049/ip-syb:20050064 [DOI] [PubMed] [Google Scholar]
- 49. Gillespie DT. Stochastic simulation of chemical kinetics. Annu Rev Phys Chem. 2007;58:35–55. 10.1146/annurev.physchem.58.032806.104637 . [DOI] [PubMed] [Google Scholar]
- 50. Ball DA, Adames NR, Reischmann N, Barik D, Franck CT, Tyson JJ, et al. Measurement and modeling of transcriptional noise in the cell cycle regulatory network. Cell Cycle. 2013;12(19):3203–18. 10.4161/cc.26257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sanchez A, Choubey S, Kondev J. Stochastic models of transcription: from single molecules to single cells. Methods. 2013;62(1):13–25. 10.1016/j.ymeth.2013.03.026 . [DOI] [PubMed] [Google Scholar]
- 52. Dong D, Shao X, Deng N, Zhang Z. Gene expression variations are predictive for stochastic noise. Nucleic Acids Res. 2011;39(2):403–13. 10.1093/nar/gkq844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ribeiro AS. Stochastic and delayed stochastic models of gene expression and regulation. Math Biosci. 2010;223(1):1–11. 10.1016/j.mbs.2009.10.007 . [DOI] [PubMed] [Google Scholar]
- 54. Blackford JA, Guo C, Zhu R, Dougherty EJ, Chow CC, Simons SS. Identification of Location and Kinetically Defined Mechanism of Cofactors and Reporter Genes in the Cascade of Steroid-regulated Transactivation. J Biol Chem. 2012;287(49):40982–95. 10.1074/jbc.M112.414805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rondelez Y. Competition for catalytic resources alters biological network dynamics. Phys Rev Lett. 2012;108(1):018102 . [DOI] [PubMed] [Google Scholar]
- 56. Zhang Z, Sun Y, Cho Y-W, Chow CC, Simons SS. PA1: a New Competitive Decelerator Acting at More Than One Step To Impede Glucocorticoid Receptor-Mediated Transactivation. J Biol Chem. 2012;288(1):42–58. 10.1074/jbc.M112.427740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dilworth FJ, Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20(24):3047–54. 10.1038/sj.onc.1204329 . [DOI] [PubMed] [Google Scholar]
- 58. Kang J, Xu B, Yao Y, Lin W, Hennessy C, Fraser P, et al. A dynamical model reveals gene co-localizations in nucleus. PLoS Comput Biol. 2011;7(7):e1002094 10.1371/journal.pcbi.1002094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Meyer ME, Gronemeyer H, Turcotte B, Bocquel MT, Tasset D, Chambon P. Steroid hormone receptors compete for factors that mediate their enhancer function. Cell. 1989;57(3):433–42. . [DOI] [PubMed] [Google Scholar]
- 60. Zhang Z, Teng CT. Estrogen receptor alpha and estrogen receptor-related receptor alpha1 compete for binding and coactivator. Mol Cell Endocrinol. 2001;172(1–2):223–33. . [DOI] [PubMed] [Google Scholar]
- 61. Min G, Kim H, Bae Y, Petz L, Kemper JK. Inhibitory cross-talk between estrogen receptor (ER) and constitutively activated androstane receptor (CAR). CAR inhibits ER-mediated signaling pathway by squelching p160 coactivators. J Biol Chem. 2002;277(37):34626–33. 10.1074/jbc.M205239200 . [DOI] [PubMed] [Google Scholar]
- 62. Charlier TD. Importance of steroid receptor coactivators in the modulation of steroid action on brain and behavior. Psychoneuroendocrinology. 2009;34 Suppl 1:S20–9. 10.1016/j.psyneuen.2009.05.004 . [DOI] [PubMed] [Google Scholar]
- 63. Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335(6192):683–9. 10.1038/335683a0 . [DOI] [PubMed] [Google Scholar]
- 64. Powis M, Celius T, Matthews J. Differential ligand-dependent activation and a role for Y322 in aryl hydrocarbon receptor-mediated regulation of gene expression. Biochem Biophys Res Commun. 2011;410(4):859–65. Epub 2011/06/28. 10.1016/j.bbrc.2011.06.079 . [DOI] [PubMed] [Google Scholar]
- 65. Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–34. Epub 2003/01/24. 10.1146/annurev.pharmtox.43.100901.135828 100901.135828 [pii]. . [DOI] [PubMed] [Google Scholar]
- 66. Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci. 2011;124(1):1–22. 10.1093/toxsci/kfr218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dere E, Lo R, Celius T, Matthews J, Zacharewski TR. Integration of genome-wide computation DRE search, AhR ChIP-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC Genomics. 2011;12:365 10.1186/1471-2164-12-365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dere E, Lee AW, Burgoon LD, Zacharewski TR. Differences in TCDD-elicited gene expression profiles in human HepG2, mouse Hepa1c1c7 and rat H4IIE hepatoma cells. BMC Genomics. 2011;12:193 10.1186/1471-2164-12-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lo R, Celius T, Forgacs AL, Dere E, MacPherson L, Harper P, et al. Identification of aryl hydrocarbon receptor binding targets in mouse hepatic tissue treated with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Appl Pharmacol. 2011;257(1):38–47. Epub 2011/09/06. doi: S0041-008X(11)00314-0 [pii] 10.1016/j.taap.2011.08.016 . [DOI] [PubMed] [Google Scholar]
- 70.Lo R, Matthews J. High-resolution genome-wide mapping of AHR and ARNT binding sites by ChIP-Seq. Toxicol Sci. 2012. 10.1093/toxsci/kfs253 . [DOI] [PubMed]
- 71. Kimura H, Tao Y, Roeder RG, Cook PR. Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol Cell Biol. 1999;19(8):5383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Karchner SI, Jenny MJ, Tarrant AM, Evans BR, Kang HJ, Bae I, et al. The active form of human aryl hydrocarbon receptor (AHR) repressor lacks exon 8, and its Pro 185 and Ala 185 variants repress both AHR and hypoxia-inducible factor. Mol Cell Biol. 2009;29(13):3465–77. Epub 2009/04/22. doi: MCB.00206-09 [pii] 10.1128/MCB.00206-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. MacPherson L, Ahmed S, Tamblyn L, Krutmann J, Forster I, Weighardt H, et al. Aryl hydrocarbon receptor repressor and TiPARP (ARTD14) use similar, but also distinct mechanisms to repress aryl hydrocarbon receptor signaling. International journal of molecular sciences. 2014;15(5):7939–57. 10.3390/ijms15057939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tigges J, Weighardt H, Wolff S, Gotz C, Forster I, Kohne Z, et al. Aryl hydrocarbon receptor repressor (AhRR) function revisited: repression of CYP1 activity in human skin fibroblasts is not related to AhRR expression. The Journal of investigative dermatology. 2013;133(1):87–96. 10.1038/jid.2012.259 . [DOI] [PubMed] [Google Scholar]
- 75. Bernshausen T, Jux B, Esser C, Abel J, Fritsche E. Tissue distribution and function of the Aryl hydrocarbon receptor repressor (AhRR) in C57BL/6 and Aryl hydrocarbon receptor deficient mice. Archives of toxicology. 2006;80(4):206–11. 10.1007/s00204-005-0025-5 . [DOI] [PubMed] [Google Scholar]
- 76. Ribeiro AS, Smolander O-P, Rajala T, Häkkinen A, Yli-Harja O. Delayed stochastic model of transcription at the single nucleotide level. J Comput Biol. 2009;16(4):539–53. 10.1089/cmb.2008.0153 . [DOI] [PubMed] [Google Scholar]
- 77. Murugan R. Stochastic transcription initiation: Time dependent transcription rates. Biophys Chem. 2006;121(1):51–6. 10.1016/j.bpc.2005.12.010 . [DOI] [PubMed] [Google Scholar]
- 78. Voliotis M, Cohen N, Molina-París C, Liverpool TB. Fluctuations, pauses, and backtracking in DNA transcription. Biophys J. 2008;94(2):334–48. 10.1529/biophysj.107.105767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gorski SA, Snyder SK, John S, Grummt I, Misteli T. Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Mol Cell. 2008;30(4):486–97. 10.1016/j.molcel.2008.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Grünwald D, Singer RH, Rout M. Nuclear export dynamics of RNA-protein complexes. Nature. 2011;475(7356):333–41. 10.1038/nature10318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hocine S, Singer RH, Grünwald D. RNA processing and export. Cold Spring Harb Perspect Biol. 2010;2(12):a000752 10.1101/cshperspect.a000752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ya Ben-Ari, Brody Y, Kinor N, Mor A, Tsukamoto T, Spector DL, et al. The life of an mRNA in space and time. J Cell Sci. 2010;123(Pt 10):1761–74. 10.1242/jcs.062638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, et al. In vivo dynamics of RNA polymerase II transcription. Nature Structural & Molecular Biology. 2007. ; 14(9):796–806. 10.1038/nsmb1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Darzacq X, Singer RH. The dynamic range of transcription. Mol Cell. 2008;30(5):545–6. 10.1016/j.molcel.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mathworks T. Simbiology Documentation [Webpage]. Natick, MA: The Mathworks; 2015; 2015a:[Simbiology Documentation]. Available: http://www.mathworks.com/help/simbio/index.html Accessed 10 March 2015. [Google Scholar]
- 86. McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci U S A. 1997;94(3):814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sand S, von Rosen D, Victorin K, Filipsson AF. Identification of a critical dose level for risk assessment: developments in benchmark dose analysis of continuous endpoints. Toxicol Sci. 2006;90(1):241–51. 10.1093/toxsci/kfj057 . [DOI] [PubMed] [Google Scholar]
- 88. Simon TW, Simons SS, Preston RJ, Boobis AR, Cohen SM, Doerrer NG, et al. The use of mode of action information in risk assessment: Quantitative key events/dose-response framework for modeling the dose-response for key events. Crit Rev Toxicol. 2014;44 Suppl 3:17–43. 10.3109/10408444.2014.931925 . [DOI] [PubMed] [Google Scholar]
- 89. Reif DM, Martin MT, Tan SW, Houck KA, Judson RS, Richard AM, et al. Endocrine profiling and prioritization of environmental chemicals using ToxCast data. Environ Health Perspect. 2010;118(12):1714–20. 10.1289/ehp.1002180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Becskei A, Kaufmann BB, van Oudenaarden A. Contributions of low molecule number and chromosomal positioning to stochastic gene expression. Nat Genet. 2005;37(9):937–44. 10.1038/ng1616 . [DOI] [PubMed] [Google Scholar]
- 91. Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76(1):17–27. . [DOI] [PubMed] [Google Scholar]
- 92. Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008;321(5892):1081–4. 10.1126/science.1158013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, et al. Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013;26(6):878–95. 10.1021/tx400021f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Patlewicz G, Simon T, Goyak K, Phillips RD, Rowlands JC, Seidel S, et al. Use and validation of HT/HC assays to support 21st century toxicity evaluations. Regul Toxicol Pharmacol. 2013;65(2):259–68. 10.1016/j.yrtph.2012.12.008 . [DOI] [PubMed] [Google Scholar]
- 95. Wang S, Houtman R, Melchers D, Aarts J, Peijnenburg A, van Beuningen R, et al. A 155-plex high-throughput in vitro coregulator binding assay for (anti-)estrogenicity testing evaluated with 23 reference compounds. ALTEX. 2013;30(2):145–57. . [DOI] [PubMed] [Google Scholar]
- 96. Aarts JMMJG, Wang S, Houtman R, van Beuningen RMGJ, Westerink WMA, Van De Waart BJ, et al. Robust array-based coregulator binding assay predicting ERα-agonist potency and generating binding profiles reflecting ligand structure. Chem Res Toxicol. 2013;26(3):336–46. 10.1021/tx300463b . [DOI] [PubMed] [Google Scholar]
- 97.Rotroff DM, Dix DJ, Houck KA, Knudsen TB, Martin MT, McLaurin KW, et al. Using in Vitro High Throughput Screening Assays to Identify Potential Endocrine-Disrupting Chemicals. Environ Health Perspect. 2012. 10.1289/ehp.1205065 . [DOI] [PMC free article] [PubMed]
- 98. Ling Z-Q, Wang Y, Mukaisho K, Hattori T, Tatsuta T, Ge M-H, et al. Novel statistical framework to identify differentially expressed genes allowing transcriptomic background differences. Bioinformatics. 2010;26(11):1431–6. 10.1093/bioinformatics/btq163 . [DOI] [PubMed] [Google Scholar]
- 99. Isella C, Renzulli T, Corà D, Medico E. Mulcom: a multiple comparison statistical test for microarray data in Bioconductor. BMC Bioinformatics. 2011;12:382 10.1186/1471-2105-12-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Shaik JS, Yeasin M. A unified framework for finding differentially expressed genes from microarray experiments. BMC Bioinformatics. 2007;8:347 10.1186/1471-2105-8-347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Xu L, Cheng C, George EO, Homayouni R. Literature aided determination of data quality and statistical significance threshold for gene expression studies. BMC Genomics. 2012;13 Suppl 8:S23 10.1186/1471-2164-13-S8-S23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Solaimani P, Damoiseaux R, Hankinson O. Genome-wide RNAi high-throughput screen identifies proteins necessary for the AHR-dependent induction of CYP1A1 by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2013;136(1):107–19. 10.1093/toxsci/kft191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD. Introducing the "TCDD-inducible AhR-Nrf2 gene battery". Toxicol Sci. 2009;111(2):238–46. Epub 2009/05/29. doi: kfp115 [pii] 10.1093/toxsci/kfp115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wang L, He X, Szklarz GD, Bi Y, Rojanasakul Y, Ma Q. The aryl hydrocarbon receptor interacts with nuclear factor erythroid 2-related factor 2 to mediate induction of NAD(P)H:quinoneoxidoreductase 1 by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Arch Biochem Biophys. 2013;537(1):31–8. 10.1016/j.abb.2013.06.001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hayes JD, Dinkova-Kostova AT, McMahon M. Cross-talk between transcription factors AhR and Nrf2: lessons for cancer chemoprevention from dioxin. Toxicol Sci. 2009;111(2):199–201. Epub 2009/07/25. doi: kfp168 [pii] 10.1093/toxsci/kfp168 . [DOI] [PubMed] [Google Scholar]
- 106. Wakabayashi N, Slocum SL, Skoko JJ, Shin S, Kensler TW. When NRF2 talks, who's listening? Antioxid Redox Signal. 2010;13(11):1649–63. 10.1089/ars.2010.3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lo R, Matthews J. The aryl hydrocarbon receptor and estrogen receptor alpha differentially modulate nuclear factor erythroid-2-related factor 2 transactivation in MCF-7 breast cancer cells. Toxicol Appl Pharmacol. 2013;270(2):139–48. 10.1016/j.taap.2013.03.029 . [DOI] [PubMed] [Google Scholar]
- 108. Vogel CFA, Khan EM, Leung PSC, Gershwin ME, Chang WLW, Wu D, et al. Cross-talk between Aryl Hydrocarbon Receptor and the Inflammatory Response: A ROLE FOR NUCLEAR FACTOR-κB. J Biol Chem. 2014;289(3):1866–75. 10.1074/jbc.M113.505578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. MacPherson L, Tamblyn L, Rajendra S, Bralha F, McPherson JP, Matthews J. 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 2013;41(3):1604–21. 10.1093/nar/gks1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Greive SJ, Dyer BA, Weitzel SE, Goodarzi JP, Main LJ, von Hippel PH. Fitting experimental transcription data with a comprehensive template-dependent modular kinetic model. Biophys J. 2011;101(5):1166–74. 10.1016/j.bpj.2011.07.043 PubMed Central PMCID: PMCPMC3164171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Greive SJ, Goodarzi JP, Weitzel SE, von Hippel PH. Development of a "modular" scheme to describe the kinetics of transcript elongation by RNA polymerase. Biophys J. 2011;101(5):1155–65. 10.1016/j.bpj.2011.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Aitken S, Robert M-C, Alexander RD, Goryanin I, Bertrand E, Beggs JD. Processivity and coupling in messenger RNA transcription. PLoS One. 2010;5(1):e8845 10.1371/journal.pone.0008845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Blake WJ, KAErn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422(6932):633–7. 10.1038/nature01546 . [DOI] [PubMed] [Google Scholar]
- 114. Ko MS. A stochastic model for gene induction. J Theor Biol. 1991;153(2):181–94. [DOI] [PubMed] [Google Scholar]
- 115. Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4(10):e309 10.1371/journal.pbio.0040309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Gradin K, McGuire J, Wenger RH, Kvietikova I, fhitelaw ML, Toftgård R, et al. Functional interference between hypoxia and dioxin signal transduction pathways: competition for recruitment of the Arnt transcription factor. Mol Cell Biol. 1996;16(10):5221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Brunnberg S, Pettersson K, Rydin E, Matthews J, Hanberg A, Pongratz I. The basic helix-loop-helix-PAS protein ARNT functions as a potent coactivator of estrogen receptor-dependent transcription. Proc Natl Acad Sci U S A. 2003;100(11):6517–22. 10.1073/pnas.1136688100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Rüegg J, Swedenborg E, Wahlström D, Escande A, Balaguer P, Pettersson K, et al. The transcription factor aryl hydrocarbon receptor nuclear translocator functions as an estrogen receptor beta-selective coactivator, and its recruitment to alternative pathways mediates antiestrogenic effects of dioxin. Mol Endocrinol. 2008;22(2):304–16. 10.1210/me.2007-0128 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.United States Environmental Protection Agency (USEPA). State of the Science Evaluation: Nonmonotonic Dose Responses as They Apply to Estrogen, Androgen, and Thyroid Pathways and EPA Testing and Assessment Procedures. Washington, DC: 2013 June 6. Report No.
- 120. National Research Council NRC. Review of the Environmental Protection Agency's State-of-the-Science Evaluation of Nonmonotonic Dose-Response Relationships as they Apply to Endocrine Disrupters Washington, DC: The National Academies Press; 2014. [Google Scholar]
- 121. Staršíchová A, Hrubá E, Slabáková E, Pernicová Z, Procházková J, Pěnčíková K, et al. TGF-β1 signaling plays a dominant role in the crosstalk between TGF-β1 and the aryl hydrocarbon receptor ligand in prostate epithelial cells. Cell Signal. 2012;24(8):1665–76. 10.1016/j.cellsig.2012.04.008 . [DOI] [PubMed] [Google Scholar]
- 122. Lipniacki T, Paszek P, Marciniak-Czochra A, Brasier AR, Kimmel M. Transcriptional stochasticity in gene expression. J Theor Biol. 2006;238(2):348–67. 10.1016/j.jtbi.2005.05.032 . [DOI] [PubMed] [Google Scholar]
- 123. Volfson D, Marciniak J, Blake WJ, Ostroff N, Tsimring LS, Hasty J. Origins of extrinsic variability in eukaryotic gene expression. Nature. 2006;439(7078):861–4. 10.1038/nature04281 . [DOI] [PubMed] [Google Scholar]
- 124. Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280(5363):585–90. . [DOI] [PubMed] [Google Scholar]
- 125. Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462(7269):58–64. 10.1038/nature08497 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains: Code A, MATLAB Script for Model Runs. Code B, SBML Code for the Model. Equation A, Calculation of Percent Recruitment from the model results. Equation B, Calculation of Time-Averaged Numbers of ligand-bound AHR. Equation C, Calculation of Time-Averaged Numbers of cofactor bound to AHR-ARNT. Equation D, Equations used for Baseline Projection to determine Transitional Dose Values from Simon et al., (2014). Fig A, Dose-response to the number of ligand-bound AHR molecules as a function of TCDD concentration and fitted Hill functions. Fig B, Histograms of CYP1A1 mRNA copy number induced by TCDD. Table A, Compartments and Species. Table B, Reactions and Rate Constants. Table C, Comparison of classical rate constants, molecular rate constants and propensities. Table D, Hill Model fits of the Percentage of Cells with one or more CYP XRE bound. Table E, Goodness of fit of Poisson distribution for modeled CYP1A1 mRNA copy number.
(DOCX)
Data Availability Statement
The MATLAB script used to run the model is included in the Supporting Information.