Abstract
Context:
Primary aldosteronism is one of the leading causes of secondary hypertension, causing significant morbidity and mortality. A number of genetic defects have recently been identified in primary aldosteronism, whereas we identified mutations in ARMC5, a tumor-suppressor gene, in cortisol-producing primary macronodular adrenal hyperplasia.
Objective:
We investigated a cohort of 56 patients who were referred to the National Institutes of Health for evaluation of primary aldosteronism for ARMC5 defects.
Methods:
Patients underwent step-wise diagnosis, with measurement of serum aldosterone and plasma renin activity followed by imaging, saline suppression and/or oral salt loading tests, plus adrenal venous sampling. Cortisol secretion was also evaluated; unilateral or bilateral adrenalectomy was performed, if indicated. DNA, protein, and transfection studies in H295R cells were conducted by standard methods.
Results:
We identified 12 germline ARMC5 genetic alterations in 20 unrelated and two related individuals in our cohort (39.3%). ARMC5 sequence changes in 6 patients (10.7%) were predicted to be damaging by in silico analysis. All affected patients carrying a variant predicted to be damaging were African Americans (P = .0023).
Conclusions:
Germline ARMC5 variants may be associated with primary aldosteronism. Additional cohorts of patients with primary aldosteronism and metabolic syndrome, particularly African Americans, should be screened for ARMC5 sequence variants because these may underlie part of the known increased predisposition of African Americans to low renin hypertension.
Cardiovascular disease is the leading cause of death worldwide. It is estimated that by 2030, over 23 million people will die from cardiovascular diseases each year (1). Recognition of primary aldosteronism (PA), a major cause of secondary hypertension, and its appropriate treatment may lead to a significant reduction of morbidity associated with cardiovascular diseases. PA may account for up to 10–15% of secondary hypertension (2, 3). The most common causes of PA are bilateral adrenal hyperplasia (60%) and aldosterone-producing adenomas (30%) (2, 4).
Genetic causes of PA are becoming more evident. Somatic mutations in KCNJ5 have been described as a common cause of PA (5–9). Germline mutations in KCNJ5 and CACNA1D cause familial hyperaldosteronism (FH) type III (9–13), whereas as yet unidentified gene(s) on chromosome 7p22 may harbor additional defects for FH type II (14, 15). Glucocorticoid-remediable hyperaldosteronism (also known as FH type I) is caused by a chimeric gene (made by the fusion of the 5′-end of CYP11B1 to the 3′-end of CYP11B2) (15–17).
Glucocorticoid cosecretion in PA has been previously documented (18–24). We and others recently described germline and somatic mutations in an armadillo repeat containing 5 (ARMC5) gene that causes hypercortisolism in primary macronodular adrenal hyperplasia (PMAH) (25–29). ARMC5 maps to 16p11.2 and is likely to be a tumor-suppressor gene (27). Tumors caused by mutations in ARMC5 are likely to be polyclonal: both alleles of ARMC5 carry mutations at the somatic and germline levels. Different nodules on the same adrenal carry different variations of the ARMC5 gene. Although the function of the gene is still under investigation, it appears that ARMC5 inactivation affects steroidogenesis (25, 26).
In this study, we investigated glucocorticoid hormone secretion in patients with PA and queried whether genetic alterations in ARMC5 were involved. This retrospective clinical and genetic study was conducted at the National Institutes of Health (NIH) Clinical Research Center (CRC).
Patients and Methods
Clinical studies and patient samples
A total of 56 patients were evaluated for PA at the NIH CRC in the last 10 years (2004–2013). Age, duration of disease, and vital signs were recorded at the time of initial presentation. All research subjects signed an informed consent. The Institutional Review Boards of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) (until 2010) and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (2010 to present), NIH, approved the research protocol (Clinical Trial Registration no. NCT00005927).
PA testing
All patients underwent a step-wise diagnosis, as previously described (30). Subjects had varying levels of hyperaldosteronism. The aldosterone-to-renin ratio (ARR) was used as an initial screening test to identify potential patients with PA (30). Because the ARR is dependent on the actual renin value, we considered ARR >15 positive for PA and ARR = 10–15 indeterminate. Several patients were taking antihypertensive medications that may have interfered with the testing at screening in other institutions. Eplerenone and spironolactone were discontinued 2–4 and 6–8 weeks before testing, respectively. If necessary, patients on these medications were switched to an α-blocker (doxazosin or prazosin), calcium channel blocker (usually verapamil), and/or hydralazine before all testing at the NIH CRC.
Subjects underwent a saline suppression test (SST) and/or an oral salt-loading test for confirmation of PA (30). SST was performed in the morning with a continuous infusion of 2 L of 0.9% normal saline over 4 hours. Aldosterone was measured at baseline and hourly. Postinfusion plasma aldosterone levels >10 ng/dL were considered strongly positive for PA, whereas values between 5 and 10 ng/dL were considered indeterminate (30, 31). An oral salt-loading test was performed as previously described (30). Briefly, patients took 2-g sodium tablets three times daily with meals for 3 days. Urinary sodium and aldosterone were measured in the 24-hour urine collection from the morning of day 3 until the morning of day 4. Urine aldosterone > 12 μg/24 h along with urine sodium >200 mmol/d confirmed the diagnosis of PA.
All research subjects underwent adrenal computed tomography scanning with contrast. Adrenal venous sampling (AVS) was performed in 50 patients. Some protocol participants refused adrenal sampling or were not surgical candidates. AVS was performed under cosyntropin stimulation, as previously described (32). Good surgical candidates with lateralizing AVS results underwent unilateral adrenalectomy. Patient ADT178.02, who had bilateral adrenocortical tumors upon imaging, was also diagnosed with hypercortisolism and underwent bilateral adrenalectomy; histopathology confirmed bilateral hyperplasia in this patient. All patients that were not surgical candidates were started on mineralocorticoid receptor antagonists.
Other hormonal measurements and dexamethasone (Liddle's) test
Diurnal plasma ACTH and serum cortisol levels were measured, as previously described (26, 33). Briefly, plasma ACTH and serum cortisol levels were measured in the morning (7:30 and 8 am) and late night (11:30 pm and midnight); averages of samples were used in all analyses. Urinary 24-hour 17-hydroxysteroids (17OHS) and urinary free cortisol (UFC) were measured and corrected by gram creatinine (Cr) and body surface area (BSA), respectively, as previously described (26, 34). The 6-day Liddle's (low-dose and high-dose dexamethasone) test was performed, as previously described (26, 35, 36), with the first 2 days as baseline.
ARMC5 and KCNJ5 sequencing and in silico analysis
ARMC5 analysis was performed in peripheral blood leukocytes and in available tumor specimens, as previously described (26). In silico predictions for mutations leading to alternative splicing were made using Alamut version 2.3 (Interactive Biosoftware). We followed guidelines for investigating causality of sequence variants in human disease (37). ARMC5 variants predicted to be damaging by in silico were defined as variants that may alter the normal levels or biochemical function of a gene or gene product. The PolyPhen analysis tool (http://genetics.bwh.harvard.edu/pph/) was used for the predictions. ARMC5 variants predicted to be damaging by in silico analysis were called “predicted damaging,” whereas ARMC5 variants predicted to be benign were called “benign.” Available adrenal tissue from two patients with predicted to be damaging ARMC5 variants were screened for KCNJ5 mutations, as previously described (6).
Immunohistochemistry, Western blot, and H295R cell culture and transfection
Immunohistochemistry for ARMC5 1/10 (rabbit antihuman NBP1-94024; Novus Biologicals) was performed on tissues embedded in paraffin, as previously described (26). Five-micrometer sections were cut and deparaffinized in Histo-Clear (Nationals Diagnostics). These were rehydrated through serial ethanol dilutions. All epitopes were retrieved by 20 minutes of boiling in Vector Antigen Retrieval Solution (H3300; Vector Labs). Primary antibody-antigen was detected with the rabbit secondary antibody (MP-7401; Vector Labs), and horseradish peroxidase activity was detected with 3,3′-diaminobenzidine tetrahydrochloride (SK-4105; Vector Labs).
Forty micrograms of proteins extracted from tumor tissues obtained during surgery were analyzed by Western blot using the ARMC5 antibody and a commercially available glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (rabbit antihuman GAPDH 14C10; Cell signaling), as previously described (26).
Human adrenocortical cancer cells (H295R) were cultured and transfected with control or ARMC5 small interfering RNA (siRNA) to investigate the effect of ARMC5 inactivation on the expression of CYP11B2, as previously described (25). CYP11B2 expression has been analyzed by the reverse transcription quantitative PCR. The list of primer sequences used in the experiment is recorded in Supplemental Table 1.
In vitro analysis of the ARMC5 c.583+26 G>T mutation for alternative splicing
Wild-type ARMC5 exon 2, intron 2, and exon 3, together with flanking intronic regions, were amplified from the genomic DNA of a healthy donor (primers: forward, 5′-caggccacattctccaggtt-3′; reverse, 5′-agacagaataggggtggaggt-3′). The purified fragment was ligated into pGEMTeasy (Promega) and then subcloned into pSPL3 (Life Technologies). The mutation (c.583+26G>T, rs9921490) was introduced by site-directed mutagenesis using the QuikChange mutagenesis kit (Agilent).
Sequence and orientation of all constructs was confirmed by Sanger sequencing. The pSPL3 plasmids containing either wild-type or mutated inserts were transiently transfected into HEK293 cells and H295R cells using Lipofectamine 2000 (Life Technologies). After 24 hours, RNA was extracted (RNeasy kit; QIAGEN) and reverse transcribed (Superscript III; Life Technologies). The SD6 and SA2 primers (binding to the pSPL3 exonic regions flanking the insert) were used for RT-PCR, and products were analyzed for size differences on an agarose gel. Bands were gel-purified using the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel) and analyzed by Sanger sequencing.
Statistical analysis
Data are described as frequencies and percentages or mean ± SD, unless otherwise indicated, and were analyzed using SAS version 9.2 (SAS Institute, Inc). Continuous data were compared between the patients with ARMC5 variants predicted to be damaging and patients without and/or with benign ARMC5 variants using two-sample Student's t tests or Wilcoxon rank-sum test, as appropriate. Log-transformation of data was carried out as necessary. Categorical data were compared using Fisher's exact tests. Mixed models were used for repeated-measures analysis of 17OHS/Cr, UFC/BSA, and aldosterone levels during saline suppression between the patient's groups. Percentage change was computed based on the change from baseline to the last 24 hours of the Liddle's test.
Fisher's exact test and χ2 test with Yates correction were used to compare the distribution of allele frequencies in ARMC5 mutations between our study samples and the general population from the 1000 Genomes and between African Americans in our study samples and African Americans from the National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project (NHLBI GO ESP). A P value <.05 was considered statistically significant.
Results
Demographics
The mean age of our subjects was 50.7 ± 9.6 years at the time of presentation to the NIH CRC. Six (10.7%) patients carried germline ARMC5 variants that were predicted damaging. Most subjects were male (60.7%), and 44.6% were Caucasian. Interestingly, all patients carrying predicted damaging ARMC5 mutations were African Americans (P = .0023) (Table 1). There were significant differences in allele frequencies of ARMC5 variants between our study cohort and the general population from both the 1000 Genomes and the ESP. Indeed, the allele frequencies were significantly higher for all five predicted damaging single nucleotide polymorphisms (SNPs). In addition, we observed statistically significant differences in the same ARMC5 variant frequencies in our African American sample compared to the African American population from the NHLBI GO ESP (Table 2). Available tumor specimens from 2 patients with predicted damaging ARMC5 variants had no somatic KCNJ5 gene defects.
Table 1.
Demographic Characteristics
| Demographic Characteristics | All Cohort | Patients With Predicted Damaging ARMC5 Variants | Patients Without and/or With Benign ARMC5 Variants |
|---|---|---|---|
| Predicted damaging ARMC5 variants | 56 | 6 (10.7) | 50 (89.3) |
| Age at the time of presentation to NIH, y | 50.7 ± 9.6 | 48.5 ± 7.6 | 51.0 ± 9.9 |
| Sex | |||
| Females | 22 (39.3) | 1 (16.7) | 21 (42.0) |
| Males | 34 (60.7) | 5 (83.3) | 29 (58.0) |
| Racea | |||
| Asian | 4 (7.1) | 0 | 4 (8.0) |
| Black/African American | 22 (39.3) | 6 (100.0) | 16 (32.0) |
| Multiracial | 2 (3.6) | 0 | 2 (4.0) |
| Unknown race | 3 (5.4) | 0 | 3 (6.0) |
| White | 25 (44.6) | 0 | 25 (50.0) |
| Ethnicity | |||
| Hispanic or Latino | 5 (8.9) | 0 | 5 (10.0) |
| Not Hispanic or Latino | 51 (91.1) | 6 (100.0) | 45 (90.0) |
Data are expressed as number (percentage) or mean ± SD.
Reflects the comparison of simplified racial groups of African American vs All Others. The P value for the detailed group comparison was <.05.
Table 2.
Allele Frequency (Minor Allele) of Sequence Variations in ARMC5 in Patients With PA and in Controls From the 1000 Genomes Project (Using All Population as a Base) and in African American Controls in the ESP6500
| Protein Change | SNP ID | Prediction | All Patients With PA vs ALL in the 1000 Genomes |
AA Patients With PA vs AAs in ESP6500 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ARMC5 SNPs in Patients (n = 56) | ARMC5 SNPs in 1000 Genomes Sample (n = 1092) | χ2c | P Valued | ARMC5 SNPs in AA Patients (n = 22)f | ARMC5 SNPs in AA ESPS6500 Sample (n = 2203)e | χ2 | P Value | |||
| p.F14Y | rs151069962 | Benign | 3 | 63 | 0.03 | NS | 2 | 48 | 2.11 | NS |
| p.L156F | rs114930262 | Damaging | 4 | 17 | 6.41 | .016 | 4 | 81 | 8.84 | .009 |
| p.I170V | rs35923277 | Benign | 4 | 54 | 0.18 | NS | 0 | 36 | 0.37 | NS |
| Splice | rs9921490 | Benign | 4 | 82 | 0.02 | NS | 4 | 504 | 0.07 | NS |
| p.G323A | rs35461188 | Benign | 1 | 3 | 0.50 | NS | 0 | 0 | N/A | N/A |
| Splice | rs200655247 | Damaging | 1 | 0 | 4.39 | .049 | 1 | 4 | 4.16 | .049 |
| p.P507L | rs142376949 | Benign | 2 | 19 | 0.24 | NS | 0 | 20 | 0.20 | NS |
| p.T643M | rs370836071 | Damaging | 1 | 0 | 4.39 | .049 | 1 | 0 | 24.55 | .010 |
| p.G798A | rs115611533 | Benign | 1 | 17 | 0.17 | NS | 1 | 94 | 0.00 | NS |
| p.P826Ha | Novel | Damaging | 1 | 0 | 4.39 | .049 | 1 | 0 | 24.55 | .010 |
| p.R898Wa | Faucz et alb | Damaging | 1 | 0 | 4.39 | .049 | 1 | 0 | 24.55 | .010 |
| Splice | rs200289596 | Benign | 1 | 0 | 4.39 | .049 | 0 | 1 | 0.01 | NS |
Abbreviations: N/A, not available; NS, nonsignificant; ALL, all individuals from 1000 Genomes database are being considered; AA, African Americans.
Variation newly identified in PA patients, comparing with the database.
Variation previously described in patients with macronodular adrenal hyperplasia, but still not present in the National Center for Biotechnology Information Genome database (26).
χ2 is calculated after Yates correction for continuity (Yates correction was applied for all calculations having a number <10 in any cell of the contingency table).
The Fisher's exact test was used for the P values because the numbers are small.
The total number of analyzed samples differs among the variations (but they are generally 2203).
African American data from NHLBI GO ESP (Exome Variant Server, NHLBI GO Exome Sequencing Project [ESP], Seattle, WA). http://evs.gs.washington.edu/EVS/. Accessed July 25, 2014.
Clinical parameters and baseline biochemical data
Baseline clinical signs and biochemistry are presented in Supplemental Table 2. There was no statistical significance in vital signs between the group carrying predicted damaging ARMC5 variants and the group without and/or with benign variants. Importantly, average fasting serum glucose in the entire patient cohort was 115.71 ± 38.03 mg/dL. Eight patients (14.3%) with PA had impaired fasting glucose, and 19 others (33.9%) met the criteria for the diagnosis of diabetes mellitus.
PA screening, confirmatory testing, and adrenal venous sampling
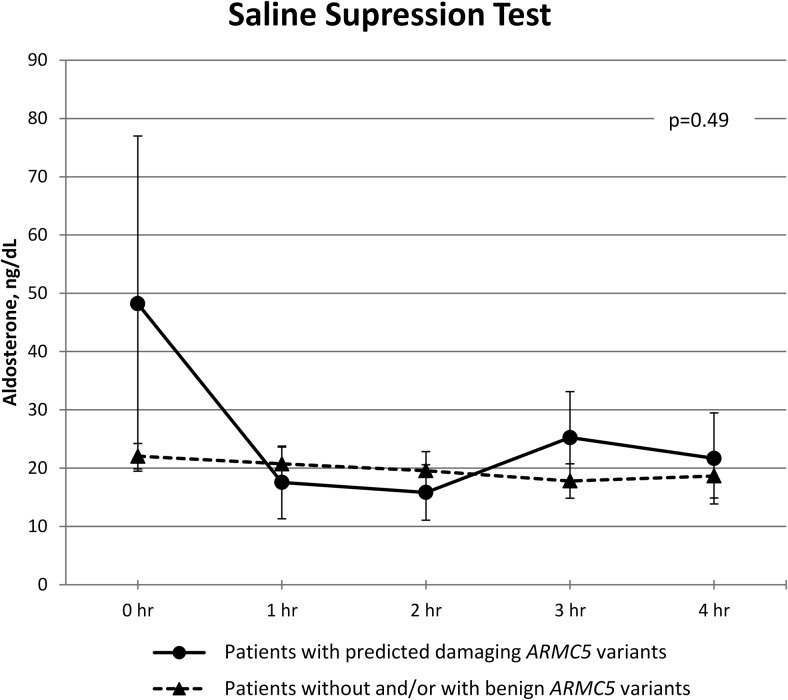
The results of the PA screening and confirmatory testing are shown in Supplemental Table 2. Data from the SST are shown in Figure 1. There was no difference in serum aldosterone during the SST between patients carrying predicted damaging ARMC5 variants and patients without and/or with benign ARMC5 variants across time (P = .49). Additional clinical characteristics are described in the Supplemental Table 3. Adrenal venous sampling data from five patients with predicted damaging ARMC5 variants are presented in Supplemental Figures 1–5.
Figure 1.
SST. Aldosterone was checked at baseline and hourly. Data are expressed as mean ± SEM. P = .49 from repeated measures analysis. Although patients with damaging ARMC5 variants started with higher aldosterone values, the difference was not significant.
Hypothalamic-pituitary-adrenal axis testing
Baseline plasma ACTH, average of diurnal serum cortisol, and average baseline urinary 17OHS/Cr and UFC/BSA are presented in Supplemental Table 2. There were no statistically significant differences between the groups. Notably, late-night serum cortisol in the whole cohort was 4.06 ± 2.28 μg/dL.
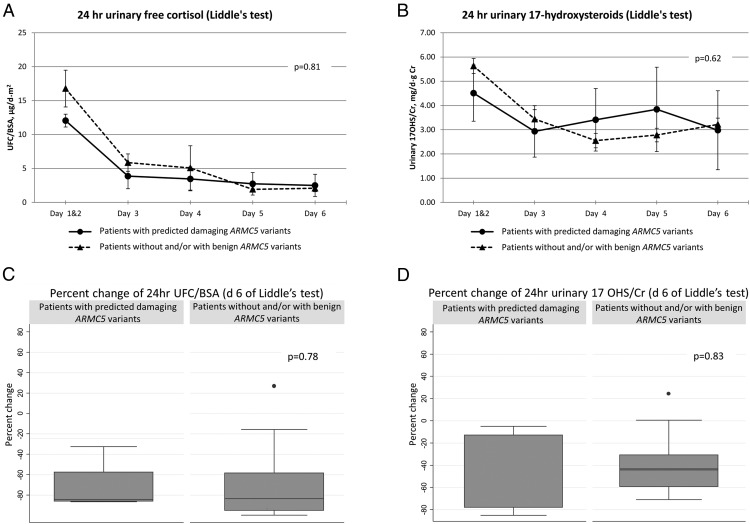
Dexamethasone (Liddle's) test
UFC/BSA measurements were not different between the groups (P = .81) (Figure 2A). Subjects with predicted damaging ARMC5 variants also did not have significantly different urinary 17OHS/Cr levels throughout the high- and low-dose dexamethasone administration during the Liddle's test (P = .62) (Figure 2B). There was no difference in the percentage change of 17OHS/Cr (P = .83) or UFC/BSA (P = .78) when baseline values (days 1–2) were compared to the day 6 values at the end of the Liddle's test (Figure 2, C and D).
Figure 2.
Liddle's test. A, 24-hour UFC, corrected by BSA (P = .81 from repeated measures analysis). B, 24-hour urinary 17OHS, corrected by gram creatinine (P = .62 from repeated measures analysis). C, Percentage change of 24-hour UFC, corrected for BSA (d 6 of Liddle's test) (P = .78). D, Percentage change of 17OHS, corrected by gram creatinine (P = .83).
ARMC5 mutations
We identified 12 ARMC5 genetic alterations in 20 unrelated and two related individuals from our cohort of 56 patients (39.3%). Nine of the germline variations were missense and resulted in an amino acid substitution, and three were intronic variations near the donor or dinucleotide acceptor. Seven of the missense and all three of the intronic variants were previously described in public databases (38) (Table 2). Two missense variants that are not present in the database were identified: the mutation c.2692C>T (p.R898W) that was previously described by our group in patients with PMAH (26), and a novel missense mutation c.2476C>A (p.P826H). Both of them are predicted as damaging. All sequence variations were found in the heterozygote state in germline DNA; tumor DNA sequencing confirmed the germline mutations found in these patients (p.T643M, p.L156F, and c.1370-3C>A) but failed to identify additional somatic mutations in ARMC5 exons. There were also no KCNJ5 defects in these samples.
ARMC5 in silico analysis
In silico analysis predicted a likely benign effect on the ARMC5 protein function for the five (of nine) previously described missense variants found in our cohort. The variants p.L156F, p.T643M, and p.R898W (previously found in patients with PMAH) were predicted to significantly impair protein function as well as the novel mutation found in our cohort (p.P826H) (Table 3). From the three intronic variants that were found (c.583+26G>T [rs9921490], c.1370-3C>A [rs200655247], and c.475+11C>T [rs200289596]), two were predicted (by in silico analysis) to affect splicing: c.583+26G>T and c.1370-3C>A.
Table 3.
In Silico Modelling of the Effect of ARMC5 Missense Substitution on the Protein Function in Patients With ARMC5 Variantsa
| DNA Change | Protein Change | SNP ID | Domains | In Silico Modelling |
Interspecies Alignment |
||||
|---|---|---|---|---|---|---|---|---|---|
| Prediction | Scorea | Mus musculus | Dasypus novemcinctus | Xenopus tropicalis | Petromyzon marinus | ||||
| c.41T>A | p.F14Y | rs151069962 | - x - | Likely benign | 0.255 | F | - | D | A |
| c.466C>T | p.L156F | rs114930262 | Armadillo | Possible damaging | 0.527 | L | - | - | - |
| c.508A>G | p.I170V | rs35923277 | Armadillo | Likely benign | 0.311 | I | I | I | I |
| c.968G>C | p.G323A | rs35461188 | Armadillo | Likely benign | 0.001 | G | G | A | - |
| c.1520C>T | p.P507L | rs142376949 | - x - | Likely benign | 0.001 | P | L | L | R |
| c.1928C>T | p.T643M | rs370836071 | - x - | Probably damaging | 1.000 | T | T | T | N |
| c.2393G>C | p.G798A | rs115611533 | BTB/POZ-like | Likely benign | 0.015 | G | S | C | A |
| c.2476C>A | p.P826H | Novel | - x - | Probably damaging | 0.895 | P | P | S | - |
| c.2692C>T | p.R898W | Faucz et al (26) | - x - | Probably damaging | 1.000 | R | R | - | R |
Letters in interspecies alignment columns are relative to the amino acid present in the position and the absence of a letter means there is no amino acid at that specific position: F, phenylalanine; D, aspartic acid; A, alanine; L, leucine; I, isoleucine; G, glycine; P, proline; R, arginine; T, threonine; N, asparagine; S, serine; C, cysteine; dash, no amino acid present.
PolyPhen-2 was used as standard. Scores go from 0.000 to 1.000. Greater score indicates higher probability to impair the protein function. The main factors taken into account for the calculation of the score are: 1) difference in the thermo-physical properties of the wild-type and mutant protein; and 2) evolutionary preservation of the residue in the corresponding position.
ARMC5 expression, cell culture, and transfection
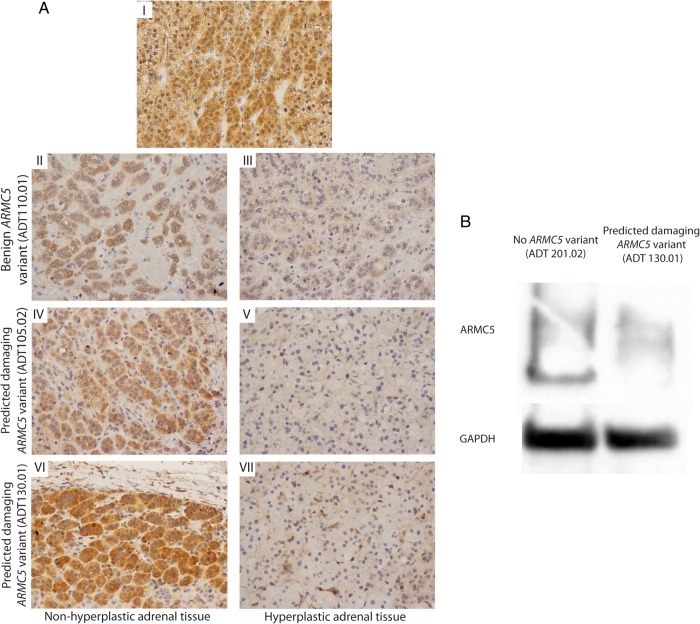
ARMC5 immunohistochemistry was performed on three adrenal tumors from three nonrelated patients and one nonhyperplastic adjacent tissue (Figure 3A). As previously described (25), ARMC5 was strongly expressed in the nonhyperplastic adjacent adrenal tissue (Figure 3A-I). Patient ADT110.01 had a benign germline ARMC5 variant, and two other tumors were from patients with predicted damaging defects (ADT105.02 and ADT130.01). For the ADT110.01, even if a small area had a concentration of some cytoplasmic ARMC5 staining (Figure 3A-II), the expression of ARMC5 was overall lower (Figure 3A-III). In both tumors with predicted damaging variants, ARMC5 expression was stronger in the adjacent tissue (Figure 3, A-IV and A-V) than in the nodular area where its expression was almost completely abolished (Figure 3, A-VI and A-VII). This has been confirmed by a Western blot experiment (in patient with predicted damaging ARMC5 variant, ADT130.01). Indeed, ARMC5 expression is clearly reduced in the adrenal adenoma from this patient compared to the expression in one adenoma without ARMC5 variant (ADT201.02) (Figure 3B). Sequencing of the ARMC5 gene in these tumors failed to identify any additional somatic alterations in ARMC5 exons. Transfection of H295R cells showed that ARMC5 silencing down-regulated the expression of CYP11B2 (Supplemental Table 1 and Supplemental Figure 6).
Figure 3.
ARMC5 expression in adrenocortical nodules. A, Immunohistochemistry of adrenocortical nodules stained for ARMC5. A-I, Nonhyperplastic adrenal tissue, surrounding aldosterone-producing adenoma (no ARMC5 mutations). A-II and A-III, ADT110.01 (with a benign ARMC5 variant) is a 51-year-old male with a 5-year history of hypertension who underwent left adrenalectomy, which revealed a dominant left adrenal nodule 1.1 × 1.0 × 0.7 cm and surrounding hyperplasia. A-IV and A-V, ADT105.2 (with a predicted damaging ARMC5 variant) is a 63-year-old male with a 35-year history of hypertension who underwent left adrenalectomy, revealing a dominant nodule 1.3 × 0.8 × 0.8 cm, surrounded by macro- and micronodular adrenal hyperplasia. A-VI and A-VII, ADT130.01 is 44-year-old male (with a predicted damaging ARMC5 variant) with a 12-year history of hypertension. Computed tomography revealed bilateral adrenal hyperplasia; left adrenalectomy revealed a 1.4-cm dominant nodule with surrounding hyperplasia. B, Western blot analysis of ARMC5 protein in normal adrenal (N) and aldosterone-producing adrenal adenoma from a patient without ARMC5 mutations (ADT201.02) and from a patient with a predicted damaging ARMC5 variant (ADT130.01).
Analysis of alternative splicing of ARMC5 c.583 +26G>T mutation
The ARMC5 c.583 +26G>T variant (rs9921490), which was found in four patients in our cohort (Table 2), was predicted in silico to create a splice acceptor site at c.583+31 that could lead to intron retention of 69 3′ bases of intron 2. This prediction was tested in a minigene assay in which exon 2, intron 2, and exon 3 of wild type compared to mutant ARMC5 and the flanking intronic sequences were cloned into the pSPL3 vector. The plasmids were transfected into HEK293 cells and the adrenal carcinoma cell line H295R in which mRNA synthesis from the plasmids using the cells' own transcription and splicing machinery would lead to mRNA products with exon 2 and exon 3 of ARMC5 flanked by two exons from the pSPL3 vector. The presence or absence of alternative splicing in the mutant compared to the wild type was assessed by RT-PCR using primers binding to the flanking pSPL3 exonic regions. RT-PCR products from the wild-type and mutant ARMC5 plasmids in cDNA from both cell lines were of equal apparent size, and the presence of both complete exons 2 and 3 and the absence of intronic sequences was confirmed by sequencing of the gel purified bands. Hence, there was no evidence that the ARMC5 c.583+26G>T variant led to alternative splicing in those cell lines, although a different effect on splicing in the adrenal tissue of PA patients carrying this variant cannot be excluded (Supplemental Figure 7).
Discussion
Our group and others previously described ARMC5 inactivating mutations as a frequent cause of macronodular adrenal hyperplasia and Cushing syndrome (25–29). In the present study, we also identified predicted damaging germline ARMC5 gene mutations in 10.7% of patients with PA (six of 56 patients). Two available tumors from patients with predicted damaging ARMC5 variants were sequenced, and no additional somatic changes were found. Notably, all subjects with predicted damaging ARMC5 variants were African American (P = .0023). To our knowledge, this is the first published report showing that germline predicted damaging ARMC5 mutations may play a role in the development of PA.
PA caused by identifiable germline mutations is rare. A series of somatic mutations confined to the adrenal cortex have been reported recently, which may be responsible for about half of the aldosterone-producing adenomas (12). Genes encoding components of the Kir 3.4 (GIRK4) potassium channel (KCNJ5), sodium/potassium and calcium ATPases (ATP1A1 and ATP2B3), and a voltage-dependent C-type calcium channel (CACNA1D) have mutations causing PA (7, 10, 39). FH type I (or glucocorticoid-remediable hyperaldosteronism) is caused from a chimeric gene (5′-end of CYP11B1 fused to 3′-end of CYP11B2) and accounts for up to 1% of PA (40). Bilateral expression of mutant KCNJ5 results in florid hyperaldosteronism requiring early bilateral adrenalectomy and causes FH type III (13, 41). FH type II, the most common form of hereditary PA, representing approximately 6% of PA, is likely to be caused by as yet unknown gene(s) (42).
Could germline ARMC5 gene alterations in patients with PA be a new form of PA, perhaps, FH type IV? ARMC5 appears to function as a tumor-suppressor gene with incomplete autosomal dominant penetrance (25). Preliminary data in the animal models suggests that inactivation of the ARMC5 gene may lead to severe developmental abnormalities of the adrenal glands in mouse and in zebra fish inter-renal tissue (43). The ARMC5 gene appears to be expressed throughout the adrenal cortex (including zona glomerulosa). Inactivation of ARMC5 in the present study was associated with decreased expression of CYP11B2, the gene encoding aldosterone synthase enzyme. This is consistent with the effects of ARMC5 on other steroidogenic enzymes (25). We speculate that whereas aldosterone-secretory capacity of individual zona glomerulosa cells may be somewhat reduced, the overall increased adrenocortical mass leads to higher steroid hormone secretion including hyperaldosteronism, as seen with hypercortisolism in patients with PMAH (25, 26).
African Americans are known to have higher rates of hypertension than Caucasians, although the exact mechanism is not clear (44–47). Kidambi et al (46) found that plasma aldosterone was correlated overall with blood pressure and was higher in the hypertensive subjects, compared to normotensive adults. Plasma renin activity was inversely related to blood pressure. Similar findings were reported in a prospective observational study of children (44). Spence (45) highlighted the importance of the measurement of aldosterone and plasma renin activity in African Americans. Could there be a selective advantage for people of African descent to excrete water more slowly to survive in hot climates (48)? Could ARMC5 thus be a genetic cause of resistant hypertension in African Americans? Obviously, larger studies are needed to answer these important questions.
Metabolic abnormalities, such as obesity, insulin resistance, and cortisol cosecretion in patients with PA, have been documented (46, 49–52). Our cohort had an elevated late-night serum cortisol (mean = 4.06 ± 2.28 μg/dL; normal, <1.8 μg/dL) and high fasting serum glucose levels (mean = 115.71 ± 38.03 mg/dL); however, it was not statistically significantly different in subjects with predicted damaging ARMC5 variants. We speculate that adrenal cells producing aldosterone in zona glomerulosa may have similar origin to cells producing cortisol in zona fasciculata, as suggested by others (53, 54). This may be the reason why some patients with PA may be cosecretors. We also speculate that obesity may be the cause of fasting hyperglycemia in our patients.
To conclude, we found that 10.7% of our cohort with PA had predicted damaging ARMC5 gene defects, which may be a novel cause of PA. Interestingly, ethnic background may play a role in this condition, given that 100% of affected subjects were African Americans. Selected family members of the patients with ARMC5 defects may be screened and counseled accordingly—a study that is ongoing in our center. This novel finding should be confirmed in larger cohorts of patients with low-renin hypertension and especially those involving African Americans.
Acknowledgments
We thank Diane Cooper, MSLS, NIH Library, for providing assistance in writing this manuscript.
This research was supported in part by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (protocol 00-CH-0160; Clinical and Molecular Analysis of ACTH-Independent Steroid Hormone Production in Adrenocortical Tissue). These organizations had no further role in the collection, analysis, and interpretation of data, in the writing of the report, and in the decision to submit the paper for publication. The principal investigator had full access to all the data in the case and takes responsibility for the integrity of the data and the accuracy of the data interpretation.
Clinical Trial Registration No.: NCT00005927.
Disclosure Summary: M.Z., P.X., F.R.F., A.G., A.B., M.H.S.-R., M.B., N.S., M.M.Q., M.M., A.H., S.B.A., R.L., G.A., S.E., L.D., B.R., A.D., S.Y.G., R.N., E.K., M.B.L., and C.A.S. have nothing to declare. J.B. received honoraria from Atterocor, grant support from Novartis Pharmaceuticals, and fees from Ipsen.
Footnotes
- ARMC5
- armadillo repeat containing 5
- ARR
- aldosterone-to-renin ratio
- AVS
- adrenal venous sampling
- BSA
- body surface area
- Cr
- creatinine
- FH
- familial hyperaldosteronism
- 17OHS
- 17-hydroxysteroid
- PA
- primary aldosteronism
- PMAH
- primary macronodular adrenal hyperplasia
- SNP
- single nucleotide polymorphism
- SST
- saline suppression test
- UFC
- urinary free cortisol.
References
- 1. Finegold JA, Asaria P, Francis DP. Mortality from ischaemic heart disease by country, region, and age: statistics from World Health Organisation and United Nations. Int J Cardiol. 2013;168:934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mulatero P, Stowasser M, Loh KC, et al. Increased diagnosis of primary aldosteronism, including surgically correctable forms, in centers from five continents. J Clin Endocrinol Metab. 2004;89:1045–1050. [DOI] [PubMed] [Google Scholar]
- 3. Rossi GP, Bernini G, Caliumi C, et al. A prospective study of the prevalence of primary aldosteronism in 1,125 hypertensive patients. J Am Coll Cardiol. 2006;48:2293–2300. [DOI] [PubMed] [Google Scholar]
- 4. Schirpenbach C, Reincke M. Primary aldosteronism: current knowledge and controversies in Conn's syndrome. Nat Clin Pract Endocrinol Metab. 2007;3:220–227. [DOI] [PubMed] [Google Scholar]
- 5. Torpy DJ, Stratakis CA, Gordon RD. Linkage analysis of familial hyperaldosteronism type II–absence of linkage to the gene encoding the angiotensin II receptor type 1. J Clin Endocrinol Metab. 1998;83:1046. [DOI] [PubMed] [Google Scholar]
- 6. Xekouki P, Hatch MM, Lin L, et al. KCNJ5 mutations in the National Institutes of Health cohort of patients with primary hyperaldosteronism: an infrequent genetic cause of Conn's syndrome. Endocr Relat Cancer. 2012;19:255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choi M, Scholl UI, Yue P, et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011;331:768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boulkroun S, Beuschlein F, Rossi GP, et al. Prevalence, clinical, and molecular correlates of KCNJ5 mutations in primary aldosteronism. Hypertension. 2012;59:592–598. [DOI] [PubMed] [Google Scholar]
- 9. Charmandari E, Sertedaki A, Kino T, et al. A novel point mutation in the KCNJ5 gene causing primary hyperaldosteronism and early-onset autosomal dominant hypertension. J Clin Endocrinol Metab. 2012;97:E1532–E1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scholl UI, Goh G, Stölting G, et al. Somatic and germline CACNA1D calcium channel mutations in aldosterone-producing adenomas and primary aldosteronism. Nat Genet. 2013;45:1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scholl UI, Nelson-Williams C, Yue P, et al. Hypertension with or without adrenal hyperplasia due to different inherited mutations in the potassium channel KCNJ5. Proc Natl Acad Sci USA. 2012;109:2533–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Funder JW. Genetics of primary aldosteronism. Front Horm Res. 2014;43:70–78. [DOI] [PubMed] [Google Scholar]
- 13. Monticone S, Bandulik S, Stindl J, et al. A case of severe hyperaldosteronism caused by a de novo mutation affecting a critical salt bridge Kir3.4 residue. J Clin Endocrinol Metab. 2015;100:E114–E118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lafferty AR, Torpy DJ, Stowasser M, et al. A novel genetic locus for low renin hypertension: familial hyperaldosteronism type II maps to chromosome 7 (7p22). J Med Genet. 2000;37:831–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stowasser M, Gordon RD, Tunny TJ, Klemm SA, Finn WL, Krek AL. Familial hyperaldosteronism type II: five families with a new variety of primary aldosteronism. Clin Exp Pharmacol Physiol. 1992;19:319–322. [DOI] [PubMed] [Google Scholar]
- 16. Gordon RD, Klemm SA, Tunny TJ, Stowasser M. Primary aldosteronism: hypertension with a genetic basis. Lancet. 1992;340:159–161. [DOI] [PubMed] [Google Scholar]
- 17. Gordon RD, Stowasser M, Klemm SA, Tunny TJ. Primary aldosteronism–some genetic, morphological, and biochemical aspects of subtypes. Steroids. 1995;60:35–41. [DOI] [PubMed] [Google Scholar]
- 18. Späth M, Korovkin S, Antke C, Anlauf M, Willenberg HS. Aldosterone- and cortisol-co-secreting adrenal tumors: the lost subtype of primary aldosteronism. Eur J Endocrinol. 2011;164:447–455. [DOI] [PubMed] [Google Scholar]
- 19. Adachi J, Hirai Y, Terui K, et al. A report of 7 cases of adrenal tumors secreting both cortisol and aldosterone. Intern Med. 2003;42:714–718. [DOI] [PubMed] [Google Scholar]
- 20. Willenberg HS, Späth M, Maser-Gluth C, et al. Sporadic solitary aldosterone- and cortisol-co-secreting adenomas: endocrine, histological and genetic findings in a subtype of primary aldosteronism. Hypertens Res. 2010;33:467–472. [DOI] [PubMed] [Google Scholar]
- 21. Allan CA, Kaltsas G, Perry L, et al. Concurrent secretion of aldosterone and cortisol from an adrenal adenoma - value of MRI in diagnosis. Clin Endocrinol (Oxf). 2000;53:749–753. [DOI] [PubMed] [Google Scholar]
- 22. Fujii H, Kamide K, Miyake O, et al. Primary aldosteronism combined with preclinical Cushing's syndrome in an elderly patient. Circ J. 2005;69:1425–1427. [DOI] [PubMed] [Google Scholar]
- 23. Guthrie GP, Jr, Kotchen TA. Hypertension and aldosterone overproduction without renin suppression in Cushing's syndrome from an adrenal adenoma. Am J Med. 1979;67:524–528. [DOI] [PubMed] [Google Scholar]
- 24. Honda T, Nakamura T, Saito Y, Ohyama Y, Sumino H, Kurabayashi M. Combined primary aldosteronism and preclinical Cushing's syndrome: an unusual case presentation of adrenal adenoma. Hypertens Res. 2001;24:723–726. [DOI] [PubMed] [Google Scholar]
- 25. Assié G, Libé R, Espiard S, et al. ARMC5 mutations in macronodular adrenal hyperplasia with Cushing's syndrome. N Engl J Med. 2013;369:2105–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Faucz FR, Zilbermint M, Lodish MB, et al. Macronodular adrenal hyperplasia due to mutations in an armadillo repeat containing 5 (ARMC5) gene: a clinical and genetic investigation. J Clin Endocrinol Metab. 2014;99:E1113–E1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berthon A, Stratakis CA. From beta-catenin to ARM-repeat proteins in adrenocortical disorders. Horm Metabol Res. 2014;46:889–896. [DOI] [PubMed] [Google Scholar]
- 28. Gagliardi L, Schreiber AW, Hahn CN, et al. Armc5 mutations are common in familial bilateral macronodular adrenal hyperplasia. J Clin Endocrinol Metab. 2014;99:E1784–E1792. [DOI] [PubMed] [Google Scholar]
- 29. Elbelt U, Trovato A, Kloth M, et al. Molecular and clinical evidence for an ARMC5 tumor syndrome: concurrent inactivating germline and somatic mutations are associated with both primary macronodular adrenal hyperplasia and meningioma. J Clin Endocrinol Metab. 2015;100:E119–E128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Funder JW, Carey RM, Fardella C, et al. Case detection, diagnosis, and treatment of patients with primary aldosteronism: an Endocrine Society Clinical Practice Guideline. J Clin Endocrinol Metab. 2008;93:3266–3281. [DOI] [PubMed] [Google Scholar]
- 31. Rossi GP, Belfiore A, Bernini G, et al. Prospective evaluation of the saline infusion test for excluding primary aldosteronism due to aldosterone-producing adenoma. J Hypertens. 2007;25:1433–1442. [DOI] [PubMed] [Google Scholar]
- 32. Webb R, Mathur A, Chang R, et al. What is the best criterion for the interpretation of adrenal vein sample results in patients with primary hyperaldosteronism? Ann Surg Oncol. 2012;19:1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Papanicolaou DA, Yanovski JA, Cutler GB, Jr, Chrousos GP, Nieman LK. A single midnight serum cortisol measurement distinguishes Cushing's syndrome from pseudo-Cushing states. J Clin Endocrinol Metab. 1998;83:1163–1167. [DOI] [PubMed] [Google Scholar]
- 34. Gomez MT, Malozowski S, Winterer J, Vamvakopoulos NC, Chrousos GP. Urinary free cortisol values in normal children and adolescents. J Pediatr. 1991;118:256–258. [DOI] [PubMed] [Google Scholar]
- 35. Hsiao HP, Kirschner LS, Bourdeau I, et al. Clinical and genetic heterogeneity, overlap with other tumor syndromes, and atypical glucocorticoid hormone secretion in adrenocorticotropin-independent macronodular adrenal hyperplasia compared with other adrenocortical tumors. J Clin Endocrinol Metab. 2009;94:2930–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liddle GW. Tests of pituitary-adrenal suppressibility in the diagnosis of Cushing's syndrome. J Clin Endocrinol Metab. 1960;20:1539–1560. [DOI] [PubMed] [Google Scholar]
- 37. MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. 1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beuschlein F, Boulkroun S, Osswald A, et al. Somatic mutations in ATP1A1 and ATP2B3 lead to aldosterone-producing adenomas and secondary hypertension. Nat Genet. 2013;45:440–444, 444e1–e2. [DOI] [PubMed] [Google Scholar]
- 40. Carvajal CA, Stehr CB, González PA, et al. A de novo unequal cross-over mutation between CYP11B1 and CYP11B2 genes causes familial hyperaldosteronism type I. J Endocrinol Invest. 2011;34:140–144. [DOI] [PubMed] [Google Scholar]
- 41. Chomette G, Malki B, Auriol M, Karkouche B. Pregnancy toxemia and hyperplasia of the juxtaglomerular apparatus [in French]. Arch Anat Cytol Pathol. 1985;33:175–178. [PubMed] [Google Scholar]
- 42. Jeske YW, So A, Kelemen L, et al. Examination of chromosome 7p22 candidate genes RBaK, PMS2 and GNA12 in familial hyperaldosteronism type II. Clin Exp Pharmacol Physiol. 2008;35:380–385. [DOI] [PubMed] [Google Scholar]
- 43. Zilbermint M, Faucz FR, Lodish MB, et al. Macronodular adrenal hyperplasia due to mutations in an armadillo repeat containing 5 (ARMC5) gene: a clinical and genetic investigation. In: Proceedings from the Annual Meeting and Expo of The Endocrine Society; June 21–24, 2014; Chicago, IL Abstract OR14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tu W, Eckert GJ, Hannon TS, et al. Racial differences in sensitivity of blood pressure to aldosterone. Hypertension. 2014;63:1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spence JD. Lessons from Africa: the importance of measuring plasma renin and aldosterone in resistant hypertension. Can J Cardiol. 2012;28:254–257. [DOI] [PubMed] [Google Scholar]
- 46. Kidambi S, Kotchen JM, Grim CE, et al. Association of adrenal steroids with hypertension and the metabolic syndrome in blacks. Hypertension. 2007;49:704–711. [DOI] [PubMed] [Google Scholar]
- 47. Grim CE, Cowley AW, Jr, Hamet P, et al. Hyperaldosteronism and hypertension: ethnic differences. Hypertension. 2005;45:766–772. [DOI] [PubMed] [Google Scholar]
- 48. Weder AB, Gleiberman L, Sachdeva A. Whites excrete a water load more rapidly than blacks. Hypertension. 2009;53:715–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peppa M, Pikounis V, Papaxoinis G, et al. Adrenocortical carcinoma secreting cortisol, androgens and aldosterone: a case report. Cases J. 2009;2:8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamada M, Nakajima Y, Taguchi R, et al. KCNJ5 mutations in aldosterone- and cortisol-co-secreting adrenal adenomas. Endocr J. 2012;59:735–741. [DOI] [PubMed] [Google Scholar]
- 51. Goodfriend TL, Egan BM, Kelley DE. Aldosterone in obesity. Endocr Res. 1998;24:789–796. [DOI] [PubMed] [Google Scholar]
- 52. Rocchini AP, Katch VL, Grekin R, Moorehead C, Anderson J. Role for aldosterone in blood pressure regulation of obese adolescents. Am J Cardiol. 1986;57:613–618. [DOI] [PubMed] [Google Scholar]
- 53. Chang SP, Morrison HD, Nilsson F, Kenyon CJ, West JD, Morley SD. Cell proliferation, movement and differentiation during maintenance of the adult mouse adrenal cortex. PLoS One. 2013;8:e81865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Freedman BD, Kempna PB, Carlone DL, et al. Adrenocortical zonation results from lineage conversion of differentiated zona glomerulosa cells. Dev Cell. 2013;26:666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]