Abstract
Actinomycetes and filamentous fungi produce a wide range of bioactive compounds, with applications as antimicrobials, anticancer agents or agrochemicals. Their genomes contain a far larger number of gene clusters for natural products than originally anticipated, and novel approaches are required to exploit this potential reservoir of new drugs. Here, we show that co-cultivation of the filamentous model microbes Streptomyces coelicolor and Aspergillus niger has a major impact on their secondary metabolism. NMR-based metabolomics combined with multivariate data analysis revealed several compounds that correlated specifically to co-cultures, including the cyclic dipeptide cyclo(Phe-Phe) and 2-hydroxyphenylacetic acid, both of which were produced by A. niger in response to S. coelicolor. Furthermore, biotransformation studies with o-coumaric acid and caffeic acid resulted in the production of the novel compounds (E)-2-(3-hydroxyprop-1-en-1-yl)-phenol and (2E,4E)-3-(2-carboxy-1-hydroxyethyl)-2,4-hexadienedioxic acid, respectively. This highlights the utility of microbial co-cultivation combined with NMR-based metabolomics as an efficient pipeline for the discovery of novel natural products.
Filamentous microorganisms have an enormous potential for the production of a wide range of bioactive compounds, with application as among others antibiotic, anticancer or antifungal drugs1,2,3. In particular, the emergence of infectious diseases involving multi-drug resistant (MDR) bacterial pathogens since the 1980s has reinforced the screening efforts using filamentous microorganisms. It is likely that in terms of the biosynthetic potential of microbes we have only seen the tip of the iceberg4. However, the return of investment on high-throughput screening campaigns has dropped alarmingly, resulting in an almost complete withdrawal of BigPharma from the antibiotic scene5. Genome sequencing has now revealed that prolific antibiotic producers, and in particular fungi6,7,8 and actinomycetes9,10,11, which had been intensively studied for decades, harbour far more biosynthetic gene clusters for natural products than originally anticipated. The challenge scientists now face is to elicit the expression of these potential treasures, via chemical, genetic or biological triggers12,13. Strategies that have been employed include heterologous expression14, screening or selecting for antibiotic resistant strains15,16, and the use of chemical elicitors17,18. Besides the activation of cryptic gene clusters in known species, a second major reservoir for natural products is formed by microorganisms that have not yet been cultivated, which recently led to the discovery of novel antibiotics in unexpected microbial sources19,20.
Growth in microbial communities or interactions between different microorganisms is the next logical step in the search for new molecules, as many natural products can be expected to be activated specifically by competitors or symbionts in the natural habitat. Microbial co-cultures have been shown to elicit the biosynthesis of novel metabolites, thus increasing the pre-existent chemical diversity21,22. The model microorganisms that are studied intensively in our laboratory are Streptomyces coelicolor, a Gram-positive bacterium with a complex multicellular lifestyle23, and the fungus Aspergillus niger. It was shown previously that interaction with bacteria may have profound effects on the producing ability of Aspergillus species24,25. Here, we report the application of co-cultivation of S. coelicolor and A. niger for the discovery of novel natural products. In addition, the structurally related o-coumaric acid and caffeic acid were specifically converted into the novel molecules (E)-2-(3-hydroxyprop-1-en-1-yl)-phenol and (2E,4E)-3-(2-carboxy-1-hydroxyethyl)-2,4-hexadienedioxic acid, respectively. We thereby apply NMR-based metabolomics, a technology that is particularly effective in identifying molecules in complex biological mixtures, which therefore facilitates the direct biochemical analysis of the community metabolism26. While routinely applied in plant metabolomics, its application in microbial research is rare27.The combination of microbial co-cultivation with NMR-based metabolomics is presented as an efficient way to discover new molecules.
Results and discussion
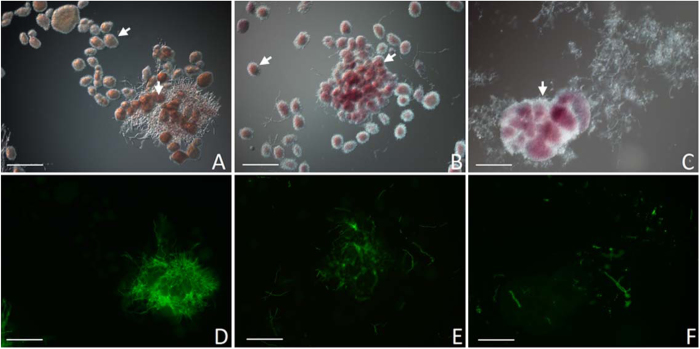
The interaction between the two filamentous model microbes Streptomyces coelicolor A3(2) M145 and Aspergillus niger N402 was studied in co-cultivations in submerged NMMP cultures. Preliminary experiments indicated that the pH of the medium had contrasting effects on the growth of both microbes: whereas Streptomyces species grow well at neutral pH, those of Aspergillus grow optimally at lower pH values. A phosphate buffer of pH 5 was optimal for obtaining sufficient biomass of both microorganisms for a mutual interaction to occur. As the growth rate of A. niger was higher under these conditions, the fungal spores were added to the culture flasks 72 h after inoculation of S. coelicolor. To allow the ready visualization of the interaction between the two microorganisms, we used a test setup with A. niger AR19#1 which expresses eGFP28. A. niger AR19#1 formed pellets with a diameter of 100-500 μm that were rapidly colonized by S. coelicolor within 24 h after addition of A. niger to the Streptomyces culture. The colonization initiated with the adhesion of S. coelicolor to the A. niger biomass (Fig. 1A,D), eventually leading to pellets that were largely covered with Streptomyces biomass after 72 h of co-cultivation (Fig. 1B,E). Notably, loose fungal hyphae were evident in the culture broth in the following 24 h, inferring that the fungal mycelium was almost completely degraded by the bacterium (Fig. 1C,F).
Figure 1. Mycelial interactions between Aspergillus niger AR19#1 (large open mycelial structures) and Streptomyces coelicolor A3(2) M145 (small red-pigmented pellets indicated by arrow heads) during co-culture.
Strain AR19#1 is a derivative of A. niger N402 which expresses eGFP. Samples were observed following inoculation of A. niger at 24 h (A,D), 72 h (B,E), and 96 h (C,F). The reduction in fluorescence intensity in the eGFP-positive hyphae of A. niger (D-F) highlights the strong decline in viable fungal biomass in later stages of the co-culture. Bar, 250 μm (A,B) or 125 μm (C).
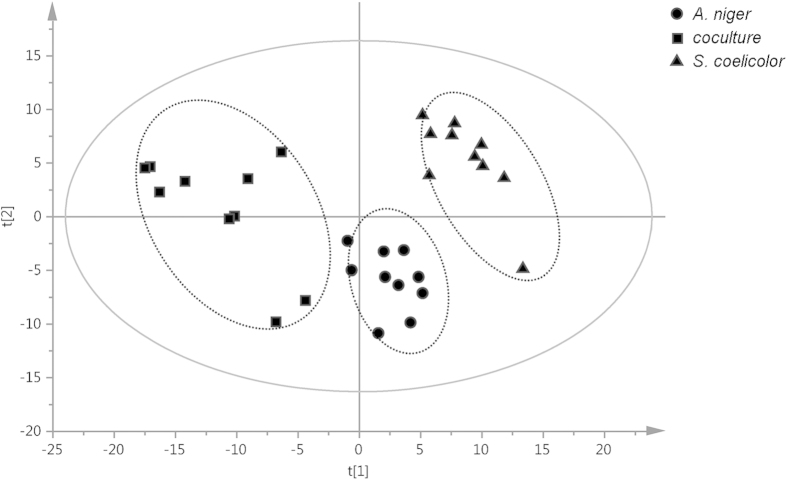
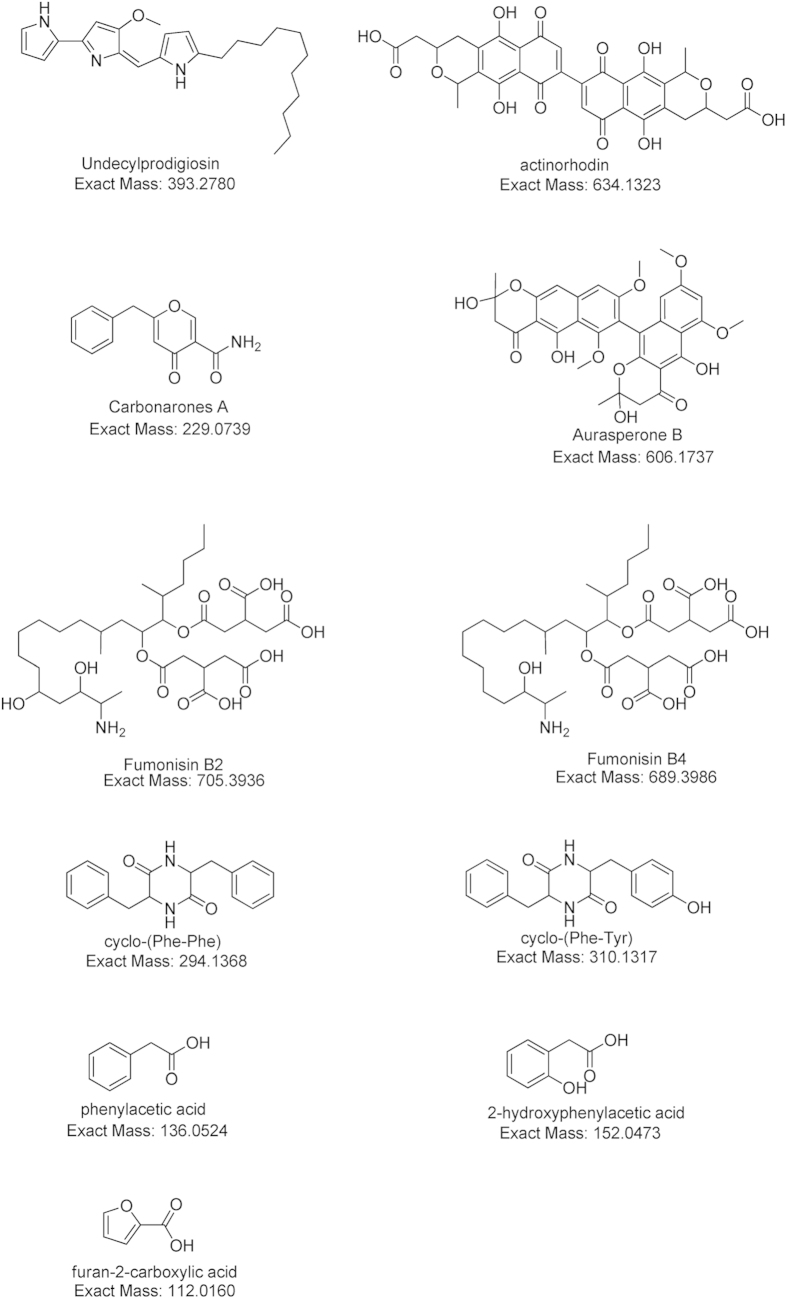
Previous studies have shown that evident metabolic changes occur when different organisms are grown together29. An NMR-based metabolomics approach was employed to study such effects in the Aspergillus-Streptomyces co-cultivations, using S. coelicolor A3(2) M145 and A. niger N402 (the plasmid-free parent of strain AR19#1, i.e. not expressing eGFP). The method of NMR-based metabolomics was performed according to our previously published protocol, including sample preparation, NMR analysis, metabolite identification and multivariate data analysis26. In the current study, a total of 30 liquid cultures (10 for each monoculture and 10 for the coculture) were separately extracted followed by global NMR-profiling of the secondary metabolites (Figure S1). The 1H NMR spectra were then subjected to statistical analysis. An unsupervised principal component analysis (PCA) revealed clustering of the samples in three groups, indicating that the co-culture fingerprints did not overlap with the two corresponding monoculture clusters (Fig. 2). This showed that the dataset contained information that allowed the discrimination of the chemical composition of the co-culture from that of the monocultures. Features exclusively related to the co-culture were found within the dataset of induced compounds (see below), implying that microbial interactions modulated the biosynthetic pathways for the production of secondary metabolites. The PCA loading plot (not shown) allowed detecting biomarkers responsible for the discrimination among the three groups. The compounds were identified with different NMR techniques (1D and/or 2D experiments) and UHPLC-TOF-MS analysis (positive and/or negative modes) (Table 1) and the results were compared with spectroscopic data from literature. The major discriminators for the S. coelicolor culture were the well-studied pigmented antibiotics undecylprodigiosin and actinorhodin30,31, while the A. niger monoculture was abundant in the γ-pyrone derivative carbonarone A32, as well as several minor components, namely naphtho-γ-pyrone aurasperone B, and fumonisins B2 and B433,34. Notably, some compounds accounting for the PCA separation of the metabolites in the co-culture were identified as the previously described compounds cyclo-(Phe-Phe)35, cyclo-(Phe-Tyr)36, phenylacetic acid37, 2-hydroxyphenylacetic acid37, and furan-2-carboxylic acid38 (Fig. 3).
Figure 2. Unsupervised (PCA) multivariate data analysis of the 1H NMR fingerprint data included in Figure S1.
Circles represent the S. coelicolor monoculture, squares the A. niger monoculture, while the co-culture of S. coelicolor and A. niger is represented by triangles.
Table 1. Spectral data assignments for the compounds displayed in Fig. 3. All compounds were summarized according to their corresponding producers, namely S. coelicolor monoculture, A. niger monoculture, and referred co-culture. Compound identification was based on 1H NMR and/or high resolution mass spectrometry, and compared with literature. Proton coupling constants (J in Hz) are given in parentheses.
| Producer | Compounds | Molecular Formula | Characteristic 1H NMR Chemical shifts | Exact Mass | Experimental high resolution mass | Reference |
|---|---|---|---|---|---|---|
| S. coelicolor | Undecylprodigiosin | C25H35N3O | 7.32 (dd, J = 2.4, 1.2); 6.37 (dd, J = 4.2, 1.2); 6.18 (d, J = 4.2); 2.78 (m) | 393.2780 | 394.2854 [M + H]+ | 31 |
| Actinorhodin | C32H26O14 | 7.38 (s) | 634.1323 | 657.3169 [M + Na]+; 633.1256 [M – H]– | 30 | |
| A. niger | Carbonarones A | C13H11NO3 | 8.88 (s); 7.24 (d, J = 8.4); 7.37 (m); 7.31 (m); 6.38 (s); 3.88 (s) | 229.0739 | 230.0809 [M + H]+; 252.0630 [M + Na]+; 269.1353 [M + K]+; | 32 |
| Fumonisin B2 | C34H59NO14 | 705.3936 | 706.4009 [M + H]+ | 34 | ||
| Fumonisin B4 | C34H59NO13 | 689.3986 | 690.4063 [M + H]+; 688.3917 [M – H]– | 33 | ||
| Aurasperone B | C32H30O12 | 6.64 (d, J = 2.4); 3.93 (s); 3.72 (s); 3.60 (s); 3.13 (d, J = 17.4); 3.12 (d, J = 17.4) | 606.1737 | 607.1808 [M + H]+; 629.1605 [M + Na]+; 605.1665 [M – H]– | 33 | |
| Co-culture | Cyclo-(Phe-Phe) | C18H18N2O2 | 7.33 (t, J = 7.8); 7.25 (m); 7.10 (brd, J = 7.8); 4.08 (dd, J = 7.2, 4.2); 2.78 (dd, J = 13.2, 4.2); 2.14 (dd, J = 13.2, 7.2) | 294.1368 | 295.1429 [M + H]+; 317.1238 [M + Na]+ | 35 |
| Cyclo-(Phe-Tyr) | C18H18N2O3 | 7.33 (t, J = 7.8); 7.25 (m); 7.10 (brd, J = 7.8); 6.88 (d, J = 8.4); 6.68 (d, J = 8.4) | 310.1317 | 311.1379 [M + H]+; 333.1185 [M + Na]+ | 35 | |
| Phenylacetic acid | C8H8O2 | 3.58 (s); 6.77 (m); 7.09 (m) | 136.0524 | 135.0442 [M − H]– | 37 | |
| 2-hydroxyphenylacetic | C8H8O3 | 3.47 (s) | 152.0473 | 153.0551 [M + H]+ | 40 | |
| Furan-2-carboxylic acid | C5H4O3 | 7.61 (dd, J = 1.8, 1.2); 7.06 (dd, J = 3.6, 0.6); 6.51 (dd, J = 3.6, 1.8) | 112.0160 | 135.0413 [M + Na]+; 111.0090 [M − H]– | 38 |
Figure 3. Major discriminating compounds responsible for the PCA separation (Fig. 2) of the S. coelicolor and A. niger monocultures from their co-culture.
S. coelicolor monoculture: undecylprodigiosin and actinorhodin; A. niger monoculture: carbonarones A, aurasperone B, fumonisin B2 and fumonisin B4; Co-culture: cyclo(Phe-Phe), cyclo(Phe-Tyr), phenylacetic acid, 2-hydroxyphenylacetic acid, and furan-2-carboxylic acid. The shown theoretical exact mass of each compound was calculated with ChemDraw Ultra 12.0 software. The structure elucidation was done on the basis of NMR and/or experimental UHPLC-TOF-MS high resolution mass, and the spectral data assignments were summarized in Table 1.
Recently, the implementation of the co-cultivation strategy attracted considerable interest and application, since it proved to be an effective way to harvest unique structures with pronounced biological activities29. It may trigger biosynthetic pathways for ‘cryptic’ natural products that would otherwise remain silent under standard laboratory culture conditions. However, often it has not been established which of the co-cultivated microorganisms is actually responsible for manufacturing these metabolites. Here, we focused on the production of cyclo(Phe-Phe) that was produced abundantly in the co-culture of A. niger with S. coelicolor. Since Aspergilli produce 2,5-diketopiperazine-type compounds29,39, A. niger was the most obvious candidate. To test this hypothesis, A. niger was inoculated into the culture filtrate (supernatant without bacterial biomass) of a 5-day-old S. coelicolor culture and incubated for another two days. 1H NMR metabolic profiling (Figure S2) unambiguously demonstrated that the culture filtrate of S. coelicolor effectively elicited the production of cyclo(Phe-Phe) and phenylacetic acid by A. niger. We hypothesize that A. niger metabolizes phenylalanine into phenylacetic acid, which is then further oxidized to 2-hydroxyphenylacetic acid40, while cyclo(Phe-Phe) can be synthesized by condensation and cyclization of two molecules of phenylalanine41,42.
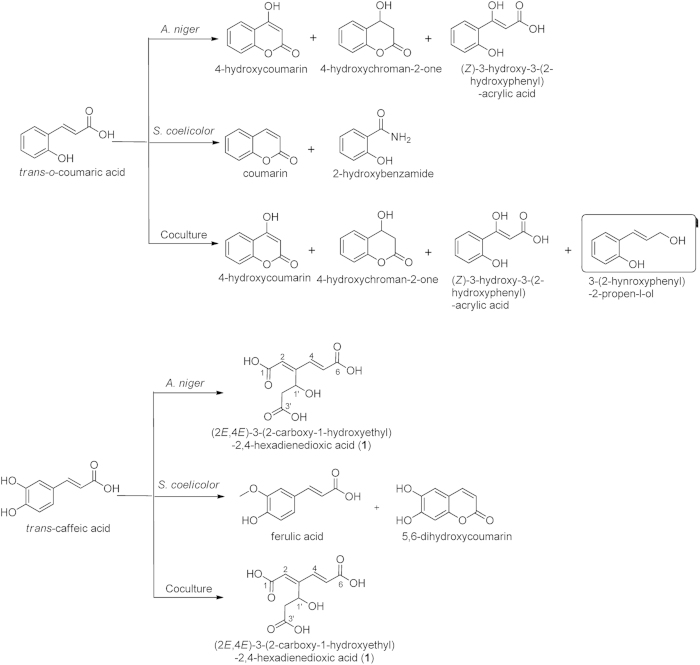
Thus, compounds in the culture fluid derived from S. coelicolor allowed the production of secondary metabolites by A. niger, without the need for physical interaction between them. In contrast, direct contact was required for the biosynthesis of the aromatic polyketides orsellinic acid, lecanoric acid, F-9775A and F-9775B in a co-culture of Aspergillus nidulans and Streptomyces hygroscopicus43. We then investigated whether exogenously added compounds could also act as starter material for bioconversion to new compounds. For this, co-cultivation was done in the presence of one of a variety of hydroxycinnamic acids. Seven structurally related molecules, namely cinnamic acid, o-, m- and p- coumaric acid, caffeic acid, ferulic acid, and sinapinic acid, were fed as substrates to the co-culture and to each of the monocultures. After incubation, the cultures were extracted with ethyl acetate and subsequently subjected to 1H NMR and UHPLC-TOF-MS analysis (positive and/or negative modes) without further chromatographic fractionation. All of the compounds obtained by bioconversion by the three types of cultures were unambiguously identified on the basis of spectral data (Table S1-S7) and summarized (Figure S3-S9). All structures could be readily elucidated based on spectra of known compounds, except for the novel compound 1 (see below). Under these culturing conditions, the majority of substrates were almost completely consumed by A. niger, whereas S. coelicolor exhibited a limited capacity to convert these substrates, yielding at best trace amounts of biotransformation products. However, although the biotransformation process in co-cultures was dominated by A. niger, both qualitative and quantitative differences were found between the mono- and co-cultures in terms of the biotransformation compounds. The addition of o-coumaric acid to the co-culture yielded (E)-2-(3-hydroxyprop-1-en-1-yl)-phenol, which was not produced in either of the two monocultures (Fig. 4).
Figure 4. Biotransformation products of o-coumaric acid and caffeic acid by S. coelicolor and A. niger monocultures and their co-culture.
The boxed compound (E)-2-(3-hydroxyprop-1-en-1-yl)-phenol was exclusively detected in co-culture. The structurally novel molecule (2E,4E)-3-(2-carboxy-1-hydroxyethyl)-2,4-hexadienedioxic acid was derived from caffeic acid biotransformation by fungus, while conversion rate was around 3.5 times in co-culture
Unexpectedly, caffeic acid was converted by A. niger in both mono- and co-culture into a previously undescribed compound (1). Its molecular formula, C9H10O7, was determined using UHPLC-TOF-2Q high-resolution mass analysis with an ion at m/z 211.0254 [M – H2O – H]– (calculated mass for C9H8O6 is 211.0254). The 1H NMR spectrum exhibited a characteristic signal corresponding to a trans-substituted olefinic bond, δH 6.42 (d, J = 16.2 Hz), 7.47 (dt, J = 16.2, 0.6 Hz), which indicated the acrylic acid side chain of caffeic acid resisted the biodegradation. The disappearance of the typical ABX system proton signals for 1,2,4-trisubstituted benzene moiety implied that this aromatic ring had been opened. With the aid of 2D NMR experiments (1H-1H COSY, HSQC, HMBC, Figure S10-S14), the planar structure of compound 1 was unambiguously identified as (2E,4E)-3-(2-carboxy-1-hydroxyethyl)-2,4-hexadienedioxic acid (Table 2) shown in Fig. 1. The formation of 1 was likely due to oxygenation of caffeic acid resulting in cleavage of the aromatic ring between two adjacent hydroxyl groups, thus producing two carboxyl groups at the site of ring opening44. We propose that the resulting product 4-carboxymethylene-2,5-heptadienoic acid44 reacts with one molecule of water to generate the final adduct (2E,4E)-3-(2-carboxy-1-hydroxyethyl)-2,4-hexadienedioxic acid (Figure S7). In view of the oxidative cleavage of the benzene ring, the geometry at theΔ2,3 double bond was established as E. Because the nucleophilic addition of olefin with H2O tends to generate racemates, the stereochemistry at C-1’ was assigned to be both R and S. The (2E,4E)-3-(2-carboxy-1-hydroxyethyl)-2,4-hexadienedioxic acid bioconversion rate was accelerated when A. niger was co-cultivated with S. coelicolor, to around 3.5 times the rate of that occurring in the fungal monoculture.
Table 2. 1H and 13C NMR data assignment for new compound (2E,4E)-3-(2-carboxy-1-hydroxyethyl)-2,4-hexadienedioxic acid (1) in CD3OD. Proton coupling constants (J in Hz) are given in parentheses. 1H NMR and 13C NMR spectra were recorded at 600 MHz. All chemical shift assignments were done on the basis of 1D- and 2D -NMR techniques.
| NO. | δC | δH (J in Hz) | HMBC (1H→13C) | COSY (1H→1H) |
|---|---|---|---|---|
| 1 | 172.4 | |||
| 2 | 120.6 | 6.42 (d, J = 0.6) | C-1, C-4, C-1' | H-1' |
| 3 | 162.5 | |||
| 4 | 132.0 | 7.47 (dt, J = 16.2, 0.6) | C-1’, C-2, C-6, C-5, C-3 | H-5, H-1' |
| 5 | 128.5 | 6.42 (d, J = 16.2) | C-6, C-3, C-4 | H-4 |
| 6 | 167.1 | |||
| 1' | 79.0 | 5.68 (dddd, J = 7.2, 3.6, 1.8, 0.6) | H-2, H-4, H-2' | |
| 2' | 37.6 | 3.07 (dd, J = 16.8, 3.6); 2.58 (dd, J = 16.8, 3.6) | C-3’, C-3 | H-1' |
| 3' | 171.1 |
In conclusion, co-cultivation of S. coelicolor and A. niger allowed the production of the compounds cyclo(Phe-Phe), 2-hydroxyacetic acid and phenylacetic acid by A. niger that were not produced by its monoculture. The fact that the bacterial cell-free filtrate was sufficient for cyclo(Phe-Phe) and phenylacetic acid production by A. niger proves that the change in fungal phenylalanine metabolism was induced by secreted compounds that act as starter molecules or as elicitors of silent biosynthetic pathways, rather than by cell-to-cell contact with or nutrient depletion by S. coelicolor. NMR-based metabolomics coupled to eliciting compounds by microbial co-cultivation is thereby a very effective strategy to dereplicate known molecules and effectively highlighted the compounds induced specifically by co-cultivation. In addition, chemical modification of hydroxycinnamic acids was achieved by Aspergillus and/or Streptomyces biotransformation, whereby only the co-culture specifically converted o-coumaric acid into (E)-2-(3-hydroxyprop-1-en-1-yl)-phenol. Importantly, besides novel metabolic activity, the biotransformation also allowed the discovery of the novel compound (2E,4E)-3-(2-carboxy-1-hydroxyethyl)-2,4-hexadienedioxic acid, which was produced following addition of caffeic acid. Thus, implementation of co-cultivation and biotransformation has the capacity to increase chemical diversity, which offers new leads for drug-discovery efforts.
Methods
Bacterial strains and culturing conditions
Streptomyces coelicolor A3(2) M14545 was obtained from the John Innes Centre strain collection, Aspergillus niger N402 (cspA1) is a derivative of A. niger ATCC9029. A. niger AR19#128 expresses eGFP under control of the A. nidulans gpdA promoter. NMMP45 was used as liquid minimal media for all co-cultivation experiments. Growth curves of S. coelicolor M145 were set up in 50 ml NMMP with glucose (0.5% w/v) as described46, with an inoculum of 106 spores in 250 ml flasks equipped with a spring, and grown at 30 °C with constant shaking at 160 rpm. After 72 h, 105 spores of A. niger N402 were added to the culture, and cultivation continued for another 72 h. In parallel, monocultures of S. coelicolor and A. niger were grown for 144 h and 72 h, respectively, corresponding to the cultivation time of each strain in the co-culture. Ten replicates of each experiment, namely Aspergillus niger monoculture, Streptomyces coelicolor monoculture, and the coculture (so 30 samples in total), were prepared for the metabolomics studies. Fluorescence and corresponding light micrographs were obtained with a Zeiss Axioscope A1 upright fluorescence microscope (with an Axiocam Mrc5 camera at a resolution of 37.5 nm/pixel) as described47. The green fluorescent images were created using 470/40 nm band pass (bp) excitation and 525/50 bp detection.
Biotransformation of hydroxycinnamic acids
Seven hydroxycinnamic acid congeners (cinnamic acid, o-coumaric acid, m-coumaric acid, p-coumaric acid, caffeic acid, ferulic acid, and sinapinic acid) were selected as substrates for the biotransformation study. For this, 10 mg of each substrate dissolved in 200 μl DMSO were added to the co-culture, 24 h after addition of A. niger to the S. coelicolor culture. In parallel, the substrates were added to monocultures of S. coelicolor (after 96 h) and A. niger (after 24 h), corresponding to the cultivation time of each strain in the co-culture. The biotransformation products were analyzed 48 h after adding the substrates. All biotransformation experiments were conducted in triplicate.
Extraction of metabolites
Culture replicates were harvested by centrifugation at 4000 rpm for 10 min. The culture broth (50 ml) was extracted twice with 20 ml of ethyl acetate. The organic phase was washed with 30 ml of water and subsequently dried with 5 g of anhydrous Na2SO4. Finally, the EtOAc was removed under vacuum at 38 °C and the residue was dissolved in 2.0 ml of EtOAc in a microtube (Eppendorf type-5415C, Hamburg, Germany). The solvent was then evaporated at room temperature under nitrogen gas, and subsequently dipped into liquid nitrogen and lyophilized using a freeze dryer (Edwards Ltd., Crawley, England).
NMR measurements
NMR sample preparation and measurements were performed according to a protocol that was published previously26. Briefly, 500 μl of methanol-d4 were added to freeze-dried samples, and the resultant mixtures were vortexed for 10 sec and sonicated for 20 min at 42 kHz using an Ultrasonicator 5510E-MT (Branson, Danbury, CT, USA), followed by centrifugation at 16,000x g at room temperature for 5 min. The supernatant (300 μl) was transferred to a 3 mm micro NMR tube and analyzed. The 1H NMR spectra were recorded at 25 °C on a 600 MHz Bruker DMX-600 spectrometer (Bruker, Karlsruhe, Germany) operating at a proton NMR frequency of 600.13 MHz. Deuterated methanol was used as the internal lock. Each 1H NMR spectrum consisted of 128 scans using the following parameters: 0.16 Hz/point, pulse width (PW) = 30 (11.3 ls) and relaxation delay (RD) = 1.5 s. Free induction decays (FIDs) were Fourier transformed with a line broadening (LB) = 0.3 Hz. The resulting spectra were manually phased and baseline corrected, and calibrated to methanol at 3.30 ppm, using XWIN NMR (version 3.5, Bruker).
UHPLC-TOF mass spectrometry
The UHPLC-TOF-MS analyses were performed on an Ultimate 3000 UHPLC system (Thermoscientific, USA) coupled to a micro-ToF-2Q mass spectrometer from Bruker Daltonics (Bremen, Germany) with an electrospray (ESI) interface48. In separate runs, detection was achieved in both positive and negative ion modes. The m/z range was set to be 100–1000 and the ESI conditions were as follows: capillary voltage of 3500 V, source temperature of 250 °C, desolvation temperature of 250 °C, dry gas low of 10.0 l/min and the nebulizer at 2.0 Bar. For positive ionization, all conditions were the same and the capillary voltage was 4000 V. Internal calibration was performed using a 10 mM sodium formate solution from Sigma–Aldrich (Steinheim, Germany). Samples of 3 μl were injected onto a Kinetex C18, 150 × 2.0 mm column, packed with 1.7 μm particles (Phenomenex, USA), and eluted in gradient mode at a flow rate of 0.3 ml/min with the following solvent system: (A) 0.1% formic acid (v/v) in water; (B) 0.1% (v/v) formic acid in acetonitrile. Analysis began with a gradient of 5% to 90% B to in 19.5 min, followed by an isocratic step of 90% B for 1 min and a re-equilibration of 1 minute with 5% B. The total run time was 21.5 min. The temperature was maintained at 30 °C.
Multivariate Data analysis
The 1H NMR data files were processed as described26,49. The 1H NMR spectra were converted to an ASCII file using AMIX software (Bruker Biospin GmbH), with total intensity scaling. Bucketing or binning was performed and the spectral data were reduced to include regions of equal width (0.04 ppm) equivalent to the region of δ 0.30–10.02. The regions of δ 4.85 – 4.95 and δ 3.25 – 3.35 were removed in the analysis because of the remnant signals of the solvents, HDO and CD3OD, respectively. Principal component analysis (PCA) was performed with the SIMCA-P+ software (Version 13.0, Umetrics, Umeå, Sweden). Pareto-scaling method was used for PCA.
Additional Information
How to cite this article: Wu, C. et al. Expanding the chemical space for natural products by Aspergillus-Streptomyces co-cultivation and biotransformation. Sci. Rep. 5, 10868; doi: 10.1038/srep10868 (2015).
Supplementary Material
Acknowledgments
The work was supported by grants 10379 and 12957 from the Netherlands Technology Foundation to GVW and DC, respectively, and by a grant from the Chinese Scholarship Council (CSC) to CSW.
Footnotes
Author Contributions CSW performed the experiments with the help of BZ; AFJR, GPvW, DC and YHC designed the study; GPvW, DC and YHC and CSW wrote the paper with the help of AFJR. All authors discussed the results and their interpretation and commented on the manuscript at all stages.
References
- Harvey A. L., Edrada-Ebel R. & Quinn R. J. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 14, 111–129 (2015). [DOI] [PubMed] [Google Scholar]
- Hopwood D. A. Streptomyces in nature and medicine: the antibiotic makers. (Oxford University Press, New York; 2007). [Google Scholar]
- Newman D. J. & Cragg G. M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75, 311–335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimm S. L., Russell G. J., Gittleman J. L. & Brooks T. M. The future of biodiversity. Science 269, 347–350 (1995). [DOI] [PubMed] [Google Scholar]
- Payne D. J., Gwynn M. N., Holmes D. J. & Pompliano D. L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6, 29–40 (2007). [DOI] [PubMed] [Google Scholar]
- Bergmann S. et al. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol 3, 213–217 (2007). [DOI] [PubMed] [Google Scholar]
- Pel H. J. et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25, 221–231 (2007). [DOI] [PubMed] [Google Scholar]
- van den Berg M. A. et al. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat Biotechnol 26, 1161–1168 (2008). [DOI] [PubMed] [Google Scholar]
- Bentley S. D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002). [DOI] [PubMed] [Google Scholar]
- Cruz-Morales P. et al. The genome sequence of Streptomyces lividans 66 reveals a novel tRNA-dependent peptide biosynthetic system within a metal-related genomic island. Genome Biol Evol 5, 1165–1175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y. et al. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol 190, 4050–4060 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezel G. P., McKenzie N. L. & Nodwell J. R. Chapter 5. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol 458, 117–141 (2009). [DOI] [PubMed] [Google Scholar]
- Zhu H., Sandiford S. K. & van Wezel G. P. Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol 41, 371–386 (2014). [DOI] [PubMed] [Google Scholar]
- Schumann J. & Hertweck C. Advances in cloning, functional analysis and heterologous expression of fungal polyketide synthase genes. J Biotechnol 124, 690–703 (2006). [DOI] [PubMed] [Google Scholar]
- Ochi K., Tanaka Y. & Tojo S. Activating the expression of bacterial cryptic genes by rpoB mutations in RNA polymerase or by rare earth elements. J Ind Microbiol Biotechnol 41, 403–414 (2014). [DOI] [PubMed] [Google Scholar]
- Thaker M. N. et al. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat Biotechnol 31, 922–927 (2013). [DOI] [PubMed] [Google Scholar]
- Craney A., Ozimok C., Pimentel-Elardo S. M., Capretta A. & Nodwell J. R. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem Biol 19, 1020–1027 (2012). [DOI] [PubMed] [Google Scholar]
- Zhu H. et al. Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbiology 160, 1714–1725 (2014). [DOI] [PubMed] [Google Scholar]
- Ling L. L. et al. A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. C. et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62 (2014). [DOI] [PubMed] [Google Scholar]
- Angell S., Bench B. J., Williams H. & Watanabe C. M. Pyocyanin isolated from a marine microbial population: synergistic production between two distinct bacterial species and mode of action. Chem Biol 13, 1349–1359 (2006). [DOI] [PubMed] [Google Scholar]
- Oh D. C., Kauffman C. A., Jensen P. R. & Fenical W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J Nat Prod 70, 515–520 (2007). [DOI] [PubMed] [Google Scholar]
- Claessen D., Rozen D. E., Kuipers O. P., Sogaard-Andersen L. & van Wezel G. P. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol 12, 115–124 (2014). [DOI] [PubMed] [Google Scholar]
- Benoit I. et al. Bacillus subtilis attachment to Aspergillus niger hyphae results in mutually altered metabolism. Environ Microbiol (2014), in press. [DOI] [PubMed] [Google Scholar]
- Konig C. C. et al. Bacterium induces cryptic meroterpenoid pathway in the pathogenic fungus Aspergillus fumigatus. Chembiochem 14, 938–942 (2013). [DOI] [PubMed] [Google Scholar]
- Kim H. K., Choi Y. H. & Verpoorte R. NMR-based metabolomic analysis of plants. Nat Protoc 5, 536–549 (2010). [DOI] [PubMed] [Google Scholar]
- Wu C. S., Kim H. K., van Wezel G. P. & Choi Y. K. Metabolomics in the natural products field – a gateway to novel antibiotics. Drug Disc Today Technol. In press (2015). [DOI] [PubMed] [Google Scholar]
- Vinck A. et al. Hyphal differentiation in the exploring mycelium of Aspergillus niger. Mol Microbiol 58, 693–699 (2005). [DOI] [PubMed] [Google Scholar]
- Bertrand S. et al. Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery. Biotechnol Adv 32, 1180–1204 (2014). [DOI] [PubMed] [Google Scholar]
- Wright L. F. & Hopwood D. A. Actinorhodin is a chromosomally-determined antibiotic in Streptomyces coelicolor A3(2). J Gen Microbiol 96, 289–297 (1976). [DOI] [PubMed] [Google Scholar]
- Rudd B. A. & Hopwood D. A. A pigmented mycelial antibiotic in Streptomyces coelicolor: control by a chromosomal gene cluster. J Gen Microbiol 119, 333–340 (1980). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Carbonarones A and B, new bioactive gamma-Pyrone and alpha-Pyridone derivatives from the marine-derived fungus Aspergillus carbonarius. J Antibiot (Tokyo) 60, 153–157 (2007). [DOI] [PubMed] [Google Scholar]
- Nielsen K. F., Mogensen J. M., Johansen M., Larsen T. O. & Frisvad J. C. Review of secondary metabolites and mycotoxins from the Aspergillus niger group. Anal Bioanal Chem 395, 1225–1242 (2009). [DOI] [PubMed] [Google Scholar]
- Frisvad J. C., Smedsgaard J., Samson R. A., Larsen T. O. & Thrane U. Fumonisin B2 production by Aspergillus niger. J Agric Food Chem 55, 9727–9732 (2007). [DOI] [PubMed] [Google Scholar]
- Wang F. W. Bioactive metabolites from Guignardia sp., an endophytic fungus residing in Undaria pinnatifida. Chin J Nat Med 10, 72–76 (2012). [DOI] [PubMed] [Google Scholar]
- Nishanth Kumar S., Dileep C., Mohandas C. B. N. & Jayaprakas C. A. Cyclo(D-Tyr-D-Phe): a new antibacterial, anticancer, and antioxidant cyclic dipeptide from Bacillus sp. N strain associated with a rhabditid entomopathogenic nematode. J Peptide Sci 20, 173–185 (2014). [DOI] [PubMed] [Google Scholar]
- Kishore G., Sugumaran M. & Vaidyanathan C. S. Metabolism of DL-(+/−)-phenylalanine by Aspergillus niger. J Bacteriol 128, 182–191 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T. et al. Chemical constituents of Pseudolarix kaempferi Gord. J Chin Pharm Sci 21, 428–435 (2012). [Google Scholar]
- Wang F. et al. Seven new prenylated indole diketopiperazine alkaloids from holothurian-derived fungus Aspergillus fumigatus. Tetrahedron 64, 7986–7991 (2008). [Google Scholar]
- Faulkner J. K. & Woodcock D. The metabolism of phenylacetic acid by Aspergillus niger. Phytochem 7, 1741–1742 (1968). [Google Scholar]
- Guo C. J. et al. Biosynthetic pathway for the epipolythiodioxopiperazine acetylaranotin in Aspergillus terreus revealed by genome-based deletion analysis. J Am Chem Soc 135, 7205–7213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauguet L. et al. Cyclodipeptide synthases, a family of class-I aminoacyl-tRNA synthetase-like enzymes involved in non-ribosomal peptide synthesis. Nucleic Acids Res 39, 4475–4489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeckh V. et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci U S A 106, 14558–14563 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkle B. J., Nozaki M. & Fujisawa H. Ring cleavage of plant catechols by crystalline oxygenases. Phytochemistry 10, 235–242 (1971). [Google Scholar]
- Kieser T., Bibb M. J., Buttner M. J., Chater K. F. & Hopwood D. A. Practical Streptomyces genetics. (John Innes Foundation, Norwich, U.K.; 2000). [Google Scholar]
- van Wezel G. P. et al. A new piece of an old jigsaw: glucose kinase is activated posttranslationally in a glucose transport-dependent manner in Streptomyces coelicolor A3(2). J Mol Microbiol Biotechnol 12, 67–74 (2007). [DOI] [PubMed] [Google Scholar]
- Colson S. et al. The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology 154, 373–382 (2008). [DOI] [PubMed] [Google Scholar]
- Li N. et al. Relative quantification of proteasome activity by activity-based protein profiling and LC-MS/MS. Nat Protoc 8, 1155–1168 (2013). [DOI] [PubMed] [Google Scholar]
- Wu C. S., van Wezel G. P. & Choi Y. Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66. J Antibiotics (Tokyo) in press (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.