Abstract
Genome-wise association studies (GWAS) identify risk variants and modifiers that can influence the pathophysiological processes involved in colorectal cancer (CRC) and thus are important to detect associations between disease phenotypes. Our literature review, performed as per PRISMA statement indicates a significant lack of GWAS functional studies in Saudi Arabia. Therefore, studies on sequencing and mapping are needed to identify gene variants that play a role in the pathophysiology of CRC in this specific population. Because it is not apt to generalize disease associations found in other racial and/or ethnic groups to the Arabic or Middle Eastern population, it is very important to conduct GWAS taking into account multiple ethnicities in this region. In addition, linkage studies and case–control studies that include the various confounding and epigenetic factors are needed for appropriate diagnosis of CRC. We recommend that studies in this region be conducted to understand the role of gene–environment interactions across the various ethnic groups, stages of cancer, tumor type, clinical variables, and the population risk to CRC.
Keywords: Colorectal cancer, GWAS, Saudi Arabia
Colorectal cancer (CRC) is the third and fourth most common cancer in women and men worldwide, respectively, and the fourth most common cause of cancer death.[1] CRC exhibits global geographic variations in its incidence with multiple factors (social, demographic, environmental, and genetic) playing different roles in its pathogenesis. Diet rich in fat and low in fiber, high levels of triglycerides, physical inactivity, diabetes, alcohol, obesity, and smoking are the identified risk factors of colorectal cancer. Hereditary factors play a definite role, but gene–environment interactions are also important in the pathogenesis.[2] Approximately 70% of the risk of colorectal cancer can be related to environmental factors, and identification of these may help prevent the development of the disease. CRC is generally sporadic but approximately 25% of the patients have a genetic predisposition. Instability in chromosomes, CpG island methylation, and microsatellite instability have been reported in key genes leading to the developing of CRC.[3] The disease-specific mortality in development countries for CRC has been reported to be approximately 33%.[4]
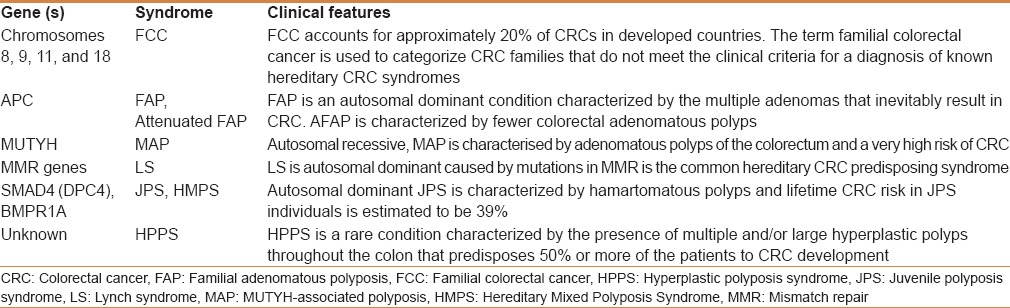
Histopathologically, CRC is manifested in the host as crypt lesions, adenomatous polyps, and carcinomas, which are malignant. CRC develops through a series of clinical processes including inactivation of adenomatous polyposis coli (APC) and mutations in tumor suppressor genes and oncogenes, which are involved in carcinogenesis and due to alterations.[5] Alterations in the levels of gene expressions along with epigenetic modifications in the promoter regions of these genes. Some of the genes and associated syndromes, which have a potential risk in CRC are presented in Table 1. Polymorphisms causing genetic susceptibility and associated CRC risk were found in GSTT1, GSTM1, COX 2, MTHFR, NATs, MTR, and TGF-beta1 genes.[3] The gene polymorphisms in these genes and gene–environment interactions in these genes were found to be associated with increased risk of CRC.
Table 1.
Genes and chromosomes that cause syndromes associated with risk of colorectal cancer
The current article is presented as per the PRISMA statement (www.prisma-statement.org). Published genome-wide association studies (GWAS) in CRC from Saudi Arabia were searched using the search engines PubMed, MEDLINE, EMBASE, and Cochrane Collaboration databases up to May 2014 using the search strategy: (“colorectal cancer” OR “colon cancer”) AND (“genetic studies” OR “genome wide association studies,” OR “gene polymorphisms”) AND (“Saudi Arabia” OR “Kingdom of Saudi Arabia”). The references within the selected articles were manually searched for any relevant literature within this topic. Only studies that included information of GWAS within CRC and conducted in Saudi Arabia were narrowed down to retain the focus of this article. The diagnosis of CRC within the selected studies was as per the internationally accepted criteria. The data were collected independently by two reviewers and any conflicts were resolved through consensus.
Role of pathophysiology and gene polymorphisms in CRC
Tumor progression from normal epithelium to adenoma and carcinoma involves a lot of cellular and molecular events, including genetic alterations, chromosomal instability, hypermethylation of genes, microsatellite instability.[5,6,7] Chromosomal instability leads to aneuploidy and loss of important segments in chromosomes, which was detectable in chromosomes 5, 18, and 17. These mutations may cause changes that influence tumor growth and progression, including characteristic histological changes.[7] Family history of CRC also increases the risk in close relatives, but the magnitude of risk is dependent on the age of diagnosis and the extent of relationship among individuals. It is thus very important to analyze the genetic loci vis-à-vis environmental factors and family history and focusing on candidate genes of biologic relevance to CRC pathogenesis.[7,8] Some studies have used a genome-wide approach to evaluate pattern of gene polymorphisms throughout the genome, based on the International HapMap Project.[9,10] This project helped to identify alleles that may confer an increased or decreased association with CRC risk and help in stratification of at-risk individuals thus helping to devise appropriate screening and treatment methods. The absolute risk of different syndromes in CRC has been estimated in many studies with values ranging from 90% by 45 years of age for FAP, 69% by 80 years for attenuated FAP, 40%–80% by 75 years for LS, 35%–53% for MYH-associated polyposis, 39% by 70 years for PJS and 17%–86% by 60 years for JPS.[11,12,13,14,15,16] The identification of biologically relevant alleles in terms of susceptibility with CRC will need further functional and molecular characterization studies, which should be collectively analyzed for firm conclusions to be drawn. From the above it is understood that genetic testing for susceptibility genes is important to assess germline mutations to formulate appropriate intervention strategies, screening programs and risk analyses. Palles et al.[17] described the transmission pattern in the families with CRCS10, which showed autosomal dominant inheritance. They identified a heterozygous mutation in the germline POLD1 gene and somatic POLE mutation by linkage analysis and sequencing. In addition, tumors showed microsatellite stability. Collectively, this study showed that replication errors may have increased the rate of mutations in CRC. In addition to germline POLD1 mutations, Palles et al.[17] identified somatic POLE mutations in five colorectal cancers from a large database. All of these tumors had additional somatic mutations. These findings suggested that the mechanism of tumorigenesis in POLD1-mutated tumors is decreased fidelity of replication-associated polymerase proofreading, leading to an increased mutation rate.
Genome-wide association studies in CRC
GWAS have been used as important tools to identify and understand disease gene loci and their role in genetic susceptibility, carcinogenesis, and disease mechanisms.[6,18,19] GWAS are used for screening, disease prevention, and risk identification in cancers. It has been widely understood that cancer can show familial gene clustering and in colorectal cancer mutations in mismatch repair genes have been identified. However, genetic linkage studies cannot confirm susceptibility due to the existence of alleles with lower penetrance [Figure 1]. Genetic association studies previusly involved genetic polymorphism analyses, studies on pathways involved in carcinogenesis, DNA repair, hormone biosynthesis, carcinogen metabolism, and cell cycle control.[6] Later, studies involved assessments of functional single nucleotide polymorphisms (SNPs) and gene sequencing.
Figure 1.
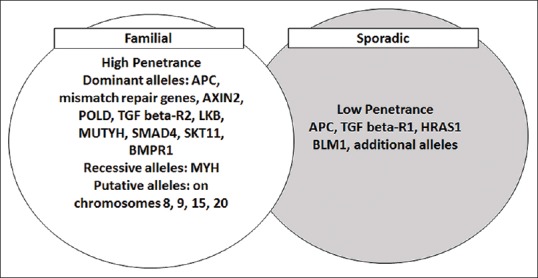
Familial and sporadic alleles with a risk for colorectal cancer
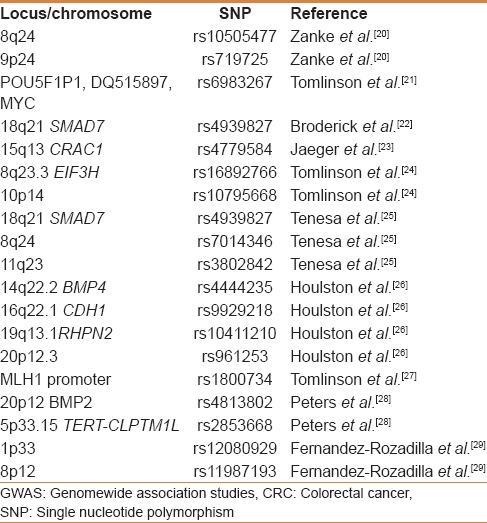
GWAS processes multiple SNPs simultaneously through genotyping platforms by tagging variants in the genome by scanning for associations. GWAS have been conducted in colorectal cancer, and five genetic predisposition loci have been identified in the western population [Table 2]. Pande et al. 2010 in a retrospective study genotyped three risk variants: 8q24 (rs10505477: T >C and rs6983627: T >G) and 9p24 (rs719725: A >C) to analyze the association between each of the risk variants and CRC risk, but none of these variants were found to be associated with CRC risk in their study group.
Table 2.
Cancer susceptibility loci identified through GWAS in CRC
It is important to determine the effects of genetic variation on individual gene expression and cell signaling pathways to understand slight perturbations at the molecular and cellular levels, thus going a long way in public health implications. It is also important to identify populations at higher risk of CRC for genetic stratification of subjects. Improved genetic surveillance programs will help to predict risk of CRC at the level of genotypes within populations. Studies of gene–environmental interactions may help provide plausible explanations to disease variance within treatment groups and overall disease risks. It is important to understand that role of GWAS in especially Arab world will provide important clues and can have high predictive values.
Recently, a new consortium called COlorectal cancer GENeTics (COGENT)[18] has been established for enhancing rigorous research in many countries. This consortium includes research groups from Europe, Australia, the Americas, China, and Japan actively working on CRC genetics. The consortium has recommended that Saudi Arabia takes part in this consortium and specifically work toward better understanding of CRC-related low-penetrance alleles in this population. Using GWAS tagging SNPs (tagSNPs) new, independent CRC predisposition SNPs close to BMP4 (rs1957636) and BMP2 (rs4813802) and near GREM1 between tagSNP rs4779584 were found to be associated with CRC risk. This technique used genetic fine-mapping studies through tagSNP with more than one functional SNP.[30]
Early detection and appropriate prevention by excision of nonmalignant polyps has been found to reduce mortality and thus improve survival rates in subjects with CRC.[31] In addition, subjects who have been classified by genetic surveillance as low, moderate, and high-risk groups could also be prognostically taken care of through disease prevention programs.[32,33]
Scenario in the Kingdom of Saudi Arabia
According to Globocan, in Saudi Arabia, the number of cancer deaths are 9,100 in which CRC cancer incidences are 1168 (14.1%). It has been reported that in Saudi Arabia, the risk for colorectal cancer is low with a crude estimated incidence rate of 6 and a crude mortality rate of close to 4 as compared with the global estimates. The Globocan 2008 data reports an annual incidence rate of CRC of 14.3 per 100,000 men and 9.8 per 100,000 women with annual death rates of 10.1 in men and 6.9 in women.[23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38] A retrospective analyses conducted by Mosli and Alahwal[39] at the National Saudi Cancer Registry from 2001 to 2006 recorded an increasing incidence of CRC with a total of 4201 reported cases and a mean age of diagnosis being 58 years and slightly higher rates reported in males. Colon was the most common site of cancer followed by rectum. Approximately 23% subjects had localized disease, whereas 24% patients had distant metastasis at the time of diagnosis. The remaining patients presented with varying degrees of regional extension and/or an unknown stage of cancer. A study conducted by Isbister[40] that analyzed data from King Faisal Specialist Hospital and Research Centre Tumour registry reported the incidence of CRC below 40 years of age in Saudi Arabia—also suggesting that CRC is more aggressive in young age subjects with metastases being more common in older age subjects. Most importantly, there was an increasing incidence of CRC with lower age of diagnosis among Saudis necessitating the need for more stringent guidelines for CRC screening in this population. The clinical and pathological features of CRC in Saudi also mimic the Western population in terms of left-sided subsite dissemination and delayed appearance of the disease.[41]
An approach that can be used is presented in Figure 2. Tracking the genes and mutations that can influence CRC, identify variants and SNPs, sequencing and re-sequencing, and develop individual patient approaches for treatment through GWAS, all in larger patient populations is the best approach. It has also been shown that GWAS can identify germline mutations, but the genetic risk of adenomas for CRC has been presented in a latest study by Joshi et al. 2013 who have reported a probable association between the 1q31.1 locus and risk of advanced adenoma. This particular study also used an in silico analysis of GWAS data and observed the association of CRC susceptibility SNPs with adenoma risk [Figure 2].
Figure 2.
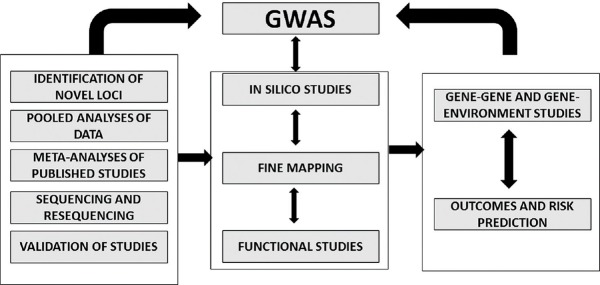
The role of genome-wide association studies (GWAS) in identifying the risk factors in a particular disease
In patients with chronic inflammatory bowel disease, a higher risk of CRC has been observed with risk increasing with family history, colitis, and severity of bowel inflammation. Therefore, early detection of CRC in these patients using molecular and genetic approaches, including DNA damage studies, changes in inflammatory mediators and oxidative stress have been proposed by Azer.[42] The geographic variations in CRC is influenced by diet as demonstrated by Nashar and Al-Murshed.[43] The study demonstrates that an increased consumption of meat and fat from animal sources could predispose an individual to an increased risk of CRC. It is therefore important to characterize as many biomarkers as possible to help in early detection of CRC. Also, biomarkers that serve as prognostic and diagnostic tools are essential in appropriate management of CRC. The development of multidrug resistance mechanisms has hindered the treatment strategies in colon cancer attributing to limited drug effects and overexpression of some oncogenes. The nonspecific action of P-gp in terms of their ability to distribute drugs to nontarget organs can cause decreased elimination and enhanced cytotoxicity of anticancer agents. This leads to initiation of studies that use gene silencing approaches for P-gp–mediated multidrug resistance. Binkhathlan and Alshamsan[44] have suggested that nanomedicine including inhibition by low molecular weight agents and RNAi technology can provide new avenues to eliminate or overcome drug resistance in cancer treatment. Defective glycosylation of galactosaminyltransferase enzymes have also been described to alter the pathology of many cancers. In silico analyses conducted on many genes that are involved in causing biochemical and molecular alterations in the etiology of cancers including colorectal cancers specific to the Saudi population may help in understanding cancers. In fact, in silico analyses of R297W-GALNT12 by researchers at King AbdulAziz University helped in the prediction of harmful effects and disruption of ionic interactions with consequent reduction of associated enzymatic activity reported in CRC.[45]
Key concepts that need to be focused on, include the following
Identifying new gene variants
Understanding the physiological role of novel gene variants at the level of cells
Developing diagnostic biomarkers for easy and early detection of CRC
Understanding the association between different loci at the level of gene expression and its impact on the generation of pathophysiological processes
Categorising risk alleles and their potential role in gene–gene and gene–environment interactions.
SUMMARY
GWAS have enhanced our understanding of the role of genetic variation in CRC risk and the possibility that target-specific drugs that suit a particular subject can be put forward for appropriate treatment by clinicians. GWAS specific to CRC have been performed in England, Scotland, and Canada. These studies recruited moderate sample sizes and provided very important results but also highlighted the need for many more such large scale population studies for the identification of new variants. Genetic population-based studies to identify new cancer predisposition genes through identification of low penetrance alleles in CRC are needed in Saudi Arabia. Published literature has so far confirmed the existence of at least 11 susceptibility loci; however, these are not enough for risk prediction necessitating the need for additional clinical studies in CRC in Saudi Arabia.
ACKNOWLEDGMENTS
We thank the reviewers who helped in narrowing the studies on CRC and GWAS conducted in Saudi Arabia.
The authors thank Chair, Medical and Molecular Genetics, Department of Clinical Lab Sciences, King Saud University for support and encouragement.
Footnotes
Source of Support: Nil
Conflict of Interest: The authors report no conflicting interests with respect to this publication.
REFERENCES
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA. Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Almurshed KS. Colorectal cancer: Case–control study of sociodemographic, lifestyle and anthropometric parameters in Riyadh. East Mediterr Health J. 2009;15:817–26. [PubMed] [Google Scholar]
- 3.Migliore L, Migheli F, Spisni R, Coppedè F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol 2011. 2011 doi: 10.1155/2011/792362. 792362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, et al. Colorectal cancer. Lancet. 2010;375:1030–47. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 5.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 6.Easton DF, Eeles RA. Genome-wide association studies in cancer. Human Mol Genet. 2008;17:R109–15. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 7.Lindblom A. Different mechanisms in the tumorigenesis of proximal and distal colon cancers. Curr Opin Oncol. 2001;13:63–9. doi: 10.1097/00001622-200101000-00013. [DOI] [PubMed] [Google Scholar]
- 8.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 9.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 10.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15:1592–3. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burt RW, Leppert MF, Slattery ML, Samowitz WS, Spirio LN, Kerber RA, et al. Genetic testing and phenotype in a large kindred with attenuated familial adenomatous polyposis. Gastroenterology. 2004;127:444–51. doi: 10.1053/j.gastro.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Vasen HF, Wijnen JT, Menko FH, Kleibeuker JH, Taal BG, Griffioen G, et al. Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology. 1996;110:1020–7. doi: 10.1053/gast.1996.v110.pm8612988. [DOI] [PubMed] [Google Scholar]
- 13.Stoffel E, Mukherjee B, Raymond VM, Tayob N, Kastrinos F, Sparr J, et al. Calculation of risk of colorectal and endometrial cancer among patients with Lynch syndrome. Gastroenterology. 2009;137:1621–7. doi: 10.1053/j.gastro.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aretz S, Uhlhaas S, Goergens H, Siberg K, Vogel M, Pagenstecher C, et al. MUTYH-associated polyposis: 70 of 71 patients with biallelic mutations present with an attenuated or atypical phenotype. Int J Cancer. 2006;119:807–14. doi: 10.1002/ijc.21905. [DOI] [PubMed] [Google Scholar]
- 15.Hearle N, Schumacher V, Menko FH, Olschwang S, Boardman LA, Gille JJ, et al. Frequency and spectrum of cancers in the Peutz-Jeghers syndrome. Clin Cancer Res. 2006;12:3209–15. doi: 10.1158/1078-0432.CCR-06-0083. [DOI] [PubMed] [Google Scholar]
- 16.Coburn MC, Pricolo VE, DeLuca FG, Bland KI. Malignant potential in intestinal juvenile polyposis syndromes. Ann Surg Oncol. 1995;2:386–91. doi: 10.1007/BF02306370. [DOI] [PubMed] [Google Scholar]
- 17.Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nature Genet. 2013;45:136–44. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houlston RS members of COGENT. COGENT (COlorectal cancer GENeTics) revisited. Mutagenesis. 2012;27:143–51. doi: 10.1093/mutage/ger059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Marchand L. Genome-wide association studies and colorectal cancer. Surg Oncol Clin N Am. 2009;18:663–8. doi: 10.1016/j.soc.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 21.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 22.Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–7. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 23.Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, Broderick P, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40:26–8. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 24.Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–30. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 25.Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–7. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–35. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomlinson IP, Houlston RS, Montgomery GW, Sieber OM, Dunlop MG. Investigation of the effects of DNA repair gene polymorphisms on the risk of colorectal cancer. Mutagenesis. 2012;27:219–23. doi: 10.1093/mutage/ger070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters U, Hutter CM, Hsu L, Schumacher FR, Conti DV, Carlson CS, et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Human Genet. 2012;131:217–34. doi: 10.1007/s00439-011-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez-Rozadilla C, Cazier JP, Tomlinson IP, Carvajal-Carmona LG, Palles C, Lamas MJ, et al. A colorectal cancer genome-wide association study in a Spanish cohort identifies two variants associated with colorectal cancer risk at 1p33 and 8p12. BMC Genomics. 2013;14:55. doi: 10.1186/1471-2164-14-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomlinson IP, Carvajal-Carmona LG, Dobbins SE, Tenesa A, Jones AM, Howarth K, et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet. 2011;7:e1002105. doi: 10.1371/journal.pgen.1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10:353–8. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 32.Towler BP, Irwig L, Glasziou P, Kewenter J, Weller D, Silagy C. Screening for colorectal cancer using the faecal occult blood test, hemoccult. Cochrane Database Syst Rev. 2007;24:CD001216. doi: 10.1002/14651858.CD001216.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarvinen HJ, Aarnio M, Mustonen H, Aktan-Collan K, Aaltonen LA, Peltomäki P, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 2000;118:829–34. doi: 10.1016/s0016-5085(00)70168-5. [DOI] [PubMed] [Google Scholar]
- 34.Pisani P, Bray P, Parkin F. Estimates of the worldwide prevalence of cancer for twenty-five sites in the adult population. Int J Cancer. 2002;97:72–81. doi: 10.1002/ijc.1571. [DOI] [PubMed] [Google Scholar]
- 35.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. Lyon: IARC Press; 2004. GLOBOCAN 2002. Cancer incidence, mortality and prevalence worldwide, version 2.0. (IARC Cancer Base No. 5) [Google Scholar]
- 36.Isbister WH. Malignant neoplasia of the colon, rectum and anus at the King Faisal Specialist Hospital and Research Center. Ann Saudi Med. 1992;12:429–33. doi: 10.5144/0256-4947.1992.429. [DOI] [PubMed] [Google Scholar]
- 37.El-Sheikh MA, Al-Karawi MA, Koreich OM. Incidence of colorectal cancer and colonic polyps in Saudi patients. Ann Saudi Med. 1990;10:19–21. [Google Scholar]
- 38.Age-Adjusted Colorectal Cancer Incidence and Mortality Rates for 2008 for 32 countries, Organized by Region of the World, Participating in the ICSN. Globocan 2008, International Agency for Research on Cancer [Google Scholar]
- 39.Mosli M, Alahwal M. The epidemiology of colorectal cancer in the Kingdom of Saudi Arabia: A retrospective analysis of data from the National Saudi Cancer Registry (SCR) from 2001-2006. CDDW Abstracts. 2012:A214. [Google Scholar]
- 40.Isbister WH. Colorectal Cancer below age 40 in the Kingdom of Saudi Arabia. Aust N Z J Surg. 1992;62:468–72. doi: 10.1111/j.1445-2197.1992.tb07227.x. [DOI] [PubMed] [Google Scholar]
- 41.Ayyub MI, Al-Radie AO, Khazeindart AM, Nagio AH, Maniyare IA. Clinicopathological trends in colorectal cancer in a tertiary care hospital. Saudi Med J. 2002;2:160–3. [PubMed] [Google Scholar]
- 42.Azer SA. Overview of molecular pathways in inflammatory bowel disease associated with colorectal cancer development. Eur J Gastroenterol Hepatol. 2013;25:271–81. doi: 10.1097/MEG.0b013e32835b5803. [DOI] [PubMed] [Google Scholar]
- 43.Nashar RM, Almurshed KS. Colorectal cancer: A case control study of dietary factors, King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia. J Family Community Med. 2008;15:57–64. [PMC free article] [PubMed] [Google Scholar]
- 44.Binkhathlan Z, Alshamsan A. Emerging nanodelivery strategies of RNAi molecules for colon cancer therapy: Preclinical developments. Ther Deliv. 2012;3:1117–30. doi: 10.4155/tde.12.89. [DOI] [PubMed] [Google Scholar]
- 45.Hussain MR, Nasir J, Al-Aama JY. Clinically significant missense variants in human GALNT3, GALNT8, GALNT12 and GALNT13 genes: Intriguing in silico findings. J Cell Biochem. 2014;115:313–27. doi: 10.1002/jcb.24666. [DOI] [PubMed] [Google Scholar]


