Abstract
This Matters Arising paper is in response to Guo et al (2013) in Neuron. See also the Matters Arising by Eckler et al. published concurrently. Using genetic fate-mapping with Cux2-Cre and Cux2-CreERT2 mice we demonstrated that the neocortical ventricular zone (VZ) contains radial glial cells (RGCs) with restricted fate potentials (Franco et al., 2012). Using the same mouse lines, Guo et al. (2013) concluded that the neocortical VZ does not contain lineage restricted RGCs. We now show that the recombination pattern in Cux2-Cre/CreERT2 mice depends on genetic background and breeding strategies. We provide evidence that Guo et al. likely reached different conclusions because they worked with transgenic sublines with drifted transgene expression patterns. In Cux2-Cre and Cux2-CreERT2 mice that recapitulate the endogenous Cux2 expression pattern, the vast majority of fate-mapped neurons express Satb2 but not Ctip2, confirming that a restricted subset of all neocortical projection neurons belongs to the Cux2 lineage.
INTRODUCTION
We identified an RGC lineage in the neocortex that expresses the Cux2 gene and is fate-restricted (Franco et al., 2012). Using Cux2-Cre mice for cumulative lineage-tracing studies, we reported that 75% of all neurons in the Cux2-Cre lineage are found in upper neocortical cell-layers and 25% in lower layers (Franco et al., 2012). Most neurons of the Cux2-Cre lineage expressed Satb2 (Franco et al., 2012), which is used as a marker for callosal projection neurons in upper and lower layers and for locally projecting neurons in layer 4 (Alcamo et al., 2008; Arlotta et al., 2005; Britanova et al., 2008). We will refer to these neurons as corticocortical projection neurons. Some cells in the Cux2-Cre lineage expressed the interneuron marker Gad65/67 and few cells were positive for Ctip2 (Franco et al., 2012), which is expressed in interneurons and in corticofugal projection neurons (Arlotta et al., 2005; Franco et al., 2012). Similar observations were made when we used Cux2-CreERT2 mice and tamoxifen injections at E10.5 for temporal genetic fate-mapping (Franco et al., 2012), indicating that progenitors expressing Cux2-CreERT2 at E10.5 are fate-restricted.
Using similar strategies, Guo et al. (2013) found no evidence for fate-restricted RGCs. Here we have addressed this discrepancy and provide a likely explanation why Guo et al. reached a conclusion different from ours. We show that the recombination pattern in Cux2-Cre/CreERT2 mice depends on genetic background and breeding strategies. Specifically, repeated sibling interbreedings of mice carrying the transgene on the C57BL/6 genetic background lead to progressive changes in the expression pattern of transgenes from the Cux2 locus that no longer reflects endogenous Cux2 expression. Changes in the expression pattern of the transgene are also observed on different genetic backgrounds. Notably, Cux2-Cre/CreERT2 mice obtained by the Chen laboratory originally came from colonies that were maintained for over 10 generations (>3 years) by interbreeding mice homozygous for the transgene, which we show here affects the Cre expression pattern. Analysis of the results presented in Eckler et al. (this issue) suggests that the Chen laboratory is working with a Cux2-CreERT2 subline with a recombination pattern that no longer recapitulates the expression pattern of the endogenous Cux2 locus. Importantly, by breeding mice with the aberrant transgene expression pattern onto different genetic backgrounds, the recombination pattern that recapitulates the expression pattern of the endogenous Cux2 genetic locus can be reestablished. Using these “recovered” mice as well as additional fate mapping strategies, we provide further evidence supporting the conclusion that the neocortical VZ contains fate-restricted progenitors.
RESULTS
The Cux2 genetic locus exhibits variable activity that depends on genetic background, and is active in the developing germline
Cux2-Cre and Cux2-CreERT2 mice were generated on a C57BL/6 background (Franco et al., 2012; 2011). For experimentation we routinely used heterozygous Cux2+/Cre and Cux2+/CreERT2 mice maintained by breeding to wild-type C57Bl/6 mice. When crossed to different Cre reporter lines on a congenic C57Bl/6J background, Cux2+/Cre mice consistently exhibited a recombination pattern that recapitulated the upper-layer biased expression pattern of the endogenous Cux2 gene (Fig. 1A).
Figure 1. The Cux2 genetic locus exhibits variable activity that depends on genetic background.
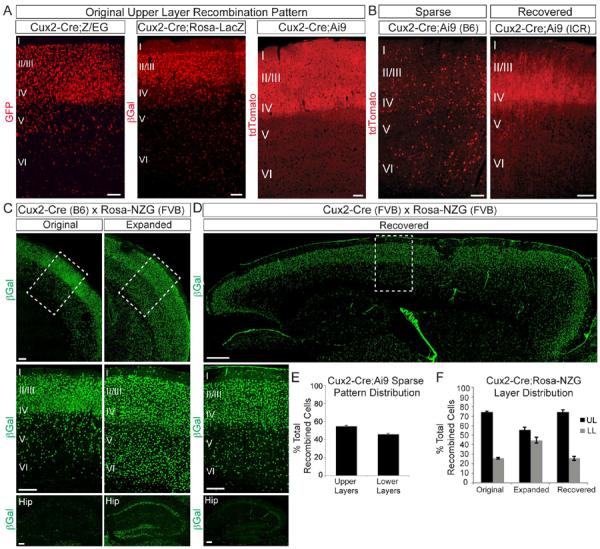
(A) Coronal sections from Cux2-Cre mice crossed to Cre reporter lines (all on a congenic C57BL/6J background). Note recombination primarily in upper neocortical layers, with some scattered cells in lower layers. B) Coronal sections from Cux2-Cre mice crossed to the Ai9 reporter showing sparse recombination in the homozygous inbred line (left panel) and the recovered original pattern after outbreeding onto the ICR strain (right panel). (C) Coronal sections from Cux2-Cre;Rosa26-NZG mice showing the original (left) and expanded (right) recombination patterns in the neocortex (top, middle) and hippocampus (bottom). Middle panels: enlarged images of boxed areas in top panels. (D) Sagittal section from a Cux2-Cre;Rosa26-NZG mouse showing the recovered original recombination pattern in the neocortex (top, middle) and hippocampus (bottom) after outbreeding Cux2-Cre onto an FVB/NJ background for 5 generations. Middle panel is enlarged image of boxed area in top panel. (E) Quantification (mean ± SEM) of layer distribution of tdTomato+ cells in sparsely recombined Cux2-Cre;Ai9 mice (502 cells from 3 animals). (F) Quantification (mean ± SEM) of the layer distribution of βGal+ cells in Cux2-Cre;Rosa26-NZG mice exhibiting the different recombination patterns in (C) and (D). Scale bars: (A,B) 100 μm; (C) 200 μm; (D) 500 μm (top) and 200 μm (middle, bottom).
To facilitate maintenance of the lines for frequent shipments, we generated homozygous Cux2Cre/Cre or CuxCreERT2/CreERT2 mice. Mice that were ultimately obtained by the Chen laboratory were maintained for more than 10 generations of interbreeding in our homozygous colony. Significantly, when we crossed these inbred Cux2Cre/Cre mice to the Ai9fl/fl reporter, their offspring often exhibited sparse recombination patterns (Fig. 1B; “Sparse”) that spanned all neocortical cell layers equally (Fig. 1B,E). This was in stark contrast to the expression pattern of the endogenous Cux2 genetic locus and the recombination pattern in Cux2-Cre mice that were not maintained by breeding homozygous littermates (Fig. 1A) (Franco et al., 2012). We observed this shifted recombination pattern with increasing magnitude and frequency upon prolonged inbreeding of Cux2Cre/Cre mice. The aberrant recombination pattern was stably inherited even when the mice were subsequently crossed to C57BL/6J wild-type mice to generate Cux2+/Cre heterozygotes. This suggests that once established, the epigenetic changes at the genetically modified Cux2 locus are stably inherited in the C57BL/6J genetic background. To test whether the expression pattern could be reset on a different genetic background, we outcrossed inbred Cux2Cre/Cre mice with the outbred strain ICR for 3 generations. We then crossed the resulting Cux2+/Cre mice to Ai9fl/fl mice and found that we recovered the original upper layer-biased pattern (Fig. 1B; “Recovered”).
We also observed that crossing inbred Cux2Cre/Cre mice to the Rosa26-NZG reporter line on an FVB/NJ background resulted in a recombination pattern that was much more broad than the original pattern (Fig. 1C). This expanded pattern was seen in all brain regions, including the neocortex and hippocampus (Fig. 1C) and included all neocortical cell layers (Fig. 1F). The original pattern was recovered when Cux2Cre/Cre mice were outbred onto the FVB/NJ background for 5 generations (Fig. 1D,F). These data indicate that the recombination pattern in Cux2-Cre mice is subject to differences in genetic background of the parents carrying the Cre and/or Cre-reporter transgenes.
We also collaborated with The Jackson Laboratory and the Allen Institute for Brain Science to characterize Cux2-Cre and Cux2-CreERT2 mice for Allen Brain Atlas (ABA) website (brain-map.org). Mice that were shipped to The Jackson Laboratory and Allen Institute had been maintained by homozygous inbreeding for 3–4 generations. Cux2-Cre mice maintained by homozygous breedings for a few generations exhibited layer-specific recombination throughout different functional domains of the neocortex (Fig. S1A,C), although some brains showed a somewhat broadened expression pattern especially in the lateral cortex (not shown). This broadened expression was likely caused by genetic drift caused by inbreeding of the Cux2-Cre homozygous mice for a few generations. Surprisingly, some brains on the ABA website displayed uniform recombination throughout the whole brain including of all neocortical cell layers (Fig. S1D, not shown). Mice with this broad recombination pattern inherited Cux2-Cre and the Cre-reporter allele from the same parent (Fig. S1B; Breeding Strategy 2). We hypothesized that Cux2-Cre is expressed in the developing F1 germline where it drives recombination of the reporter, thus leading to inheritance of the recombined reporter allele in F2 progeny (Fig. S1B). Indeed, breeding Cux2-Cre;Ai9 double heterozygous mice to WT mice led to ubiquitous reporter gene expression throughout the entire animal in F2 progeny (Fig. S1E–F). This was true even in F2 animals that did not inherit Cux2-Cre (Fig. S1E–F), indicating F1 germline recombination. Not all F2 embryos exhibited ubiquitous recombination (Fig. S1D–E), indicating mosaic germline recombination. We confirmed that breeding Cux2-Cre;Rosa-NZG double heterozygous F1 mice to WT animals produced similar results, with some F2 embryos exhibiting ubiquitous recombination even in the absence of the Cre allele (Fig. S1G). Future releases of the ABA website will include information to specify the breeding scheme and inheritance pattern of the Cux2-Cre and Cre-reporter alleles.
We conclude that the expression pattern of the Cre transgene in Cux2-Cre/CreERT2 mice is dependent on genetic background and breeding scheme.
Cux2-Cre mice mark a subset of neocortical projection neurons
To confirm our original lineage mapping results (Franco et al., 2012), we performed additional analyses using Cux2-Cre mice that were continuously maintained by breedings of heterozygous Cux2-Cre mice with C57BL/6 wild-type mice, a breeding scheme that maintains the same expression pattern for Cre as for the endogenous Cux2 gene as determined by in situ hybridization (Franco et al., 2012; Nieto et al., 2004; Zimmer et al., 2004). We analyzed recombination in the mediolateral part of the neocortex along its rostrocaudal extent at embryonic days (E) 10.5, E12.5, and E14.5. As reported (Franco et al., 2012), Cux2-Cre induced recombination in a subset of Pax6 positive RGCs that increased in numbers from E10.5 to E14.5 (Fig. S2A,B). Cre protein was present in the recombined progenitors (Fig. S2C). We then analyzed brains at postnatal day (P) 30 along the rostrocaudal extent of the brain. A Nex/Neurod6-Cre control transgene that induces recombination in nearly all neocortical projection neurons (Belvindrah et al., 2007a; Wu et al., 2005), labeled neurons equally in all cell layers (Fig. S2D). Cux2-Cre mice exhibited recombination extensively in upper neocortical cell layers and much less in lower layers (Fig. S2D). Some signal in lower layers is expected because Cux2-Cre-mediated recombination labels interneurons that reside in all cell layers, and Satb2+ projection neurons that reside in upper and, to a lesser extent, lower layers (Franco et al., 2012). In addition, the Ai9 reporter exhibited a diffuse signal in lower layers consistent with the fact that tdTomato is expressed in the processes of upper layer projection neurons that arborize in lower layers. However, fluorescence in cell bodies was distinguished from that in the neuropil by confocal microscopy, which revealed no fluorescence in the majority of pyramidal neurons in lower layers (Fig. S2E). These data are consistent with our earlier publication (Franco et al., 2012) and confirm that when genetic background is carefully controlled, Cux2-Cre mice are a useful tool to determine the full complement of cells that express Cre from the Cux2 locus at any point during their development and maturation.
To define the subtype of neurons fate-mapped with the Cux2-Cre line, we analyzed Cux2-Cre;Rosa26R-LacZ for co-expression of βGal with Satb2, a marker for corticocortical projection neurons (Alcamo et al., 2008; Britanova et al., 2008) and with Ctip2 and Tle4, markers for subsets of corticofugal projection neurons (Arlotta et al., 2005; Koop et al., 1996). β-gal protein in the Rosa26R-LacZ reporter is strongly expressed in cell bodies thus facilitating the identification of individual cells. Consistent with our previous study (Franco et al., 2012), the vast majority of cells fate mapped by Cux2-Cre expressed Satb2 (Fig. 2A), but not Ctip2 (Fig. 2B) or Tle4 (not shown). We next extended our analyses to the Ai9 tdTomato reporter (Madisen et al., 2010) for better comparison with Guo et al. (2013). We used Satb2 and Ctip2 as markers of corticocortical and corticofugal projection neurons, respectively (Fig. 2C–D). There was strong overlap between Satb2 and tdTomato in upper and lower layers (Fig. 2D). Since essentially all projection neurons in upper layers are Satb2+ and Ctip2− (Alcamo et al., 2008; Britanova et al., 2008), we performed quantitative analysis on recombined cells in lower layers in serial sections. We analyzed 2902 cells in 3 animals, focusing on the medio-lateral part of the neocortex that primarily comprises the somatosensory cortex. We counted total numbers of Satb2+/Ctip2−, Satb2+/Ctip2+ and Satb2−/Ctip2+ cells and determined within each category the percentage that were recombined (Fig. 2E). 92 ± 4% of all lower layer Satb2+/Ctip2− cells belonged to the Cux2-Cre lineage. Only 12 ± 6% of all Ctip2+/Satb2− cells in lower layers were recombined. 24 ± 7% of the Satb2+/Ctip2+ population belonged to the Cux2-Cre lineage; we speculate that this is a poorly characterized projection neuron population that cannot be defined as corticocortical or corticofugal by staining for Satb2 and Ctip2.
Figure 2. Cux2-Cre cumulative fate mapping labels Satb2+ corticocortical projection neurons.
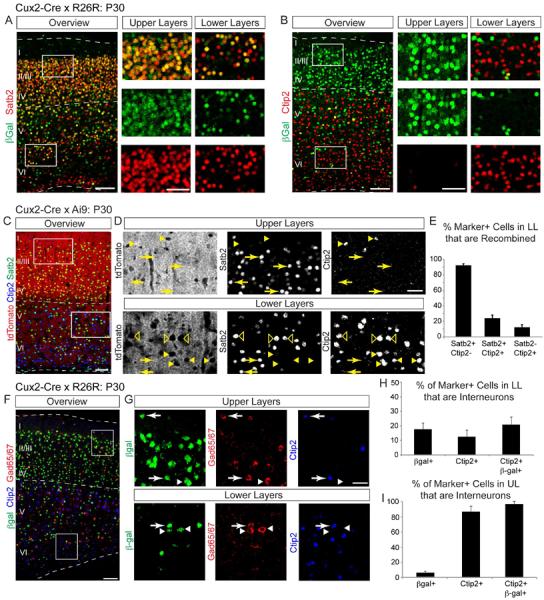
(A,B) Sagittal sections from Cux2-Cre;Rosa-LacZ adult brains immunostained for βGal (green) to reveal recombined cells, and for two projection neuron markers (red): Satb2, corticocortical; Ctip2, corticospinal. Note the high degree of co-localization between βGal and Satb2 (A) and modest co-localization of βGal with Ctip2 (B). (C) Coronal section from a Cux2-Cre;Ai9 adult neocortex immunostained for Satb2 (green) and Ctip2 (blue). tdTomato fluorescence (red) marks recombined cells. (D) Higher magnification views of boxed regions in (C). Labeled are examples of Satb2+(arrows), Ctip2+ (arrowheads) and Satb2+/Ctip2+ cells (empty arrowheads). (E) Percentage of recombined lower layer cells that express Satb2 and/or Ctip2 in the adult neocortex (mean ± SEM). 2902 cells from 3 animals were quantified. (F) Sagittal section from a Cux2-Cre;Rosa-LacZ adult neocortex immunostained for βGal to reveal recombined cells (green), Gad65/67+ interneurons (red) and Ctip2 (blue). (G) Higher magnification views of the boxed regions in (H). Some recombined cells are Gad65/67+ (arrows and arrowheads) and a subset of these are Ctip2+ (arrows). (I–J) Percentage of interneurons expressing indicated markers (mean ± SEM). 1240 cells from 2 animals were quantified. Scale bars: (A–C) 100 μm; 50 μm in boxed insets; (D, G) 100 μm; (E, H) 50 μm.
During development, Cux2 is expressed in some migrating interneurons (Zimmer et al., 2004). Consistent with this and with our earlier findings (Franco et al., 2012), we found that 10% of all cells in the Cux2-Cre lineage expressed the interneuron marker Gad65/67 (Fig. 2F–I, S2F; 6% in upper layers and 17% in lower layers). Since Ctip2 is expressed in a subset of interneurons (Fig. 2F–I; 86% of Ctip2+ cells in upper layers and 12% in lower layers are Gad65/67+), some recombined Ctip2+ cells might be interneurons. We quantified the number of Ctip2+ interneurons within the Cux2-Cre lineage (1240 cells analyzed in 2 animals). 21% of recombined Ctip2+ cells in lower layers were interneurons (Fig. 2H). In upper layers, nearly all recombined Ctip2+ cells were interneurons (Fig. 2I).
Our Cux2-Cre data were in stark contrast to the patterns in Neurod6-Cre and Emx1-Cre, which drive recombination in all neocortical projection neurons but not interneurons (Belvindrah et al., 2007b; Gorski et al., 2002; Wu et al., 2005). 90% and 88% of all Ctip2+ cells belonged to the Neurod6-Cre and Emx1-Cre lineages, respectively, whereas only 7% of all Ctip2+ cells belonged to the Cux2-Cre lineage (many of which are interneurons) (Fig. S2G). Thus, unlike in fate mappings with Neurod6-Cre and Emx1-Cre very few cells in the Cux2 lineage are Ctip2+/Satb2− corticofugal projection neurons.
We reported that astrocytes were not detected in the Cux2-CreERT2 fate-mapped lineage at P10 (Franco et al., 2012). If Cux2+ progenitors generate astrocytes at the end of upper layer neurogenesis, P10 may be too early a time point to observe late-born, morphologically mature astrocytes. We therefore analyzed later time points in Cux2-Cre;Ai9 animals using an antibody against the astrocyte marker Aldh1L1. Only a minor fraction of Aldh1L1+ astrocytes were within the Cux2-Cre fate-mapped lineage (Fig. S2H).
These data confirm our published findings (Franco et al., 2012), which concluded that the vast majority of excitatory neurons in the Cux2-Cre lineage are Satb2+ projection neurons that are abundant in upper neocortical cell layers but can also be found in lower layers. In agreement with published findings (Franco et al., 2012; Zimmer et al., 2004), Cux2-Cre also labels a subset of interneurons, and a minor fraction of Ctip2+ projection neurons and neocortical astrocytes. In light of our discovery that expression of transgenes from the Cux2 locus is dependent on genetic background and breeding strategies, the discrepancy between our study (Franco et al., 2012) and that of the Chen laboratory (Guo et al., 2013) is likely explained by altered recombination patterns caused by differences in genetic background and breeding strategies used to maintain the Cux2-Cre and Cux2-CreERT2 lines.
Temporal genetic fate-mapping using Cux2-CreERT2 mice identifies a population of fate-restricted RGCs
Like other mice that constitutively express Cre, Cux2-Cre mice do not inform about the time point when recombination occurs. Inducible genetic fate-mapping using CreERT2 allows for temporal control over recombination by activating recombination with tamoxifen at defined time points to investigate progenitor-offspring relationships (Hayashi and McMahon, 2002; Zervas et al., 2004). We showed previously that an early E10.5 injection of tamoxifen into Cux2-CreERT2;Ai9 mice labeled RGCs that gave rise to neurons in the P10 neocortex that predominantly resided in upper neocortical layers (Franco et al., 2012). To extend these findings, we injected the faster-acting and shorter-lived tamoxifen metabolite 4-hydroxy-tamoxifen (4-OHT) (Guenthner et al., 2013) into Cux2-CreERT2;Ai9 animals at E10.5 and E11.5; time-points when intermediate progenitors (IPs) and differentiated neurons, including interneurons, are largely absent from the emerging neocortex (Fig. 3A). As shown for tamoxifen induction (Franco et al., 2012), a subpopulation of Pax6+ RGCs in the VZ expressed tdTomato one day after injection of 4-OHT (Fig. 3B), confirming recombination in progenitors. CreERT2 was active in the 4-OHT-injected animals for only a short time period. Two days after 4-OHT injection into Cux2-CreERT2 pregnant dams, we used in utero electroporation to introduce a Cre-responsive EGFP-expression plasmid into the VZ of the embryos. When we analyzed the brains two days later, we did not observe reporter activation (Fig. S3A–C). In controls, the same Cre-responsive EGFP-expression plasmid was activated in the presence of constitutively active Cre (data not shown; Franco et al., 2012). Thus, in experiments with Cux2-CreERT2 mice recombination was induced in progenitors within one day of 4-OHT admininistration but not in differentiated neurons that emerge several days later.
Figure 3. Temporal genetic fate-mapping using Cux2-CreERT2 mice.
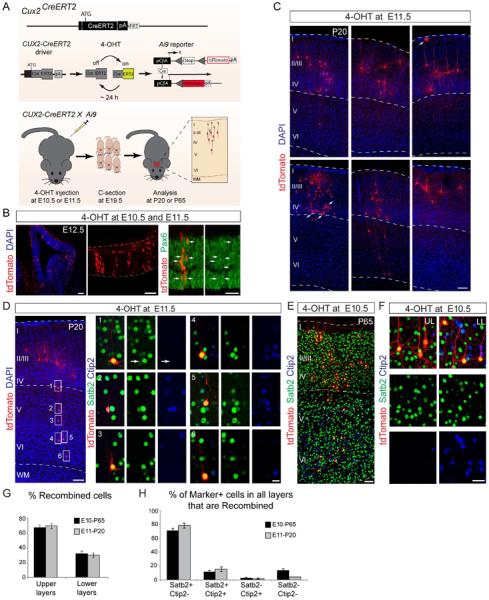
(A) Targeting strategy for Cux2-CreERT2 mice and fate-mapping strategy. Pregnant dams were injected with 4-OHT at the indicated times. Pups were delivered by C-section at E19.5 and transferred to foster mothers. Brains were analyzed at the indicated times. (B) Confocal images showing the recombination pattern in the Cux2-CreERT2;Ai9 neocortex at E12.5 after 4-OHT injections at E10.5 and E11.5. Recombined cells in the VZ express Pax6 (arrows). (C) Neocortex from Cux2-CreERT2;Ai9 animals injected with 4-OHT at E11.5 and analyzed at P20. Most recombined tdTomato+ cells (red) are located in upper layers. Astrocytes were occasionally observed (arrows). Nuclei are stained with DAPI (blue). (D) Example of a 4-OHT injection in which an isolated “clone” (red) contains neurons that reside in upper and lower layers. Note that all boxed cells in lower layers V-VI are Satb2+ (green), whereas none express exclusively Ctip2 (blue). (E) Neocortex from Cux2-CreERT2;Ai9 animals injected with 4-OHT at E10.5 and analyzed at P65 for Satb2 (green) and Ctip2 (blue) expression in recombined tdTomato+ cells (red). (F) Higher-magnification view of the neocortex from Cux2-CreERT2;Ai9 animals analyzed as in (E). (G–H) Quantification of the layer distribution (G) and molecular marker expression (H) in isolated recombined “clones” in serial sections from injections at E10.5 and E11.5, analyzed at P65 and P20, respectively (mean ± SEM). 1745 cells from 9 animals were analyzed in “clones” that spanned several cell layers in serial sections. Scale bars: (B) left to right: 100, 100 and 50 μm; (C, E) 100 μm; (D) 100 μm; 25 μm for boxed insets; (F) 50 μm.
We next analyzed the distribution and molecular identities of fate-mapped cells in mature brains at P20 and P65. A large fraction of cells derived from the Cux2-CreERT2+ progenitor lineage were located in upper layers, with a smaller number of recombined cells settling in lower layers (Fig. 3C–F). The distribution of cells in upper and lower layers (Fig. 3G) was similar to the distribution of cells cumulatively fate-mapped with Cux2-Cre (Franco et al., 2012). The identities of the cells in the Cux2-CreERT2 lineage were determined using molecular markers to quantify 1745 cells in serial sections from 9 animals (Fig. 3D–F, H). Even among cells positioned in lower layers, nearly all recombined cells were Satb2+ (Fig. 3H; >80% for E10.5 injections analyzed at P65; >90% for E11.5 injections analyzed at P20). Some recombined cells expressed Ctip2, but most of these co-expressed Satb2 (Fig. 3H). Only 2–3% of recombined cells were Ctip2+ and Satb2−. In agreement with cumulative fate-mapping results, some of the recombined cells in lower layers were interneurons (Fig. S3D–H).
Consistent with earlier findings (Franco et al., 2012), we rarely found astrocytes in the Cux2-CreERT2 lineage in P10 brains following 4-OHT injections at E10.5 (Fig. S2I). Recombined astrocytes were detected at P20 (Fig. 3C, arrows) and more frequently at P65 (Fig. S2I). This is consistent with the late postnatal local proliferation of astrocytes in the neocortex (Ge et al., 2012) and suggests that the small pool of astrocytes in the Cux2-CreERT2 fate-mapped lineage is amplified postnatally by proliferation.
In summary, our temporal genetic fate-mapping results provide evidence that neocortical progenitors of the Cux2-CreERT2 lineage that are labeled around E10.5–E11.5 generate predominantly Satb2+ projection neurons, as well as interneurons and a minor population of astrocytes.
DISCUSSION
The study by Guo et al. (2013) has raised concerns regarding the fate-restriction of Cux2-Cre+ progenitors. Although the interpretation of their data was confounded by the fact that they analyzed cell lineage in the immature cortex, a further likely explanation for this discrepancy is that the Chen laboratory used Cux2-Cre/CreERT2 mice that no longer expressed Cre in a pattern reflecting that of the endogenous Cux2 locus, as a result of breeding strategies and genetic background variations that likely can cause epigenetic changes at the genetically modified Cux2 locus. The mice used by the Chen laboratory were derived from colonies in our laboratory that had been maintained by homozygous sibling matings for over 10 generations (the mice were first shipped to Stanford from where they were imported to the Chen laboratory), a breeding scheme that we now show affects the expression pattern of the Cre transgenes. Once established, the shifted recombination patterns (Fig. 1B,D) were maintained even in mice that were bred to C57BL/6J wild-type mice to obtain mice heterozygous for the transgenic locus, indicating that the epigenetic changes are stably inherited on the C57BL/6J background (but not following outbreeding to ICR mice). The shifted expression patterns exhibited equal distributions between upper and lower layers that closely resembled the distribution reported by Guo et al. (2013). Additionally, in their “Matters Arising” article the Chen laboratory achieved a very sparse “clone”-like recombination pattern in Cux2-CreERT2 mice by injecting 4mg tamoxifen per 40g body weight, which is twice the amount that we used to induce robust widespread recombination throughout upper neocortical cell layers (Franco et al., 2012). Together these data suggest that the Chen laboratory is working with Cux2-Cre/CreERT2 mice with shifted recombination patterns. Notably, at the time of shipment we were not aware of the effects of breeding scheme and genetic background on transgene expression from the Cux2 locus.
By analyzing mice that exhibited reproducible recombination patterns recapitulating the expression pattern of the endogenous Cux2 gene as determined by in situ hybridization, we provide further evidence that the VZ of the neocortex contains a subpopulation of RGCs that is fate restricted. Only a subset of RGCs express the Cux2 gene and cumulative genetic fate-mapping experiments using Cux2-Cre mice demonstrate that the vast majority of the lineage-traced cells in the adult neocortex are Satb2+ corticocortical projection neurons that are abundant in upper neocortical layers, but can also be found in lower layers (Alcamo et al., 2008; Britanova et al., 2008). Using temporal genetic fate mapping, we demonstrate that cells derived from Cux2-CreERT2+ VZ progenitors generate predominantly Satb2+ projection neurons, whereas the vast majority of Ctip2+ neurons is derived from progenitors that do not belong to the Cux2+ RGC lineage. Notably, although we used an unbiased approach to quantify cells in serial sections along the rostrocaudal axis from sparsely labeled brains, we cannot claim that we analyzed true clones derived from single progenitors. In this regard, we are concerned about the claim by Eckler et al. (this issue) that they carried out a clonal analysis. Cell clones in their study spanned more than 0.5 mm along the rostrocaudal axis and the authors analyzed thousands of cells for each brain. This cell spread and density appears far too high to represent clones. This is a particular important point for additional lineage tracing studies carried out by Guo et al. (2013) and Eckler et al. (this issue) using FezF2-CreERT2 mice. The authors observed that in these mice cells in upper and lower layers were lineage traced. However, it is unclear from their analysis whether the cells in upper or lower layers were derived from single progenitors.
Guo et al. (2013) concluded that most cells in the Cux2-Cre lineage in the VZ are interneurons and that Cux2-Cre is not expressed in progenitors because they could not detect Cux2 protein in the VZ. However, we show that Cux2-Cre and Cux2-CreERT2 induce recombination in a subset of Pax6+ RGCs, and that Cre protein is present in VZ progenitors. These data are consistent with in situ hybridization studies for Cux2 that revealed expression of Cux2 mRNA in RGCs and IPs in the VZ and SVZ (Franco et al., 2012; Nieto et al., 2004; Zimmer et al., 2004). Translation of the Cux2 mRNA may be initiated later, as reported for other genes in the VZ/SVZ (Yang et al., 2014), or available antibodies may not be sensitive enough for detecting Cux2. Significantly, the number of proliferating IPs in the neocortex is increased in mice lacking Cux2, suggesting that Cux2 protein is expressed in progenitors and acts to restrain their proliferation (Cubelos et al., 2008). In this regard, we are surprised that interneurons appear absent in the lineage analysis carried out by Eckler et al (this issue), since they reported in their previous study (Guo et al. 2013) that interneurons are abundantly recombined with Cux2-CreERT2 mice. Perhaps, Cre was no longer expressed in interneurons in the mice used in the recent study (Eckler et al. this issue) due to changes in gene expression caused by breeding strategies.
We also show that the vast majority of neocortical neurons in the Cux2-Cre lineage of adult mice are Satb2+ corticocortical projection neurons, some are interneurons and only a very minor percentage are Ctip2+ /Satb2− projection neurons. These data provide important insights into the total cell population that expresses Cre from the Cux2 locus at any time during their development and provide evidence for restriction within the Cux2-Cre fate-mapped lineage. Notably, additional support for expression of Cux2 in a restricted subset of progenitors comes from functional studies. The dab-1 gene regulates migration of projection neurons of all neocortical cell layers from the VZ into the cortical wall. When dab1 is inactivated with Cux2-Cre, migration of upper layer neurons is disrupted, while deep layer neurons are largely unaffected (Franco et al. 2011).
Our temporal genetic fate mapping studies indicate that Cux2-CreERT2+ progenitors in the neocortical VZ at E10.5–11.5 generate a restricted subset of neocortical projection neurons. Although we observed some variations in the number of fate-mapped projection neurons in upper layers ranging from 75–89%, which differed between functional cortical areas, the vast majority of neurons in the Cux2-CreERT2 lineage in all cases expressed Satb2, a marker for corticocortical projection neurons (Fame et al., 2011; Molyneaux et al., 2009; Sohur et al., 2014). Variations in layer distribution may depend on functional domains of the neocortex and differences in experimental protocol (e.g. kinetics of tamoxifen versus 4-OHT), but do not change the conclusion that Cux2-CreERT2+ progenitors are restricted in their fate potential.
A study using Mosaic Analysis with Double Markers (MADM) concluded that single RGCs in the Emx1-CreERT2 lineage produced clones containing neurons with upper- and lower-layer identities (Gao et al., 2014). Notably, MADM depends on Cre-mediated interchromosomal mitotic recombination during the G2 phase of the cell cycle (Gao et al., 2014). Recombination efficiency will depend on cell cycle length and on the genetic locus of the MADM reporter and thus likely does not capture all progenitor types with equal probability. Perhaps Emx1-CreERT2 and MADM preferentially label a slowly proliferating multipotent progenitor subtype that subsequently generates various lineage-restricted progenitors with faster proliferation kinetics, including those labeled by Cux2-CreERT2. Surprisingly, Gao et al. (2014) concluded that during an asymmetric neurogenic division, a single RGC produces 8–9 neurons. This result is difficult to reconcile with studies using Tbr2-Cre to lineage-trace the output of IP cells (Vasistha et al., 2014). IPs are generated from RGCs and proliferate before generating neurons (Miyata et al., 2004; Noctor et al., 2004). Vasistha et al. (2014) concluded that IPs predominantly generate clones comprising >16 cells. The observation that single Emx1-CreERT2+ RGCs produce clones that are smaller than those traced by their IP offspring is difficult to explain, but may reflect preferential labeling of a specific RGC sub-lineage with MADM.
In summary, we conclude that the neocortical VZ contains lineage-restricted progenitors around E10.5–11.5 that can be traced with Cux2-CreERT2 mice. We further conclude that Cux2-Cre mice are a useful tool to trace the entire Cux2-lineage. It is critical when using these tools to control for genetic background, while also avoiding breeding schemes that allow for germline recombination.
EXPERIMENTAL PROCEDURES
All experimental procedures, genetic background of mice and breeding schemes used in this article are presented in full in the supplemental information.
Supplementary Material
Highlights.
-
-
Transgene expression patterns from the Cux2 genetic locus depend on genetic background
-
-
Cux2 is expressed in a subset of radial glial cells
-
-
The vast majority of excitatory neocortical neurons fate mapped by Cux2-Cre are Satb2+
-
-
Cux2-CreERT2+ progenitors in the E11.5 dorsal ventricula zone generate Satb2+ neurons
ACKNOWLEDGEMENTS
This work was supported by the NIH (SJF, NS060355; UM, NS046456, MH078833, HD070494), the Dorris Neuroscience Center (UM), the Skaggs Institute for Chemical Biology (UM), Children's Hospital Colorado Program in Pediatric Stem Cell Biology (SJF), CIRM (AE, C-GS, CLC, IM-G), a TSRI Stem Cell Postdoctoral Fellowship (KKB), the Ministerio de Educacion (CG-S, EX2009-0416; IM-G, FU-2006-1238), and the Generalitat Valenciana (CG-S, APOSTD/2010/064).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Fariñas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Belvindrah R, Graus-Porta D, Goebbels S, Nave K-A, Müller U. Beta1 integrins in radial glia but not in migrating neurons are essential for the formation of cell layers in the cerebral cortex. J Neurosci. 2007a;27:13854–13865. doi: 10.1523/JNEUROSCI.4494-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvindrah R, Hankel S, Walker J, Patton BL, Müller U. Beta1 integrins control the formation of cell chains in the adult rostral migratory stream. J Neurosci. 2007b;27:2704–2717. doi: 10.1523/JNEUROSCI.2991-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Cubelos B, Sebastián-Serrano A, Kim S, Moreno-Ortiz C, Redondo JM, Walsh CA, Nieto M. Cux-2 controls the proliferation of neuronal intermediate precursors of the cortical subventricular zone. Cereb Cortex. 2008;18:1758–1770. doi: 10.1093/cercor/bhm199. [DOI] [PubMed] [Google Scholar]
- Fame RM, MacDonald JL, Macklis JD. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 2011;34:41–50. doi: 10.1016/j.tins.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Gil-Sanz C, Martinez-Garay I, Espinosa A, Harkins-Perry SR, Ramos C, Müller U. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Müller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011;69:482–497. doi: 10.1016/j.neuron.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Postiglione MP, Krieger TG, Hernandez L, Wang C, Han Z, Streicher C, Papusheva E, Insolera R, Chugh K, et al. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell. 2014;159:775–788. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W-P, Miyawaki A, Gage FH, Jan Y-N, Jan LY. Local generation of glia is a major astrocyte source in postnatal cortex. Nature. 2012;484:376–380. doi: 10.1038/nature10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JLR, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron. 2013;78:773–784. doi: 10.1016/j.neuron.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Eckler MJ, McKenna WL, McKinsey GL, Rubenstein JLR, Chen B. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron. 2013;80:1167–1174. doi: 10.1016/j.neuron.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Koop KE, MacDonald LM, Lobe CG. Transcripts of Grg4, a murine groucho-related gene, are detected in adjacent tissues to other murine neurogenic gene homologues during embryonic development. Mech Dev. 1996;59:73–87. doi: 10.1016/0925-4773(96)00582-5. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Fame RM, MacDonald JL, MacQuarrie KL, Macklis JD. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29:12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Götz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Sohur US, Padmanabhan HK, Kotchetkov IS, Menezes JRL, Macklis JD. Anatomic and molecular development of corticostriatal projection neurons in mice. Cereb Cortex. 2014;24:293–303. doi: 10.1093/cercor/bhs342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasistha NA, García-Moreno F, Arora S, Cheung AFP, Arnold SJ, Robertson EJ, Molnár Z. Cortical and Clonal Contribution of Tbr2 Expressing Progenitors in the Developing Mouse Brain. Cereb Cortex. 2014 doi: 10.1093/cercor/bhu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-X, Goebbels S, Nakamura K, Nakamura K, Kometani K, Minato N, Kaneko T, Nave K-A, Tamamaki N. Pyramidal neurons of upper cortical layers generated by NEX-positive progenitor cells in the subventricular zone. Proc Natl Acad Sci USA. 2005;102:17172–17177. doi: 10.1073/pnas.0508560102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Smibert CA, Kaplan DR, Miller FD. An eIF4E1/4E-T complex determines the genesis of neurons from precursors by translationally repressing a proneurogenic transcription program. Neuron. 2014;84:723–739. doi: 10.1016/j.neuron.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Zervas M, Millet S, Ahn S, Joyner AL. Cell behaviors and genetic lineages of the mesencephalon and rhombomere 1. Neuron. 2004;43:345–357. doi: 10.1016/j.neuron.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Zimmer C, Tiveron M-C, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb. Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


