ABSTRACT
Analysis of the genome sequence of Pseudomonas aeruginosa PA14 revealed the presence of an operon encoding an ABC-type transporter (NppA1A2BCD) showing homology to the Yej transporter of Escherichia coli. The Yej transporter is involved in the uptake of the peptide-nucleotide antibiotic microcin C, a translation inhibitor that targets the enzyme aspartyl-tRNA synthetase. Furthermore, it was recently shown that the Opp transporter from P. aeruginosa PAO1, which is identical to Npp, is required for uptake of the uridyl peptide antibiotic pacidamycin, which targets the enzyme translocase I (MraY), which is involved in peptidoglycan synthesis. We used several approaches to further explore the substrate specificity of the Npp transporter. Assays of growth in defined minimal medium containing peptides of various lengths and amino acid compositions as sole nitrogen sources, as well as Biolog Phenotype MicroArrays, showed that the Npp transporter is not required for di-, tri-, and oligopeptide uptake. Overexpression of the npp operon increased susceptibility not just to pacidamycin but also to nickel chloride and the peptidyl nucleoside antibiotic blasticidin S. Furthermore, heterologous expression of the npp operon in a yej-deficient mutant of E. coli resulted in increased susceptibility to albomycin, a naturally occurring sideromycin with a peptidyl nucleoside antibiotic. Additionally, heterologous expression showed that microcin C is recognized by the P. aeruginosa Npp system. Overall, these results suggest that the NppA1A2BCD transporter is involved in the uptake of peptidyl nucleoside antibiotics by P. aeruginosa PA14.
IMPORTANCE One of the world's most serious health problems is the rise of antibiotic-resistant bacteria. There is a desperate need to find novel antibiotic therapeutics that either act on new biological targets or are able to bypass known resistance mechanisms. Bacterial ABC transporters play an important role in nutrient uptake from the environment. These uptake systems could also be exploited by a Trojan horse strategy to facilitate the transport of antibiotics into bacterial cells. Several natural antibiotics mimic substrates of peptide uptake routes. In this study, we analyzed an ABC transporter involved in the uptake of nucleoside peptidyl antibiotics. Our data might help to design drug conjugates that may hijack this uptake system to gain access to cells.
INTRODUCTION
Newly developed antimicrobial compounds that show strong in vitro activity are often inactive under in vivo conditions. A possible reason why these compounds fail as practically useful antibiotics is that they do not reach their intracellular targets because of the impermeability of bacterial membranes. The wide range of nutrient uptake systems harbors immense potential for the delivery of antibiotics into bacterial cells (1). Uptake systems could be hijacked for drug delivery by the Trojan horse strategy, in which the antibiotic mimics either the structure of the natural substrate of the transporter or is covalently linked to the substrate. In this context, it is essential to understand the different nutrient uptake mechanisms and entry routes to develop new drug delivery strategies.
ABC (ATP-binding cassette) transporters play an important role in the nutritional uptake of substrates from the environment. They typically consist of two permease domains and various ATP-binding domains that are responsible for the energy supply (2). Translocation of molecules across membranes is achieved by the two hydrophobic transmembrane domains upon ATP hydrolysis. The substrate specificity of ABC importers is determined by their substrate-binding protein(s) (SBP[s]), which scavenges solutes in the periplasm and delivers them to the translocator permease (2).
ABC transporters connected to the uptake of various nutrients have been associated with the virulence of pathogenic bacteria (3). For example, ABC transporter mutants defective in the uptake of amino acids or oligopeptides have shown attenuated virulence in multiple animal models (3, 4). Moreover, oligopeptide transporters have also been connected to genetic competence, cell wall metabolism, sporulation, and adherence (5, 6). The diversity of the peptide uptake machinery makes it a viable target for Trojan horse strategy compounds by improving drug internalization into the cytoplasm.
Analysis of the genome sequence of Pseudomonas aeruginosa PA14 revealed the presence of three putative peptide ABC transporters belonging to the peptide-opine-nickel uptake transporter (PepT) family. We have previously characterized the transporter system DppBCDF (PA14_58440-PA14_58490), which is responsible for the utilization of di- or tripeptides (7). Available RNA sequencing data (8) have revealed that the operon encoding the second member of the PepT family, PA14_37840-PA14_37880, is not expressed during in vitro growth. The objective of the present study was to characterize the third member of the PepT family of P. aeruginosa PA14, NppA1A2BCD (PA14_41110-PA14_41160). A recent study showed that in P. aeruginosa PAO1, this transporter, previously designated OppABCDE, is involved in the translocation of the uridyl peptide antibiotic pacidamycin (9). In the present study, we demonstrate that pacidamycin is also a substrate of the Npp permease in P. aeruginosa PA14 and further show that three other antibiotics, blasticidin S, albomycin, and microcin C (McC), are also transported by this uptake system. The four Npp permease substrates identified have diverse chemical structures, mechanisms of antibacterial action, and intracellular targets; however, they are all peptidyl nucleosides, which we propose is a feature specifically recognized by the Npp transport system.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa strains were cultured at 37°C in double yeast tryptone (dYT) medium or minimal medium P (MMP) (10). For growth assays, MMP was modified by omitting NH4Cl (MMP-N) so that various peptides could be used as sole nitrogen sources. Selection for transformed pseudomonads was achieved on King's B medium. Escherichia coli strains were cultured in dYT or M9 minimal medium (11), and cells were routinely maintained at 37°C, except when strains were used that contained the FLP or λ Red recombinase (30°C). E. coli XL-1 Blue was used as the cloning host, E. coli W3110 was used for heterologous expression of transporter genes from PA14, and E. coli ST18 was used for biparental mating where the medium was supplemented with 50 μg/ml 5-aminolevulinic acid.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) or genotypea | Reference or source |
|---|---|---|
| Strains | ||
| Pseudomonas aeruginosa | ||
| PA14 | Wild type | 51 |
| PA14 ΔdppBCDF | dppBCDF deletion mutant | 7 |
| PA14 ΔnppBCD | nppBCD deletion mutant | This study |
| PA14 ΔdppBCDF ΔnppBCD | dppBCDF nppBCD deletion mutant | This study |
| PA14 pscC::MAR2×T7 | pscC transposon mutant, Gmr | 52 |
| Escherichia coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 lac [F′ proAB lacIqZDM15 Tn10(Tcr)] | Stratagene |
| ST18 | pro thi hsdR+ Tpr Smr; chromosome::RP4-2 Tc::Mu-Kan::Tn7/λpir ΔhemA | 53 |
| W3110 | F− λ− rph-1 IN(rrnD rrnE) | 54 |
| W3110 ΔyejABEF | yejABEF deletion mutant | This study |
| BW28357 | F− λ− ΔlacZ4787(::rrnB-3) hsdR514 Δ(araD-araB)567 Δ(rhaD-rhaB)568 | 55 |
| P. syringae pv. phaseolicola 6/0 | Wild type from bush bean, producer of phaseolotoxin | 56 |
| Plasmids | ||
| pGEM-T Easy | Apr, high-copy-number cloning vector | Promega |
| pPS858-Eco | Apr Gmr, source of Gmr-GFP-FRT cassette | 57 |
| pEX18Ap | Apr, gene replacement vector | 57 |
| pEX18Ap.nppBCD-ko | Apr Gmr, contains 2.9-kb fusion fragment of PA14_41130, Gmr-GFP-FRT, and PA14_41170 | This study |
| pBBR1MCS-5 | Gmr, broad-host-range cloning vector | 58 |
| pBBR5.npp | Gmr, contains 7.5-kb fragment carrying nppA1A2BCD operon under lac promoter control | This study |
| pFLP2 | Apr, source of FLP recombinase | 57 |
| pKD46 | Apr, lambda Red recombinase expression plasmid | 14 |
| pKD3 | Apr Cmr, source of Cmr cassette flanked by FRT sequences | 14 |
| pUHAB | Apr, pUC19 carrying mccABCDE genes from E. coli R51 on 11.2-kb fragment | 59 |
Antibiotic resistance: Apr, ampicillin; Gmr, gentamicin; Kan, kanamycin; Smr, streptomycin, Tc, tetracycline; Tpr, trimethoprim.
Cultures harboring individual vectors were supplemented with 50 μg/ml ampicillin (Ap), 25 μg/ml chloramphenicol (Cm), and 25 μg/ml gentamicin (Gm) for E. coli or 500 μg/ml carbenicillin (Cb) and 100 μg/ml Gm for PA14. Bacterial growth was monitored by measuring optical density at 600 nm (OD600) with a spectrophotometer.
PCR amplifications and DNA modifications.
For the PCR primers used in this study, see Table S1 in the supplemental material. All primer sequences were based on the genome of P. aeruginosa UCBPP-PA14 (GenBank accession no. NC_008463.1). Screening PCRs were carried out with the DreamTaq DNA polymerase (Thermo Scientific) in accordance with the manufacturer's instructions at the optimal annealing temperature for each primer set. For screening PCRs performed with PA14, bacterial cells were boiled at 95°C for 5 min and subsequently pelleted at 13,000 rpm for 1 min. Phusion DNA polymerase (Thermo Scientific) was used for high-fidelity PCRs (supplemented with 5% dimethyl sulfoxide). Restriction digestions were performed with Thermo Scientific restriction enzymes according to the manufacturer's instructions at the appropriate temperature. All ligation reactions were carried out at room temperature with Thermo Scientific T4 DNA ligase. DNA purifications were performed with either the GeneJET PCR purification or the GeneJET gel extraction kit (Thermo Scientific) by following the manufacturer's instructions.
Construction of the PA14 nppBCD knockout mutant.
The construction of the knockout vector was based on the protocol described by Zumaquero et al. (12). Briefly, approximately 500-bp sequences flanking the 5′ and 3′ regions of the nppBCD operon were PCR amplified with primers 41130-A1 and 41130-A2 and primers 41170-B1 and 41170-B2. The T7 primer sequence, as well as a KpnI restriction site, was incorporated into primers A2 and B1 to provide homology between the two fragments. After amplification, the fragments obtained were gel purified and approximately 40 ng of each fragment was used in a PCR with primers A1 and B2. The resulting fusion product was gel purified, further ligated into the pGEM-T easy vector, and verified by sequencing.
A Gmr cassette, coupled to green fluorescent protein (GFP) and flanked by Flp-FRT sites, was cut from plasmid pPS858-Eco and subsequently inserted into the KpnI-digested pGEM construct. The deletion allele was cut and further ligated into EcoRI-digested pEX18Ap, yielding the final replacement vector pEX18Ap.nppBCD-ko.
The generation of the PA14 nppBCD mutant was based on the site-specific insertional mutagenesis strategy of Schweizer and Hoang (13) and carried out as described previously (7). In order to confirm the transporter deletion, locus-specific primers that bind up- and downstream of the operon were designed (41130_out1 and 41170_out2) and used in combination with primers binding within the Gmr-GFP cassette (Gm-GFP_out_F and Gm-GFP_out_R).
Construction of the nppA1A2BCD overexpression plasmid.
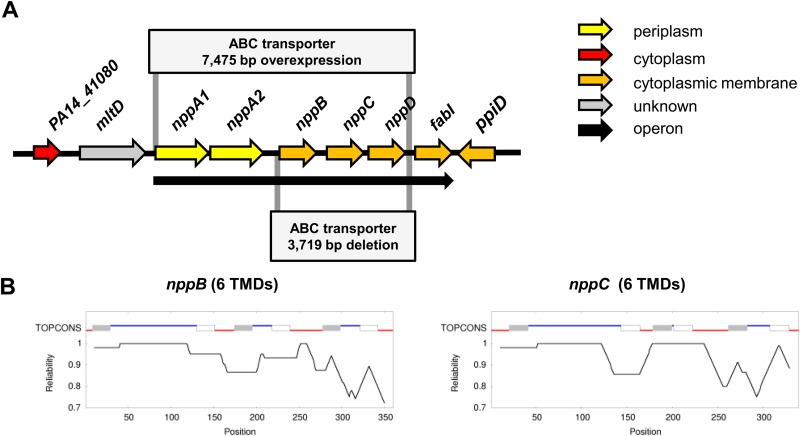
The 7,475-bp operon comprising the ABC transporter NppA1A2BCD was PCR amplified (Fig. 1A), cloned into pBBR1MCS-5 to obtain expression under Plac, and subsequently sequenced to rule out mutations that might have occurred during the PCR. The construct obtained was mobilized into PA14 via biparental conjugation as described earlier (7).
FIG 1.
Overview of the P. aeruginosa PA14 nucleoside transport machinery. (A) Schematic representation of the genomic region surrounding the transporter operon nppA1A2BCD-fabI. The 3,719-bp deletion of the ABC transporter nppBCD operon, as well as the overexpressed 7,475-bp ABC transporter region, is indicated. The subcellular locations of the proteins are indicated by different colors. The black arrow indicates an operon structure. (B) Transmembrane domain analysis of ABC transporter permeases NppB and NppC. The upper line indicates the predicted topology from TOPCONS based on amino acid sequences. Red lines indicate an inner membrane orientation, blue lines indicate an outer membrane orientation, gray boxes indicate transmembrane helices spanning from the inside to the outside, and white boxes indicate transmembrane helices spanning from the outside to the inside. Below the line is a graphic interpretation of the reliability of the prediction for each amino acid.
Construction of the W3110 yejABEF knockout mutant.
The construction of the yejABEF deletion mutant was based on the protocol described by Datsenko and Wanner (14). This protocol utilizes linear DNA fragments for the recombination event. Briefly, primers yej-ko_fwd and yej-ko_rev containing 50-nucleotide extensions that were homologous to the region surrounding the yejABEF operon were used to amplify the Cmr cassette of plasmid pKD3. The resulting PCR product was digested with DpnI and gel purified.
E. coli W3110 cells, harboring the λ Red recombinase on plasmid pKD46, were grown in dYT medium at 30°C. When the culture reached an OD600 of 0.4, l-arabinose was added to a final concentration of 0.3% to induce the expression of RecET recombinase. The culture was shifted to 37°C and further incubated for 1 h. Cells were harvested by centrifugation, washed with ice-cold water, and stored on ice until electroporation.
A 500-ng sample of the purified PCR product was transformed by electroporation into E. coli W3110 cells with an Eppendorf Electroporator 2510 at 1,350 V. Following the recovery of electrotransformants for 2 h at 37°C in dYT medium, bacterial cell suspensions were plated on dYT plates containing half of the standard antibiotic concentration. Homologous recombination events were screened by colony PCR with primers yej_out1 and yej_out2 flanking the knockout region. PCR products were verified by sequencing.
Phenotype MicroArrays.
We utilized PM6, PM7, and PM8 Biolog Phenotype MicroArray plates to monitor the catabolism of the di- or tripeptides used as nitrogen sources by PA14 and its mutant strains. Preparation of PM plates was done as described previously (7).
All experiments were performed with a minimum of three replicates. Outliers among all replicates were identified by means of their standard deviation. On the basis of the difference between the standard deviation and the average value, we defined a maximum deviation threshold of 20% to be a good replicate. A deviation of more than 50% disqualified the sample from further analysis. Samples with deviations between 20 and 50% were considered acceptable.
The respiratory activity of bacterial cells in the wells of PM plates was quantified by a kinetic plot of color formation against time. The resulting area of each plot was represented by a specific value. The threshold of this area value was set to 2,000 (approximately 15 to 20% of the average area value of the positive control for all of the plates). In order to assist visualization of the data, we created a heat map based on values representing the area under the respiration curve (see Fig. S1 in the supplemental material).
Growth experiments with various peptides as sole nitrogen sources.
We performed growth experiments with selected peptides as sole nitrogen sources (Table 2). Therefore, bacteria grown overnight on KB agar plates were scraped from the plates, resuspended in MMP-N, washed by centrifugation, and adjusted to an OD600 of 0.05. For the growth assay, MMP-N was individually supplemented with various peptides at 0.2 mM to serve as sole sources of nitrogen. Cell cultures (1 ml of culture in a 2-ml Eppendorf tube) were incubated in an incubator shaking at 200 rpm at 37°C. After 24 h, an endpoint measurement (OD600) was taken. All experiments were repeated at least three times.
TABLE 2.
Effect of nppBCD and dppBCDF mutations on the use of peptides as nitrogen sourcesa
| Nitrogen source | Length (aa) | Avg OD600 ± SD |
|||
|---|---|---|---|---|---|
| PA14 | ΔnppBCD | ΔdppBCDF | ΔnppBCD ΔdppBCDF | ||
| Controls | |||||
| None | 0.02 ± 0.00 | 0.01 ± 0.00 | 0.02 ± 0.00 | 0.02 ± 0.00 | |
| 0.2 mM NH4Cl | 0.35 ± 0.01 | 0.38 ± 0.02 | 0.35 ± 0.02 | 0.36 ± 0.03 | |
| Oligopeptidesb | |||||
| Gly-Pro-Ile-Ser | 4 | 0.07 ± 0.01 | 0.09 ± 0.00 | 0.06 ± 0.01 | 0.05 ± 0.00 |
| Gly-Pro-Arg-Pro | 4 | 0.08 ± 0.01 | 0.06 ± 0.00 | 0.07 ± 0.00 | 0.06 ± 0.00 |
| Gly-Arg-Gly-Asp | 4 | 0.14 ± 0.02 | 0.13 ± 0.01 | 0.14 ± 0.01 | 0.13 ± 0.00 |
| Val-Ile-His-Asn | 4 | 0.14 ± 0.01 | 0.13 ± 0.00 | 0.14 ± 0.02 | 0.13 ± 0.01 |
| Lys-Thr-Thr-Lys-Ser | 5 | 0.18 ± 0.03 | 0.19 ± 0.02 | 0.17 ± 0.02 | 0.16 ± 0.02 |
| Phe-Leu-Glu-Glu-Val | 5 | 0.05 ± 0.00 | 0.05 ± 0.00 | 0.05 ± 0.00 | 0.04 ± 0.01 |
| Phe-Tyr-Gly-Pro-Val | 5 | 0.14 ± 0.01 | 0.11 ± 0.03 | 0.14 ± 0.01 | 0.13 ± 0.00 |
| Tyr-Gly-Gly-Phe-Leu | 5 | 0.15 ± 0.00 | 0.16 ± 0.02 | 0.06 ± 0.00 | 0.04 ± 0.01 |
| Tyr-Gly-Gly-Phe-Met | 5 | 0.17 ± 0.02 | 0.15 ± 0.02 | 0.11 ± 0.01 | 0.11 ± 0.02 |
| Gly-Arg-Gly-Asp-Asn-Pro | 6 | 0.07 ± 0.02 | 0.05 ± 0.00 | 0.08 ± 0.02 | 0.05 ± 0.01 |
| Lys-Lys-Thr-Pro-Glu-Glu | 6 | 0.09 ± 0.02 | 0.07 ± 0.01 | 0.09 ± 0.02 | 0.08 ± 0.01 |
| Val-Tyr-Ile-His-Pro-Phe | 6 | 0.16 ± 0.01 | 0.16 ± 0.02 | 0.16 ± 0.02 | 0.13 ± 0.02 |
| Asp-Arg-Val-Tyr-Ile-His-Pro | 7 | 0.10 ± 0.01 | 0.09 ± 0.01 | 0.09 ± 0.01 | 0.10 ± 0.01 |
| Ile-Ile-Asn-Phe-Glu-Lys-Leu | 7 | 0.08 ± 0.03 | 0.08 ± 0.01 | 0.09 ± 0.02 | 0.10 ± 0.03 |
| Ser-Gln-Asn-Tyr-Pro-Ile-Val | 7 | 0.36 ± 0.07 | 0.32 ± 0.05 | 0.30 ± 0.03 | 0.32 ± 0.03 |
| Asp-Arg-Val-Tyr-Ile-His-Pro-Phe | 8 | 0.34 ± 0.04 | 0.34 ± 0.03 | 0.31 ± 0.01 | 0.32 ± 0.02 |
| Asp-Arg-Val-Tyr-Val-His-Pro-Phe | 8 | 0.39 ± 0.06 | 0.38 ± 0.03 | 0.36 ± 0.04 | 0.38 ± 0.01 |
| Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys | 8 | 0.12 ± 0.03 | 0.10 ± 0.02 | 0.13 ± 0.04 | 0.11 ± 0.02 |
| Ser-Tyr-Ser-Met-Glu-His-Phe-Arg-Trp-Gly | 10 | 0.39 ± 0.03 | 0.38 ± 0.02 | 0.36 ± 0.03 | 0.35 ± 0.04 |
| Ala-Pro-Gly-Asp-Arg-Ile-Tyr-Val-His-Pro-Phe | 11 | 0.05 ± 0.00 | 0.07 ± 0.01 | 0.07 ± 0.01 | 0.05 ± 0.00 |
| AMPsb | |||||
| HHC36 | 9 | 0.05 ± 0.03 | 0.06 ± 0.02 | 0.07 ± 0.03 | 0.04 ± 0.02 |
| Bac2a | 12 | 0.06 ± 0.01 | 0.06 ± 0.00 | 0.05 ± 0.01 | 0.06 ± 0.00 |
| Indolicidin | 13 | 0.09 ± 0.01 | 0.10 ± 0.01 | 0.14 ± 0.03 | 0.12 ± 0.01 |
| LL37 | 37 | 0.14 ± 0.00 | 0.14 ± 0.00 | 0.13 ± 0.01 | 0.12 ± 0.01 |
| Cell wall peptides (0.1 mg/ml) | |||||
| B. subtilis PGN | 0.18 ± 0.01 | 0.18 ± 0.00 | 0.16 ± 0.02 | 0.15 ± 0.02 | |
| Tri-DAP | 0.08 ± 0.00 | 0.07 ± 0.01 | 0.08 ± 0.01 | 0.06 ± 0.02 | |
| Miscellaneous | |||||
| Phe-Arg-β-naphthylamide (0.2 mM) | 0.24 ± 0.02 | 0.22 ± 0.02 | 0.24 ± 0.01 | 0.21 ± 0.03 | |
| Aminolevulinic acid (0.2 mM) | 0.06 ± 0.01 | 0.06 ± 0.01 | 0.07 ± 0.01 | 0.06 ± 0.00 | |
| Aminopterin (0.2 mM) | 0.08 ± 0.01 | 0.04 ± 0.02 | 0.05 ± 0.02 | 0.04 ± 0.01 | |
| Pacidamycin D (0.1 mg/ml) | 0.05 ± 0.00 | 0.04 ± 0.00 | 0.04 ± 0.01 | 0.04 ± 0.00 | |
| Glutathione (0.2 mM)c | 0.41 ± 0.01 | 0.38 ± 0.04 | 0.42 ± 0.01 | 0.38 ± 0.02 | |
Bacterial growth was measured in triplicate by determining the increase in OD600. Boldface values are significantly different as determined by a two-tailed t test (P < 0.05).
The concentration of peptides in growth assays was 0.2 mM.
Glutathione was tested in growth assays as a nitrogen or sulfur source with similar results.
Drug susceptibility tests.
The MICs of drugs for P. aeruginosa strains were determined by a 2-fold dilution assay in a 96-well plate with MMP. MIC determination for E. coli cultures was done with Mueller-Hinton broth. All tests were performed at least in triplicate in accordance with Clinical and Laboratory Standards Institute recommendations (15). Growth of bacteria at 37°C was examined by visual inspection after 24 h of incubation. The MIC was defined as the lowest concentration of an antibiotic that completely prevented visible cell growth.
McC toxicity and competition assay.
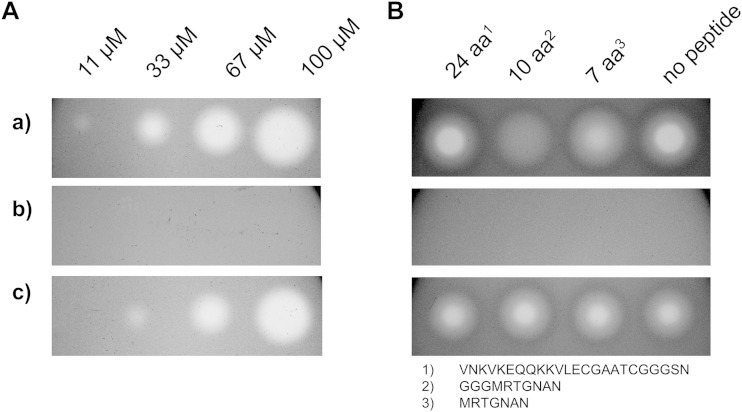
McC was purified from E. coli BW28357 harboring plasmid pUHAB as described previously (16). Chemically synthesized peptides MRTGNAN (7 amino acids [aa]), GGGMRTGNAN (10 aa), and VNKVKEQQKKVLECGAATCGGGSN (24 aa) were purchased from GeneScript USA. McC toxicity and transporter competition assays were performed with E. coli strain W3110 and its corresponding ΔyejABEF mutant harboring either empty plasmid pBBR1MCS-5 or a plasmid overexpressing the PA14 npp operon. Overnight cultures were diluted 1:100 into 15 ml of M9 soft agar (0.6%) supplemented with 0.5% glycerol, 0.1% Casamino Acids, 0.01% thiamine, and Gm at 25 μg/ml.
The toxicity assay was performed with a 3-fold dilution of 100 μM McC, where 2.5 μl of McC was dropped onto an agar plate and subsequently incubated at 37°C for 16 h. The transporter competition assay contained a mixture of 30 μM McC and one of the aforementioned peptides at 3 mM.
Agar diffusion assay with toxic tripeptides.
The toxic tripeptide phaseolotoxin [Nδ(N′-sulfodiaminophosphinyl)-ornithyl-alanyl-homoarginine] was obtained from a culture of Pseudomonas syringae pv. phaseolicola as previously described (7). The tripeptide bialaphos was obtained from Alfa Aesar.
For preparation of the test plates, P. aeruginosa strains grown overnight were scraped from agar plates, resuspended in sterile water, and adjusted to an OD600 of 1.0. A 500-μl volume of the cell suspension was added to 50 ml of MMP agar warmed to 50°C. After the plates was prepared, 7-mm holes were punched into the agar and 50-μl volumes of the tripeptides were added to the holes. Plates were incubated for 24 h at 37°C and subsequently visually analyzed in terms of growth inhibition zones on the bacterial lawn.
Human bronchial epithelial cells.
The immortalized human bronchial epithelial cell line 16HBE14o− was obtained from Dieter C. Gruenert (University of California, San Francisco, CA). Cells were cultured in coated T25 cell culture flasks at 37°C in a humidified 5% CO2 atmosphere. The coating solution contained 10% bovine serum albumin (Sigma-Aldrich), 1% bovine collagen I (Invitrogen), and 1% human fibronectin (Sigma-Aldrich) in LHC basal medium (Invitrogen) and was applied to the T25 flasks 1 day prior to seeding of the cells. The culture medium was minimum essential medium (Invitrogen) supplemented with 10% fetal bovine serum (Sigma-Aldrich), 2 mM l-glutamine (Invitrogen), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen).
Cytotoxicity assay.
16HBE14o− cells were cultured in T25 flasks until 90% confluent (5 days). Bacteria were grown aerobically in LB medium to an OD600 of 1.0. Bacterial densities were adjusted in infection medium (minimal essential medium with 2 mM l-glutamine) to infect epithelial cells at a ratio of 50:1. After the 16HBE14o− cell layer was washed twice with phosphate-buffered saline, epithelial cells were infected with P. aeruginosa PA14 or its transporter mutants. Samples from the supernatant of the cell cultures were drawn 3, 6, and 9 h postinfection. The lactate dehydrogenase (LDH) activity released into the medium by damaged cells was determined with the Pierce LDH cytotoxicity kit (Thermo Scientific). Absorbance of the samples was detected with a microplate reader (FLUOstar Omega; BMG Labtech) at wavelengths of 490 and 680 nm, respectively. LDH activity was calculated by subtracting the 680-nm absorbance value (background signal from the instrument) from the 490-nm absorbance value. All experiments were repeated in triplicate.
Multiple-sequence alignment and phylogenetic analysis.
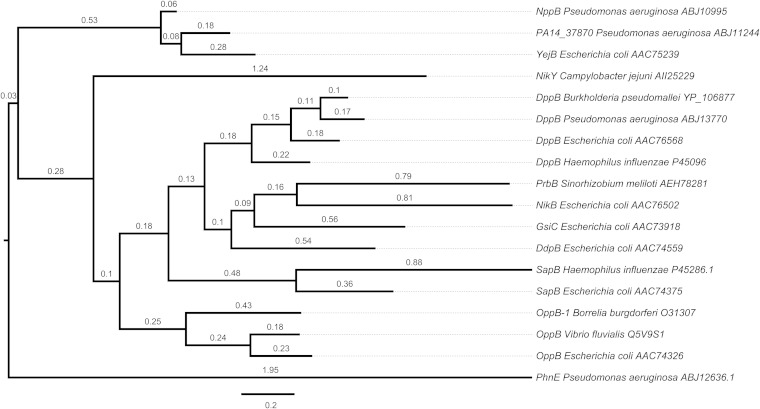
Protein sequence data were obtained from NCBI and the Pseudomonas Genome Database (17). Putative peptide ABC transporter proteins from P. aeruginosa PA14 were identified on the basis of homology to already described transporters from other organisms found in the Transporter Classification Database (18) and respective BLASTP searches. The phylogenetic tree reconstruction was performed by the Phylogeny.fr web service (19) with the phosphonate ABC permease PhnE of P. aeruginosa PA14 included as the outgroup. The tree obtained was subsequently visualized by FigTree (20).
RESULTS
Computational analysis of the ABC transporter NppA1A2BCD from P. aeruginosa PA14.
It was previously shown that inactivation of an ABC transporter mediates high-level resistance of P. aeruginosa PAO1 to the uridyl peptide antibiotic pacidamycin. Mistry et al. (9) named this transporter Opp because of its sequence similarity to the oligopeptide permease of Helicobacter pylori. However, phylogenetic analysis shows that this transporter and known oligopeptide permeases are clearly separated. On the basis of this observation and functional findings reported below, we propose that this ABC transporter be renamed Npp (nucleoside peptide permease).
In P. aeruginosa strain PA14, the gene for the Npp transporter is located in an operon-like structure containing two genes for SBPs PA14_41110 (nppA1) and PA14_41130 (nppA2), two genes for membrane-associated permeases PA14_41140 (nppB) and PA14_41150 (nppC), a gene for hydrophilic ATP-binding protein PA14_41160 (nppD), and a gene for NADH-dependent enoyl-acyl carrier protein reductase PA14_41170 (fabI) (Fig. 1A). Analysis of the genome sequence upstream of the ABC transporter genes (Fig. 1A) revealed the presence of the gene for lytic murein transglycosylase D (MltD), which is involved in bacterial cell wall degradation (21). The mltD gene is not part of the npp operon, which is expressed as a single polycistronic nppA1A2BCD-fabI transcript (8, 9, 17) from three promoters (σ32, σ38, and σ70) located in the intergenic region between mltD and nppA (22). The nppA1A2BCD-fabI operon is expressed at a low level in cells of P. aeruginosa PA14 during growth in LB medium, as previously shown by RNA sequencing (8). The transcriptional profile correlates with mass spectrometry-based proteomic data showing that the Npp transporter appears to be present at approximately 30 copies per cell (D. Bumann, personal communication).
Analysis of NppB and NppC topology with the TOPCONS prediction software (23) revealed the typical six α-helical transmembrane-spanning domains of ABC-type permeases (Fig. 1B). Using the NppB protein sequence, we identified homologs of this permease in P. aeruginosa PA14 (PA14_37870, 68% identity) and E. coli (YejB, 64% identity). Phylogenetic analysis showed that these three permeases form a cluster, whereas other ABC-type peptide uptake transporters appeared with greater distance (<35% identity), as depicted in the phylogenetic tree in Fig. 2.
FIG 2.
Phylogenetic relationships among ABC transporter permeases. The tree reconstruction shown was performed by the Phylogeny.fr web service, and the result was visualized by FigTree. Distance is expressed as the number of amino acid substitutions per 100 positions.
Role of the Npp transporter in peptide utilization.
It has previously been reported that oligopeptide permeases are responsible for the uptake of cell wall peptides in E. coli and Salmonella enterica serovar Typhimurium (24). These data, together with the presence of the aforementioned gene for lytic MltD immediately upstream of the npp operon, led us to the initial hypothesis that the Npp transporter might be involved in the uptake of cell wall peptides such as peptidoglycan (PGN). In order to test this hypothesis, we performed batch culture growth assays with PGN from Bacillus subtilis or Tri-DAP (l-Ala-γ-d-Glu-meso-diaminopimelic acid), a compound present in PGN, as a sole source of nitrogen (Table 2). Although we could demonstrate that PA14 is able to utilize PGN as a nitrogen source, we could not demonstrate its uptake by the Npp transporter.
Next, we used PM6 to PM8 Biolog Phenotype MicroArray plates to test whether the Npp transporter is involved in the utilization of di- and tripeptides. We have previously demonstrated that the ABC transporter Dpp of P. aeruginosa PA14 is an uptake system for these peptides (7). In contrast, analysis of Biolog data showed that the Npp transporter does not contribute to the utilization of di- or tripeptides by PA14 (see Fig. S1 in the supplemental material).
Recently, Mistry et al. (9) reported that bialaphos (l-alanyl-l-alanyl-phosphinothricin), a tripeptide with antibacterial activity, is a substrate of the Opp system of P. aeruginosa PAO1, which is identical to the Npp system of PA14. This prompted us to investigate whether the Npp transporter is involved in the uptake of bialaphos and another toxic tripeptide, phaseolotoxin. However, our results show that, in PA14, these peptides are taken up only by the dipeptide permease DppBCDF (see Fig. S2 in the supplemental material).
To elucidate whether the Npp transporter is responsible for the uptake of longer peptides, we performed growth assays with defined minimal medium containing peptides of various lengths, ranging from 4 to 11 aa, as the sole nitrogen source (Table 2). We found that the wild-type PA14 strain grew well on 4 of the 20 oligopeptides tested (OD600 of >0.3 after 24 h), growth was moderate with 9 oligopeptides (OD600 of ≥0.1 after 24 h), and the remaining 7 peptides could not be utilized by PA14. It should be noted that analysis of culture supernatants by thin-layer chromatography revealed that some peptides were partially degraded (data not shown). However, as we observed no difference in the growth of the wild type and the nppBCD-deficient mutant in this assay, the results suggest that Npp of P. aeruginosa PA14 is not involved in the utilization of oligopeptides. Interestingly, in the course of these studies, we found that the dipeptide transporter DppBCDF appears to be involved in the uptake of two pentapeptides (Table 2).
The Yej transporter of S. enterica serovar Typhimurium, which is homologous to the Npp transporter of PA14, is able to confer resistance to antimicrobial peptides (AMPs) (25). However, antimicrobial susceptibility tests showed that mutational inactivation of nppBCD had no effect on the susceptibility of PA14 to four AMPs (Bac2a, HHC-36, indolicidin, and LL37). Thus, we tested whether PA14 is able to utilize these AMPs as nitrogen sources. Two of the four AMPs tested allowed slight growth (OD600 of ≥0.1 after 24 h) of the PA14 wild-type strain, as well as of the nppBCD-deficient mutant, suggesting that the Npp permease is not involved in the translocation of these AMPs (Table 2).
Contribution of the Npp transporter to P. aeruginosa PA14 antibiotic resistance.
Recently, Mistry et al. (9) demonstrated that the Npp transporter of P. aeruginosa PAO1 is required for uptake of the uridyl peptide antibiotic pacidamycin across the inner membrane. In order to investigate the role of the Npp transporter in P. aeruginosa PA14 antibiotic resistance and to identify additional substrates, wild-type PA14 and peptide transporter mutants were tested for antimicrobial susceptibility. Deletion of the nppBCD genes caused the expected increase in the pacidamycin MIC, as previously described for PAO1 (Table 3) (9). In addition, we observed increases in resistance to blasticidin S, another peptidyl nucleoside antibiotic, in the nppBCD mutant (2-fold) and the nppBCD dppBCDF double mutant (4-fold) (Table 3).
TABLE 3.
Antimicrobial susceptibility profiles of P. aeruginosa strains
| Drug | MIC (μg/ml)a for: |
|||||||
|---|---|---|---|---|---|---|---|---|
| PA14 | PA14 ΔnppBCD | PA14 ΔdppBCDF | PA14 ΔnppBCD ΔdppBCDF | PA14(pBBR5) | PA14(pBBR5.npp) | PA14 ΔnppBCD(pBBR5) | PA14 ΔnppBCD(pBBR5.npp) | |
| Peptidyl nucleoside antibiotics | ||||||||
| Pacidamycin D | 125 | 500 | 125 | 500 | 125 | <3.9 | 500 | <3.9 |
| Blasticidin S | 125 | 250 | 125 | 500 | 125 | 15.6 | 250 | 15.6 |
| Nikkomycin | >500 | >500 | >500 | >500 | >500 | >500 | >500 | >500 |
| Aminonucleoside antibiotic: puromycin | 625 | 625 | 625 | 625 | 625 | 625 | 625 | 625 |
| Fatty acyl nucleoside antibiotic: tunicamycin | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| AMPs | ||||||||
| LL37 | 12.5 | 12.5 | 6.2 | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 |
| Protamine | 3.1 | 3.1 | 3.1 | 3.1 | 3.1 | 3.1 | 3.1 | 3.1 |
| Polymyxins | ||||||||
| Colistin | 1.2 | 1.2 | 1.2 | 1.2 | 1.2 | 1.2 | 1.2 | 1.2 |
| Polymyxin B | 1.2 | 1.2 | 1.2 | 1.2 | 1.2 | 1.2 | 1.2 | 1.2 |
| Glycopeptide: vancomycin | 500 | 250 | 500 | 250 | 500 | 500 | 500 | 500 |
| Heavy metal: nickel chloride | 125 | 125 | 125 | 125 | 125 | 15.6 | 250 | 31.2 |
MICs were determined by 2-fold dilution assay in three or more independent experiments with similar results. Boldface values are more than 2-fold higher or lower MICs.
To identify additional substrates of the ABC transporter, the nppA1A2BCD operon was overexpressed from plasmid pBBR1MCS-5. Consistent with the results of the nppBCD deletion mutant analysis, overproduction of the Npp transporter increased the susceptibility of PA14 to blasticidin S (8-fold) and pacidamycin (>32-fold). In addition, we detected a 16-fold increase in the susceptibility of PA14 overexpressing npp to nickel chloride, which has previously been described as a substrate of the NikABCDE transporter of E. coli, another member of the ABC superfamily of transporters belonging to the peptide-opine-nickel uptake transporter family (Transporter Classification Database TC number 3.A.1.5) (26) (Table 3). Chivers et al. (26) found that the NikABCDE system transports nickel ions in the presence of exogenously added l-histidine in the form of a Ni-(l-His)2 complex. In the case of Npp, the form in which nickel ions are transported remains to be established.
Heterologous expression of the Npp transporter in E. coli.
The Yej transporter is the closest homolog of the Npp system in E. coli (Fig. 2). It is involved in the uptake of the peptide nucleotide antibiotic McC, a protein synthesis inhibitor that targets the enzyme aspartyl-tRNA synthetase (27). In order to test whether the P. aeruginosa Npp and E. coli Yej transporter systems are similar in substrate specificity, we constructed a yejABEF-negative mutant of E. coli W3110 and complemented it with the nppA1A2BCD genes from P. aeruginosa PA14.
To determine if McC is a substrate of the Npp system, we used an agar diffusion assay and applied various concentrations of McC to lawns of cells being tested. Clear zones of inhibition of the E. coli wild-type strain and a complete lack of growth inhibition of the yejABEF-deficient mutant were observed, as expected. Interestingly, clear growth inhibition zones were detected on lawns of the yej mutant complemented with the nppA1A2BCD genes (Fig. 3A), indicating that McC is efficiently recognized by the Npp system. Moreover, heterologous expression of the Npp transporter in a yejABEF-negative E. coli background increased susceptibility to pacidamycin D, a uridyl peptide antibiotic, and albomycin, a sideromycin composed of a thioribosyl pyrimidine antibiotic conjugated to an iron-complexing moiety as found in ferrichrome (Table 4), indicating that these compounds are also recognized by Npp.
FIG 3.
Heterologous expression of the Npp transporter in E. coli W3110. (A) McC toxicity assay with a 3-fold dilution of McC (right to left). (B) Peptide transporter competition assay with 30 μM McC and the peptides indicated at 3 mM. Rows: a, W3110(pBBR1MCS-5); b, W3110 ΔyejABEF(pBBR1MCS-5); c, W3110 ΔyejABEF(pBBR5.npp).
TABLE 4.
Antimicrobial susceptibility profiles of E. coli strains
| Drug | MIC (μg/ml)a for: |
|||
|---|---|---|---|---|
| W3110(pBBR5) | W3110(pBBR5.npp) | W3110 ΔyejABEF(pBBR5) | W3110 ΔyejABEF(pBBR5.npp) | |
| Peptidyl nucleoside antibiotics | ||||
| Pacidamycin D | >500 | 250 | >500 | 125 |
| Blasticidin S | 15.6 | 15.6 | 15.6 | 15.6 |
| Albomycin | NDb | ND | 250 | 15.6 |
| AMPs | ||||
| LL37 | >500 | >500 | >500 | >500 |
| Protamine | >500 | >500 | >500 | >500 |
| Cyclic polypeptides | ||||
| Colistin | <3.9 | <3.9 | <3.9 | <3.9 |
| Bacitracin | >2,500 | >2,500 | >2,500 | >2,500 |
| Glycopeptide: vancomycin | 312 | 312 | 312 | 312 |
| Heavy metals | ||||
| Nickel chloride | 1,250 | 625 | 1,250 | 625 |
| Copper sulfate | 1,250 | 1,250 | 1,250 | 1,250 |
| Cadmium acetate | 31.2 | 31.2 | 31.2 | 31.2 |
MICs were determined by 2-fold dilution assay in three or more independent experiments with similar results. Boldface values are more than 2-fold higher or lower MICs.
ND, not determined.
The peptide moiety of McC is responsible for facilitated transport of the toxic aspartate-adenylate inside the target cell. Vondenhoff et al. (28) previously showed that peptides ranging from 7 to 13 aa in length outcompete McC uptake by E. coli, decreasing the susceptibility to the antibiotic. In order to test whether the Npp system is also able to recognize unmodified peptides, a competition assay with McC and peptides of various lengths was performed. In the E. coli wild-type strain, the effect of McC was impaired by simultaneous incubation with peptides 7 or 10 aa in length (Fig. 3B), in agreement with earlier observations. Interestingly, when cells overexpressing the Npp system were tested, no comparable inhibition of McC uptake was observed. This finding is in line with our previous results, where we were not able to associate peptide utilization with the Npp transport machinery in P. aeruginosa.
Roles of peptide ABC transporters in P. aeruginosa PA14 cytotoxicity.
Kiely et al. (29) demonstrated that the dipeptidase MdpA contributes to P. aeruginosa PAO1 cytotoxicity. This prompted us to investigate the roles of the dipeptide ABC transporter Dpp and the nucleoside peptide transporter Npp in P. aeruginosa PA14 cytotoxicity to cells of a human bronchial epithelial line. The cytotoxicity of wild-type PA14 and the corresponding transporter mutants was measured by quantifying the cytosolic enzyme LDH release by damaged cells in culture supernatants. The data showed that none of the transporters is involved in P. aeruginosa PA14 cytotoxicity (see Fig. S3 in the supplemental material).
DISCUSSION
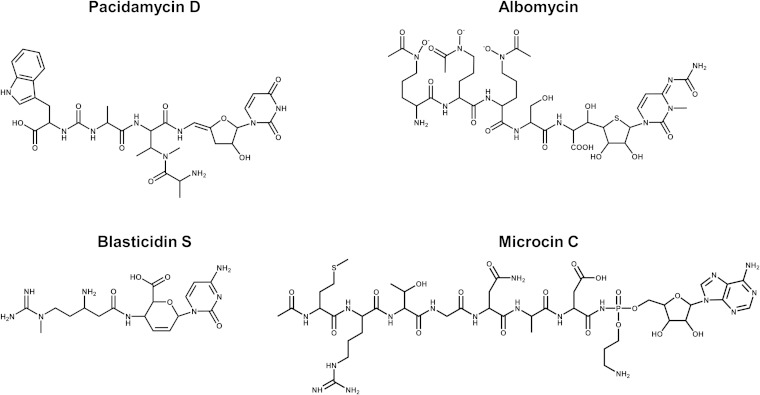
In the present study, we show that the ABC-type transporter NppA1A2BCD from P. aeruginosa PA14 is involved in the uptake of four diverse antibiotics with antifungal and antibacterial activities acting on various intracellular targets (30, 31). While these antibiotics have very different chemical structures (Fig. 4), their scaffold consists of peptides joined to nucleosides, and apparently, this common scaffold is required for Npp-mediated transport. Two of the Npp transporter substrates identified are ineffective against Pseudomonas.
FIG 4.
Chemical structures of the nucleoside peptidyl antibiotics recognized by the Npp transporter of P. aeruginosa PA14.
McC.
McC is produced by some E. coli strains carrying plasmids with the mccABCDEF gene cluster (32). McC consists of an MRTGNAD peptide with a modified AMP attached to the α-carboxyl group of aspartate through an N-acyl phosphoramidate linkage (33). The N-terminal methionine is formylated, and the phosphate group is decorated with a propylamine group (33). McC is processed within the target cell to a toxic nonhydrolyzable aspartyl-adenylate analogue that inhibits translation by preventing the synthesis of aminoacylated tRNAAsp (27, 34). In E. coli, McC uptake through the outer membrane occurs via the porin OmpF and other unknown transport mechanisms (28). Once in the periplasmic space, McC is recognized via its peptide moiety by the inner membrane ABC-type transporter YejABEF (27, 28). The Npp system of P. aeruginosa PA14 is homologous to the Yej transporter of E. coli (Fig. 2). Heterologous overexpression of Npp in an E. coli strain lacking the yej operon showed that the Npp transporter is able to replace the Yej system and translocate McC through the inner membrane. Curiously, McC showed no antibiotic activity against P. aeruginosa. Since proteolytic processing is rather nonspecific, while the target of processed McC is highly conserved, we surmise that the natural resistance of P. aeruginosa to McC is probably due to the lack of a transport system moving the nucleoside heptapeptide across the outer membrane or is caused by the presence of highly active enzymatic systems that detoxify processed McC, such as Rim family acetyltransferases (35) or efflux systems. Similarly, while the Yej transporter of Salmonella confers resistance to AMPs (25), mutational inactivation of nppBCD had no effect on the susceptibility of P. aeruginosa PA14 to these peptides. The reason for this difference remains to be established.
Previous studies have shown that peptides with a minimal chain length of 6 aa can compete with McC for uptake by the Yej transporter (28). Interestingly, the Pseudomonas Npp transporter seems to work in a different way since the peptides tested could not compete with the uptake of McC into E. coli cells by the heterologously expressed Npp transporter. This finding seems to indicate that the Npp transporter specifically “senses” the presence of nucleotide modification on transported peptides. The substrate specificity of ABC importers from Gram-negative bacteria is defined by periplasmic SBPs that capture substrates with high affinity and specificity and deliver them to the transporter (36). Whether structural variations of the SBPs YejA, NppA1, and NppA2 are responsible for the substrate specificity differences observed in the respective transporters will be the subject of future studies.
Albomycin.
Albomycin, a naturally occurring peptidyl nucleoside sideromycin structurally related to the hydroxamate siderophore ferrichrome, consists of an aminoacyl-nucleotide antibiotic moiety linked to a ferrichrome siderophore (37, 38). The high specific activity of albomycin against E. coli comes from its ability to use the ferric hydroxamate import system for its uptake across both the outer membrane and the cytoplasmic membrane. Albomycin is actively transported across the outer membrane of E. coli by the TonB-dependent transporter FhuA (39). Transport across the cytoplasmic membrane is mediated by the ABC transporter FhuBCD (37). Upon release of the active antibiotic moiety (aminoacyl-thioribosyl pyrimidine) from the iron-chelating group (40), it interferes with the loading of serine to the seryl-tRNA by seryl-tRNA synthetases (41). Interestingly, we found that the Npp permease of P. aeruginosa is also able to translocate albomycin when heterologously expressed in E. coli. Again, as is the case with Npp-mediated McC transport discussed above, it is tempting to speculate that the binding proteins of the Npp transporter recognizes the peptidyl nucleoside moiety of albomycin, whereas the binding protein of the Fhu transporter recognizes the siderophore moiety. P. aeruginosa possesses two TonB-dependent transporters, FiuA and FoxA, that are responsible for the uptake of hydroxamate siderophores, including ferrichrome, across the outer membrane (42, 43). Albomycin displays only weak bactericidal activity against P. aeruginosa (44, 45). This allows speculation that cleavage of albomycin inside the cell and/or detoxification of its toxic payload is the limiting factor that modulates its bactericidal activity in P. aeruginosa.
Pacidamycin D.
Pacidamycin D belongs to the large group of uridyl peptide antibiotics. These antibiotics have a structural feature in common, namely, a 3′-deoxyuridine with an enamide linkage at the 5′ position that is attached to a tetrapeptide moiety via a central diaminobutyric acid that connects the N-terminal amino acid, the ureadipeptide, and the 3′-deoxyuridine moiety (46). Pacidamycins exhibit antimicrobial activity inhibiting translocase I (MraY), an enzyme located on the cytoplasmic face of the inner membrane (47). MraY catalyzes the formation of lipid I from UDP-N-acetylmuramoyl-pentapeptide and undecaprenyl phosphate during bacterial cell wall biosynthesis in most Gram-positive and Gram-negative bacteria (31). However, treatment of most bacteria with pacidamycins fails because of their intrinsic resistance, caused by a lack of antibiotic uptake and extrusion via efflux pumps. Intriguingly, pacidamycin is very effective against P. aeruginosa, although its therapeutic use is limited by the fact that resistant mutants emerge with high frequency (9). Recently, the uptake of this peptide antibiotic through the inner membrane has been associated with the putative peptide ABC transporter Opp of P. aeruginosa PAO1 (9). This transporter is identical to the Npp transporter of P. aeruginosa PA14. In the present study, we showed that heterologous expression of the Npp system renders E. coli sensitive to pacidamycin. This result suggests that E. coli is resistant to pacidamycin because of a lack of import through the inner membrane. Furthermore, our data show that the Yej transporter of E. coli, which is a close homolog of the Npp transporter that used the same McC substrate, is not able to recognize uridyl peptide antibiotics.
Blasticidin S.
Blasticidin S is a peptidyl nucleoside antibiotic produced by Streptomyces griseochromogenes (48). Unlike other peptidyl nucleoside antibiotics that typically contain nucleosides with a five-carbon sugar, the structure of blasticidin S features a glucuronic acid-derived hexose that is coupled to cytosine (Fig. 4) (49). The cytidine of blasticidin S can bind tightly to the peptidyl transferase center of the large ribosomal subunit of the ribosome, which makes it a potent inhibitor of protein synthesis in both prokaryotic and eukaryotic cells (50). Interestingly, it appears that several transporters are involved in the uptake of blasticidin S through the inner membrane of P. aeruginosa PA14. Deletion of the NppBCD transporter leads to a 2-fold increase in PA14 resistance to blasticidin S, whereas the double mutant defective in the Npp permease and the dipeptide ABC transporter Dpp showed a 4-fold increase in resistance.
In summary, we have demonstrated that the NppA1A2BCD transporter of P. aeruginosa PA14 is involved in the uptake of the peptidyl nucleoside antibiotics McC, albomycin, pacidamycin, and blasticidin S. Two of these antibiotics are ineffective against P. aeruginosa, suggesting that they do not pass through the outer membrane of the bacterial cell. Furthermore, our results show that the Npp transporter recognizes peptidyl nucleosides with widely different structures. This observation might help to design new AMP-drug conjugates that hijack this specific entry route, thereby increasing our arsenal of pathogen-specific antibiotics.
A question that remains unresolved is the natural substrate of the Npp transporter system. Since we found that the Npp permease is involved in the translocation of nucleoside peptidyl antibiotics, we tested whether the system is involved in the uptake of nucleosides and nucleotide sugars (UPD-glucose and UDP-GlcNAc). However, under our experimental conditions, PA14 was not able to utilize the tested compounds as sole carbon sources (data not shown).
Furthermore, we found that the Npp transporter of PA14 is not involved in the translocation of peptides. Our data suggest that the previously described di- or tripeptide permease Dpp of PA14 (7) is also responsible for the uptake of several short-chain peptides composed of 5 aa. We identified another as-yet-uncharacterized putative peptide transporter (PA14_12980) in the genome of PA14. This inner membrane transporter belongs to the oligopeptide transporter family (Transporter Classification Database TC number 2.A.67). However, under our experimental conditions, this transporter was not involved in oligopeptide uptake in PA14 (data not shown).
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Volkmar Braun for sending us albomycin, and we appreciate Robert E. W. Hancock for providing AMPs. We thank Dieter C. Gruenert for the 16HBE14o− cell line. We also thank Sebastian Springer for providing several peptides. Furthermore, we acknowledge Mathias Winterhalter and Roland Benz for generous laboratory support and useful discussions.
The research leading to these results was conducted as part of the Translocation consortium (www.imi.europa.eu/content/translocation) and received support from the Innovative Medicines Joint Undertaking under grant agreement no. 115525, resources which are composed of a financial contribution from the European Union seventh framework program (FP7/2007-2013) and an EFPIA companies in-kind contribution. Research in the lab of K.S. is supported by grants from the Russian Academy of Sciences Molecular and Cell Biology and Nanotechnology program.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00234-15.
REFERENCES
- 1.Mislin GL, Schalk IJ. 2014. Siderophore-dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa. Metallomics 6:408–420. doi: 10.1039/c3mt00359k. [DOI] [PubMed] [Google Scholar]
- 2.Higgins CF. 2001. ABC transporters: physiology, structure and mechanism—an overview. Res Microbiol 152:205–210. doi: 10.1016/S0923-2508(01)01193-7. [DOI] [PubMed] [Google Scholar]
- 3.Garmory HS, Titball RW. 2004. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun 72:6757–6763. doi: 10.1128/IAI.72.12.6757-6763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coulter SN, Schwan WR, Ng EY, Langhorne MH, Ritchie HD, Westbrock-Wadman S, Hufnagle WO, Folger KR, Bayer AS, Stover CK. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol 30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 5.Podbielski A, Pohl B, Woischnik M, Körner C, Schmidt KH, Rozdzinski E, Leonard BA. 1996. Molecular characterization of group A streptococcal (GAS) oligopeptide permease (Opp) and its effect on cysteine protease production. Mol Microbiol 21:1087–1099. doi: 10.1046/j.1365-2958.1996.661421.x. [DOI] [PubMed] [Google Scholar]
- 6.LeDeaux JR, Solomon JM, Grossman AD. 1997. Analysis of non-polar deletion mutations in the genes of the spo0K (opp) operon of Bacillus subtilis. FEMS Microbiol Lett 153:63–69. doi: 10.1111/j.1574-6968.1997.tb10464.x. [DOI] [PubMed] [Google Scholar]
- 7.Pletzer D, Lafon C, Braun Y, Köhler T, Page MG, Mourez M, Weingart H. 2014. High-throughput screening of dipeptide utilization mediated by the ABC transporter DppBCDF and its substrate-binding proteins DppA1-A5 in Pseudomonas aeruginosa. PLoS One 9:e111311. doi: 10.1371/journal.pone.0111311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, Greenberg EP, Sorek R, Lory S. 2012. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog 8:e1002945. doi: 10.1371/journal.ppat.1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mistry A, Warren MS, Cusick JK, Karkhoff-Schweizer RR, Lomovskaya O, Schweizer HP. 2013. High-level pacidamycin resistance in Pseudomonas aeruginosa is mediated by an opp oligopeptide permease encoded by the opp-fabI operon. Antimicrob Agents Chemother 57:5565–5571. doi: 10.1128/AAC.01198-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luckett JC, Darch O, Watters C, Abuoun M, Wright V, Paredes-Osses E, Ward J, Goto H, Heeb S, Pommier S, Rumbaugh KP, Camara M, Hardie KR. 2012. A novel virulence strategy for Pseudomonas aeruginosa mediated by an autotransporter with arginine-specific aminopeptidase activity. PLoS Pathog 8:e1002854. doi: 10.1371/journal.ppat.1002854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 12.Zumaquero A, Macho AP, Rufian JS, Beuzon CR. 2010. Analysis of the role of the type III effector inventory of Pseudomonas syringae pv. phaseolicola 1448a in interaction with the plant. J Bacteriol 192:4474–4488. doi: 10.1128/JB.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schweizer HP, Hoang TT. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15–22. doi: 10.1016/0378-1119(95)00055-B. [DOI] [PubMed] [Google Scholar]
- 14.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. CLSI document M7-A7 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Metlitskaya A, Kazakov T, Vondenhoff GH, Novikova M, Shashkov A, Zatsepin T, Semenova E, Zaitseva N, Ramensky V, Van Aerschot A, Severinov K. 2009. Maturation of the translation inhibitor microcin C. J Bacteriol 191:2380–2387. doi: 10.1128/JB.00999-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39:D596–600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saier MH Jr, Reddy VS, Tamang DG, Vastermark A. 2014. The transporter classification database. Nucleic Acids Res 42:D251–258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonnet GH, Hallett MT, Korostensky C, Bernardin L. 2000. Darwin v. 2.0: an interpreted computer language for the biosciences. Bioinformatics 16:101–103. doi: 10.1093/bioinformatics/16.2.101. [DOI] [PubMed] [Google Scholar]
- 21.Bateman A, Bycroft M. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J Mol Biol 299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- 22.de Jong A, Pietersma H, Cordes M, Kuipers OP, Kok J. 2012. PePPER: a webserver for prediction of prokaryote promoter elements and regulons. BMC Genomics 13:299. doi: 10.1186/1471-2164-13-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernsel A, Viklund H, Hennerdal A, Elofsson A. 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res 37:W465–468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodell EW, Higgins CF. 1987. Uptake of cell wall peptides by Salmonella typhimurium and Escherichia coli. J Bacteriol 169:3861–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eswarappa SM, Panguluri KK, Hensel M, Chakravortty D. 2008. The yejABEF operon of Salmonella confers resistance to antimicrobial peptides and contributes to its virulence. Microbiology 154:666–678. doi: 10.1099/mic.0.2007/011114-0. [DOI] [PubMed] [Google Scholar]
- 26.Chivers PT, Benanti EL, Heil-Chapdelaine V, Iwig JS, Rowe JL. 2012. Identification of Ni-(L-His)2 as a substrate for NikABCDE-dependent nickel uptake in Escherichia coli. Metallomics 4:1043–1050. doi: 10.1039/c2mt20139a. [DOI] [PubMed] [Google Scholar]
- 27.Novikova M, Metlitskaya A, Datsenko K, Kazakov T, Kazakov A, Wanner B, Severinov K. 2007. The Escherichia coli Yej transporter is required for the uptake of translation inhibitor microcin C. J Bacteriol 189:8361–8365. doi: 10.1128/JB.01028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vondenhoff GH, Blanchaert B, Geboers S, Kazakov T, Datsenko KA, Wanner BL, Rozenski J, Severinov K, Van Aerschot A. 2011. Characterization of peptide chain length and constituency requirements for YejABEF-mediated uptake of microcin C analogues. J Bacteriol 193:3618–3623. doi: 10.1128/JB.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiely PD, O'Callaghan J, Abbas A, O'Gara F. 2008. Genetic analysis of genes involved in dipeptide metabolism and cytotoxicity in Pseudomonas aeruginosa PAO1. Microbiology 154:2209–2218. doi: 10.1099/mic.0.2007/015032-0. [DOI] [PubMed] [Google Scholar]
- 30.Merino P. 2013. Chemical synthesis of nucleoside analogues. John Wiley & Sons, Inc., Hoboken, NJ. [Google Scholar]
- 31.Walsh CT, Zhang W. 2011. Chemical logic and enzymatic machinery for biological assembly of peptidyl nucleoside antibiotics. ACS Chem Biol 6:1000–1007. doi: 10.1021/cb200284p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Severinov K, Nair SK. 2012. Microcin C: biosynthesis and mechanisms of bacterial resistance. Future Microbiol 7:281–289. doi: 10.2217/fmb.11.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guijarro JI, González-Pastor JE, Baleux F, San Millán JL, Castilla MA, Rico M, Moreno F, Delepierre M. 1995. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J Biol Chem 270:23520–23532. doi: 10.1074/jbc.270.40.23520. [DOI] [PubMed] [Google Scholar]
- 34.Metlitskaya A, Kazakov T, Kommer A, Pavlova O, Praetorius-Ibba M, Ibba M, Krasheninnikov I, Kolb V, Khmel I, Severinov K. 2006. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic microcin C. J Biol Chem 281:18033–18042. doi: 10.1074/jbc.M513174200. [DOI] [PubMed] [Google Scholar]
- 35.Novikova M, Kazakov T, Vondenhoff GH, Semenova E, Rozenski J, Metlytskaya A, Zukher I, Tikhonov A, Van Aerschot A, Severinov K. 2010. MccE provides resistance to protein synthesis inhibitor microcin C by acetylating the processed form of the antibiotic. J Biol Chem 285:12662–12669. doi: 10.1074/jbc.M109.080192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berntsson RP, Smits SH, Schmitt L, Slotboom DJ, Poolman B. 2010. A structural classification of substrate-binding proteins. FEBS Lett 584:2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 37.Braun V. 1999. Active transport of siderophore-mimicking antibacterials across the outer membrane. Drug Resist Updat 2:363–369. doi: 10.1054/drup.1999.0107. [DOI] [PubMed] [Google Scholar]
- 38.Zeng Y, Kulkarni A, Yang Z, Patil PB, Zhou W, Chi X, Van Lanen S, Chen S. 2012. Biosynthesis of albomycin delta(2) provides a template for assembling siderophore and aminoacyl-tRNA synthetase inhibitor conjugates. ACS Chem Biol 7:1565–1575. doi: 10.1021/cb300173x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferguson AD, Braun V, Fiedler HP, Coulton JW, Diederichs K, Welte W. 2000. Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA. Protein Sci 9:956–963. doi: 10.1110/ps.9.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braun V, Günthner K, Hantke K, Zimmermann L. 1983. Intracellular activation of albomycin in Escherichia coli and Salmonella typhimurium. J Bacteriol 156:308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pramanik A, Braun V. 2006. Albomycin uptake via a ferric hydroxamate transport system of Streptococcus pneumoniae R6. J Bacteriol 188:3878–3886. doi: 10.1128/JB.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Llamas MA, Sparrius M, Kloet R, Jiménez CR, Vandenbroucke-Grauls C, Bitter W. 2006. The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J Bacteriol 188:1882–1891. doi: 10.1128/JB.188.5.1882-1891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hannauer M, Barda Y, Mislin GL, Shanzer A, Schalk IJ. 2010. The ferrichrome uptake pathway in Pseudomonas aeruginosa involves an iron release mechanism with acylation of the siderophore and recycling of the modified desferrichrome. J Bacteriol 192:1212–1220. doi: 10.1128/JB.01539-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wencewicz TA, Mollmann U, Long TE, Miller MJ. 2009. Is drug release necessary for antimicrobial activity of siderophore-drug conjugates? Syntheses and biological studies of the naturally occurring salmycin “Trojan horse” antibiotics and synthetic desferridanoxamine-antibiotic conjugates. Biometals 22:633–648. doi: 10.1007/s10534-009-9218-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braun V, Pramanik A, Gwinner T, Köberle M, Bohn E. 2009. Sideromycins: tools and antibiotics. Biometals 22:3–13. doi: 10.1007/s10534-008-9199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamoto K, Sakagami M, Feng F, Togame H, Takemoto H, Ichikawa S, Matsuda A. 2011. Total synthesis of pacidamycin D by Cu(I)-catalyzed oxy enamide formation. Org Lett 13:5240–5243. doi: 10.1021/ol202124b. [DOI] [PubMed] [Google Scholar]
- 47.Brandish PE, Kimura KI, Inukai M, Southgate R, Lonsdale JT, Bugg TD. 1996. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: inhibition of phospho-N-acetylmuramyl-pentapeptide translocase from Escherichia coli. Antimicrob Agents Chemother 40:1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeuchi S, Hirayama K, Ueda K, Sakai H, Yonehara H. 1958. Blasticidin S, a new antibiotic. J Antibiot (Tokyo) 11:1–5. [PubMed] [Google Scholar]
- 49.Isono K. 1988. Nucleoside antibiotics: structure, biological activity, and biosynthesis. J Antibiot (Tokyo) 41:1711–1739. [DOI] [PubMed] [Google Scholar]
- 50.Hansen JL, Moore PB, Steitz TA. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol 330:1061–1075. doi: 10.1016/S0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- 51.He J, Baldini RL, Deziel E, Saucier M, Zhang Q, Liberati NT, Lee D, Urbach J, Goodman HM, Rahme LG. 2004. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc Natl Acad Sci U S A 101:2530–2535. doi: 10.1073/pnas.0304622101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thoma S, Schobert M. 2009. An improved Escherichia coli donor strain for diparental mating. FEMS Microbiol Lett 294:127–132. doi: 10.1111/j.1574-6968.2009.01556.x. [DOI] [PubMed] [Google Scholar]
- 54.Bachmann BJ. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev 36:525–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou L, Lei XH, Bochner BR, Wanner BL. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J Bacteriol 185:4956–4972. doi: 10.1128/JB.185.16.4956-4972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arndt H, Henning C, Völksch B, Fritsche W. 1989. Beziehung zwischen Virulenz und Phaseolotoxinbildungsvermögen bei verschiedenen Pseudomonas syringae pv. phaseolicola-Stämmen. Arch Phytopathol Plant Protect 25:347–357. [Google Scholar]
- 57.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. [DOI] [PubMed] [Google Scholar]
- 58.Kovach ME, Phillips RW, Elzer PH, Roop RM II, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802. [PubMed] [Google Scholar]
- 59.Kurepina NE, Basyuk EI, Metlitskaya AZ, Zaitsev DA, Khmel IA. 1993. Cloning and mapping of the genetic determinants for microcin C51 production and immunity. Mol Gen Genet 241:700–706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.