Abstract
Our understanding of the genus Natrinema is presently limited due to the lack of available genetic tools. Auxotrophic markers have been widely used to construct genetic systems in bacteria and eukaryotes and in some archaeal species. Here, we isolated four auxotrophic mutants of Natrinema sp. J7-2, via 1-methyl-3-nitro-1-nitroso-guanidin mutagenesis, and designated them as J7-2-1, J7-2-22, J7-2-26, and J7-2-52, respectively. The mutant phenotypes were determined to be auxotrophic for leucine (J7-2-1), arginine (J7-2-22 and J7-2-52), and lysine (J7-2-26). The complete genome and the biosynthetic pathways of amino acids in J7-2 identified that the auxotrophic phenotype of three mutants was due to gene mutations in leuB (J7-2-1), dapD (J7-2-26), and argC (J7-2-52). These auxotrophic phenotypes were employed as selectable makers to establish a transformation method. The transformation efficiencies were determined to be approximately 103 transformants per µg DNA. And strains J7-2-1 and J7-2-26 were transformed into prototrophic strains with the wild type genomic DNA, amplified fragments of the corresponding genes, or the integrative plasmids carrying the corresponding genes. Additionally, exogenous genes, bgaH or amyH gene, were expressed successfully in J7-2-1. Thus, we have developed a genetic manipulation system for the Natrinema genus based on the isolated auxotrophic mutants of Natrinema sp. J7-2.
1. Introduction
Archaea were recognized as an independent domain distinct from eukarya and bacteria by Woese in the late 1970s [1, 2]. Unique biological features of archaea that help to adapt to harsh environment could provide opportunities to gain insight into a number of fundamental problems in biology. While so far, literatures about construction of genetic system in archaea are still relatively limited, in 1987, Cline and Ford Doolittle first demonstrated that Halobacterium halobium could uptake the naked genomic DNA of temperate halovirus φH by PEG- (polyethylene glycol-) mediated spheroplast transfection method and produce plaques successfully [3], which shows the possibility to setup a genetic operation system for these organisms. To establish genetic manipulation system in archaea, selectable marker and shuttle vector based on the marker are critical. In terms of systems based on antibiotic resistance, mevinolin which inhibits 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) [4, 5] and novobiocin which inhibits DNA gyrase [6–8] are two main antibiotics that have successfully been applied in halophiles. In 1989, a resistance gene for mevinolin was isolated to construct the shuttle vector pWL102 for E. coli and Haloferax volcanii. Hfx. volcanii could be transformed with a shuttle vector by the PEG-mediated method with selection of mevinolin [9]. Later in 1990, shuttle vectors pUBP1 and pUBP2 were developed for the transformation of Halobacterium halobium, using the replicon region from halobacterial phage φH and the mevinolin resistance as the selectable marker. Resistance gene for the gyrase inhibitor novobiocin was selected from the genus Haloferax and cloned into the endogenous plasmid pHK2 of Haloferax strain Aa2.2 to form pMDS2. And a shuttle vector pMDS1 was developed and efficiently transformed into Hfx. volcanii by using novobiocin resistance as a selectable marker [6, 7]. Some other vectors were also developed for these two halophiles; for example, pMPK35, pMPK52 based on pGRB1, pUBP4, and pUBP8 based on pHH1 [5] were developed for Hbt. Halobium; pWL101 based on pHV2 [4] and pMDS1 based on pHK2 [7] were developed for Hfx. volcanii. And, in 2010, a transformation system for Pyrococcus furiosus was described based on the shuttle vector system pYS2 from Pyrococcus abyssi, in which the pyrE gene was replaced by the HMG-CoA as a selectable marker conferring resistance to the antibiotic simvastatin [10]. Mayrhofer-Iro et al. developed a transformation/vector system for halo(alkali)philic members by using a genomic elements of Natrialba magadii virus φCh1 and a novobiocin resistance cassette as a selectable marker [11].
Besides the antibiotic resistance and shuttle vectors described above, auxotrophic markers are also useful selectable markers for construction of genetic system. Auxotrophs are a group of organisms that lost the ability to synthesize certain substances required for their growth owing to the presence of mutations. Compared to the wild type strain, the auxotrophic mutants cannot grow in minimal medium if the corresponding nutrients are not supplied. Auxotrophic strains have been widely employed for genetic investigation in bacteria and eukaryotes, but only a few cases have been documented in archaea. In 1987, several auxotrophic strains of Methanococcus voltae PS were obtained by irradiation with UV and gamma rays. These mutants were determined to be auxotrophic for histidine or purine and were used to demonstrate the occurrence of transformation with homologous wild type DNA by using the auxotrophic markers [12]. In 1990, auxotrophs of Methanococcus maripaludis created by ethyl methanesulfonate treatment were used to investigate the role of the acetyl-CoA pathway of autotrophic CO2 fixation in Methanococcus maripaludis [13]. Recently, a Sulfolobus solfataricus pyrEF/lacS mutant was used to construct a reporter gene system with integrative vectors containing the beta-galactosidase gene [14]. In the case of halophilic archaea, auxotrophy is an important tool since only limited selectable markers can be used in them. Several haloarchaeal auxotrophs obtained from mutagenesis or gene knockout have been employed in genetic studies. In 1985, for instance, auxotrophic mutants of Hfx. volcanii generated by chemical mutagenesis were used to demonstrate a native genetic transfer system in this extremely halophilic autotroph [15]. In 1990, Conover and Doolittle demonstrated that cosmid DNA prepared from Escherichia coli can be used to transform the histidine auxotroph Hfx. volcanii to a prototrophy [16]. Uracil auxotrophs based on the pyrE, pyrF, or ura3 gene were later used to develop a gene knockout system for Hfx. volcanii [17, 18], Halobacterium salinarum [19], Haloferax mediterranei, and Haloarcula hispanica [20], respectively. Aromatic amino acid auxotrophy of Hbt. salinarum was obtained by in-frame deletions of OE1477R [21]. And, in 2014, Choi et al. reported a novel integrative expression vector which contained a pyrE gene as an auxotrophic selection marker for Sulfolobus species [22]. However, it should be noted that the employment of auxotrophic mutants is presently mainly confined to genetically tractable model organisms; thus similar research in halophiles is important for establishing their respective genetic manipulation system.
The halobacterial genus Natrinema gen. nov., which requires a medium with at least 10% salt concentration for cell growth (optimum 19.8%–25.1%), is a genus of halophilic archaea first proposed by McGenity et al. according to the phylogenetic analysis of 16S rRNA gene sequences and taxonomic properties [23]. This genus now contains seven species, Natrinema altunense, Natrinema ejinense, Natrinema gari, Natrinema pallidum, Natrinema pellirubrum, Natrinema versiforme, and Natrinema salaciae. Studies of the genus Natrinema have mainly focused on their phenotypic, physiological, biochemical, and taxonomic properties, and no studies analyzing their genetics or genetic engineering potential have yet been reported.
Natrinema sp. strains J7-1 and J7-2 were isolated from a salt mine in Yingcheng, Hubei Province, China [24]. The genomes of the two strains were sequenced (GenBank ID: PRJNA157873 and PRJNA89473), revealing that they were genetically identical excluding the fact that J7-1 contains a plasmid named pHH205 and J7-2 contains a plasmid named pJ7-I [25]. Interestingly, the 16,341 bp endogenous plasmid pHH205 included in J7-1 [24, 26] was proven to be the proviral genome of a temperate membrane-containing halophilic virus called SNJ1, which could be induced from J7-1 with mitomycin C and produces plaques on lawns of J7-2 [25, 27]. In addition, another provirus named SNJ2 has recently been found on the chromosomes of these two strains, but the susceptible host of this virus has not yet been identified (unpublished data). Thus the two strains could be valuable models to study the interaction of the proviruses and their host strains. These two strains have also been employed to conduct studies of the halophilic protease SptA [28, 29], heat shock protein 70 [30], and DNA fragments conferring promoter activity in three domains [31, 32]. However, the lack of genetic tools is delaying further studies of these strains. Fortunately, the availability of the genome sequence and the amino acid biosynthetic pathways of strain J7-2 [33] allow us to isolate and characterize auxotrophic mutants of these strains, which may be used to establish a genetic manipulation system for the genus Natrinema.
In the present study, we report the isolation of four auxotrophs of Natrinema sp. J7-2 via 1-methyl-3-nitro-1-nitroso-guanidin (NTG) mutagenesis. Their auxotrophic phenotypes and genes responsible for the phenotypes were determined. The auxotrophic phenotypes were further used to establish a DNA transformation protocol suitable for the strains. In addition, we showed that both the bgaH gene from Haloferax alicantei [34, 35] and amyH gene from Har. hispanica DSM 4426 [36, 37] could be integrated and expressed in strain J7-2-1 by using auxotrophy as a selectable marker.
2. Materials and Methods
2.1. Strains and Culture Conditions
Strains and plasmids used in this study are listed in Table 1. Natrinema sp. strains were grown in Halo-2 (H-2) medium prepared as previously described [27]. Modified growth media (MGM) contained 18% (w/v) salt water (SW) [38]; minimal medium (MM) used to isolate and identify Natrinema sp. J7-2 auxotrophs contained, per liter, 188 g NaCl, 43 g MgSO4·7H2O, 2.5 g KCl, 5 mM Tris HCl (pH 7.5), 5 mL 1 M NH4Cl solution, 5 mL 10% disodium succinic acid, 2 mL 0.05 M K2HPO4, 2 g glucose, 4.865 mL 1 M CaCl2, and 1 mL of trace element solution. The trace element solution contained, per 100 mL, 47 mg MnCl2·4H2O, 44 mg ZnSO4·7H2O, 5 mg CuSO4·5H2O, and 42 mg FeSO4·7H2O. If required, the corresponding amino acid was added to the MM medium to make a final concentration of 50 μg mL−1. Solid agar was made with the addition of Bacto Agar (BD, USA) to the final concentration of 1.5%. Soluble starch was applied to the MGM and MM media at a final concentration of 1%-2%.
Table 1.
Strains, plasmids, and amplification fragments used in this work.
| Strains, plasmids, and amplification fragments | Description | Source or reference |
|---|---|---|
| Strain | ||
| Natrinema sp. J7-2 | Haloarchaeal strain isolated from a salt mine | CCTCC AB91141, China [24, 25, 27] |
| Natrinema sp. J7-2-1 | Auxotrophic strain from J7-2, Leu− | This work |
| Natrinema sp. J7-2-22 | Auxotrophic strain from J7-2, Arg− | This work |
| Natrinema sp. J7-2-26 | Auxotrophic strain from J7-2, Lys− | This work |
| Natrinema sp. J7-2-52 | Auxotrophic strain from J7-2, Arg− | This work |
| Escherichia coli DH5α | F − endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(r K − m K + ), λ− | [42] |
| Escherichia coli JM110 | rpsL thr leu thi lacY galK galT ara tonA tsx dam dcm glnV44 Δ(lac-proAB) e14-[F′ traD36 proAB + lacI q lacZΔM15] hsdR17(r K − m K +) | [43] |
| Plasmid | ||
| pUC19 | Cloning vector | Lab stock |
| pMD19T-simple | Sequence vector | Purchased from TaKaRa |
| pUC19-KM1752 | pUC19 containing leuB gene with KpnI and MfeI from J7-2 | This work |
| pUC19-HM1752 | pUC19 containing the ORF of leuB gene with HindIII and MfeI from J7-2 | This work |
| pUC19-HM4049 | pUC19 containing dapD gene with HindIII and MfeI from J7-2 | This work |
| pUC19-HM1752-amyH | pUC19-HM1752 containing amyH gene from Haloarcula hispanica DSM 4426 | This work |
| pUC19-HM1752-bgaH | pUC19-HM1752 containing bgaH gene from Haloferax alicantei | This work |
| Amplification fragments | ||
| KM1752 | leuB gene containing promoter from J7-2 with KpnI and MfeI | This work |
| HM1752 | Amplification fragments of leuB gene from J7-2 with HindIII and MfeI | This work |
| HM4049 | Amplification fragments of dapD gene from J7-2 with HindIII and MfeI | This work |
E. coli strains were grown in Luria-Broth (LB) medium supplemented with antibiotics when necessary [39]. Among them, DH5α was used for plasmid construction and propagation, and JM110 was employed to prepare unmethylated plasmid DNA for efficient transformation into Natrinema sp. J7-2.
2.2. Molecular Manipulation
Restriction endonuclease digestion, agarose gel electrophoresis, and additional molecular cloning techniques were carried out according to standard protocols [39, 40] or to the manufacturer's instructions. Small amounts of plasmid DNA were prepared using TIANprep Mini Plasmid Kit from TIANGEN (China). The primers used in this study are listed in Table 2. Restriction enzymes and other DNA modifying enzymes were purchased from TaKaRa or Fermentas and used according to the manufacturer's directions. Oligonucleotides were purchased from Sangon (Shanghai, China). DNA sequencing was performed by BGI (China).
Table 2.
Primers used in this work.
| Primer name | Sequence (5′-3′) | Restriction site |
|---|---|---|
| KpnI-1752 | TAAGGTACCTCGCTGCGATCTTGCAGGGACT | KpnI |
| 1752-MfeI | TAACAATTGCGGCGGTCGCGGGCCACTAAA | MfeI |
| HindIII-1752 | TTAAAGCTTATGACTCACGAAATCGCCGTCATT | HindIII |
| HindIII-4049 | TTAAAGCTTATGAGCGCACTCGAAA | HindIII |
| 4049-MfeI | TATCAATTGCCGGTTACGCCACCCCGCG | MfeI |
| MfeI-amyH | AATCAATTGATCATATATGCCAACTATTTA | MfeI |
| amyH-PstI | AAACTGCAGTAGTGGAAAGCGAGCCAGC | PstI |
| leuA-F | ATGGCACGTTATCCGCCATCT | / |
| leuA-R | TCAGTCGTCGGCCGGCGT | / |
| leuC-F | ATGAGCGCGGAAACGCTG | / |
| leuC-R | TCATACCGAGGTCACCTCCT | / |
| leuD-F | ATGACGGACGAGGTCGAGATT | / |
| leuD-R | TACTCCGCTTCGGGAATCGC | / |
| leuB-F | GTGAGTATGACTCACGAAAT | / |
| leuB-R | TTACAGTCGGTCGATGAT | / |
| dapA-F | GCGACCCAAGAGACCACA | / |
| dapA-R | TCATCGCTCATCCTCCG | / |
| dapF-F | ATGAATATCCCATTTCAGAAGT | / |
| dapF-R | TCACGGCTCGGCCGGGGATT | / |
| lysA-F | ATGAACGAAACCCAAACGTT | / |
| lysA-R | TATCGGTCGTATTCGTTGTC | / |
| dapD-F | ATGAGCGCACTCGAAACCGA | / |
| dapD-R | TTACTCCCGCAGCGCGTCCT | / |
| dapB-F | ATGACGACGCGAATCGGCGTCAC | / |
| dapB-R | TCATTCGCTGATCACGTCTGCGAA | / |
| argF-F | ATGACGACCGATTCCGATCC | / |
| argF-R | TCACTCGAGCACCCAACTCA | / |
| argE-F | ATGAGTGCAACCATGGACGACA | / |
| argE-R | TCATTCGTCGTCCTCGCGAA | / |
| argD-F | ATGAGCGACCTCGATTTCGT | / |
| argD-R | TCATGCGTTGGAGTCGGT | / |
| argB-F | GGGCACCCTAACATGACGAC | / |
| argB-R | TCATTGGGCCACCTCC | / |
| argC-F | ATGGCGGTCGGGACCGAG | / |
| argC-R | TTAGGGTGCCCCCACGGG | / |
| argXW-F | ATGACCGAATGCGTCGAGTG | / |
| argXW-R | TCAGGCGGTCACCTC | / |
| argH-F | ATGACCGAGGAGAGCGCTCAC | / |
| argH-R | TCAGACATAGCCGTTCACCT | / |
| argG-F | GATATGACCCGCGTGGCACT | / |
| argG-R | TTACTGGTCGTCTCCCGTCCC | / |
2.3. Mutagenesis and Isolation of Auxotrophs from Strain J7-2
Strain J7-2 was grown in H-2 medium at 42°C for approximately 36 h. The cells were collected after centrifugation at 12,857 ×g for 2 min and resuspended in an equal volume of fresh medium. Mutagenesis was carried out using the following three methods: (1) NTG was added to the cell culture with final concentrations of 0.5, 0.75, 1.0, and 1.5 mg mL−1. The culture was kept at 37°C with shaking (180 rpm) for 1.5 h. (2) The concentrated cell cultures were directly spread on the solid H-2 plate. An NTG particle was then placed on the edge region of the plate. The plate was kept at 42°C until the appearance of the lawn. There was a large inhibition zone around the NTG particle. Cells collected from the edge of this zone were transferred to liquid H-2 medium and cultured for approximately 36 h at 42°C. (3) Under sterile conditions, the cell cultures were stirred using a magnetic stirrer under ultraviolet radiation (UV) for around 1.5 h. After the mutagenic treatment described above, six successive 10-fold dilutions of the cell cultures were taken. The 10−4, 10−5, and 10−6 dilutions of the cultures were spread on H-2 plates and incubated at 42°C for 7–10 days until colonies appeared. Subsequently, the colonies were inoculated in parallel on the MGM and MM plates and incubated at 42°C for 7–10 days. Strains that showed growth on MGM but failed to grow on the MM plate were considered as candidate auxotrophic mutants. The candidates were subjected to single colony isolation and inoculation in parallel on the MGM and MM plate experiments again, until the auxotrophic phenotype was confirmed.
2.4. Phenotypic and Molecular Characterization of NTG-Induced Auxotrophs
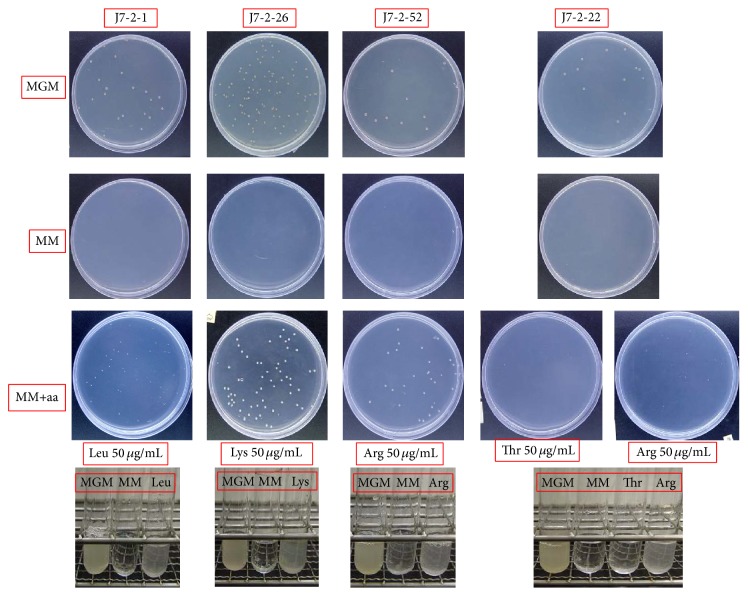
To identify the phenotypes of the auxotrophic mutants, different nutrients (amino acids, vitamins, purine, and pyrimidine) were combined into different groups (Table 3). Auxotrophic mutants were spread on the MM plates. Small filter papers (approximately 5 mm in diameter) soaked in different types of liquid nutrition were placed on separate regions of the agar surface. Filter paper soaked with MGM medium was used as control. The plates were kept at 42°C for 7–15 days. According to their growth status, the nutrients required for the growth of the auxotrophic mutants were determined. Subsequently, the auxotrophic mutants were cultured again in both solid and liquid media of MGM, MM, and MM with added nutrients to reaffirm their phenotypes (Figure 1).
Table 3.
Different combination groups of growth factors used in this work to determine the auxotrophic phenotype of the four mutation strains.
(a) Single growth factors used in this work and groups containing all of the growth factors belonged to the same type
| Group 1: containing amino acids from 2–24 | 2: Asp | 3: Cys | 4: Ile |
| 5: Gly | 6: Glu | 7: Val | 8: Ser |
| 9: Ala | 10: His | 11: Tyr | 12: Phe |
| 13: Met | 14: Thr | 15: Met | 16: Trp |
| 17: Pro | 18: Leu | 19: Lys | 20: cystine |
| 21: Arg | 22: Asp | 23: Asn | 24: Gln |
|
| |||
| Group 25: containing vitamins from 26 to 35 | 26: vitamin H | 27: vitamin C | 28: vitamin B6 |
| 29: vitamin B3 | 30: vitamin B5 | 31: vitamin B4 | 32: vitamin B2 |
| 33: vitamin B9 | 34: vitamin B12 | 35: vitamin B1 | |
|
| |||
| Group 36: containing dNTPs from 37 to 43 | 37: dATP | 38: dGTP | 39: dTTp |
| 40: dCTP | 41: dUTP | 42: xanthine | 43: hypoxanthine |
(b) Different combination groups containing amino acids
| Group A | Group B | Group C | Group D | Group E | |
|---|---|---|---|---|---|
| Group F | 9: Ala | 21: Arg | 23: Asn | 22: Asp | 3: Cys |
| Group G | 6: Glu | 24: Gln | 5: Gly | 10: His | 4: Ile |
| Group H | 18: Leu | 19: Lys | 15: Met | 12: Phe | 17: Pro |
| Group I | 8: Ser | 14: Thr | 16: Trp | 11: Tyr | 7: Val |
(c) Different combination groups containing vitamins
| Group I | Group II | Group III | ||
|---|---|---|---|---|
| Group IV | 32: vitamin B2 | 33: vitamin B9 | 31: vitamin B4 | 35: vitamin B1 |
| Group V | 28: vitamin B6 | 27: vitamin C | 29: vitamin B3 | 35: vitamin B1 |
| Group VI | 30: vitamin B5 | 34: vitamin B12 | 26: vitamin H | 35: vitamin B1 |
(d) Different combination groups containing purine and pyrimidine
| Group X1 | Group X2 | Group X3 | |
|---|---|---|---|
| Group X4 | 41: dUTP | 37: dATP | 39: dTTP |
| Group X5 | 42: xanthine | 43: hypoxanthine | 40: dCTP |
| 38: dGTP | 38: dGTP | 38: dGTP |
(e) The growth of the four auxotrophic strains on MM medium supplied with different nutrients
| Supplied nutrients in MM medium | J7-2-1 | J7-2-22 | J7-2-26 | J7-2-52 |
|---|---|---|---|---|
| MM medium | Na | N | N | N |
| MGM medium | Yb | Y | Y | Y |
| Group ALL | Y | Y | Y | Y |
| Group 1 | Y | Y | Y | Y |
| Group 25 | N | N | N | N |
| Group 36 | N | N | N | N |
| Group A | Y | N | N | N |
| Group B | N | Y | Y | Y |
| Group F | N | Y | N | Y |
| Group H | Y | N | Y | N |
| Group I | N | Ic | N | N |
| 18 | Y | N | N | N |
| 19 | N | N | Y | N |
| 21 | N | Y | N | Y |
| Other nutrients | N | N | N | N |
aN: the growth of the auxotrophs was not detected. bY: the growth of the auxotrophs was detected. cI: an inhibition zone was detected.
Figure 1.
Growth behavior of the auxotrophic strains in different media. The method used to determine the nutrients required for the four auxotrophic mutants is described in Materials and Methods, after which the phenotypes of the mutants were confirmed. We picked out a single colony of each of the four mutants and cultivated it in a liquid MGM medium at 42°C. The cultures of each strain were obtained, centrifuged, and resuspended in MM medium. Then, the suspended cultures were inoculated in liquid media of MGM, MM, and MM with the corresponding nutrients added (as shown in the figure, leucine was added to the MM medium to reconfirm the phenotype of J7-2-1; lysine was added for reconfirming J7-2-26, arginine for J7-2-52, and threonine and arginine for J7-2-22). In corresponding solid media, the suspended cultures were diluted with MM media before spreading on the corresponding media plates. Their growth states were recorded and pictures were taken after storage at 42°C for 7–15 days. MGM: a complete medium that could support the growth of all J7-2 auxotrophic strains; MM: a minimal medium that could only support the growth of the J7-2 prototrophic strain; MM+aa: MM supplied with the relevant single amino acid to a final concentration of 50 μg/mL.
To find significant mutations in the auxotrophic mutants, genes important in the pathways of the corresponding amino acid synthesis were analyzed by PCR and DNA sequencing. The mutant genes were determined by comparison of the corresponding DNA sequences between the wild type strain and the auxotroph. Information about these genes and their primers is shown in Tables 2 and 4.
Table 4.
Molecular characterization of the auxotrophic mutants by analyzing relevant genes in the corresponding amino acid biosynthetic pathways.
| Auxotrophic strains | Phenotype | Gene | Gene number | Notation | Length (bp) | Mutation status | |
|---|---|---|---|---|---|---|---|
| J7-2-1 | Leu− | leuA | NJ7_G1272 | 2-Isopropylmalate synthase | 831 | None | |
| leuC | NJ7_G1748 | 3-Isopropylmalate dehydratase, large subunit | 1388 | None | |||
| leuD | NJ7_G1749 | 3-Isopropylmalate dehydratase, small subunit | 837 | None | |||
| leuB | NJ7_G1752 | 3-Isopropylmalate dehydrogenase | 768 | 12 bp fragment deletion from 690 to 701 bp | |||
|
| |||||||
| J7-2-26 | Lys− | dapA | NJ7_G4051 | Dihydrodipicolinate synthase | 951 | None | |
| dapF | NJ7_G4046 | Diaminopimelate epimerase | 1639 | None | |||
| lysA | NJ7_G4047 | Diaminopimelate decarboxylase | 1422 | None | |||
| dapD | NJ7_G4049 | 2,3,4,5-Tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | 635 | 18 bp deletion from 583 to 600 bp | |||
| dapB | NJ7_G4050 | Dihydrodipicolinate reductase | 987 | None | |||
|
| |||||||
| Auxotrophic strains | Phenotype | Gene | Gene number | Notation | Length (bp) | Mutation Status | |
| J7-2-22 | J7-2-52 | ||||||
|
| |||||||
| J7-2-22 J7-2-52 |
Arg− | argF | NJ7_G0258 | Ornithine carbamoyltransferase | 921 | None | None |
| argE | NJ7_G0259 | N-Acetyl-ornithine/N-acetyl-lysine deacetylase | 1110 | None | None | ||
| argD | NJ7_G0260 | Acetylornithine aminotransferase | 1155 | None | None | ||
| argB | NJ7_G0261 | Acetylglutamate kinase | 870 | None | None | ||
| argC | NJ7_G0262 | N-Acetyl-gamma-glutamyl-phosphate reductase | 1077 | None | 1051 bp: G-A(GGG-GAG) | ||
| argX | NJ7_G0263 | Ribosomal protein S6 modification protein | 1043 | None | None | ||
| argW | NJ7_G0264 | Putative biosynthetic carrier protein | |||||
| argH | NJ7_G0265 | Argininosuccinate lyase | 1539 | None | None | ||
| argG | NJ7_G0266 | Argininosuccinate synthase | 1242 | None | None | ||
2.5. Isolation of Natrinema sp. J7-2 Total Genomic DNA
High molecular weight Natrinema sp. J7-2 genomic DNA was prepared according to the protocol of TIANamp Bacteria DNA Kit from TIANGEN (China). DNA samples prepared were examined by agarose gel electrophoresis and then used directly for transformation.
2.6. Transformation
All procedures were performed at room temperature unless indicated otherwise. The main transformation process of the auxotrophic mutants of Natrinema sp. J7-2 was performed based on the transformation protocol used for Hfx. volcanii [41] with several modifications: the standing time was modified from 10 min to 15 min to form spheroplasts; regeneration time was altered from 20–30 min at room temperature (RT) to 2–5 hours at 37°C and the transformation solutions were changed from 1 M NaCl to 2 M NaCl, a higher saline concentration suitable to Hbt. salinarum [41]. Finally, the resultant mixture was spread on the selected solid medium and stored at 45°C for 7–10 days to detect the transformants.
2.7. Molecular Identification of the Transformants
Target gene fragments were amplified from the genomic DNA of the wild type strain, auxotrophic strain, and the corresponding transformant. The DNA sequences were comparatively analyzed to determine whether the mutation sites had been complemented by the transformation.
2.8. β-Galactosidase Detection
The β-galactosidase activity of Natrinema sp. J7-2-1 transformants on plates was determined as described previously [44]. Briefly, agar plates were sprayed with X-gal (20 mg mL−1); blue colonies indicated the existence of β-galactosidase activity.
2.9. Amylase Activity Assay
The amylase activity of Natrinema sp. J7-2-1 transformants on plates was determined as described previously [37, 44] using 18% MGM supplemented with 2% (w/v) soluble starch. After incubation at 42°C for 5–7 days, plates were flooded with 0.3% I2/0.6% KI solution; a clear zone around the growth indicated starch hydrolysis.
3. Results and Discussion
3.1. Isolation of Auxotrophic Mutants of Natrinema sp. J7-2 by Mutagenesis
The halophilic archaea Natrinema sp. J7-2 could be a useful model to investigate the interactions between provirus and host, but such studies have so far been prevented by a lack of genetic tools. As genetic manipulation systems based on auxotrophic selectable markers have been developed in other halophilic archaea, such as Hb. salinarum NRC-1, Haloferax mediterranei, and Haloarcula hispanica, we decided to isolate auxotrophic mutants of Natrinema sp. J7-2 and develop a genetic system based on them.
Before mutagenesis, Natrinema sp. J7-2 was tested that it could grow on MM plate (Figure S1 in Supplementary Material available online at http://dx.doi.org/10.1155/2015/483194). The cultures of Natrinema sp. J7-2 having undergone NTG or UV mutagenesis were spread on the MGM plate to obtain individual colonies; randomly selected colonies were inoculated on the MGM and MM plates in parallel. Colonies which appeared on the MGM plates but not on the corresponding MM plates were considered as candidate auxotrophic mutants. Ten colonies from approximately 7000 randomly selected candidates were able to grow on the MGM plates, but not on the corresponding MM plates. These candidate auxotrophic mutants were purified using the spread plate method on MGM agar media, and the resultant individual colonies were again inoculated on the MGM and MM plates in parallel for further identification. Finally, four auxotrophic mutant strains were confirmed and were named as J7-2-1, J7-2-22, J7-2-26, and J7-2-52 (Figures S2 and S3). All were obtained from the NTG (0.75 mg mL−1) mutagenesis treatment, which has previously been used to isolate mutants in Sulfolobus acidocaldarius [45] and other species [46, 47].
3.2. Phenotypic and Molecular Characterization of Auxotrophic Mutants of Natrinema sp. J7-2
The phenotypes of the isolated auxotrophic strains were determined by restoration of their growth on the MM plate by supplying them with a range of nutrient combinations (as shown in Tables 3(a), 3(b), 3(c), and 3(d)). The results indicate that all were auxotrophic for specific amino acids. As shown in Table 3(e), strain J7-2-1 was a leucine auxotroph, since it could grow in MM medium supplied with groups ALL, 1, A, or H, all of which contain leucine. Strain J7-2-26 was determined to be a lysine auxotroph because it exhibited growth only on MM plates supplied with groups ALL, 1, B, or H, all of which contain lysine. Strain J7-2-52 was an arginine auxotroph as it could grow in MM medium supplied with groups ALL, 1, B, or F, all of which contain arginine. However, strain J7-2-22 could be auxotrophic for either arginine or threonine (Table 3(e)) because it could grow not only in MM medium supplied with groups ALL, 1, B, or F, but also on MM plates supplied with group I. The auxotrophic phenotypes of the four mutant strains were confirmed by assessment of their ability to grow in MM medium supplied with the corresponding single amino acid (Figure 1). The results reaffirmed that strain J7-2-1 is auxotrophic for leucine, strain J7-2-26 is auxotrophic for lysine, and both strains J7-2-22 and J7-2-52 are auxotrophic for arginine.
After the phenotypes of the auxotrophic strains were determined, we wanted to identify the mutations responsible for them. Each of the genes predicted to be involved in the corresponding amino acid biosynthesis was amplified from the genomes of the auxotrophs and the wild type strain (Figure S4). The mutation sites were determined by comparative analysis of the gene sequences (Table 4). The results showed that strain J7-2-1 (Leu−) has a 12 bp fragment deletion from 690 bp to 701 bp in leuB, whereas strain J7-2-26 (Lys−) has an 18 bp fragment deletion from 583 bp to 600 bp in dapD (Figure S5). We believe that these deletions might be the cause of their auxotrophic phenotypes. Consistent with its arginine auxotrophic phenotype, a single nucleotide substitution at position 1051 bp of the argC gene was identified in strain J7-2-52. This substitution resulted in a change of the codon from GGG to GAG. However, no mutations were identified in the coding regions of the genes involved in arginine biosynthesis in the other arginine auxotroph strain J7-2-22, indicating that its phenotype might be the result of mutations in other regulatory genes for arginine biosynthesis.
3.3. Establishment of a Genetic Transformation System for the Natrinema Genus
In 1987, Cline and Ford Doolittle developed a PEG-mediated transformation protocol for Hfx. volcanii [3]. In this protocol, cells are first treated with EDTA, which converts the cell to a spheroplast by removing its paracrystalline glycoprotein surface layer. DNA is then introduced into the spheroplasts with the help of PEG600, after which the cells recover in a regenerate solution or rich media before plating on a selective medium [48]. Although this transformation protocol has been used in a number of archaea, especially haloarchaea, it is only applicable for species in which a spheroplast can be generated readily [49].
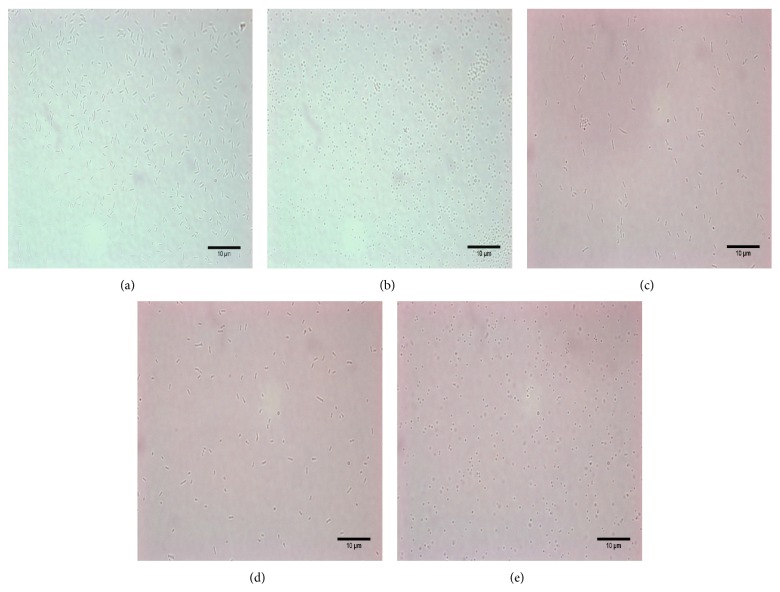
As there was no transformation protocol available for the Natrinema genus, we aimed to develop a transformation method for Natrinema based on the above transformation protocol for Hfx. volcanii. We determined whether Natrinema J7-2 cells could become spheroplasts after EDTA treatment and secondly if the spheroplast could recover in regenerate solution. The standing time in the process was modified to 15 min to form spheroplasts; regeneration time was changed as two to five hours at 37°C and the transformation solutions were also changed to 2 M NaCl, a higher saline concentration suitable for Hbt. salinarum. As shown in Figures 2(a) and 2(b) and Table 5, the morphology of the cells changed from rod shaped to spherical after EDTA treatment for 15 min, indicating the generation of the spheroplast. Three different regeneration solutions were tested to identify the most suitable for recovery of Natrinema spheroplast: H-2 medium; 18% MGM; and regeneration solution. The first two are rich media used to cultivate Natrinema sp. J7 serials strains and Hfx. volcanii, respectively. The regeneration solution, comprised of an 18% salt solution supplied with 15% sucrose, 0.05% yeast extract, 0.01% peptone, and 0.1% casamino acids, was used for transformation of Hfx. volcanii. Cell morphology could be restored to rods using all three mediums, but the cells recovered more effectively in the H-2 medium and 18% MGM than in the regeneration solution (Figures 2(c), 2(d), and 2(e) and Table 5). Thus, in the following experiments, we chose H-2 medium as the recovery solution.
Figure 2.
Morphological changes to the cells during the transformation process. As there was no transformation protocol available for the genus of Natrinema, we used the transformation process used for Hfx. volcanii. All procedures were performed at room temperature. We picked out a single colony of J7-2 and cultivated it in liquid MGM media at 42°C. Selected cultures of J7-2 were obtained and treated with spheroplasting solution for 15 min, while some were not treated to provide a control. Cell morphology was observed and recorded under the microscope. The results are shown in (a) and (b). During the recovery process, three different regeneration solutions were tested to determine whether the cell morphology could be recovered to short rods and which medium was more suitable for Natrinema: H-2 medium, 18% MGM and regeneration solution. We modified the protocol slightly. The regeneration time was changed from 20–30 min at RT to 2 to 5 hours at 37°C and the transformation solutions were also changed from 1 M NaCl to 2 M NaCl. The regeneration results were also observed and recorded under microscope. All recovered results are shown in (c), (d), and (e). (a) Cells of J7-2 grown in MGM medium before spheroplasting solution treatment. (b) Cells of J7-2 treated with spheroplasting solution. (c) Cells of J7-2 regenerated with H-2 medium treatment. (d) Cells of J7-2 regenerated with 18% MGM medium treatment. (e) Cells of J7-2 regenerated with regeneration solution treatment.
Table 5.
Percentages of spheroplasts formation and spheroplasts regenerated to wt cells.
| Percentage of Spheroplasts formation | Percentage of spheroplasts regenerated to wt cells | ||
|---|---|---|---|
| Different regeneration solutions | |||
| About 95% | H-2 medium | 18% MGM medium | Regeneration solution |
| About 90% | About 80% | About 20% | |
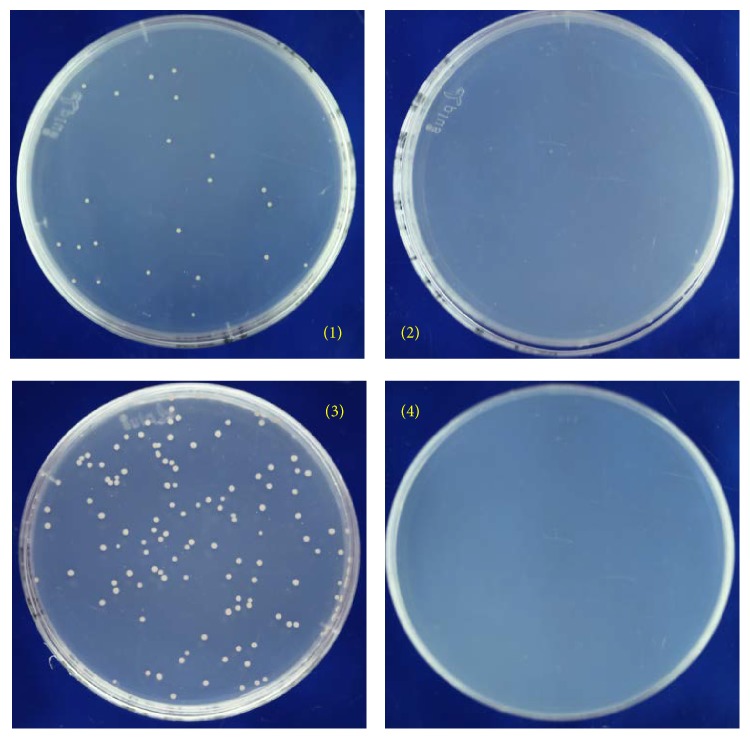
Since the spheroplast of Natrinema could be generated and recover successfully, we tested if spheroplasted J7-2 cells could be transformed by exogenous DNA using the isolated auxotrophic mutants J7-2-1 and J7-2-26 as recipients. The genomic DNA of Natrinema sp. J7-2 was prepared as the transforming DNA and applied to auxotrophic strains. As strains J7-2-1 and J7-2-26 require the presence of leucine or lysine for growth, only recombinants that restore the ability to synthesize leucine or lysine would grow on MM plates. As shown in Figure 3, strains of J7-2-1 and J7-2-26 transformed with the genomic DNA of J7-2 appeared in the MM plates (plates 1 and 3), but colonies did not appear on plates when DNA was not added to the spheroplasts of J7-2-1 and J7-2-26. The transformation efficiency is estimated to be 3.62 × 103 transformants per μg DNA. To confirm the results, the leuB gene of the transformants of J7-2-1 and the dapD gene of the transformants of J7-2-26 were amplified and subjected to sequencing. Compared to the parent strains (J7-2-1 has a 12 bp fragment deletion from 690 to 701 bp in leuB gene and J7-2-26 has an 18 bp fragment deletion from 583 to 600 bp in dapD gene), the results showed that the mutations had been corrected in all the transformants, presumably by homologous recombination (Figures S5 and S6). These results indicate that, using the protocol we developed, exogenous DNA can be transformed into Natrinema and the resultant auxotrophic strains represent useful genetic tools.
Figure 3.
Results of the transformation of J7-2-1 and J7-2-26 with J7-2 genome. The wild type Natrinema sp. J7-2 total genomic DNA was isolated and then transformed into two auxotrophic mutants, J7-2-1 and J7-2-26. The transformation mixture was spread on the MM plates and cultivated at 42°C for 15 days. The detailed transformation process was described in Materials and Methods. (1) J7-2-1 transformed with J7-2 genome. (2) Negative control, J7-2-1 transformed with salt solution without DNA. (3) J7-2-26 transformed with J7-2 genome. (4) Negative control, J7-2-26 transformed with salt solution without DNA.
To confirm that the auxotrophs were transformed to prototrophs by homologous recombination at the corresponding mutated genes, the leuB gene fragment and the dapD gene fragment from the genome of the wild type strain were amplified and transformed into J7-2-1 and J7-2-26 spheroplasts, respectively. As expected, transformants of each auxotroph transformed with the matching gene appeared on the MM plate and sequencing of leuB or dapD from the corresponding transformants confirmed that the mutation had been corrected (Figure S6). Furthermore, in the J7-2-1 transformation process, we amplified two fragments, KM1752, a leuB cloning fragment containing the promoter from J7-2, and HM1752, a leuB open reading frame (ORF) cloning fragment from J7-2 (Table 1), to identify any difference in transformation result. No obvious difference was observed (Figure S6). It is noteworthy that although the amount of leuB or dapD coding fragment should be much higher when transformed with the amplified gene fragment than that of transformation with genomic DNA, transformation efficiency of the former was slightly lower (1.02 × 103 transformants per μg DNA versus 3.62 × 103 transformants per μg DNA). We speculate that this is because the amplified fragments were short in length, resulting in a reduced efficiency of homologous recombination. However additional evidence is needed to prove this hypothesis.
3.4. Possibility of Transformation with Circular Plasmids and the Ability to Express Exogenous Genes in Auxotrophic Strains through Homologous Recombination
The above results showed that auxotrophs of Natrinema sp. J7-2 can be transformed to prototrophs by transformation with genomic DNA or amplified gene fragments of the wild type strain. However, both of them were linear DNA fragments. We wanted to know whether circular plasmids could also be transformed. Three plasmids containing the corresponding genes were constructed and named pUC19-KM1752, pUC19-HM1752, and pUC19-HM4049 (Table 1). The former two plasmids carrying the leuB gene were transformed into auxotrophic strain J7-2-1, while the latter, carrying the dapD gene, was transformed into auxotrophic strain J7-2-26. They were subsequently demethylated in E. coli strain JM110. As expected, all three circular plasmids could be transformed efficiently into auxotrophs and the prototrophic phenotype was restored on MM plates. We further quantified transformation efficiency, which was equal to approximately 2.96 × 103 transformants per μg DNA. Although the transformation efficiency is slightly increased, compared with the amplified gene fragment, it is still relatively low compared to that of Hfx. volcanii (103 transformants per μg DNA versus 106 transformants per μg DNA [7]). One possibility is that the transformation method we developed still requires improvement, though it is also possible that Natrinema sp. J7-2 is less effective than Hfx. volcanii at taking up extracellular DNA and performing homologous recombination. Nevertheless, the successful transformation of DNA into the auxotrophic strains of J7-2 shows that transformation in Natrinema is possible and the leuB and dapD genes could be used as selectable markers for the development of genetic manipulation systems.
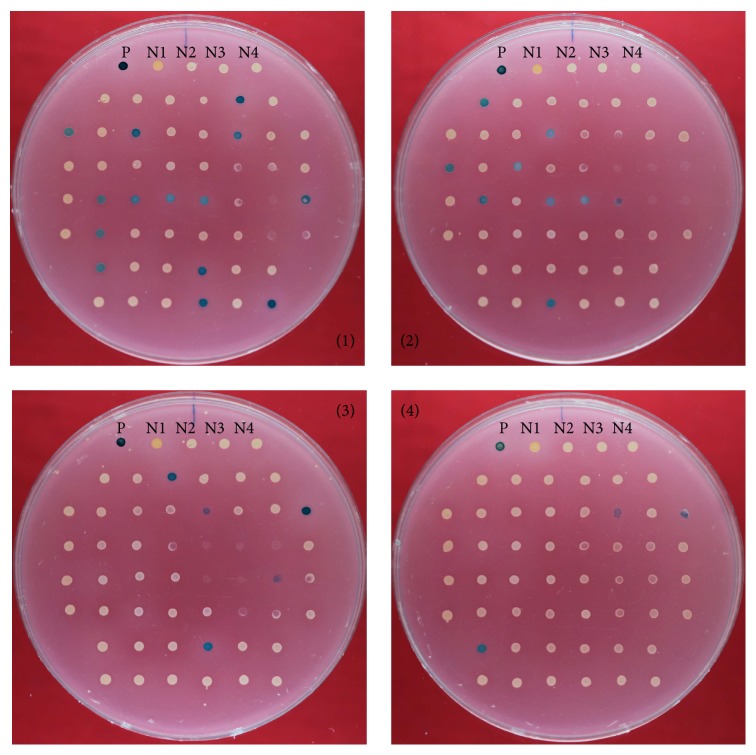
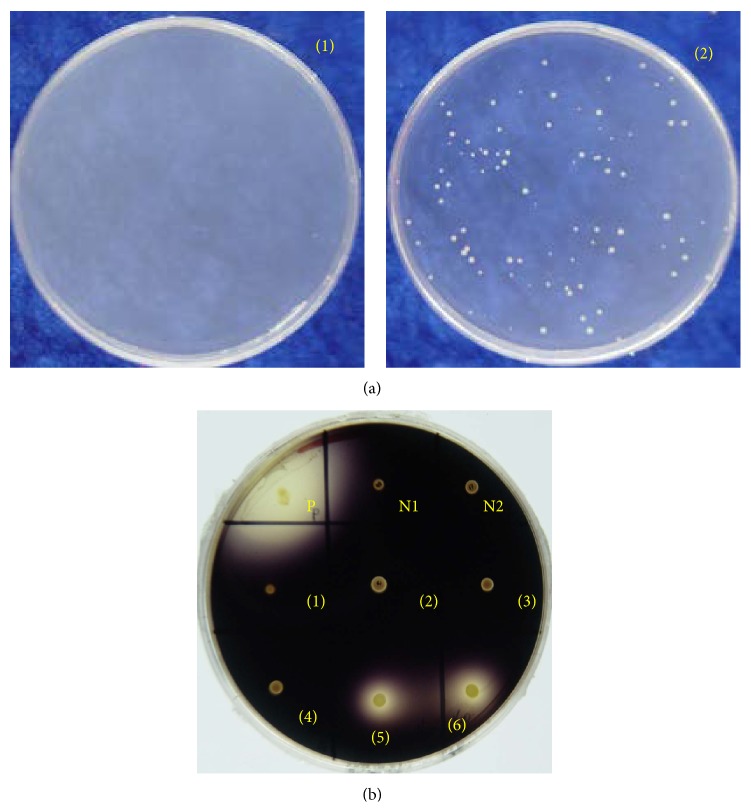
Sequencing of the genomic regions of the transformants of J7-2-1 or J7-2-26 revealed that the mutated leuB or dapD genes had been replaced with wild type leuB and dapD (Figure S6), suggesting that the restoration of growth on the MM plate was due to homologous recombination between the chromosome and the incoming plasmids. Since the circular plasmids could be transformed into auxotrophs, we attempted to use these plasmids to integrate and express exogenous genes in the auxotroph strain J7-2-1. The bgaH gene from Hfx. alicantei [34, 35] was cloned into pUC19-HM1752 to produce the plasmid pUC19-HM1752-BgaH, which was then transformed into J7-2-1 and selected on MM plates. The resultant transformants were analyzed for the expression of the β-galactosidase using MGM plates supplied with X-gal. As shown in Figure 4, 31 of 200 tested transformants (ca. 15.5% of the transformants) displayed β-galactosidase activity, detected by a color change (from pink to blue), on the MGM plates supplied with X-gal.
Figure 4.
Expression of bgaH gene in J7-2-1 via the integrative plasmid pUC19-HM1752-bgaH. The plasmid pUC19-HM1752-bgaH containing bgaH was transformed into strain J7-2-1. In total, 200 transformants were picked out randomly and cultured. The activity of β-galactosidase in these 200 transformants was detected on four MGM plates (50 transformants on each plate) supplied with X-gal (detailed process is discussed in Materials and Methods). If the transformants had β-galactosidase activity, the color of the corresponding colony would turn blue. The results were recorded and pictures taken after the four plates were stored at 42°C for 7–15 days. P: positive control. N1: negative control Hfx. volcanii DS52. N2: negative control J7-2-1. N3: negative control J7-2-1 transformed with J7-2 genome. N4: negative control J7-2-1 transformed with KM1752 fragment.
Besides the bgaH gene from Hfx. alicantei, we also tested the expression of the amyH gene from Har. hispanica DSM 4426 [36] in strain J7-2-1. Similar to bgaH, amyH was cloned into plasmid pUC19-HM1752 and the resulting plasmid pUC19-HM1752-amyH was transformed into J7-2-1. Transformants were screened on MM plates (Figure 5(a)). Twenty-nine transformants were selected and inoculated on the MGM plate containing soluble starch in order to detect the expression and activity of the amyH gene. As shown in Figure 5(b), four out of the 29 transformants exhibited amylase activity and produced a hydrolysis ring on the MGM plate. Consistent with this, the amyH gene could be amplified from two randomly selected transformants that produced the hydrolysis ring, and sequence analysis confirmed that the amyH gene was integrated into the genome by a recombination event at the leuB gene. Other elements on the integrative plasmid, such as the replication region, the antibiotic gene of ampicillin, and the lacZ gene in pUC19, were also detected by PCR amplification from the same two randomly selected transformants conferring amylase activity (Figure S7). Interestingly, we found that specific fragments within the ampicillin resistance gene and ColE1 replication region could be detected in a randomly selected transformant without amylase activity. Currently, it is unclear why the amyH gene was dropped off from the chromosome; one possibility is that some sections of pUC19-HM1752-amyH were integrated into this transformant, but not including the amyH gene or the lacZ gene. Another possibility is that a secondary recombination event occurred between the J7-2-1 genome and the integrative plasmid, resulting in regions including amyH gene or lacZ gene dropping off from genome. Additional studies are required to solve this problem and therefore increase the percentage of positive transformants.
Figure 5.
Expression of amyH gene in J7-2-1 via the integrative plasmid pUC19-HM1752-amyH. The plasmid pUC19-HM1752-amyH containing amyH was transformed into strain J7-2-1. Twenty-nine transformants were picked out randomly and cultured. Amylase activity in these transformants was detected on an MGM plate supplied with 2% (w/v) soluble starch. (detailed process is discussed in Materials and Methods). The plate was flooded with 0.3% I2/0.6% KI solution after incubation at 42°C for 7–15 days. If the transformants had amylase activity, a clear zone indicating starch hydrolysis would be observed. The results were recorded and pictures were taken. (a) (1) Negative control, J7-2-1 transformed with salt solution without DNA. (2) J7-2-1 transformed with integrative plasmid pUC19-HM1752-amyH. (b) MGM plates containing soluble starch are used to detect the expression of amyH gene in J7-2-1 by integrative plasmid pUC19-HM1752-amyH. P. Positive control. N1: negative control Haloferax volcanii DS52. N2: negative control J7-2-1. N3: negative control J7-2-1 transformed with J7-2 Genome. N4: negative control J7-2-1 transformed with KM1752 fragment. 1–29: samples from transformants of J7-2-1 transformed with integrative plasmid pUC19-HM1752-amyH.
The integration relative positions of the exogenous amyH gene on the genomes of transformants conferring amylase activity were identified through amplification experiments and sequencing with different primers and different primer combinations in the plasmid and are shown in Figure 6, confirming that the plasmid was integrated into the chromosome at the leuB locus by homologous recombination.
Figure 6.
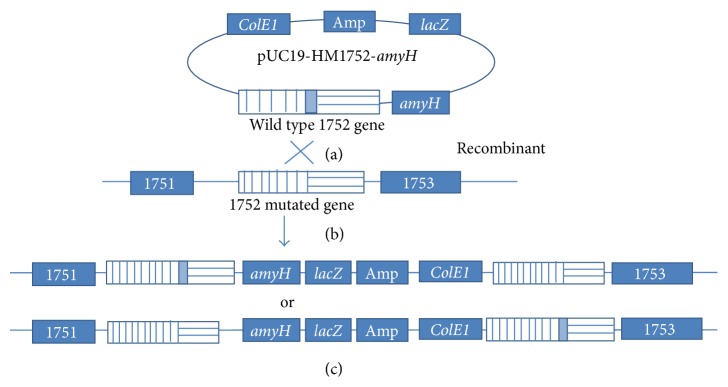
The possible integration position of exogenous DNA in the genome of transformants conferring amylase activity. Strain J7-2-1 was transformed with pUC19-HM1752-amyH and the schematic diagram shows the possible crossover event between the plasmid and J7-2-1 genome. (a) Plasmid pUC19-HM1752-amyH. (b) J7-2-1 genome with the mutated 1752 gene. (c) The possible integration position of exogenous DNA on the genome of the transformants conferring amylase activity.
Nonetheless, the auxotrophs isolated here have been proven to be useful as selectable markers for the expression of exogenous genes in Natrinema. These auxotrophs could also facilitate the development of more sophisticated genetic tools to study other proteins and their functions in the genus of Natrinema and to investigate horizontal gene transfer between strains from this genus and others. In ongoing research, we are attempting to use the elements of the halophilic virus SNJ1 to construct a shuttle vector system for Natrinema sp. J7-2 and CJ7 (another Natrinema strain without any plasmid). We believe that the above provide useful information for further study and will contribute to understanding the genetics of Natrinema genus and halophilic archaea.
Supplementary Material
Supplementary Figure S1: showed the growth of Natrinema sp. J7-2 on MM plate.
Supplementary Figure S2: showed that in the isolation process of mutants, four strains J7-2-1, J7-2-22, J7-2-26 and J7-2-52 which grow on MGM but failed on MM plate were obtained.
Supplementary Figure S3: the auxotroph of the four auxotrophic strains were reconfirmed with the same experimental procedure that they grow on MGM but failed on MM plate.
Supplementary Figure S4: amplification of important genes in the pathways of the amino acid synthesis from the four auxotrophic strains were shown.
Supplementary Figure S5: showed that the molecular characterization of the gene mutation status of the auxotrophic strains.
Supplementary Figure S6: showed the complementation of the mutation genes by auxotrophic strains' transformation and comparison the genes of these different transformants.
Supplementary Figure S7: amplification of specific fragments in J7-2-1/ pUC19-HM1752-amyH transformants conferring amylase activity were shown.
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (973 Program) (no. 2011CB808800), the National Natural Science Foundation of China (no. 30870133), the National Found for Fostering Talents of Basic Sciences (J1103513), and the Research (Innovative) Fund of Laboratory Wuhan University. The authors thank for Dr. Shishen Du (Department of Microbiology, Molecular Genetics and Immunology, University of Kansas Medical Center) for many helpful discussions and for revising the paper.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cline S. W., Ford Doolittle W. Efficient transfection of the archaebacterium Halobacterium halobium . Journal of Bacteriology. 1987;169(3):1341–1344. doi: 10.1128/jb.169.3.1341-1344.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam W. L., Doolittle W. F. Shuttle vectors for the archaebacterium Halobacterium volcanii . Proceedings of the National Academy of Sciences of the United States of America. 1989;86(14):5478–5482. doi: 10.1073/pnas.86.14.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaseio U., Pfeifer F. Transformation of Halobacterium halobium: development of vectors and investigation of gas vesicle synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(17):6772–6776. doi: 10.1073/pnas.87.17.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes M. L., Dyall-Smith M. L. A plasmid vector with a selectable marker for halophilic archaebacteria. Journal of Bacteriology. 1990;172(2):756–761. doi: 10.1128/jb.172.2.756-761.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes M. L., Nuttall S. D., Dyall-Smith M. L. Construction and use of halobacterial shuttle vectors and further studies on Haloferax DNA gyrase. Journal of Bacteriology. 1991;173(12):3807–3813. doi: 10.1128/jb.173.12.3807-3813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmes M., Pfeifer F., Dyall-Smith M. Improved shuttle vectors for Haloferax volcanii including a dual-resistance plasmid. Gene. 1994;146(1):117–121. doi: 10.1016/0378-1119(94)90844-3. [DOI] [PubMed] [Google Scholar]
- 9.Cline S. W., Lam W. L., Charlebois R. L., Schalkwyk L. C., Doolittle W. F. Transformation methods for halophilic archaebacteria. Canadian Journal of Microbiology. 1989;35(1):148–152. doi: 10.1139/m89-022. [DOI] [PubMed] [Google Scholar]
- 10.Waege I., Schmid G., Thumann S., Thomm M., Hausner W. Shuttle vector-based transformation system for Pyrococcus furiosus . Applied and Environmental Microbiology. 2010;76(10):3308–3313. doi: 10.1128/aem.01951-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayrhofer-Iro M., Ladurner A., Meissner C., et al. Transformation of Natrialba magadii: utilization of virus φCh1 elements to establish a shuttle vector system for halo(alkali)philic Archaea. Applied and Environmental Microbiology. 2013;79(8):2741–2748. doi: 10.1128/aem.03287-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertani G., Baresi L. Genetic transformation in the methanogen Methanococcus voltae PS. Journal of Bacteriology. 1987;169(6):2730–2738. doi: 10.1128/jb.169.6.2730-2738.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladapo J., Whitman W. B. Method for isolation of auxotrophs in the methanogenic archaebacteria: role of the acetyl-CoA pathway of autotrophic CO2 fixation in Methanococcus maripaludis . Proceedings of the National Academy of Sciences of the United States of America. 1990;87(15):5598–5602. doi: 10.1073/pnas.87.15.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonuscheit M., Martusewitsch E., Stedman K. M., Schleper C. A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Molecular Microbiology. 2003;48(5):1241–1252. doi: 10.1046/j.1365-2958.2003.03509.x. [DOI] [PubMed] [Google Scholar]
- 15.Mevarech M., Werczberger R. Genetic transfer in Halobacterium volcanii . Journal of Bacteriology. 1985;162(1):461–462. doi: 10.1128/jb.162.1.461-462.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conover R. K., Doolittle W. F. Characterization of a gene involved in histidine biosynthesis in Halobacterium (Haloferax) volcanii: isolation and rapid mapping by transformation of an auxotroph with cosmid DNA. Journal of Bacteriology. 1990;172(6):3244–3249. doi: 10.1128/jb.172.6.3244-3249.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bitan-Banin G., Ortenberg R., Mevarech M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. Journal of Bacteriology. 2003;185(3):772–778. doi: 10.1128/jb.185.3.772-778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allers T., Ngo H.-P., Mevarech M., Lloyd R. G. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Applied and Environmental Microbiology. 2004;70(2):943–953. doi: 10.1128/aem.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peck R. F., DasSarma S., Krebs M. P. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Molecular Microbiology. 2000;35(3):667–676. doi: 10.1046/j.1365-2958.2000.01739.x. [DOI] [PubMed] [Google Scholar]
- 20.Liu H., Han J., Liu X., Zhou J., Xiang H. Development of pyrF-based gene knockout systems for genome-wide manipulation of the archaea Haloferax mediterranei and Haloarcula hispanica . Journal of Genetics and Genomics. 2011;38(6):261–269. doi: 10.1016/j.jgg.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Gulko M. K., Dyall-Smith M., Gonzalez O., Oesterhelt D., Budisa N. How do haloarchaea synthesize aromatic amino acids? PLoS ONE. 2014;9(9) doi: 10.1371/journal.pone.0107475.e107475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi K., Hwang S., Yoon N., Cha J. A novel integrative expression vector for Sulfolobus species. Journal of Microbiology and Biotechnology. 2014;24(11):1503–1509. doi: 10.4014/jmb.1405.05060. [DOI] [PubMed] [Google Scholar]
- 23.McGenity T. J., Gemmell R. T., Grant W. D. Proposal of a new halobacterial genus Natrinema gen. nov., with two species Natrinema pellirubrum nom. nov. and Natrinema pallidum nom. nov. International Journal of Systematic Bacteriology. 1998;48(4):1187–1196. doi: 10.1099/00207713-48-4-1187. [DOI] [PubMed] [Google Scholar]
- 24.Shen P., Chen Y. Plasmid from Halobacterium halobium and its restriction map. Yi Chuan Xue Bao. 1994;21(5):409–416. [PubMed] [Google Scholar]
- 25.Zhang Z., Liu Y., Wang S., et al. Temperate membrane-containing halophilic archaeal virus SNJ1 has a circular dsDNA genome identical to that of plasmid pHH205. Virology. 2012;434(2):233–241. doi: 10.1016/j.virol.2012.05.036. [DOI] [PubMed] [Google Scholar]
- 26.Ye X., Ou J., Ni L., Shi W., Shen P. Characterization of a novel plasmid from extremely halophilic Archaea: nucleotide sequence and function analysis. FEMS Microbiology Letters. 2003;221(1):53–57. doi: 10.1016/s0378-1097(03)00175-7. [DOI] [PubMed] [Google Scholar]
- 27.Mei Y., Chen J., Sun D., et al. Induction and preliminary characterization of a novel halophage SNJ1 from lysogenic Natrinema sp. F5. Canadian Journal of Microbiology. 2007;53(9):1106–1110. doi: 10.1139/w07-072. [DOI] [PubMed] [Google Scholar]
- 28.Shi W., Tang X.-F., Huang Y., Gan F., Tang B., Shen P. An extracellular halophilic protease SptA from a halophilic archaeon Natrinema sp. J7: gene cloning, expression and characterization. Extremophiles. 2006;10(6):599–606. doi: 10.1007/s00792-006-0003-8. [DOI] [PubMed] [Google Scholar]
- 29.Xu Z., Du X., Li T., Gan F., Tang B., Tang X.-F. Functional insight into the C-terminal extension of halolysin SptA from haloarchaeon Natrinema sp. J7. PLoS ONE. 2011;6(8) doi: 10.1371/journal.pone.0023562.e23562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H., Lin L., Zeng C., Shen P., Huang Y.-P. Cloning and characterization of a haloarchaeal heat shock protein 70 functionally expressed in Escherichia coli . FEMS Microbiology Letters. 2007;275(1):168–174. doi: 10.1111/j.1574-6968.2007.00881.x. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y., Huang Y. P., Shen P. The 492-bp RM07 DNA fragment from the halophilic Archaea confers promoter activity in all three domains of life. Current Microbiology. 2003;47(5):388–394. doi: 10.1007/s00284-003-4034-4. [DOI] [PubMed] [Google Scholar]
- 32.Sun G., Jiang A. Plasmid DNA fragments from Halobacterium halobium active as eubacteria promoters in Escherichia coli . Yi Chuan Xue Bao. 1997;24(4):383–384. [PubMed] [Google Scholar]
- 33.Feng J., Liu B., Zhang Z., et al. The complete genome sequence of Natrinema sp. J7-2, a haloarchaeon capable of growth on synthetic media without amino acid supplements. PLoS ONE. 2012;7(7) doi: 10.1371/journal.pone.0041621.e41621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes M. L., Scopes R. K., Moritz R. L., et al. Purification and analysis of an extremely halophilic β-galactosidase from Haloferax alicantei . Biochimica et Biophysica Acta—Protein Structure and Molecular Enzymology. 1997;1337(2):276–286. doi: 10.1016/s0167-4838(96)00174-4. [DOI] [PubMed] [Google Scholar]
- 35.Patenge N., Haase A., Bolhuis H., Oesterhelt D. The gene for a halophilic β-galactosidase (bgaH) of Haloferax alicantei as a reporter gene for promoter analyses in Halobacterium salinarum . Molecular Microbiology. 2000;36(1):105–113. doi: 10.1046/j.1365-2958.2000.01831.x. [DOI] [PubMed] [Google Scholar]
- 36.Zeng C., Zhao Y.-Z., Cui C.-Z., et al. Characterization of the Haloarcula hispanica amyH gene promoter, an archaeal promoter that confers promoter activity in Escherichia coli . Gene. 2009;442(1-2):1–7. doi: 10.1016/j.gene.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Coronado M.-J., Vargas C., Mellado E., et al. The alpha-amylase gene amyH of the moderate halophile Halomonas meridiana: cloning and molecular characterization. Microbiology. 2000;146, part 4:861–868. doi: 10.1099/00221287-146-4-861. [DOI] [PubMed] [Google Scholar]
- 38.Nuttall S. D., Dyall-Smith M. L. HF1 and HF2: novel bacteriophages of halophilic archaea. Virology. 1993;197(2):678–684. doi: 10.1006/viro.1993.1643. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J., Russell D. W. Molecular Cloning: A Laboratory Manual. Vol. 2. CSHL Press; 2001. [Google Scholar]
- 40.Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning. 2. New York, NY, USA: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Dyall-Smith M. The Halohandbook: Protocols for Haloarchaeal Genetics. Melbourne, Australia: Haloarchaeal Genetics Laboratory; 2008. [Google Scholar]
- 42.Hanahan D. Studies on transformation of Escherichia coli with plasmids. Journal of Molecular Biology. 1983;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 43.Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 44.Lv J., Wang S., Zeng C., Huang Y., Chen X. Construction of a shuttle expression vector with a promoter functioning in both halophilic Archaea and Bacteria . FEMS Microbiology Letters. 2013;349(1):9–15. doi: 10.1111/1574-6968.12278. [DOI] [PubMed] [Google Scholar]
- 45.Bernander R., Poplawski A., Grogan D. W. Altered patterns of cellular growth, morphology, replication and division in conditional-lethal mutants of the thermophilic archaeon Sulfolobus acidocaldarius . Microbiology. 2000;146(3):749–757. doi: 10.1099/00221287-146-3-749. [DOI] [PubMed] [Google Scholar]
- 46.Harper M., Lee C. J. Genome-wide analysis of mutagenesis bias and context sensitivity of N-methyl-N′-nitro-N-nitrosoguanidine (NTG) Mutation Research. 2012;731(1-2):64–67. doi: 10.1016/j.mrfmmm.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 47.Kato J., Nagata C., Yang L., et al. Isolation and characterization of the Enterobacter cloacae cheR mutant defective in phosphate taxis. Bioscience, Biotechnology, and Biochemistry. 2001;65(2):456–458. doi: 10.1271/bbb.65.456. [DOI] [PubMed] [Google Scholar]
- 48.Leigh J. A., Albers S.-V., Atomi H., Allers T. Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales . FEMS Microbiology Reviews. 2011;35(4):577–608. doi: 10.1111/j.1574-6976.2011.00265.x. [DOI] [PubMed] [Google Scholar]
- 49.Allers T., Mevarech M. Archaeal genetics—the third way. Nature Reviews Genetics. 2005;6(1):58–73. doi: 10.1038/nrg1504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: showed the growth of Natrinema sp. J7-2 on MM plate.
Supplementary Figure S2: showed that in the isolation process of mutants, four strains J7-2-1, J7-2-22, J7-2-26 and J7-2-52 which grow on MGM but failed on MM plate were obtained.
Supplementary Figure S3: the auxotroph of the four auxotrophic strains were reconfirmed with the same experimental procedure that they grow on MGM but failed on MM plate.
Supplementary Figure S4: amplification of important genes in the pathways of the amino acid synthesis from the four auxotrophic strains were shown.
Supplementary Figure S5: showed that the molecular characterization of the gene mutation status of the auxotrophic strains.
Supplementary Figure S6: showed the complementation of the mutation genes by auxotrophic strains' transformation and comparison the genes of these different transformants.
Supplementary Figure S7: amplification of specific fragments in J7-2-1/ pUC19-HM1752-amyH transformants conferring amylase activity were shown.