SUMMARY
Heritable epigenetic factors can contribute to complex disease etiology. Here we examine the contribution of DNA methylation to complex traits that are precursors to heart disease, diabetes and osteoporosis. We profiled DNA methylation in the liver using bisulfite sequencing in 90 mouse inbred strains, genome-wide expression levels, proteomics, metabolomics and sixty-eight clinical traits, and performed epigenome-wide association studies (EWAS). We found associations with numerous clinical traits including bone density, insulin resistance, expression, protein and metabolite levels. A large proportion of associations were unique to EWAS and were not identified using GWAS. Methylation levels were regulated by genetics largely in cis, but we also found evidence of trans regulation, and we demonstrate that genetic variation in the methionine synthase reductase gene Mtrr affects methylation of hundreds of CpGs throughout the genome. Our results indicate that natural variation in methylation levels contributes to the etiology of complex clinical traits.
Keywords: EWAS, GWAS, Epigenetics, Genetics, DNA methylation, RRBS, bisulfite sequencing, complex traits, mouse genetics, metabolic syndrome, atherosclerosis, diabetes, bone, bone mineral density
INTRODUCTION
Methylation of DNA cytosine bases is evolutionarily conserved in multiple species from plants to humans. In mammalian species, DNA methylation plays an important role in imprinting, X-chromosome inactivation, cell differentiation, gene silencing and regulation of gene expression. Similar to genetic variation, epigenetic modifications are variable between individuals and regulated by genetics (Orozco et al., 2014). Methylation QTL (metQTL) studies in human adipose tissue found that 28% of CpGs were associated with nearby SNPs (Grundberg et al., 2013). DNA methylation is variable between inbred strains in Arabidopsis, rice, mice, and in human populations. Strain-specific methylation patterns were maintained across generations in mouse strains, and twin studies in humans have shown a higher concordance of methylation patterns in monozygotic twins relative to dizygotic twins (Gordon et al., 2012; McRae et al., 2014), suggesting that DNA methylation is under genetic control.
Both human populations and mouse strains show variation in multifactorial traits like heart disease and osteoporosis. In the past decade, genome-wide association studies (GWAS) have identified hundreds of genetic variants influencing clinical traits (Welter et al., 2014). DNA methylation has also been associated with gene expression (Bell et al., 2011; Grundberg et al., 2013) and complex traits including cancer (Shenker et al., 2013; Xu et al., 2013), aging (Heyn et al., 2013; Horvath, 2013), multiple sclerosis (Huynh et al., 2014), rheumatoid arthritis (Liu et al., 2013), and obesity in humans (Dick et al., 2014). However, although DNA methylation is influenced by genetics and could account for part of the heritability of clinical traits, epigenetic variation has typically not been considered in GWAS for complex traits.
In this study, we performed epigenome-wide association studies (EWAS) to determine the contribution of DNA methylation to complex clinical traits related to heart disease, diabetes, obesity, and osteoporosis. Our study integrates systems genetics data that includes SNP genotypes, DNA methylation bisulfite sequencing data, genome-wide gene expression, proteomics, metabolomics, and clinical phenotypes. Our results reveal a large number of associations between DNA methylation variants and clinical or molecular traits, many of which we could not identify using traditional GWAS. We explored the contribution of genetics to DNA methylation patterns and found that 52% of highly variable CpGs were under genetic control. The narrow sense heritability for CpG methylation levels was on average 27%, and 60% for highly variable CpGs. We also present evidence of common genetic variation affecting DNA methylation patterns in trans, and experimentally validate the role of Mtrr in regulating methylation levels of CpGs across the genome.
RESULTS
Data
We constructed reduced representation bisulfite sequencing (RRBS) libraries using liver genomic DNA from 16-week old male mice using a previously described protocol (Feng et al., 2011), corresponding to 90 mouse inbred strains from the Hybrid Mouse Diversity Panel, HMDP (Bennett et al., 2010). We sequenced the libraries using the Illumina HiSeq platform and obtained an average of 90+/-11 million reads per sample, then aligned the data to the mouse genome using BS-Seeker2 (Guo et al., 2013) for an average of 41+/-7 million uniquely aligned reads per sample (Figure S1A). This corresponded to 46% mappability, and 48x coverage per sample on average (Figure S1B). We filtered the cytosines based on 10x or more coverage for a total of 11,520,175 cytosines present in at least 90% of the samples, of which 2,047,165 were CG, 2,737,475 were CHG, and 6,735,535 were in CHH context. The mouse genome contains 21.3 million CpGs and we observed approximately 2 million (9.6%) of all CpGs using RRBS.
Global methylation levels in the adult mouse livers were 44% +/-1 for CpG cytosines, 1.1% +/-0.4 for CHG, and 0.8% +/-0.4 for CHH cytosines, where H is any base other than G (Figure S1C). Since non-CG methylation was too low to be studied in these samples (Figure S1C,E), we focused our analyses on CG cytosines only. We defined a set of 360,324 Variable CpGs, which showed a 50% absolute change (delta) in methylation levels in at least one sample. We further identified a set of 22,227 Hypervariable CpGs, which showed 50% or higher methylation delta, relative to the median methylation level of the CpG in 5 or more samples. An example of a Variable and a Hypervariable CpG can be found in Figure S2A-B. We excluded 6,993 CpGs that were also SNPs in the mouse strains, since the changes in methylation observed correspond to the loss of a CpG in strains carrying the SNP.
The liver is one of the main tissues involved in energy metabolism. Because of its roles in carbohydrate and fat metabolism, the liver has a significant impact on clinical phenotypes such as plasma glucose, cholesterol and lipid levels, body weight, adiposity and atherosclerosis. It would also be important to consider methylation levels in other metabolically relevant tissues such as adipose, muscle, pancreas and intestine in future studies. For the same mouse strains, we measured 68 clinical traits including atherosclerosis, diabetes, obesity, osteoporosis, and blood cell-related traits, as well as genome-wide expression levels in the liver (Bennett et al., 2010) using Affymetrix arrays. We obtained liver proteomics from 1,543 peptides measured by Liquid Chromatography–Mass Spectrometry (Ghazalpour et al., 2011). We also profiled 260 liver and plasma metabolites using Mass Spectrometry, comprising eight classes of molecules including lipids, carbohydrates, amino acids, peptides, xenobiotics, vitamins, cofactors and nucleotides (Ghazalpour et al., 2014).
DNA methylation has lower correlation in cis than SNPs
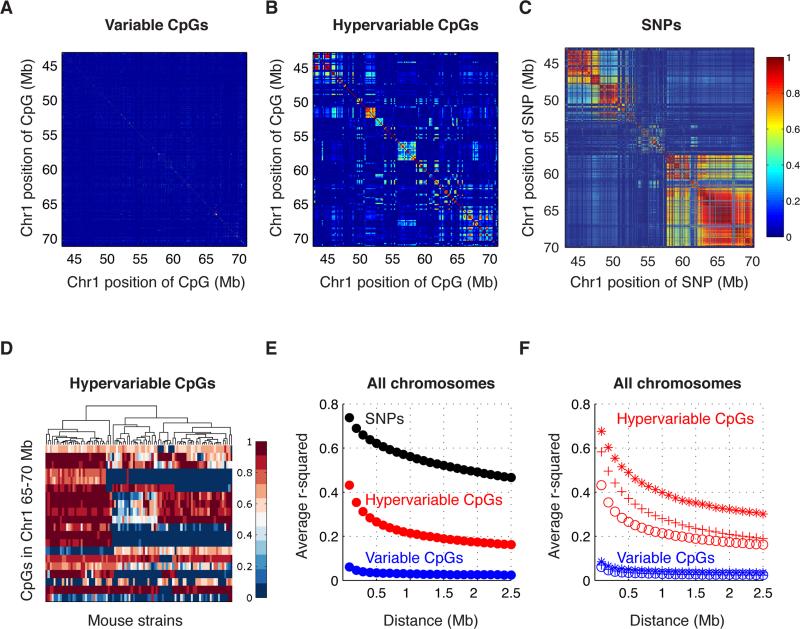
Correlations between pairs of alleles that are near each other on a chromosome, or linkage disequilibrium (LD), can result in large genomic blocks that contain multiple genes. A given association may have a few or dozens of candidate genes depending on the level of LD at that locus. We were interested in determining the correlation in pairwise CpG methylation levels, and hence the level of resolution we could achieve using CpGs in our association studies. We determined pairwise correlations of CpGs at different distances from each other. For example, we took CpGs separated by 100kb or less and estimated the correlation between the CpG methylation levels in the HMDP strains. We then estimated the average correlation between all pairs of CpGs in the genome at that distance from each other, and repeated this estimate at various distances. We compared this to the level of correlation in SNPs (LD) that we had previously calculated for the same HMDP strains (Bennett et al., 2010). Pairwise correlations in CpG methylation levels for a locus on chromosome 1 are shown in Figure 1A for Variable CpGs, Hypervariable CpGs (Figure 1B), and SNPs (Figure 1C). Methylation levels in mouse strains for a sample locus in chromosome 1 are shown in Figure 1D, where methylation levels vary between 0-100%. Correlations plots for whole chromosomes can be found on Figure S3D-I.
Figure 1. Methylation and SNP correlations.
Correlations for a locus in chromosome 1 in (A) Variable CpG methylation, (B) Hypervariable CpG methylation and (C) SNPs. The x and y axes denote the chromosome position and the color represents the correlation (r2) between CpGs, or SNPs. (D) Methylation levels of Hypervariable CpGs for a representative locus, strains are on the x-axes, CpGs are on the y-axes, and the color represents percent methylation levels. (E)-(F) Genome-wide average correlation between CpGs, or SNPs, at various distances. SNPs are shown in black, Variable CpGs are in blue, Hypervariable CpGs are in red. Each point is the average correlation at increasing 100kb bins. (E) Genome-wide average correlation between CpGs binned by their methylation level. Open circles (o) represent the average for all CpGs, asterisks (*) represent the average for CpGs with methylation levels between 0-20%, and plus symbols (+) represent the average for CpGs with methylation levels between 80-100%. See also Figure S3.
At the genome-wide level, we found that the distance-dependent correlation between CpGs was lower than that of SNPs. The average correlation across the genome between CpGs within 100kb was r2=0.06 for Variable CpGs and r2=0.43 for Hypervariable CpGs, and at 2Mb the average correlation was r2=0.03 for Variable CpGs and r2=0.17 for Hypervariable CpGs. In contrast, the average correlation across the genome was r2=0.88 for SNPs at 100kb, dropping to r2=0.49 at 2Mb. The genome-wide average of pairwise correlations is shown on Figure 1E, for CpGs or SNPs at various distances from each other in increasing 100kb bins. Since methylation levels in mammals are bimodal for a large proportion of CpGs (Figure S1D), we examined pairwise correlations between CpGs with low or high methylation levels. We found that Hypervariable CpGs with low methylation levels were generally more highly correlated with nearby CpGs, than CpGs with high methylation levels. For example, the average correlation between Hypervariable CpGs at 100kb was r2=0.68 for CpGs with 0-20% methylation levels, and r2=0.58 for CpGs with 80-100% methylation. The average correlation was r2=0.32 for CpGs with 0-20% methylation levels, and r2=0.18 for CpGs with 80-100% methylation, at a distance of 2Mb. We observed no differences in pairwise correlation levels between low and high methylated Variable CpGs (Figure 1F).
Natural variation in DNA methylation is associated with complex traits
To determine the association of epigenetic variation with complex clinical and molecular traits, we performed EWAS between CpG methylation levels and 1) 68 clinical traits, including plasma cholesterol, fatty acids, glucose and insulin, body weight, adiposity, blood cell counts, and bone mineral density phenotypes, 2) 260 plasma and liver metabolites, 3) protein levels from 1,543 peptides corresponding to 480 genes, and 4) genome-wide microarray expression levels corresponding to 12,980 genes (Figure 2). Similar to GWAS using SNPs, we used a linear mixed model (Kang et al., 2008) to determine associations between traits and CpGs, and to correct for population structure. Since we used CpG methylation as the predictors instead of SNPs, we also employed a methylation-based kinship matrix instead of a SNP kinship matrix in the model. We and others have shown that this approach corrects for false positive associations due to population structure (Bennett et al., 2010) and potential tissue heterogeneity in the methylation data (Zou et al., 2014).
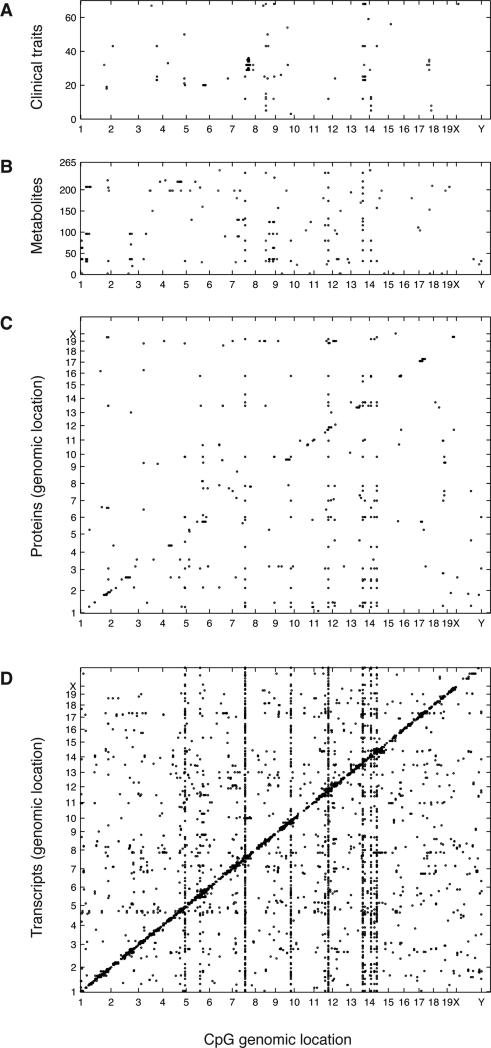
Figure 2. EWAS.
Association between CpGs methylation and (A) clinical traits, (B) metabolites, (C) protein and (D) gene expression levels. Each point is a significant EWAS at the corresponding Bonferroni thresholds across all CpGs and traits tested. The genomic position of CpGs is on the x-axes, the y-axes denote traits, and the position in the genome of the associated proteins and genes. Black points are EWAS hits for Hypervariable CpGs, and blue points are EWAS hits for Variable CpGs. For simplicity, only associations to Hypervariable CpGs are shown for the proteomics and gene expression datasets. See also Figure S4.
Each of the EWAS plots in Figure 2 summarizes associations between CpGs across the mouse genome, and traits. Due to the large number of traits in the proteomics and gene expression datasets, only associations to Hypervariable CpGs are shown for these datasets. All associations shown are significant at the Bonferroni threshold (Table S1). In summary, we found that natural variation in CpG methylation was associated with numerous complex clinical and molecular traits. A table with the number of EWAS hits and the significance threshold used can be found Table S1, and tables with the individual associations we identified for all clinical and molecular traits can be downloaded from http://ewas.mcdb.ucla.edu/download.html. We found no evidence of inflation in our EWAS results (Figure S4A-D, Supplemental Results), and no evidence of macrophage contamination in our liver samples (Figure S4E-F).
EWAS identifies both known and novel associations
We identified numerous associations between clinical traits and CpG methylation near genes known to influence those traits, including associations not identified using traditional GWAS. We previously performed GWAS for clinical traits and expression levels in the HMDP (Bennett et al., 2010). We also previously examined linkage disequilibrium (LD) for SNPs in the HMDP and estimated an average resolution of 2Mb (Bennett et al., 2010; Ghazalpour et al., 2011; Orozco et al., 2012), although LD blocks could be smaller or larger depending on the genomic region. We took the average 2Mb resolution in the HMDP and called associations in 2Mb bins, such that more than one association in a bin was considered to be the same locus. To look for the overlap between GWAS and EWAS associations, we considered associations to be overlapping if they were within 2Mb of each other. For clinical traits common to both studies we identified 266 EWAS hits and 300 GWAS hits, and 41 were identified by both EWAS and GWAS where the associated loci were within 2Mb of each other. We also observed that the overlap between EWAS and GWAS hits was much higher for expression cis-eQTL (77%) and protein (37%) cis-pQTL (Figure S5A). Clinical traits are typically more complex than molecular traits such as gene expression or protein levels. For increasingly complex traits, a smaller proportion of the variance in the phenotype can be explained by genetic or epigenetic variation. Therefore, for a given statistical power, we are likely to detect far more molecular QTL than clinical trait QTL. The low overlap between EWAS and GWAS clinical trait hits may be due to a lack of sufficient power to detect associations for complex traits, since we also observe a higher overlap between EWAS and GWAS associations for expression cis-eQTL (77%) than for clinical traits (15%).
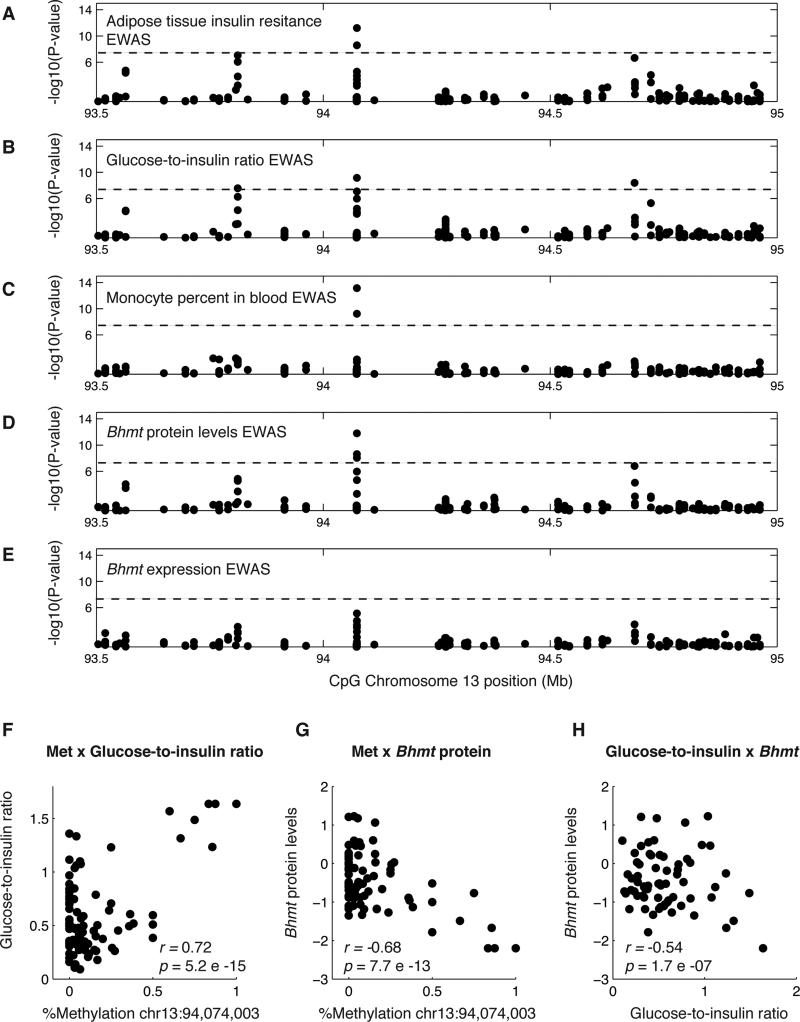
As an example, we found an EWAS hit on chromosome 13 for adipose tissue insulin resistance (ATIRI), glucose-to-insulin ratio, a measure of insulin sensitivity, and percent of monocytes in the blood (Figure 3A-C). We did not find an association using GWAS for measures of insulin resistance or monocyte levels at this locus. The gene Bhmt encoding betaine-homocysteine methyltransferase located at 94.3Mb on chromosome 13 was a candidate gene in this locus, since we also identified a cis association for protein levels of this gene using EWAS, or cis-pQTL, but no expression associations (Figure 3D-E). Methylation levels at this locus were correlated with glucose-to-insulin ratio (Figure 3F), and inversely correlated with protein levels of the gene (Figure 3G). Protein levels of Bhmt were also correlated with the trait (Figure 3H). Previous work in Bhmt knock-out mice demonstrated that Bhmt plays a role in energy metabolism, specifically in lipid synthesis, and insulin sensitivity (Teng et al., 2012). Additional examples for associations with plasma cholesterol and total bone mineral density are describe in the Supplemental Information and Figure S6.
Figure 3. Insulin resistance and Bhmt EWAS.
Manhattan plots showing association of methylation levels to (A) adipose tissue insulin resistance, (B) plasma glucose-to-insulin ratio, (C) monocyte percent in the blood, (D) liver protein levels of Bhmt, and (E) liver expression levels of Bhmt. Chromosome location is on the x-axes, the p-value for the association is on the y-axes, and each point represents a CpG. The dotted line is drawn at p<1×10−7, the Bonferroni threshold for a single phenotype. (F)-(H) Each point represents a mouse sample, showing correlation between (F) methylation levels for the peak associated CpG and glucose-to-insulin ratio levels, (G) methylation levels for the peak associated CpG and liver protein levels of Bhmt, and (H) glucose-to-insulin ratio and Bhmt protein levels. See also Figure S6.
Conditional association studies
We used conditional association studies to determine whether hits identified with both EWAS and GWAS were (1) caused by the same signal or (2) co-localizing but independent signals. To accomplish this, we performed EWAS for overlapping associations using the CpG as the predictor and the SNP genotype as a covariate. In this approach, if an EWAS hit remains significant when we use the SNP as a covariate, this would suggest that the overlapping EWAS and GWAS association was independent but co-localizing. In contrast, if the EWAS hit goes away when we use the SNP as a covariate, then we can conclude that the EWAS and GWAS overlapping association was arising from the same signal. Out of 41 overlapping clinical trait associations, only 3 (7%) remained significant when we used the SNP genotype as a covariate at the Bonferroni threshold (p<1.2×10−03), and none were significant at p<1×10−07. Similarly, 71 (4%) of overlapping cis expression associations, corresponding to 64 unique genes, remained significant after using the SNP genotype for the locus as a covariate at the Bonferroni threshold (2.7×10−05), and 30 of the associations (2%) were significant at p<1×10−07. These results suggest that the majority of associations we found using both EWAS and GWAS were likely arising from the same signal at the associated locus.
Causal inference test
To determine whether associations identified by both EWAS and GWAS were mediated by differential methylation levels, we performed causal inference tests using the R statistical package CIT (Millstein et al., 2009). The causal inference test (CIT) performs a series of conditional probability tests to determine if the association between a genetic locus (L) and a trait (T) is mediated by DNA methylation (M) in this case, by testing for the following conditions: (1) the trait is associated with the locus, L•T, (2) the trait is associated with the methylation mediator given the locus M•T | L, (3) the methylation mediator is associated with locus given the trait, L•M | T and (4) the locus is independently associated with both the mediator and the trait given the mediator, L•T | M. If the first three conditions are met, we can say that genetic variation at the locus is associated with the trait, and that this association is mediated entirely by DNA methylation, L•M•T. Alternatively, if all four conditions are met, we can say that the association is still causal and mediated by DNA methylation, but the genotype at the locus also affects methylation and the trait independently. In summary, the two causal models tested by the CIT are the causal model, where the association is mediated entirely by DNA methylation, and the causal independent model, where the association is causal but the locus is also independently associated with DNA methylation and the trait.
We performed the CIT for clinical trait associations identified by both EWAS and GWAS and found that 10 out of the 41 (24%) overlapping associations were causal and mediated by DNA methylation, and 9 of the 10 were causal independent associations (Table S2). However, it is possible that there are additional causal relationships between genetic variants, DNA methylation and traits but we lack sufficient power to detect these. Similarly, we performed the causal inference test for the gene expression associations (eQTL) we identified using both EWAS and GWAS, corresponding to 1,530 unique genes. We found 352 (22%) cis-eQTL mapping genes were mediated by variation in DNA methylation, where the locus genotype influenced methylation and DNA methylation in turn influenced the trait, L•M•T, and 321 of these were causal independent. Overall, results from the CIT indicate that a proportion (22-24%) of overlapping EWAS and GWAS hits were causal associations mediated by DNA methylation. These results in conjunction with the conditional association studies suggest that that remaining EWAS hits are likely to be secondary to the genetic associations.
Principal component analysis
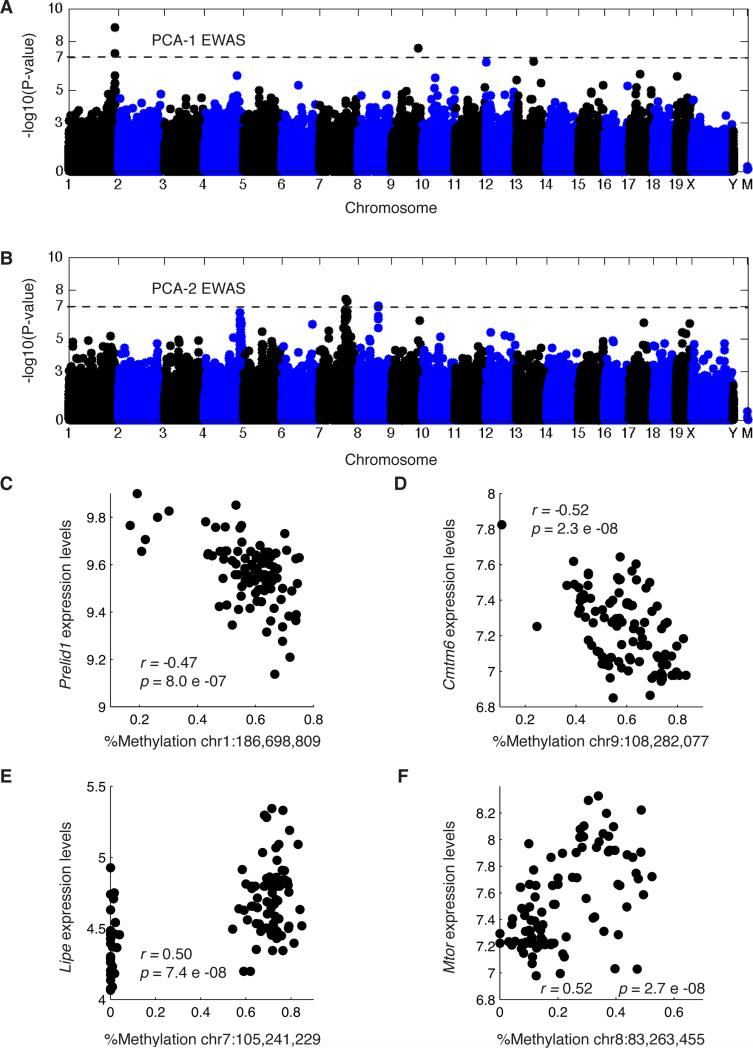
The clinical traits in our study have a complex correlation structure (Figure S5B), since several traits such as insulin resistance, plasma cholesterol levels and obesity are interrelated. To account for correlations between clinical traits, and to identify loci that drive multiple correlated traits, we performed principal component analysis on the clinical traits (Figure S5C). The first component explained 24% of the variation in the traits, and had the highest weights for glucose-to-insulin ratio and fat-related traits. The second component explained 12% of the variation in the traits, and had the highest weights for blood and fat-related traits. We performed EWAS between CpGs and the two first principal components as phenotypes, and found two significant associations with the first component on chromosomes 1 and 9 at the Bonferroni threshold (p<6.9×10−8, Figure 4A). We searched for individual methylation sites in this locus that were also associated with gene expression or protein levels. Genes associated with the chromosome 1 locus methylation levels include Prelid1 (Figure 4C), a gene involved in lipid transport, and 1110057K04Rik, a gene recently found to be involved in lipid storage (Goo et al., 2014). Methylation levels at the chromosome 9 locus were associated with Cmtm6 (Figure 4D), a gene structurally related to chemokines although its exact function is still unknown. The second component was strongly associated with a locus on chromosome 7 spanning approximately 10Mb that also coincided with the Hemoglobin beta locus (Figure 4B). Consistent with the correlation between the second principal component and fat-related traits, we found that CpGs at this locus were associated with several genes including Lipe (Figure 4E), a lipase gene involved in free fatty acid oxidation (Reid et al., 2008). In addition, we identified CpGs in chromosome 8 associated with the second principal component and with expression levels of Mtor (Figure 4F). Mtor plays a role in metabolic regulation, response to nutrients, insulin and diabetes (Zhu et al., 2013).
Figure 4. Principal component EWAS.
Association between methylation and principal components (A) one and (B) two. Each dot represents a CpG, the genomic position of CpGs is on the x-axes and the -log10 of the p-value for the association is one the y-axes, chromosomes are shown in alternating colors. (C)-(F) Each dot represents a mouse sample, showing correlation between (C) liver expression levels of Prelid1 and methylation levels of a CpG associated with PCA1, (D) expression of Cmtm6 and methylation of a CpG associated with PCA1, (E) expression of Lipe and methylation of a CpG associated with PCA2, (F) expression of Mtor and methylation of a CpG associated with PCA2. See also Figure S5.
DNA methylation can be used to infer phenotypes
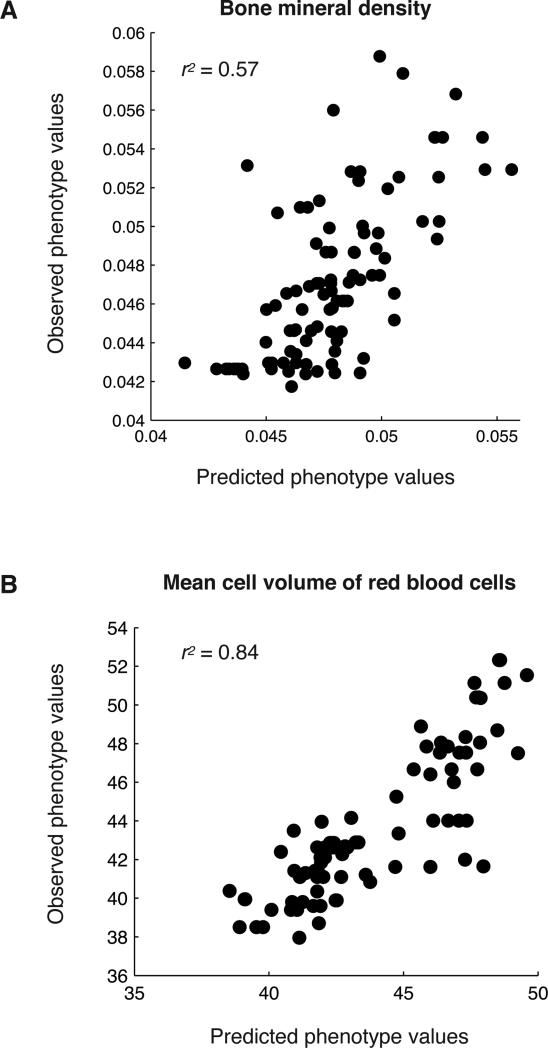
Genetics and genomics data is a highly valuable resource that can be used to model disease susceptibility and risk. An individual's genome is predominantly static, but the epigenome is variable in different tissues and is affected by transcription patterns. To determine if CpGs could be used to infer clinical traits in other individuals based on their methylation status, we built linear models with CpG sites using the generalized linear model package glmnet (Friedman et al., 2010), and tested their power to infer clinical traits in test individuals where the CpG status was known. For each trait, we randomly selected a test set of 10 mice which were kept hidden from the training set and used the remaining mice as the training set, where the methylation status was known in both sets. We used glmnet to select CpGs from the 20,000 most variable CpGs, and built a linear model for the trait based on these CpGs using the training set. We then used the resulting linear model to infer trait values on the test set of 10 mice, and determined the accuracy of the model using Pearson's correlation between the observed and inferred trait values. We found several clinical traits that could be accurately inferred on test individuals from a set of CpG methylation sites, where the trait values predicted by the model were highly correlated with the clinical trait values measured in the mice. We found eight clinical traits with r2>0.5, including plasma total cholesterol levels, plasma HDL cholesterol levels, total bone mineral density, plasma fatty acids, and red blood cell phenotypes. Examples of inferred and measured clinical trait values are shown for bone mineral density (Figure 5A), and for mean cell volume of red blood cells (Figure 5B). A list of inferred phenotypes, the correlations between predicted and observed phenotypes, and the top ten CpGs selected to model each phenotype can be found in Table S4.
Figure 5. Phenotype inference.
Phenotype predictions for (A) Bone mineral density and (B) Mean cell volume of red blood cells. The predicted phenotype value is on the x-axes and the measured phenotype value is on the y-axes. Each point is a mouse sample. See also Table S4.
Natural genetic variation influences genome-wide DNA methylation levels
To determine the extent to which genetics affects natural variation in DNA methylation, we used a linear mixed model to perform genome-wide association (GWAS) of CpG methylation levels as traits, to SNPs across the mouse genome. We and others have shown that this method reduces false-positive associations that are due to population structure (Bennett et al., 2010; Kang et al., 2008). For each CpG, we associated methylation levels to SNPs with MAF greater than 10%. We called significant associations at the Bonferroni threshold (p<1.4×10−12) by considering each CpG and SNP pair as an independent test (Table S5). We chose this stringent threshold to call significant associations in order to minimize the possibility of examining false positive associations. However, all association results at p<1×10−06 can be obtained from our website at http://ewas.mcdb.ucla.edu/download.html. We identified 3,017,453 associations between methylation levels and genetics at the Bonferroni threshold (Figure 6A), corresponding to 26,563 unique CpGs, or 7% of all methylation sites tested, and 92,959 unique SNPs. Approximately 51% of all significant associations were for Hypervariable CpGs, and 52% of all Hypervariable CpGs (11,644) were under genetic regulation. We found that 12% of the associations involved SNPs that abolished a CpG in a fraction of the mouse strains (CG-SNPs), corresponding to 2,533 of the CG-SNPs present in the strains. We note that these CG-SNPs were not used in our EWAS, since differences in methylation between strains were the result of a cytosine change to a different DNA base.
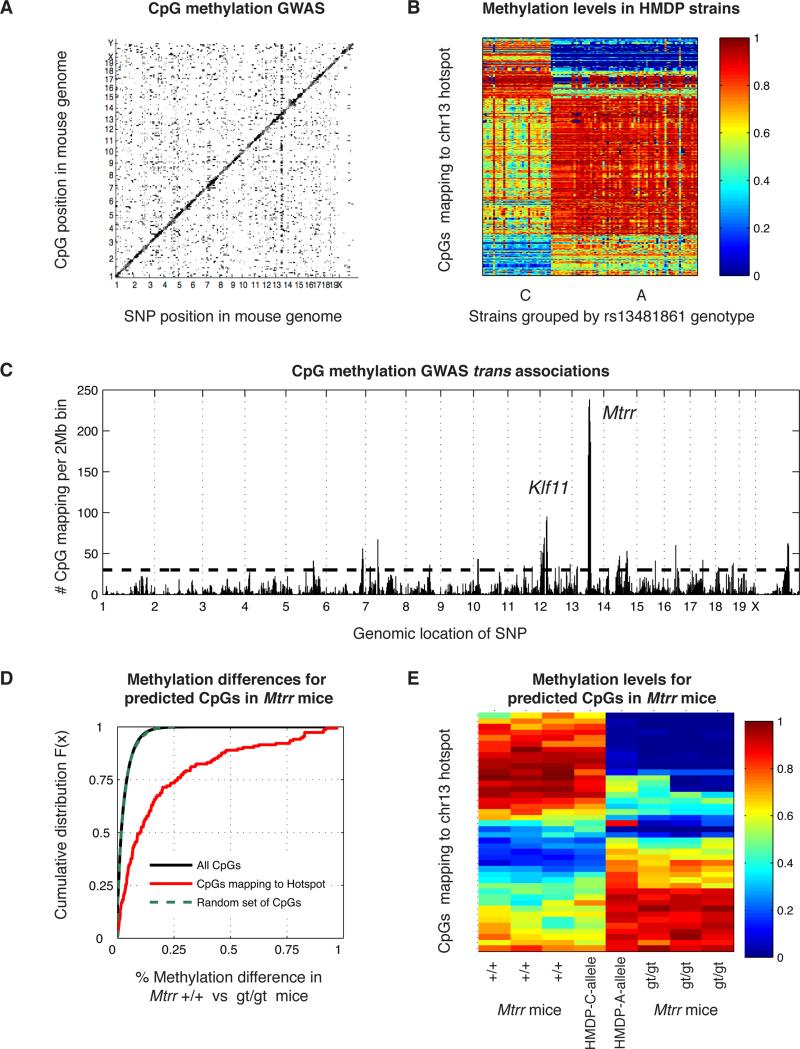
Figure 6. Natural genetic variation influences genome-wide DNA methylation.
(A) GWAS using Variable CpG methylation levels as phenotypes and SNPs as predictors. Genomic position of SNPs is on the x-axis, and the genomic position of CpGs is on the y-axis. Each point is a significant association at the Bonferroni threshold p<1.4×10−12. (B) Methylation levels for CpGs mapping to the chromosome 13 GWAS hotspot. Strains are on the x-axis grouped by their genotype of rs13481861 at the Mtrr locus, and CpGs are on the y-axis. The color is the methylation level between 0-100%. (C) CpG methylation GWAS hotspots. The number of CpGs mapping in trans to each 2Mb bin is on the y-axis. The genomic position of each bin is on the x- axis. The horizontal dotted line is the Poisson significance threshold for each hotspot bin. (D)-(E) Experimental validation of the hotspot at the Mtrr locus using RRBS in livers of wild-type mice (+/+) and mice homozygous for the Mtrr gene-trapped allele (gt/gt). (D) Distribution of methylation differences. The difference in methylation between +/+ versus gt/gt mice is on the x-axis and the cumulative distribution function is on the y-axis. The curves show the distribution of all CpGs (black), randomly sampled CpGs (green dotted), and CpGs predicted to be affected by the Mtrr genotype (red). (E) Differentially methylated CpGs between Mtrr +/+ and gt/gt mice at FDR<5%. Mice are on the x-axis and CpGs are on the y-axis. The color denotes methylation levels between 0-100%. See also Figure S7.
We estimated the variance explained by genetics, or the narrow sense heritability of DNA methylation levels for individual CpGs using an additive model. When we examined all Variable CpGs, which display variation in methylation levels in at least one strain, the variance explained by genetics was on average 27% (Figure S7A). In contrast, on average 60% of the variance was explained by genetics for Hypervariable CpGs, which display higher variation in methylation among the strains. The variance explained by genetics was on average 75% for CG-SNPs. These CG-SNPs do not show 100% heritability likely because methylation levels can still be controlled in trans for the strains with the C allele. A large proportion of the associations were local or cis. We previously estimated the GWAS mapping resolution to be 2Mb on average (Bennett et al., 2010). Here, 54% of the associations were local or cis, where SNP and CpG pairs were within 2Mb of each other. However, trans associations where SNP and CpG pairs were more than 2Mb from each other were also found in the same chromosome for approximately 79% of trans associations, suggesting that many of these associations may be cis associations caused by long range LD in SNPs. The distance between SNPs and Hypervariable CpGs was on average 1.4 times smaller than the overall distribution of SNP to CpG distances, and the distance between SNPs and CG-SNPs was on average 3.6 times smaller than the overall distribution. The distribution of the distances between SNP and CpGs pairs is shown on Figure S7B, and results of the DNA methylation GWAS are summarized in Table S5.
Mtrr influences methylation levels of CpGs across the genome
Methylation levels of hundreds of CpGs across the genome mapped to a QTL hotspot on chromosome 13, defined as a 2Mb bin at 68-70Mb, near the gene Methionine synthase reductase, Mtrr, located at 68.7Mb (Figure 6C). This methylation hotspot can also be seen as a vertical band on the GWAS plot (Figure 6A). Expression levels of Mtrr were variable among the mouse strains and were regulated in cis, since we observed both a cis-eQTL for Mtrr using GWAS (p=1.82×10−14), and an association between Mtrr expression and methylation levels 314bp from Mtrr using EWAS (p=4.97×10−14). Expression levels of Mtrr were highly correlated with methylation levels of CpGs mapping to the locus both in cis and in trans, with an average absolute Pearson's r=0.48. The distribution of these correlations was significantly different (KS-test p=4.98×10−187) from the correlation between Mtrr expression and all CpGs, which had average absolute r=0.09 (Figure S7C). Mtrr is necessary for the utilization of methyl groups from the folate cycle, which donates methyl groups to multiple cellular pathways including DNA methylation (Crider et al., 2012). MTRR was recently associated with methylation levels in autoimmune thyroid disease in humans (Arakawa et al., 2012), along with DNMT1, DNMT3A, DNMT3B and MTHFR. All these suggested that Mtrr was an ideal candidate gene for the methylation hotspot.
Based on our GWAS results, we observed 471 CpGs from the HMDP mapping to the chromosome 13 hotspot (Figure 6B). These CpGs were physically located throughout the mouse genome and we hypothesized that their methylation levels were influenced by Mtrr. To experimentally validate Mtrr as a causal gene for the chromosome 13 methylation hotspot, we measured DNA methylation levels in the livers of Mtrr wild-type (+/+) and homozygous gene-trapped mice (gt/gt) using RRBS. Mice homozygous for the gene-trapped allele display reduced expression, protein and activity of Mtrr (Elmore et al., 2007). Of the 471 CpGs mapping to the hotspot, 154 CpGs were represented in the RRBS dataset of Mtrr mice, and 42 of the 154 (27%) were differentially methylated between Mtrr wild-type and gt/gt mice at 5% FDR. Methylation levels at these differentially methylated sites are shown on Figure 6E for wild-type and gt/gt mice, as well as the average methylation levels of mouse strains with the reference allele (C) or alternate allele (T) for SNP rs13481861 located in an exon of Mtrr. The list of 154 CpGs we tested using RRBS, including the methylation delta, the p-value for differential methylation between Mtrr wild-type and gt/gt mice, and the average methylation of allele C and allele T HMDP strains can be found in Table S6. We were not able to observe all 471 CpGs because of the decrease in coverage, since we multiplexed six samples in one lane for the validation experiments and two samples per lane for all the HMDP samples.
The difference in methylation (delta) between wild-type and gt/gt mice was on average 6.7-fold higher in the 154 predicted CpGs, relative to all CpGs observed in the Mtrr mice (KS-test p=1.8×10−33). To test if the differences between Mtrr wild-type and gt/gt mice at these CpGs were due to chance, we randomly sampled sets of CpGs across the genome and calculated the delta between wild-type and gt/gt mice in 1,000 samplings, but found no significant differences between randomly sampled CpGs and all observed CpGs. Random sets of CpGs had an average delta of 0.99%, and average KS-test p=0.52 for Mtrr +/+ versus gt/gt mice across all 1,000 samplings, with a minimum KS-test p=0.001 and maximum delta of 1.1%. In contrast, the 154 CpGs that we hypothesized were regulated by Mtrr showed an average delta of 20% between Mtrr +/+ and gt/gt, with KS-test p=1.8×10−33. The distribution of methylation differences in Mtrr wild-type and gt/gt mice is shown on Figure 6D for the 154 predicted CpGs, a set of randomly sampled CpGs, and all CpGs observed in the Mtrr mouse RRBS dataset.
Online databases for the identification of candidate genes
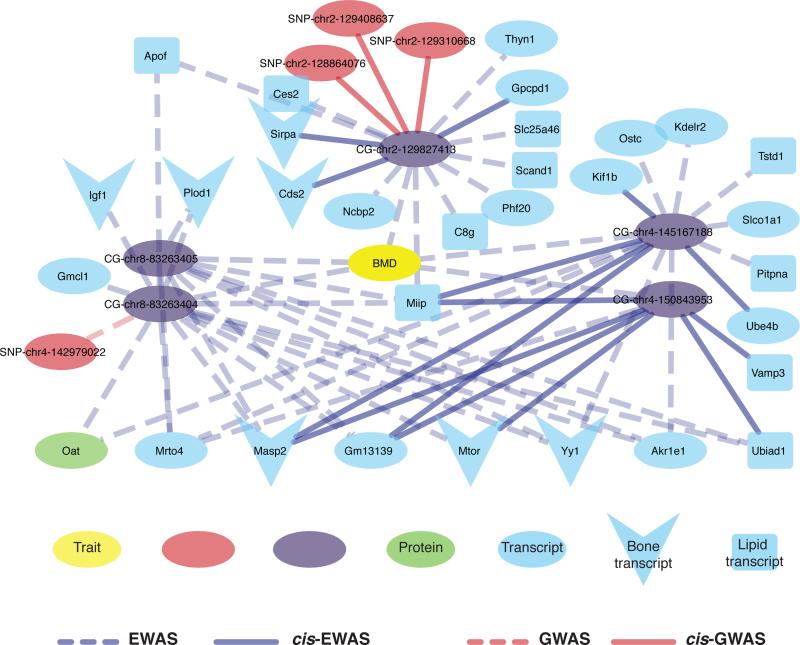
All of our EWAS and GWAS results are available through an online database to facilitate the identification of candidate genes for clinical traits using DNA methylation patterns, which can be accessed at http://ewas.mcdb.ucla.edu. We also created an different online tool that generates association graphs based on our results at http://pathways.mcdb.ucla.edu/network. A sample association graph for bone mineral density is shown in Figure 7. Further details on these online databases can also be found in the Supplemental Information.
Figure 7. Bone mineral density association graph.
Association graph of EWAS and GWAS hits. Edges are defined by EWAS=purple lines, GWAS=red lines, Cis-associations=solid lines, and Trans-associations=dotted lines. Node colors are Trait=yellow, CpG=purple, SNP=red, Gene expression=light blue, Protein levels=green. Genes implicated in bone mineral density and/or bone biology are shown with V shape, and genes implicated in fat or lipid metabolism are shown as squares.
DISCUSSION
In this study we leveraged a powerful mouse systems genetics platform to ask what is the relationship between DNA variation and methylation? What are the loci that control methylation levels? How does methylation relate to clinical traits that are precursors to heart disease and diabetes? Can methylation be incorporated into a network and causal models of complex disease? Our results demonstrate that using DNA methylation for genome-wide association studies (i.e. EWAS) complements traditional GWAS.
We identified thousands of associations between DNA methylation levels and clinical traits such as bone mineral density, adiposity, plasma cholesterol, glucose, insulin, triglyceride levels, and molecular traits such as metabolites, protein and gene expression levels (Figure 2). Roughly 15% of EWAS hits for clinical traits could also be found using GWAS in the same panel of mouse strains, but the remaining associations were unique to the EWAS (Figure S5A). The low overlap between methylation and genetic associations for clinical traits may be due a lack of statistical power to detect associations with small effects. Since molecular traits are typically less complex relative to clinical traits, we more likely to detect associations for gene expression and protein levels, and indeed we observed a much larger overlap for cis expression (77%) and cis protein associations (37%) identified with EWAS and GWAS. In cases where associations are identified by both EWAS and GWAS, we asked whether methylation was mediating the effect on the trait. We used a causal inference test (CIT) to address this question, and found evidence that DNA methylation was mediating the effect on the trait in 24% of the overlapping clinical trait associations, and 22% of the overlapping cis expression associations. However, we note that it is often difficult to establish causal relationships for either genetic or epigenetic associations for a number of reasons, including the complexity of biological pathways, linkage disequilibrium and statistical power. In addition, although the CIT examines the model where a locus affects methylation and methylation affects the trait (L•M•T), there are other possible scenarios not tested by the CIT, such as where DNA methylation changes are reactive, and are not mediators of the association.
DNA methylation levels are dynamic and can be modified in response to disease, age and environmental perturbations. In contrast, the genome remains largely static throughout an individual's lifetime, and is modified only in certain diseases such as cancer. We found that correlations in CpG methylation patterns were significantly smaller than correlations in SNP genotypes (LD), leading to a dramatic increase in our association mapping resolution (Figure 1, S3). LD blocks are typically much larger in laboratory mice than in human populations, due the breeding history of existing mouse strains. Hence, we would not expect a similar increase in mapping resolution using EWAS in human populations. The extensive LD in mice has been a major difficulty for candidate gene identification in mouse genetics studies, and we found that this can be largely overcome using EWAS in mice. We hypothesize that the plasticity in the epigenome allows methylation patterns to be at least partly decoupled from local genetic patterns, since CpG methylation can be modified by chromatin binding proteins in trans. In contrast, SNP genotypes are static and LD patterns remain fixed in the population.
In addition, DNA methylation patterns can vary across different tissues. One of the advantages of studies in mammalian model organisms such as the mouse is the ability to sample tissues, which are not readily available in human studies. A previous study identified methylation associations using blood for rheumatoid arthritis in humans(Liu et al., 2013), and in the current study we found associations for bone mineral density and methylation levels in the liver (Figure 7, S6). These findings suggest that it is possible to uncover significant associations for methylation patterns that are conserved between the tissue that is sampled and the tissue relevant to the trait of interest, but associations to methylation levels that are not conserved are likely to be missed. We believe that there is potentially a large amount of information to be gained from studying relevant tissues whenever possible.
We examined the degree to which methylation is controlled by genetics by taking methylation patterns of individual CpGs as phenotypes, and mapping them to the SNP genotypes using GWAS. We found that 7% of all CpGs and 52% of Hypervariable CpGs were under genetic control (Figure 6A). A large proportion (55%) of these associations were in cis, where the CpG and SNP were found within 2Mb of each other. Although only 7% of all CpGs we examined were significantly associated with SNPs, it is possible that we did not have sufficient power to identify additional associations, particularly for genetic variants with subtle effects on DNA methylation. In addition, we note that CpGs with minimal or no variation in methylation levels would not be significantly associated even if they were stably maintained across generations. Previous human methylation studies, or mQTL studies, found that 20% of variable CpGs were associated with genetic variation in blood leukocytes (McRae et al., 2014), and 28% in adipose tissue (Grundberg et al., 2013). We observed an average heritability of 27% for all CpGs and 60% for Hypervariable CpGs in the liver of mouse inbred strains, excluding CGSNPs. In comparison, heritability in previous human twin studies was on average 12% in cord blood mononuclear cells, 7% in human umbilical vein endothelial cells (Gordon et al., 2012), 19% in blood leukocytes (McRae et al., 2014), and median heritability of 34% in adipose tissue in the MuTHER cohort (Grundberg et al., 2013). These results support the notion that DNA methylation levels are indeed associated with genetic variation, although the extent of the heritability and genetic effects is variable across different tissues and population samples.
Although the majority of associations between CpG methylation levels and genetics were in cis, we found a trans association hotspot where methylation levels of CpGs across the genome map to a locus in chromosome 13 near Mtrr (Figure 6A,C), a gene necessary for the utilization of methyl groups from the folate cycle. This suggested that natural genetic variation in a population can influence genome-wide DNA methylation levels. The FAST kinase gene Fast3kd, was also located on chromosome 13 near Mtrr, and is another candidate gene for the hotspot. Unfortunately, Fastkd3 was not represented in either our gene expression or protein datasets and we could not observe any cis-eQTL or cis expression EWAS hits for this gene. Although Fastkd3 may also be a candidate for the hotspot, it contains a mitochondrial targeting domain and functions primarily in the mitochondria, making it a less likely candidate than Mtrr for influencing CpG methylation levels in trans. We confirmed the role of Mtrr in 27% CpGs predicted to be affected by the chromosome 13 hotspot, since these CpGs were differentially methylated between wild-type and gt/gt mice (Figure 6E). The validation results of the chromosome 13 hotspot we present here are consistent with our previous work on a gene expression hotspot on mouse chromosome 8 in primary macrophages (Orozco et al., 2012), where we experimentally validated 12% of the genes predicted to map to the chromosome 8 hotspot.
One of the most desirable applications since the advent of the human genome project has been to be able to determine a person's phenotype from their genome sequence. However, understanding how genetic variation alters cellular behavior and organismal phenotypes, and accurately inferring phenotypes from raw genotypes has proved to be an extremely difficult endeavor. A recent study demonstrated that modeling of SNPs from whole genome sequencing data could be used to infer starvation resistance and startle response in Drosophila (Ober et al., 2012). Here we show that DNA methylation patterns can be used to infer complex phenotypes in a mammalian organism, including bone mineral density, blood cell phenotypes and plasma cholesterol levels (Figure 5, Table S4). We built linear models that incorporate the DNA methylation status at specific CpGs, and used these models to infer clinical traits in other individuals in the same cohort whose methylation status was known. We note that the statistical inference approach we used to model, or explain, a phenotype is distinct from longitudinal prediction of phenotypes for a given individual at a future time.
Association studies over the past ten years have found that the majority of genetic polymorphisms associated with traits are outside protein coding regions. The associated genetic polymorphisms are thought not to alter genes themselves, but rather regulatory elements that control gene expression (Furey and Sethupathy, 2013). Our findings suggest that DNA variants can act by regulating DNA methylation, which in turn affects regulation of gene expression or protein levels of genes that function in biological mechanisms important for the phenotype expression. We hypothesize that it is the plasticity in DNA methylation which makes it ideal for quantitative trait modeling. Since DNA methylation patterns are specific to developmental stages, cell types, and can vary in response to the environment or disease, they can capture the cellular status and provide a more detailed picture of dynamic cellular behavior than our static genomes. Ultimately, our studies suggest that CpG methylation patterns are themselves under genetic control, but because they are more responsive to an organism's state, they can provide added information that cannot be obtained from the genetic sequence alone.
EXPERIMENTAL PROCEDURES
A more detailed version of the experimental procedures can be found in the Supplemental Information.
Data access
All RRBS sequencing and SNP data can be obtained from GEO XXX. The EWAS and GWAS results can be accessed in our online databases to search for candidate genes at http://ewas.mcdb.ucla.edu and to generate association graphs at http://pathways.mcdb.ucla.edu/network. Individual tables with all methylation associations can be downloaded from http://ewas.mcdb.ucla.edu/download.html. The GWAS results can also be accessed at http://systems.genetics.ucla.edu/data/hmdp.
RRBS Libraries
We prepared RRBS libraries as previously described (Feng et al., 2011), with minor modifications. We sequenced the libraries by multiplexing two libraries per lane in an Illumina HiSeq sequencer, with 100bp reads.
Alignment
We aligned the reads with BS-Seeker2 (Guo et al., 2013) to the mm9 mouse reference genome. We used Bowtie as the base aligner, trimmed adapters, allowed for up to 5 mismatches and selected uniquely aligned reads.
Linkage disequilibrium and CpG correlation studies
We computed the Pearson's r-squared between pairs of SNPs, or pairs of CpGs, excluding missing values.
EWAS
We used the linear mixed model package pyLMM (https://github.com/nickFurlotte/pylmm) to test for association and to account for population structure and relatedness among the mouse strains. This method was previously described as EMMA (Kang et al., 2008), and we implemented the model in python to allow for continuous predictors, such as CpG methylation levels that vary between 0 and 1. We applied the model: y=μ+xβ+u+e, where μ=mean, x=CpG, β=CpG effect, and u=random effects due to relatedness, with Var(u) = σg2K and Var(e) = σe2, where K=IBS (identity-by-state) matrix across all Variable CpGs. We computed a restricted maximum likelihood estimate for σg2K and σe2, and we performed association based on the estimated variance component with an F-test to test that β does not equal 0. Each phenotype was log transformed for the association test.
Inflation
We calculated the inflation factor lambda by taking the chi-squared inverse cumulative distribution function for the median of the association p-values, with one degree of freedom, and divided this by the chi-squared probability distribution function of 0.5 (the median expected p-value by chance) with one degree of freedom.
Overlap of EWAS and GWAS
We defined an overlap between EWAS and GWAS if the associations were found within 2Mb (Figure S5A). To decrease the chance of not finding an overlap based on our stringent Bonferroni EWAS thresholds, we used the per phenotype Bonferroni threshold of p<1×10−7 for EWAS, and p<4.1×10−6 for the GWAS as previously described (Bennett et al., 2010).
Conditional EWAS
We performed EWAS for clinical traits or cis expression associations identified with both EWAS and GWAS. We used the pyLMM package as described with one modification, for each EWAS we used the SNP genotype for the GWAS hit as a covariate.
Causal inference test
We performed causal inference tests using the R statistical package CIT developed by Millstein and colleagues (Millstein et al., 2009), according to the user's manual.
PCA
We performed a principal component analysis on the clinical traits. The first and second principal components explained 24% and 12% of the variation in the traits, respectively. We mapped the first two principal components as traits to CpG methylation levels across the genome using EWAS as described above.
Methylation GWAS
We tested for association between methylation levels as phenotypes, and SNPs as predictors using EMMA as previously described (Bennett et al., 2010). The difference between the EWAS model described above, and the GWAS linear mixed model is that in GWAS x=SNP, β=SNP effect, and K=IBS (identity-by-state) matrix across all SNPs.
Methylation GWAS Hotspots
We divided the genome into 2Mb bins and counted the number of all unique CpGs with a significant GWAS hit in that bin and called these “cis and trans” associations (Figure S7E). We also defined a set of trans association hotspots (Figure 6C), where we counted CpGs mapping to each bin in trans, such that the CpG was physically located at least 10Mb away from the bin. We considered CpGs to be associated at the Bonferroni threshold with p<1.4×10−12. We used the Poisson distribution to determine if individual bins had a higher than expected number of associations.
Validation of Mtrr Hotspot
We generated RRBS libraries from Mtrr gene trapped mice (Elmore et al., 2007), using three wild-type and three homozygous gene trapped (gt/gt) male mice at three months of age. We sequenced the libraries by multiplexing all six libraries in one lane and aligned the data using BS-Seeker2 as described above. We compared CpG methylation levels in +/+ and gt/gt mice using a t-test, and estimated the FDR using the Storey method (Storey, 2002). We calculated the difference in methylation levels at each CpG by taking the absolute difference in methylation between the average methylation Mtrr+/+ and the average in −/− mice, i.e. delta methylation.
Phenotype inference
We used the glmnet package in R for building linear models, which fits a generalized linear model via penalized maximum likelihood (Friedman et al., 2010).
Supplementary Material
Research Highlights.
DNA methylation is variable among individuals and is influenced by genetic variation
DNA methylation is associated with complex metabolic and molecular traits
Many associations to DNA methylation are not identified using traditional GWAS
Genetic variation in the gene Mtrr affects DNA methylation throughout the genome
ACKNOWLEDGEMENTS
We want acknowledge Joshua Millstein for his valuable advice on performing the causal inference test, and Hong Xiu Qi, Pingzi Wen and Sharda Charugundla for their valuable help in the animal experiments. LDO was supported by the Ruth L. Kirschstein National Research Service Award T32AR059033. JG was supported by the grant CIRM TB1-01183. MZ and WG were supported by the grant NBRPC 2012CB316503 and the grant NSFC 91010016. WG was supported by the China Scholarship Council. This work was supported by the National Institutes of Health grant GM095656-01A1 and HL28481.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
LDO, AJL and MP conceived the studies. LR and MM prepared the libraries. WG aligned the libraries. JG performed PCA and phenotype predictions. HS and DL created the online databases. NAF created pyLMM. RB and RR generated the Mtrr mice. BJB, CRF, AG and LDO collected the tissues, clinical traits, DNA, metabolite-, protein- and gene expression data. LDO performed analyses and wrote the manuscript. MP directed the study.
REFERENCES
- Arakawa Y, Watanabe M, Inoue N, Sarumaru M, Hidaka Y, Iwatani Y. Association of polymorphisms in DNMT1, DNMT3A, DNMT3B, MTHFR and MTRR genes with global DNA methylation levels and prognosis of autoimmune thyroid disease. Clin Exp Immunol. 2012;170:194–201. doi: 10.1111/j.1365-2249.2012.04646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate's role. Adv Nutr. 2012;3:21–38. doi: 10.3945/an.111.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S, Meduri E, Morange PE, Gagnon F, Grallert H, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014 doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- Elmore CL, Wu X, Leclerc D, Watson ED, Bottiglieri T, Krupenko NI, Krupenko SA, Cross JC, Rozen R, Gravel RA, et al. Metabolic derangement of methionine and folate metabolism in mice deficient in methionine synthase reductase. Mol Genet Metab. 2007;91:85–97. doi: 10.1016/j.ymgme.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Rubbi L, Jacobsen SE, Pellegrini M. Determining DNA methylation profiles using sequencing. Methods Mol Biol. 2011;733:223–238. doi: 10.1007/978-1-61779-089-8_16. [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Furey TS, Sethupathy P. Genetics. Genetics driving epigenetics. Science. 2013;342:705–706. doi: 10.1126/science.1246755. [DOI] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett BJ, Shih D, Che N, Orozco L, Pan C, Hagopian R, He A, Kayne P, Yang WP, et al. Genetic regulation of mouse liver metabolite levels. Mol Syst Biol. 2014;10:730. doi: 10.15252/msb.20135004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo YH, Son SH, Kreienberg PB, Paul A. Novel lipid droplet-associated serine hydrolase regulates macrophage cholesterol mobilization. Arterioscler Thromb Vasc Biol. 2014;34:386–396. doi: 10.1161/ATVBAHA.113.302448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon L, Joo JE, Powell JE, Ollikainen M, Novakovic B, Li X, Andronikos R, Cruickshank MN, Conneely KN, Smith AK, et al. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res. 2012;22:1395–1406. doi: 10.1101/gr.136598.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E, Meduri E, Sandling JK, Hedman AK, Keildson S, Buil A, Busche S, Yuan W, Nisbet J, Sekowska M, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93:876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen PY, Pellegrini M. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 2013;14:774. doi: 10.1186/1471-2164-14-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Moran S, Esteller M. Aberrant DNA methylation profiles in the premature aging disorders Hutchinson-Gilford Progeria and Werner syndrome. Epigenetics. 2013;8:28–33. doi: 10.4161/epi.23366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh JL, Garg P, Thin TH, Yoo S, Dutta R, Trapp BD, Haroutunian V, Zhu J, Donovan MJ, Sharp AJ, et al. Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat Neurosci. 2014;17:121–130. doi: 10.1038/nn.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, Reinius L, Acevedo N, Taub M, Ronninger M, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31:142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae AF, Powell JE, Henders AK, Bowdler L, Hemani G, Shah S, Painter JN, Martin NG, Visscher PM, Montgomery GW. Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol. 2014;15:R73. doi: 10.1186/gb-2014-15-5-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millstein J, Zhang B, Zhu J, Schadt EE. Disentangling molecular relationships with a causal inference test. BMC Genet. 2009;10:23. doi: 10.1186/1471-2156-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober U, Ayroles JF, Stone EA, Richards S, Zhu D, Gibbs RA, Stricker C, Gianola D, Schlather M, Mackay TF, et al. Using whole-genome sequence data to predict quantitative trait phenotypes in Drosophila melanogaster. PLoS Genet. 2012;8:e1002685. doi: 10.1371/journal.pgen.1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco LD, Bennett BJ, Farber CR, Ghazalpour A, Pan C, Che N, Wen P, Qi HX, Mutukulu A, Siemers N, et al. Unraveling inflammatory responses using systems genetics and gene-environment interactions in macrophages. Cell. 2012;151:658–670. doi: 10.1016/j.cell.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco LD, Rubbi L, Martin LJ, Fang F, Hormozdiari F, Che N, Smith AD, Lusis AJ, Pellegrini M. Intergenerational genomic DNA methylation patterns in mouse hybrid strains. Genome Biol. 2014;15:R68. doi: 10.1186/gb-2014-15-5-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BN, Ables GP, Otlivanchik OA, Schoiswohl G, Zechner R, Blaner WS, Goldberg IJ, Schwabe RF, Chua SC, Jr., Huang LS. Hepatic overexpression of hormone-sensitive lipase and adipose triglyceride lipase promotes fatty acid oxidation, stimulates direct release of free fatty acids, and ameliorates steatosis. J Biol Chem. 2008;283:13087–13099. doi: 10.1074/jbc.M800533200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P, Flanagan JM. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22:843–851. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002;64:479–498. [Google Scholar]
- Teng YW, Ellis JM, Coleman RA, Zeisel SH. Mouse betaine-homocysteine S-methyltransferase deficiency reduces body fat via increasing energy expenditure and impairing lipid synthesis and enhancing glucose oxidation in white adipose tissue. J Biol Chem. 2012;287:16187–16198. doi: 10.1074/jbc.M111.303255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Bolick SC, DeRoo LA, Weinberg CR, Sandler DP, Taylor JA. Epigenome-wide association study of breast cancer using prospectively collected sister study samples. J Natl Cancer Inst. 2013;105:694–700. doi: 10.1093/jnci/djt045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Soto J, Anderson B, Riehle C, Zhang YC, Wende AR, Jones D, McClain DA, Abel ED. Regulation of fatty acid metabolism by mTOR in adult murine hearts occurs independently of changes in PGC-1alpha. Am J Physiol Heart Circ Physiol. 2013;305:H41–51. doi: 10.1152/ajpheart.00877.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Lippert C, Heckerman D, Aryee M, Listgarten J. Epigenome-wide association studies without the need for cell-type composition. Nat Methods. 2014;11:309–311. doi: 10.1038/nmeth.2815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.