ABSTRACT
The entomopathogenic nematode Heterorhabditis bacteriophora forms a specific mutualistic association with its bacterial partner Photorhabdus temperata. The microbial symbiont is required for nematode growth and development, and symbiont recognition is strain specific. The aim of this study was to sequence the genome of P. temperata and identify genes that plays a role in the pathogenesis of the Photorhabdus-Heterorhabditis symbiosis. A draft genome sequence of P. temperata strain NC19 was generated. The 5.2-Mb genome was organized into 17 scaffolds and contained 4,808 coding sequences (CDS). A genetic approach was also pursued to identify mutants with altered motility. A bank of 10,000 P. temperata transposon mutants was generated and screened for altered motility patterns. Five classes of motility mutants were identified: (i) nonmotile mutants, (ii) mutants with defective or aberrant swimming motility, (iii) mutant swimmers that do not require NaCl or KCl, (iv) hyperswimmer mutants that swim at an accelerated rate, and (v) hyperswarmer mutants that are able to swarm on the surface of 1.25% agar. The transposon insertion sites for these mutants were identified and used to investigate other physiological properties, including insect pathogenesis. The motility-defective mutant P13-7 had an insertion in the RNase II gene and showed reduced virulence and production of extracellular factors. Genetic complementation of this mutant restored wild-type activity. These results demonstrate a role for RNA turnover in insect pathogenesis and other physiological functions.
IMPORTANCE The relationship between Photorhabdus and entomopathogenic nematode Heterorhabditis represents a well-known mutualistic system that has potential as a biological control agent. The elucidation of the genome of the bacterial partner and role that RNase II plays in its life cycle has provided a greater understanding of Photorhabdus as both an insect pathogen and a nematode symbiont.
INTRODUCTION
Members of the genus Photorhabdus are Gram-negative, motile bioluminescent bacteria that maintain two distinct life styles as insect pathogens and as symbionts of entomopathogenic heterorhabditid nematodes (for reviews, see references 1, 2, 3, 4, 5, 6, and 7). Based on molecular analysis, the genus is divided into three bacterial species: Photorhabdus luminescens, Photorhabdus temperata, and Photorhabdus asymbiotica (8, 9). Several subspecies are recognized.
The life cycle of Photorhabdus and its nematode host Heterorhabditis is best described as a cyclic association that begins and ends with infective juvenile nematodes (IJs). The bacteria are carried inside the gut of the nonfeeding third-instar infective-stage nematode. These nonfeeding IJs retain a monoculture of Photorhabdus within the anterior region of the intestine (10–12). The specific pair interaction between Photorhabdus and Heterorhabditis is highly restrictive, where one species of nematode retains only one species of bacteria.
The IJ nematodes are relatively resistant to environmental stress and provide protection to the bacteria. Since the bacteria alone are unable to penetrate the insect, the IJ serves as a vector transporting the bacteria to an insect host (12). The nematodes actively seek out and infect insect hosts by entering through natural openings or burrowing directly through the insect cuticle (13). The host range of these nematodes includes grubs, weevils, ticks, cockroaches, crickets, and the larval stage of many insects (14). Once inside the insect, the nematodes regurgitate the bacteria into the hemolymph (10). The bacteria release highly virulent toxins (15–18), which results in insect death in less than 48 h. As the bacteria enter the stationary phase of their growth cycle, they secrete extracellular enzymes such as proteases (19), lipases (20), and hemolysins (21). These enzymes aid in breaking down insect tissue, thereby providing nutrients for both the bacteria and nematodes. The bacteria also generate essential growth factors for nematode growth and development. The growth and development of Heterorhabditis nematodes have an obligate requirement for their specific bacterial symbiont (22). In addition, the bacteria release antibiotics to prevent secondary invaders and putrefaction of the insect carcass (23, 24). After several days of feeding, the nematodes and bacteria reassociate and leave in search of a new insect host.
Photorhabdus exists in two phenotypic phase-variant stages known as the primary and secondary phases (25–27). The two phase variants can be differentiated by several phenotypic traits, such as dye absorption, production of extracellular enzymes, bioluminescence, and cellular morphology (28, 29). Infective-stage nematodes harbor the primary-phase variant, which exhibits larger amounts of antibiotics, lipases, phospholipases, proteases, pigmentation, and bioluminescence than the secondary-phase variant (3, 25-27, 30). The primary-phase variant is able to swim and swarm under oxic and anoxic conditions, but the secondary-phase variant is motile only under anoxic conditions (31, 32). Although both variants are as virulent as their insect hosts, the secondary-phase variant is far less effective at providing conditions to support nematode growth, development, and reproduction (6, 9). Within the nematode, the bacteria are predominately maintained as primary-phase cells, and the nematodes grow preferentially in association with primary-phase cells. Although both phase variants have been isolated from IJs of their nematode partners (33), the numbers of IJs released from their insect host are increased with the primary-phase form. The exact molecular mechanism and biological significance of phase variation in these species remain unknown. The secondary-phase variants are defective in supporting nematode reproduction, suggesting an alternative ecological role such as survival outside the nematode and insect hosts (33, 34).
Our understanding of these bacteria has been greatly enhanced by the genome sequencing of two of the three established species: P. luminescens TT01 (35) and P. asymbiotica ATCC 43949 (36). The P. luminescens genome contains the largest number of putative insect toxin genes for any known bacterial genomes sequenced (35). This gene redundancy is thought to ensure the death of a wide range of insect hosts (37). The P. asymbiotica genome contains many genes that are responsible for virulence against human hosts. Among these genes are unique genes that may be responsible for the transition from insect pathogen to human pathogen.
Our interest in Photorhabdus is an extension of our studies on signal transduction and symbiosis. P. temperata is capable of swimming (movement in liquids) and swarming (movement on solid surfaces) motility (31, 32). Both swimming and swarming motility require the presence of additional salt (NaCl or KCl) in the medium. Motility increases the overall competitive fitness of P. luminescens during the insect infection process (38). In this study, we sequenced the genome from a third Photorhabdus species (P. temperata) and performed comparative analysis of the three Photorhabdus genomes. A genetic approach was initiated to identify mutants with altered swimming and swarming motility that were also modified in pathogenesis and/or symbiosis. We present the results of that mutant search and genome analysis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
For this study, both primary- and secondary-phase variants of Photorhabdus temperata (formerly Xenorhabdus luminescens [8, 39]) NC19 (ATCC 29304) and P. temperata Meg1 were used (40). Primary-phase cells were used to generate the mutant library that was also used in these experiments. For each subculture, phase status was identified by pigmentation and by differential dye absorption (27). Differential dye absorption was determined when the strains were grown on NBTA (nutrient agar supplemented with 25 mg of bromothymol blue and 40 μg of triphenyl 2,3,5-tetrazolium chloride per liter) or when they were grown on MacConkey agar (bromothymol blue and neutral red absorption, respectively). Primary-phase-variant colonies are blue on NBTA and red on MacConkey agar, while secondary-phase-variant colonies are red on NBTA and off-white on MacConkey agar. On LB medium, primary-phase-variant colonies of strain NC19 were pigmented (yellowish orange), while secondary-phase-variant colonies were off-white.
Cells were grown and maintained at 28°C in LB medium as described previously (31), unless otherwise noted. When appropriate, kanamycin was added to a final concentration of 25 μg/ml.
Genome sequencing.
Genomic DNA (gDNA) was isolated from overnight cultures grown in LB medium using the Qiagen genomic DNA isolation kit and following the procedure outlined for bacterial gDNA isolation. The gDNA eluted from the column was precipitated by the addition of 0.7 volume of room temperature isopropanol and recovered by spooling the precipitated DNA. The spooled gDNA was immediately transferred to a microcentrifuge tube containing 1.5 ml nuclease-free water. After the gDNA was dissolved, the quality and quantity of the gDNA were determined.
The purified gDNA was sent to the Genome Sequencing Center at the University of Indiana for genome sequencing by 454 pyrosequencing technology. One plate of genomic sequencing and one plate of 2-kb paired-end sequencing were performed to generate the 17 scaffolds for the genome. The number of genome sequencing gaps was brought down from 130 to 81 using PCR-based amplicon sequencing.
Sequence assembly and bioinformatics analysis.
The genomic sequence generated by 454 technology and PCR amplicon sequencing was assembled de novo by the Newbler assembler, which is part of the 454 platform. The orientation and order of scaffolds were determined by a combination of bioinformatic and laboratory-based methods. The scaffold arrangement was predicted by the use of the MAUVE program (41) and confirmed by pulsed-field gel electrophoresis (PFGE) analysis. The final draft assembly contained 17 scaffolds. The total size of the genome was 5.23 Mbp, and the final assembly is based on 218 Mbp of 454 data, which provides an average 39× coverage of the genome.
Genome annotation was performed within the Integrated Microbial Genomes (IMG) platform developed by the Joint Genome Institute, Walnut Creek, CA (42). Genes were identified using Prodigal (43). The predicted coding sequences (CDS) were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database and the UniProt, TIGRFam, Pfam, KEGG, COG, and InterPro databases. The tRNAscan-SE tool (44) was used to find tRNA genes, whereas rRNA genes were found by searches against models of the rRNA genes built from SILVA (45). The sequence alignment of the 16S rRNA was performed using ClustalW (46). Orthologs among the Photorhabdus genomes were determined by a modified Lerat method (47). A concatenated phylogenetic tree was generated from 1,203 conserved orthologs. For phylogenetic analysis, maximum-parsimony and neighbor-joining trees were constructed from 1,000 bootstrap replicates using MEGA 5.0 (48). To visualize their comparative analysis, the Photorhabdus genomes were aligned by use of the ARTEMIS Comparison Tool (ACT) (49). The metabolic potential of the P. temperata genome was analyzed by the use of the antiSMASH program (50, 51).
Transposon mutagenesis and generation of a mutant library.
Cells of P. temperata NC19 were mutagenized by mini-Tn5 according to the method of Ciche et al. (52). To help reduce sibling mutation, the mutant library was generated from seven different batches, and 500 to 2,800 mutants were isolated in each batch. A bank of about 10,000 transposon primary-phase mutants was isolated, grown overnight, and stored in microtiter plates with glycerol at −80°C. This bank of the mutant library consists of about 100 microtiter plates and was used in the experiments described below.
Motility assays.
Both swimming (movement in liquids) and swarming (movement on solid surfaces) behaviors were measured by swim and swarm plate migration assays as described previously (31, 32). For experiments assaying the salt requirement, NaCl was omitted from the swim growth medium.
Screen for motility mutants.
Individual colonies of transposon-induced mutants of P. temperata were screened for motility mutants by the use of the swim and swarm plate migration assays (31, 32). Swim and swarm media were poured into oversized petri dishes (150 by 15 mm; Fisher Scientific, Canada). Using a 96-well replicator, the transposants, which had been stored at −80°C, were transferred into new 96-well microtiter plates containing fresh LB medium and incubated overnight at 28°C. The freshly grown overnight culture was replica plated directly into swim medium or on top of the swarm medium. The motility plates were incubated at 28°C up to 72 h and monitored every 6 to 8 h for mutants with altered motility behavior compared to the parental wild type. Thus, the library was rapidly screened and putative mutants were selected. All preliminary mutants were retested on individual motility plates. Confirmed mutants were suspended in 30% glycerol and stored at −80°C for further study.
Phenotypic characterization.
Dye absorption with NBTA and MacConkey media was assayed as described above. In addition, EB agar (eosin Y and methylene blue at 400 and 65 mg/liter, respectively, in 2% PP3 agar [consisting of 2.0% proteose peptone 3 and 1.5% Bacto agar]) was used. Hemolytic activity was determined by observing a clearing surrounding the bacterial colonies cultured on a blood agar. Protease activity was determined by the gelatin assay (27). DNase activity was determined on DNase test agar containing methyl green. Antibiotic activity was evaluated by placing a 5-mm-diameter plug, taken at a point 5 mm from confluent growth of a 96-h culture of P. temperata on PP3 agar, onto a plate of LB medium that had been inoculated with Micrococcus luteus cells.
Growth assay.
Overnight cultures were diluted in fresh LB medium to a final optical density at 600 nm (OD600) of 0.01. The diluted cell cultures (150 μl/well) were added to wells of a 96-well polystyrene microtiter plate. The inoculated plate was placed in a Tecan plate reader with Magellan software (Tecan Group Ltd., Switzerland) and incubated at 28°C with shaking. The OD600 of the plate was determined each hour for a 24-h period. The average and standard deviation of each sample was determined from replicate wells (n = 8), and these values were plotted versus time. The doubling times were determined for the mutants and the parental primary-phase wild type.
Retrieval of DNA flanking the mini-Tn5 of the mutants.
Genomic DNA from the mutants was purified by the cetyltrimethylammonium bromide (CTAB) method (53). The purified DNA was digested with the restriction enzyme NsiI (the mini-Tn5 contains no NsiI sites) and was self-ligated. After isopropanol precipitation, the DNA was transformed by electroporation into Escherichia coli DH5α. Transformants containing the mini-Tn5 were selected by being resistant to kanamycin. The retrieved plasmid was purified and digested with NsiI and SfiI to confirm that the plasmid contained a single NsiI restriction fragment and the mini-Tn5 (determined by the presence of a 2.9-kb SfiI restriction fragment), respectively.
Sequence analysis.
The sequences of the DNA flanking the transposon insertions were obtained by using the M13 forward and reverse primers located 40 or 60 bp from the inverted repeat termini of the transposon. DNA sequencing was performed by the Hubbard Genome Center at the University of New Hampshire at Durham. Comparison of the DNA sequence to the P. temperata genome database was done using the BLAST program.
Southern hybridization.
Genomic DNA was prepared from mutant and wild-type P. temperata. Southern blot analysis was performed under high-stringency conditions using an AlkPhos direct labeling and detection system (GE Healthcare Bio-Science, Piscataway, NJ). The probes were prepared by PCR with the following primers: PT-KAN-F, GTA AAC TGG ATG GCT TTC TTG CCG, and PT-KAN-R, ATA TCA CGG GTA GCC AAC GCT ATG. Probe labeling, hybridization, washing, and detection were performed according to the manufacturer's protocol.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed as described previously (54). The DNA was cut with the restriction enzyme NotI, and the fragments were resolved and analyzed by PFGE under the appropriate conditions.
Axenic nematode cultures.
Heterorhabditis bacteriophora NC1 was used for studies with axenic nematode cultures. Nematode growth and propagation were performed according to modified methods (52). Growth of axenic nematodes was done on P. temperata Meg1. This bacterial strain was isolated from Heterorhabditis megidis, and H. bacteriophora IJs will grow and develop with P. temperata Meg1 cells. However, because P. temperata Meg1 is not specific to H. bacteriophora, the IJs do not retain the bacteria and are thus axenic (55).
Overnight cultures were plated onto lipid agar (1.6% nutrient broth, 1% yeast extract, 3% Bacto agar, 0.4% MgCl2 · 6H2O, 1.4% [vol/vol] Karo dark corn syrup, 0.8% pure corn oil) and incubated at 28°C for 24 to 48 h. Approximately 10 infective juveniles (IJs) were seeded onto the plate and incubated for 10 to 14 days at 28°C. IJs are young nematodes that usually carry the symbiotic bacterial strain in their gut. To separate IJs from adults and hermaphrodites, 10 ml of sterile distilled water (dH2O) was added to the lipid agar plate and allowed for sit at room temperature for 10 min. The liquid overlay containing nematodes was collected and washed several times. Nematodes were allowed to settle, and supernatant was removed. The remaining nematode solution was applied to a 40-mm cell filter that was placed in a petri dish filled with sterile dH2O and incubated at room temperature for 1 to 2 h without disruption. Standing water was collected, and nematodes were harvested by centrifugation at 7,000 × g for 3 min. Nematode pellets were resuspended in 2% bleach for surface sterilization. After a 15-min incubation at room temperature, the nematodes were harvested by centrifugation at 7,000 × g for 3 min. The resulting pellet was rinsed by resuspending in sterile dH2O and collected again. This step was repeated several times. After a final centrifugation step, the supernatant was removed to leave approximately 1 cm of sterile dH2O above the nematode pellet and stored at 4°C and were viable for up to 2 months. Surface sterilization was verified by a bioassay. Approximately 10 IJs were homogenized in 100 μl sterile dH2O with a micro-tissue grinder. The homogenate was plated onto PP3 agar and incubated for 48 h at 28°C.
Symbiosis assay.
The ability of motility mutants to support the growth and propagation of nematodes was determined according to modified methods (52). Overnight bacterial cultures were plated onto 24-well plates containing lipid agar and grown overnight at 28°C. After overnight growth, 10 to 12 axenic surface-sterilized IJs were added to each well. Plates were incubated at room temperature for 21 days. Primary-phase parental wild-type NC19 and Meg1 cells served as positive controls, while secondary-phase wild-type cells acted as a negative control. Following incubation, symbiosis-positive strains formed a white mass of IJs that was visible to the naked eye. Preliminary mutants exhibiting altered symbiosis capabilities were verified on individual lipid agar plates, as described previously.
Insect mortality assays.
In vitro, insect pathogenesis was tested in larvae of the greater wax moth, Galleria mellonella, according to a modification of the method of Clarke and Dowds (56). G. mellonella larvae were purchased from Grubco, Inc. (Hamilton, OH), and used within 3 weeks of arrival. Larvae were stored in wood shavings at room temperature (22°C) prior to use. P. temperata NC19 primary-phase wild-type and mutant cultures were grown in LB medium overnight with vigorous shaking. For each sample, the OD600 of the culture was measured by a spectrophotometer (Shimadzu Scientific Instruments, Columbia, MD). Each culture was standardized to an OD600 of 1.0 with LB medium and further diluted 105-fold. This dilution contained approximately 10 cells/μl, and 10 μl of diluted sample (about 100 cells) was directly injected into the insect hemocoel by use of a sterilized Hamilton syringe (Hamilton Company, Reno, NV). A replicate inoculum of 10 μl was spread onto LB medium containing kanamycin and 0.1% pyruvate to determine a viable cell count and also plated on MacConkey agar to determine the phase state of the cells. Two sets of controls, parental wild-type cells and sterile growth medium, were used in each experiment. All samples, including the controls, were injected into 20 larvae per sample. Larvae were incubated at room temperature, and their health was monitored every 6 to 8 h for the duration of the assay. At these time points, insect mortality was determined. Times to lethality were determined for each mutant and compared to those obtained with the parental wild type. LT50 was defined as the time required for 50% of the larvae to die, while LT100 was defined as the time required for 100% mortality. The effect of dosage, or multiplicity of infection (MOI), was determined by injecting different numbers of cells into G. mellonella larvae. These insects were injected as described above, but 10 μl of the 10−3 to 10−6 dilutions was used in these assays. Insect health was monitored every 3 h, and times to lethality (LT50 and LT100) were determined for each mutant and compared to those obtained with the parental wild type at each dosage.
In vivo insect pathogenesis assays were performed to whether the mutant bacterium-nematode complex was successful in killing insects in a modification of the protocol of Bilgrami et al. (57) using G. mellonella as the insect host. Approximately 1,000 IJs containing either the wild type, mutant P13-7, or HNR1307, the genetically complemented P13-7 mutant, were added to 50 g of sterile sand moistened with 7 ml of sterile water. Twenty insect larvae were added and incubated at room temperature. Insect mortality was monitored over time for 300 h, and LT50 and LT100 were determined for mutants and compared to the values obtained with the parental wild type at each dosage.
RNA extraction and DNase treatment.
Overnight cultures were subcultured in LB medium and grown to an OD600 of ∼0.7 before harvesting by centrifugation at 10,000 × g for 10 min. Pellets were frozen and stored at −80°C until RNA extraction. Thawed pellets were resuspended in 100 μl lysis solution (1 mg/ml lysozyme in 0.1% DEPC [diethyl pyrocarbonate]-treated TE buffer [1 mM EDTA, 10 mM Tris]) and incubated for 10 min at room temperature. The RNA was extracted by the use of the Qiagen RNeasy minikit (Qiagen Sciences, Valencia, CA), and the manufacturer's protocol was followed except that in the final elution step, 50 μl of nuclease-free water was added directly to the column membrane and incubated at room temperature for 10 min before elution. To remove residual DNA, RNA samples were treated with RNase-free DNase as follows: 50 μl RNA, 10 μl 10× DNase I buffer (New England BioLabs, Ipswich, MA), 5 U DNase I (2 U/μl; New England BioLabs, Ipswich, MA), 5 μl RNase Out recombinant RNase inhibitor (40 U/μl; Invitrogen, Carlsbad, CA) and RNase-free water to reach a total reaction volume of 100 μl. Samples were incubated at 37°C for 30 min. DNase activity was inactivated with the addition of 1 μl 0.5 M EDTA (pH 8.0) (Ambion, Inc., Austin, TX) and a 10-min incubation at 75°C. RNA yield and purity (260-nm/280-nm ratio of ∼2.0) was determined by a NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE). Following RNA quantification, the RNA samples were stored at −80°C until cDNA synthesis.
RT-PCR and cDNA synthesis.
Gene expression was determined by reverse transcription-PCR (RT-PCR). cDNA synthesis was performed with random hexamers by the use of a qScript cDNA synthesis kit (Quanta Biosciences, Gaithersburg, MD) according to the manufacturer's instructions with approximately 300 ng RNA. Thermal cycling parameters consisted of 22°C for 5 min, 42°C for 30 min, and 85°C for 5 min. cDNA yield and purity were determined by a NanoDrop 1000 spectrophotometer. Endpoint PCR amplification with specific primers for pte_02637 (forward, 5′-CAA TCC GCT GCT TGC ACA GCT AAA-3′, and reverse, 5′-ATT GTT GTT CGG TTC CTG CAT GGG-3′) and approximately 500 ng of cDNA template was used to identify mRNA expression. Thermal cycling parameters included 94°C for 5 min; 30 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 90 s; and a 7-min elongation at 72°C. PCR amplicons were visualized by agarose gel electrophoresis.
Genetic complementation of mutant P13-7.
The wild-type gene was first cloned by the use of a TA TOPO cloning kit according to the manufacturer's instructions (Invitrogen). The pte_02637 gene was PCR amplified with 150 ng parental wild-type gDNA as the template and designed primers (forward, 5′-CAG CGG TTA CGC TGT AGT TTG A-3′, and reverse, 5′-GTT GCT TGC TCG ACA CTG CTG ATT-3′). Fresh PCR product was ligated into the TOPO cloning vector and transformed into E. coli TOP10 cells. Colonies containing the cloned genes were identified on LB plates containing kanamycin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 20 mg/ml). Positive colonies were picked and inoculated into broth medium. Plasmid isolations were performed on cultures after an overnight incubation at 37°C. The plasmid was confirmed by its restriction pattern. The purified plasmid was digested with EcoRV, and the fragments were sized by agarose gel electrophoresis analysis. Insert orientation was determined with restriction digests and confirmed by sequencing with the primer M13F −20 (5′-GTA AAA CGA CGG CCA GTG-3′). The TOPO clone and the expression vector pBAD18-Cm (58) were cut with HindIII and XbaI according to the manufacturer's instructions (New England BioLabs). These specific enzymes enable the TOPO insert to be subcloned into the expression vector in the forward orientation. The expression vector was treated with calf intestinal alkaline phosphatase (New England BioLabs) to dephosphorylate the ends of the vector and prevent self-ligation. Manufacturer's instructions were followed except that 1 μl (1 U/μl) of calf intestinal alkaline phosphatase was added directly to the 20-μl restriction digest, and the reaction mixture was incubated at 37°C for 30 min. For ligation, the insert was added to the vector in a 5:1 insert-to-vector ratio using 400 U of T4 ligase (New England BioLabs). The mixture was incubated at room temperature overnight. The construct (4 μl) was added to 50 μl electrocompetent E. coli DH5α and transferred to a chilled 0.2-mm cuvette. The vector was introduced into the cells by electrotransformation by the use of an electroporator (E. coli Pulser; Bio-Rad, Hercules, CA) at 12,500 V/cm and 200 Ω. Immediately after electroporation, 500 μl SOC medium (2% Bacto tryptone, 0.5% yeast extract, 0.058% NaCl, 0.019% KCl, 0.2% MgCl2 · 6H2O, 0.49% MgSO4 · 7 H2O, 0.36% glucose) was added to the cuvette and transferred to a fresh tube. Bacterial suspensions were incubated at 37°C with gentle shaking for 1 h. Transformants were cultured on LB plates supplemented with chloramphenicol (25 μg/ml) at 37°C overnight. Potential transformants were isolated and retained for further use. To confirm the transformants, the plasmids were purified, digested with SfiI to linearize the plasmid, and electrophoresed on a 1% agarose gel in 1× TAE (40 mM Tris-acetate, 1 mM EDTA [pH 8.1 to 8.5]) to confirm identity. The confirmed plasmids were electroporated into the parental wild-type of P. temperata NC19 as described above. However, the electroporation recovery and selection incubation for P. temperata NC19 were performed at 28°C. Transformants were confirmed as described above. Due to difficulties directly transforming the mutant cells, the construct was first introduced into P. temperata wild-type cells. The purified construct from the wild-type cells was used for the electrotransformation experiments with the mutants.
Expression of complemented mutants.
To examine expression of complemented mutants, phenotypic assays were performed as described above, with the addition of 0.2% arabinose to all media.
Nucleotide sequence accession number.
This whole-genome project has been deposited at DDBJ/EMBL/GenBank under the accession number AYSJ00000000.1. The version described in this paper is version AYSJ01000000.
RESULTS AND DISCUSSION
Genome organization and general properties.
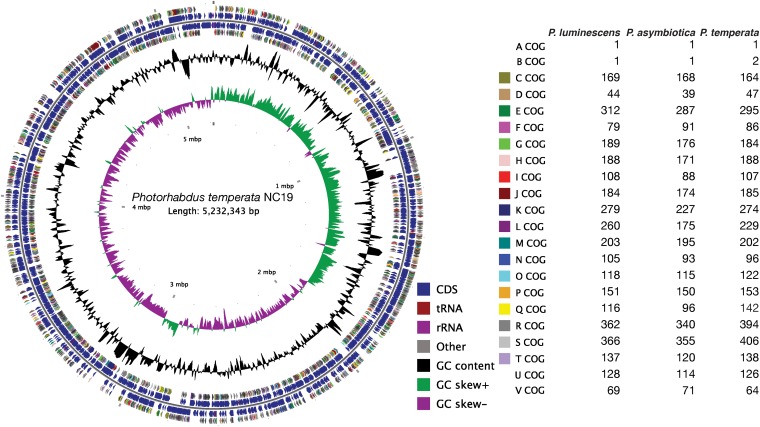
The P. temperata NC19 (ATCC 29304) genome contained a total of 5,232,343 bp and was assembled in a permanent draft sequence consisting of 17 scaffolds with an average coverage of 39×. The P. temperata genome had an average G+C content of 43.3% and contained 4,555 protein-encoding genes, 3 rRNA operons, and 69 tRNA genes. Table 1 shows the general properties of the P. temperata NC19 genome compared to the P. luminescens TT01 and P. asymbiotica genomes. Since it is a draft genome, lower numbers of rRNA operons and tRNA genes were observed in P. temperata.
TABLE 1.
Summary of genome characteristics
| Characteristic | P. temperata NC19 | P. luminescens TT01 | P. asymbiotica ATCC 43949 |
|---|---|---|---|
| Genome size (bp) | 5,232,343 | 5,688,987 | 5,064,808 |
| Scaffolds | 17 | 1 | 1 |
| GC content (%) | 43.3 | 42.8 | 42.2 |
| Total no. of protein-coding genes | 4,555 | 4,895 | 4,417 |
| No. of genes for: | |||
| rRNA | 3 | 3 | 3 |
| tRNA | 69 | 85 | 85 |
| CDS | 4,808 | 4,905 | 4,403 |
| Transposases | 129 | 111 | 61 |
| Integrases | 8 | 22 | 17 |
| Excisionase | 6 | 3 | 11 |
| Other phage related | 140 | 206 | 131 |
| Other plasmid | 0 | 0 | 1 |
| Resolvase | 6 | 3 | 2 |
| Reverse transcriptase | 5 | 1 | 0 |
| No. of CRISPRa | 4 | 5 | 6 |
| Accession no. | AYSJ00000000.1 | NC_005126.1 | NC_012962.1 |
| NC_012961.1 | |||
| Reference | 35 | 36 |
CRISPR, clustered regularly interspaced short palindromic repeats.
The arrangement of the 17 scaffolds of the P. temperata genome was predicted by MAUVE analysis (41) using the two other Photorhabdus genomes as references and confirmed by PFGE (data not shown). A singular circular map of the P. temperata genome was generated (Fig. 1). The distribution of P. temperata, P. luminescens, and P. asymbiotica genes into clusters of orthologous groups (COG) functional categories is also presented in Fig. 1. In general, these genomes showed similar profiles in their COG distribution profiles. The P. temperata and P. luminescens genomes had the most genes with COG classifications with 3,205 and 3,206, respectively, while the P. asymbiotica genome had 2,894 genes classified into COG. However, the P. temperata genome possessed an increase in the number of genes in COG Q (Secondary Metabolism Biosynthesis, Transport, and Catabolism) and the P. asymbiotica genomes had a reduction in the number of genes in COG K (Transcription), COG I (Lipid Metabolism), and COG L (Replication, Recombination, and Repair). The P. luminescens genome had the largest number of genes in COG L (Replication, Recombination, and Repair).
FIG 1.
Graphical circular map of the P. temperata NC19 genome. The circles show (from outside to inside) (i) CDS color coded with regard to COG function (plus strand), (ii) annotated ORFs (plus strand), (iii) annotated ORFs (minus strand), (iv) CDS color coded with regard to COG function (minus strand), (v) GC content, and (vi) GC skew. The distribution of CDS into COG functional groups for the three genomes is also presented.
Genomic comparison among the Photorhabdus genomes.
These three genomes represent the three identified Photorhabdus species (8, 9, 59). Based on 16S rRNA phylogeny (see Fig. S1 in the supplemental material), there was a high level of similarity between P. temperata and P. asymbiotica at 97%, and a 96% similarity between P. temperata and P. luminescens, confirming previous studies (60).
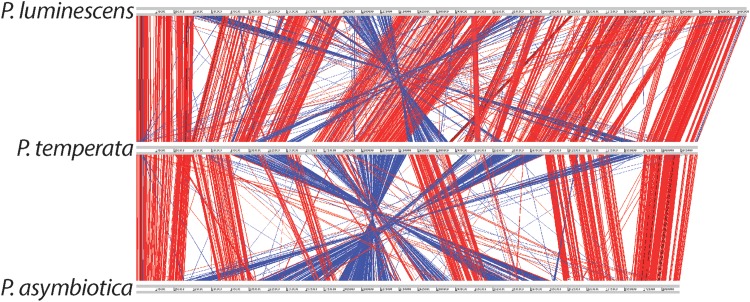
To investigate the synteny among the three genomes, they were aligned at their origins through the use of the ARTEMIS comparison tool (ACT). The P. temperata and P. luminescens genomes contained highly syntenic regions throughout their length but maintained several smaller inversions in the terminus region of the chromosome (Fig. 2). Conversely, the P. temperata and P. asymbiotica genomes had syntenic regions that were conserved near the origin region, but large-scale inversions and relocations occurred in the terminus region of the chromosome. Many of these inversions were greater than 300 kb and associated with numerous insertion or deletion events, transposases, and repeat sequences. The P. temperata chromosome also contained unique regions that were not found in the other two genomes.
FIG 2.
Alignment of the three Photorhabdus genomes by the use of the ACT. Each line represents a homologous gene. Each red line represents a gene in the same direction, while blue lines represent inverted genes.
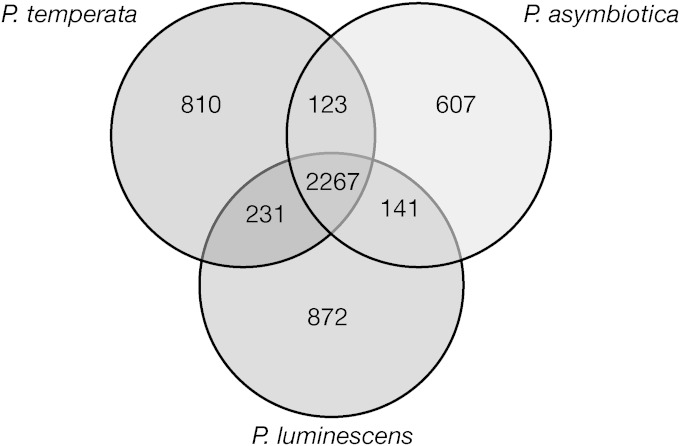
The three genomes have a conserved core of 2267 genes, which represented 50% of the P. temperata genome (Fig. 3). The number of unique genes for P. temperata, P. luminescens, and P. asymbiotica genomes were 810, 872, and 607, respectively. Among the 810 genes that were unique to P. temperata, there were 454 genes for hypothetical proteins, 44 conserved domains of unknown function, 41 phage-related genes, and 25 natural-product-related genes. Table S1 in the supplemental material presents all genes that had an assigned predicted function and were unique to P. temperata. Among some of the major classes of genes that were conserved among all three species were those related to general metabolism, cell transport and secretion systems, cell signaling, and some secondary metabolite biosynthesis. Table S2 in the supplemental material presents all genes shared among all Photorhabdus species.
FIG 3.
Pangenome among the three Photorhabdus genomes. The Venn diagram shows the numbers of orthologs among the Photorhabdus genomes.
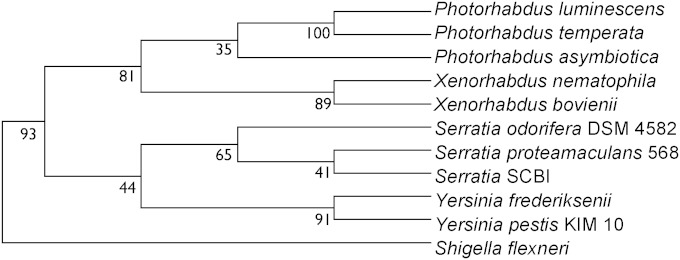
To get a clear overall picture of the phylogeny among the Photorhabdus genomes, we examined multiple genes that are common to these genomes and other related bacteria. Orthologs among Photorhabdus, Xenorhabdus, Serratia, Yersinia, and Shigella genomes were determined by a modified Lerat method (47). A concatenated maximum-likelihood phylogenetic tree was generated from 1,203 conserved orthologs (Fig. 4). Contrary to the 16S rRNA results, P. temperata and P. luminescens clustered together away from P. asymbiotica. Unlike previous studies based on a single gene, the grouping of P. temperata and P. luminescens was resolved by the use of this multigene phylogeny approach.
FIG 4.
Maximum-likelihood phylogenetic tree of the concatenated orthologs of Photorhabdus genomes and neighbors. The tree was constructed from 1,203 different maximum-likelihood phylogenetic trees for amino acid sequences of orthologs among all the genomes. The values at each branch point are the percent number of maximum-likelihood trees that displayed the same phylogeny. For each ortholog set, ClustalW (81) was used to align the sequences and a maximum-likelihood tree was created by using PHYLIP (82).
Unique group of conjugative genes.
Analysis of the P. temperata genome revealed the presence of genetic remnants for a number of genes involved in conjugative plasmid transfer (see Table S3 in the supplemental material). The proteins are responsible for transfer machinery and global regulation, which acts as an incompatibility determinant. These genes were in the P. temperata genome but not in the P. luminescens or P. asymbiotica genomes. The nearest evolutionary neighbors of these conjugative-related proteins were proteins from Xenorhabdus nematophila. In previous attempts to use conjugative genetic systems, there has been difficulty in transferring genetic material from E. coli to P. temperata (data not shown). These conjugation-related genes may account for this difficulty to move plasmids with specific origins of transfer between the two bacteria.
While examining the unique genes in P. temperata and determining the genes responsible for plasmid incompatibility, we observed a contiguous 104-kb region of the genome that consisted of 58 genes from the X. nematophila plasmid XNC1_p. This region and genes were found only in the P. temperata genome and were not present in any other Photorhabdus genomes sequenced to date (see Table S4 in the supplemental material). This region of the genome was also flanked by numerous transposases and insertion sequences. Comparative analysis of P. temperata genes to X. nematophila showed >85% identity at the protein level for the predicted amino acids and >90% identity at the nucleotide level when the whole genomic region was compared, including intergenic spaces.
Organization of the Photorhabdus secondary metabolome.
The Photorhabdus genomes encode a large number of biosynthetic gene clusters responsible for the production of natural products and secondary metabolites. Bioinformatic analysis of the P. temperata genome for biosynthetic clusters of potential secondary metabolites and other natural products was performed through the use of the antiSMASH program (50, 51). From this analysis, there were 18 potential natural product biosynthetic clusters distributed throughout the P. temperata genome (Table 2). For 10 of 18 of these clusters, the predicted chemical structure of the product was made by bioinformatic analysis alone (Table 2; also, see Fig. S2 in the supplemental material).
TABLE 2.
Distribution and predicted products of the secondary metabolite biosynthetic clusters within the P. temperata NC19 genome
| Gene cluster | Approximate location | Typea | Cluster presence in other Photorhabdus or Xenorhabdus genomesb | Predicted structurec |
|---|---|---|---|---|
| PT01 | pte_00186-pte_00208 | NRPS | plu3906-plu3935, pau_03747-pau_03784 | PT01 |
| PT02 | pte_00292-pte_00298 | Butyrolactone | pau_03747-pau_03784, pau_03338-pau_03397 | |
| PT03 | pte_00508-pte_00526 | Type I PKS | pau_01186-pau_01226, plu3516–3550 | |
| PT04 | pte_00783-pte_00787 | NRPS | plu3105-plu3144, pau_01452-pau_01511, XBJ1_1106-XBJ1_1149 | PT04 |
| PT05 | pte_00840-pte_00877 | NRPS | plu3105-plu3144, pau_01452-pau_01511 | PT05 |
| PT06 | pte_01018-pte_01031 | NRPS | Unique | PT06 |
| PT07 | pte_01583-pte_01602 | Type II PKS | plu4179–4210 | |
| PT08 | pte_02299-pte_02346 | NRPS | plu0164-plu0208 | PT08 |
| PT09 | pte_02429-pte_02439 | Bacteriocin | Unique | |
| PT10 | pte_02479-pte_02493 | Terpene | plu4322-plu4359 | |
| PT11 | pte_02783-pte_02807 | Type III PKS | pau_02342-pau_02390, plu2164-plu2205, XNC1_2270-XNC1_2323 | |
| PT12 | pte_03084-pte_03100 | NRPS-type I PKS | XBJ1_2661-XBJ1_2722 | PT12 |
| PT13 | pte_03528-pte_03541 | NRPS | XNC1_1677-XNC1_1733, XBJ1_2662-XBJ1_2722 | PT13 |
| PT14 | pte_03726-pte_03757 | NRPS | pau_01786-pau_01824, plu2716-plu2754 | |
| PT15 | pte_04021-pte_04055 | NRPS | plu0874-plu0915 | PT15 |
| PT16 | pte_04169-pte_04189 | Bacteriocin-NRPS | PT16 | |
| PT17 | pte_04198-pte_04227 | HglDE-like | pau_00901-pau_00936 | |
| PT18 | pte_04484-pte_04523 | NRPS | plu2642-plu2670 | PT18 |
NRPS, nonribosomal peptide synthetase; PKS, polyketide synthase.
Location within the other Photorhabdus and Xenorhabdus genomes based on locus tag. Plu and Pau numbers are P. luminescens TT01 and P. asymbiotica locus tags, while XBJ1 and XNC1 represent Xenorhabdus bovienii and X. nematophila.
Numbers correspond to predicted structures shown in Fig. S3 in the supplemental material.
Though Photorhabdus species are predicted to produce a wide variety of secondary metabolites, only a small number have been completely characterized. To date, the following gene clusters have been identified in Photorhabdus species: a siderophore catecholate (61), the pigment and antibiotic anthraquinone (62), the antibiotic stilbene (63, 64), GameXPeptides (65), mevalagmapeptides (65), photopyrones (66), glidobactin (67), and a carbapenem antibiotic (68). However, GameXPeptides, mevalagmapeptides, photopyrones, glidobactin, and carbapenem biosynthetic gene clusters were not present in the P. temperata genome. The 18 P. temperata biosynthetic gene clusters were compared to other Photorhabdus genomes to identify any similarities in these previously identified clusters. Clusters PT06 and PT09 identified from the antiSMASH analysis of the P. temperata genome were not found in other Photorhabdus or Xenorhabdus genomes and were completely unique to P. temperata. Clusters PT12 and PT13 shared homology with the Xenorhabdus genomes but were not found in either Photorhabdus genome. Five biosynthetic clusters (PT07, PT08, PT10, PT15, and PT18) shared homology only with the P. luminescens genome. Conversely, two biosynthetic clusters (PT02 and PT17) shared homology exclusively with the P. asymbiotica genome.
Biosynthetic cluster PT16 (pte_04184) was identified in P. temperata (previously called P. luminescens) and is responsible for the production of the catecholate siderophore photobactin (52). In our study, through BLASTP analysis, this biosynthetic cluster was also found in P. luminescens (plu3532) and P. asymbiotica (pau_00890). Photobactin has antimicrobial activity, which is activated by the phosphopantheinyl transferase NgrA (pte_04227), whose gene was present in P. temperata as the last gene of a polyketide synthase (PKS) biosynthetic (PT17) gene cluster (61). While this siderophore is important for iron sequestration, it was not involved in facilitating symbiosis (61). However, its antimicrobial activity may play a role in maintaining the monoculture environment within the insect cadaver.
Biosynthetic cluster PT07 was predicted to produce an anthraquinone, which is both a pigment and an antimicrobial. A comparative protein BLAST search within the P. temperata genome also confirmed this prediction. Furthermore, P. temperata produces a red pigment that suggests anthraquinone production. Biosynthetic cluster PT07 encodes a type II PKS (see Fig. S3 in the supplemental material), which was predicted to contain the antABCDEFGHI operon. The genes in this operon were highly homologous to the ant operon found in P. luminescens and identically organized (62). This ant operon was not found in the P. asymbiotica genome, but the remnants of 4 genes were present. At the amino acid level, the predicted products of these remnant genes had an identity of 37% or less to P. luminescens and P. temperata predicted proteins. This typical type II PKS gene-containing biosynthetic gene cluster is often found in actinomycetes and is homologous to those in Frankia sp. strain EUN1f and multiple species of Streptomyces.
Furthermore, the biosynthetic gene cluster responsible for the production of isopropylstilbenes in P. luminescens (69) was identified in the P. temperata genome. Through a comparative protein BLAST search within the P. temperata and P. asymbiotica genomes, four gene clusters responsible for stilbene biosynthesis were identified (see Fig. S4 in the supplemental material). These gene clusters were not detected through the use of antiSMASH. Many of the genes identified through the BLAST search are adjacent to predicted gene clusters from antiSMASH, but none were within these clusters. Gene cluster PT11 was flanked by two sets of genes identified from the BLAST search. In the P. temperata genome, the stlA gene (pte_02764) was located 16 genes upstream of PT11. The stlCDE (pte_02827, pte_02826, and pte_02825) genes were adjacent to gene cluster PT11, with only a single gene between PT11 and stlCDE. However, the homologous stlDE (plu2164 and plu2165) genes from P. luminescens were identified as homologs of cluster PT11 from antiSMASH. Due to this homology and organization, cluster PT11 was predicted to be involved in stilbene biosynthesis. The presence of stilbene was further confirmed with mass spectrometry (Helge Bode, personal communication).
Carbapenem biosynthesis was previously identified in P. luminescens (68). Both antiSMASH and protein searches failed to identify the presence of carbapenem biosynthetic clusters in P. temperata and P. asymbiotica. However, the remnants of a 1-carbapen-2-em-3-carboxylic acid operon were found in the P. temperata genome. Three genes were scattered within the genome.
Presence of type III and VI secretion systems.
The P. temperata genome contained two secretion systems that have been tied to pathogenesis: a type III secretion system (T3SS) and a T6SS.
All three Photorhabdus genomes contained four complete T6SS operons (see Fig. S5 to S8 in the supplemental material) and several T6SS-related genes that were not part of an operon. Previous annotations of the P. luminescens and P. asymbiotica genomes did not identify the presence of T6SS but were identified from the BLASTP comparison with the P. temperata annotation. From the BLASTP analysis, each of the four operons contained all of the genes required for a functional T6SS (70). Each of the four operons varied in structure and gene content, and they were not spatially arranged in the same way among the three species. The first T6SS operon revealed the most divergence of the four operons (see Fig. S5 in the supplemental material).
In contrast to the first T6SS operon, the three additional T6SS operons shared higher homology levels. The second T6SS operon was the most compact and gene dense of the four clusters at ∼20 kb (see Fig. S6 in the supplemental material). Nearly all the gene products in both P. luminescens and P. asymbiotica shared a sequence identity of >85% with P. temperata proteins, making it the most conserved of the four operons. T6SS operon 3 was the most expansive of the T6SS operons found in P. temperata and spanned over 30 kb (see Fig. S7 in the supplemental material). This operon contained eight central genes that were not related to any known T6SS operon and have no homology to genes in P. luminescens or P. asymbiotica. The rest of the genes that encode the T6SS are homologous to T6SS-encoding genes in P. luminescens and P. asymbiotica.
The last T6SS operon found within the P. temperata genome showed interesting properties. The gene cluster was ∼20 kb in length but contained five genes not related to known T6SS genes (see Fig. S8 in the supplemental material). Located ∼8.7 kb upstream was the TcdA/TcdB insect toxin gene (pte_02753). Upon closer examination, this TcdA/TcdB insect toxin gene was identified as mcf1 (makes caterpillars floppy) from a PSI-BLAST against the NCBI nonredundant database. This T6SS operon was not located in a similar position in the P. asymbiotica and P. luminescens genome. The fourth T6SS operon was highly conserved among the three genomes at 80 to 98% protein identity levels.
In other entomopathogenic bacteria, T6SS seem to play a role in the insect infection process (71). Furthermore, T6SS are generally known to function as a virulence factor for many pathogens that interact with eukaryotic cells, including Vibrio cholerae, Pseudomonas aeruginosa, and Burkholderia mallei (72). Besides being a virulence factor, T6SS have the potential to promote beneficial interactions with eukaryotic cells. During Helicobacter hepaticus interaction with the mouse intestinal microbiota, T6SS interacts with epithelial cells of the intestinal epithelium. This interaction limits colonization and intestinal inflammation and possibly prevents an immunological response to help promote the proper functional microbiota (73). This active suppression of the innate and adaptive immune responses may be analogous to how Photorhabdus colonizes the epithelium of its nematode partner's intestine.
At least one T3SS operon was found in all of the Photorhabdus genomes (see Table S7 in the supplemental material). The P. asymbiotica genome also contained a second novel T3SS that was not found in the other two genomes (36). The Photorhabdus T3SS contained a highly conserved type III secretion system backbone similar to those found in Yersinia pestis and Pseudomonas aeruginosa (74). Of the 30 genes found in the P. temperata T3SS operon, most gene products shared >90% identity with their P. luminescens and P. asymbiotica counterparts. Among the three genomes, these genes were spatially organized in a similar manner (see Fig. S9 in the supplemental material). Pathogens often translocate molecularly distinct effectors through the T3SS (70). The P. temperata genome contained three T3SS effectors located in the operon. The effectors were the YopT-like peptidase gene (pte_00337), YopR gene (pte_00342), and TyeA gene (pte_00360). The effector for P. asymbiotica was not located in the backbone of the T3SS. A YopT-like effector was identified by a genome-wide approach and shown to be further downstream of the backbone itself. P. temperata shared 79% and 75% identity with these same YopT-like effectors of P. luminescens and P. asymbiotica, respectively. Previous studies failed to identify the effector for P. asymbiotica in the backbone of the T3SS (75). In Photorhabdus, T3SS is necessary for insect colonization and shown to play an important role in evading the insect cellular immune reaction of nodulation (76).
Identification of potential pathogenesis genes.
Besides these secretion systems, the P. temperata genome was also analyzed for the presence of other virulence genes. Table S5 in the supplemental material lists the insect toxin complex (TC) genes identified within the P. temperata genome and their P. luminescens and P. asymbiotica homologs. There were 21 predicted TC genes present within the genome. The TC genes are essential for insect pathogenesis. Beside the TC genes, the P. temperata genome contains several other virulence factors, including three cytotoxin genes, four cytotoxin-related activating genes, and two additional homologs to RtxA-like proteins (see Table S6 in the supplemental material). Furthermore, 14 hemolysin-related genes were identified within the genome. A total of 23 protease-related genes and 16 lipase-related genes were identified. All of these gene products shared a high degree of identity with known virulence factors.
Transposon mutagenesis of P. temperata.
A genetic approach was initiated to understand the role of motility in the life cycle of Photorhabdus. The plasmid pUB394 was used to deliver a mini-Tn5 into P. temperata NC19 to construct a mutant library. To help reduce sibling mutations, the mutant bank was generated from 7 different batches of transpositions, and about 500 to 2,800 mutants were isolated in each batch. The resulting library consisted of 10,176 transposon mutants. The frequency of transposition defined as the ratio of cells resistant to kanamycin and sucrose to total cells present was 5.4 × 10−8. Southern analysis of 7 randomly chosen mutants confirmed single, random mutations of the mini-Tn5 (data not shown).
Screening and isolation of motility mutants.
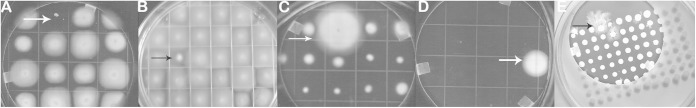
The entire transposon library was screened for mutants defective in motility by the use of the swim or swarm migration plate assays (Fig. 5). A total of 86 mutants with altered motility were identified and were classified into five different phenotypic groups. First, 23 nonmotile mutants were identified on swim medium by the absence of ring formation (Fig. 5A). These mutants formed a compact colony on this medium and did not form swim rings even after 72-h incubations. A second class of mutants was identified that exhibited aberrant motility. These mutants were motile but had defective or aberrant swim ring formation (Fig. 5B). Thus, the swim ring was smaller and/or oddly shaped. There were 12 mutants classified as this phenotype. A third class of mutants was identified as hyperswimmers (Fig. 5C). These 12 mutants formed swim rings noticeably earlier than the parental wild type.
FIG 5.
Motility mutants were identified by the use of several screens. Cells from overnight broth cultures were used to inoculate swim-migration plates as described in Materials and Methods. All motility plates were incubated at 28°C for 24 h and screened for motility mutants. (A) Nonmotile mutant; (B) reduced-swimming-motility mutant; (C) hyperswimmer mutant; (D) mutant capable of swimming in medium without NaCl; (E) hyperswarmer mutant.
P. temperata requires additional NaCl or KCl for optimal swimming motility (31). In the absence of this additional salt, the cells are nonmotile and do not produce flagella. A fourth class of mutants was isolated on swim medium lacking additional salt (Fig. 5D). Five mutants were isolated that were able to form swim rings in swim medium lacking additional salt (NaCl). The last motility mutant group consisted of 33 mutants with altered swarming behavior (movement on solid surfaces). Photorhabdus cells swarm optimally on the surface of complex media with 0.45% agar, but swarming motility is reduced at higher concentrations (above 1%) of agar (32). When the mutant library was screened for motility on swarm medium containing 1% agar, 33 mutants were identified as hyperswarmers (Fig. 5E). These mutants swarmed faster and formed larger swarm rings than the parental wild types. Many of the hyperswarmer mutants formed unique patterns on these swarm plates (see Fig. S10 in the supplemental material).
Physiological properties of the motility mutants.
All five classes of mutants were retested for motility under three different conditions (Table 3 ; also, see Table S7 in the supplemental material). As expected, all of the nonmotile mutants were nonmotile both in swim media and on swarm media. Aberrant mutants showed reduced swim ring formation with NaCl and were nonmotile without NaCl. All of these mutants were also defective in swarming motility. Hyperswimming mutants showed increased swim ring formation, and six mutants also had increased swarm ring formation (P2-H12, P10-D9, P7-E1, P62-H6, P93-A4, and P75-A4). Although these six mutants were initial classified as hyperswimmers, they can be moved into the hyperswarmer category based on the results of this secondary test (Table 3; also, see Table S7 in the supplemental material). These six mutants were probably missed in the initial screening process. However, not all of the hyperswimming mutants formed swim rings in medium without NaCl. Hyperswarmer mutants displayed increased swarm ring formation that was 4- to 10-fold greater than that of the parental wild type. These mutants also exhibited wild-type or higher levels of swim ring formation in the presence of additional NaCl. In the absence of NaCl, these mutants could be subdivided into two mutant groups: motile and nonmotile. The mutants able to swim without NaCl swam as well as the wild type with NaCl, with one mutant (P4-9) that was also a hyperswarmer.
TABLE 3.
Physiological properties of representative P. temperata NC19 mutants and their parental wild type
| Characteristic and organism | Swim ring formationa |
Swarm ring formationb | Insect mortalityc |
Symbiosisd | Dye absorption one: |
Extracellular productsf |
Pigmentation (PP3) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| +NaCl | − NaCl | LT50 | LT100 | EB | NBTA | MacConkey | Protease | DNase | Hemolytic | Antibiotic | ||||
| Wild type | ||||||||||||||
| NC19 primary (parental) | 36.9 ± 3.3 | 3.0 ± 0.0 | 10.7 ± 1.5 | 38 | 46 | + | + | + | + | ++ | ++ | + | + | Dark |
| NC19 secondary | 3.0 ± 0.0 | 3.0 ± 0.0 | ND | 35 | 43 | − | − | − | − | + | + | − | − | Light |
| Nonmotility | ||||||||||||||
| P3-C8 | 3.0 ± 0.0 | 3.0 ± 0.0 | 11.0 ± 1.1 | 38 | 46 | + | + | + | + | ++ | ++ | + | + | Dark |
| P4-27 | 3.0 ± 0.0 | 3.0 ± 0.0 | 10.0 ± 1.1 | 41 | 50 | + | + | + | + | ++ | + | + | + | Dark |
| P20 F12 | 3.0 ± 0.0 | 3.0 ± 0.0 | 9.0 ± 1.0 | 32 | 38 | + | + | + | + | ++ | ++ | − | + | Dark |
| P59-G8 | 3.0 ± 0.0 | 3.0 ± 0.0 | 8.0 ± 1.1 | 38 | 46 | + | + | + | + | ++ | + | + | + | Dark |
| P86-G6 | 3.0 ± 0.0 | 3.0 ± 0.0 | 9.0 ± 1.1 | 42 | 50 | + | + | + | + | +++ | + | + | ++ | Dark |
| Defective motility | ||||||||||||||
| P13-7 | 18.7 ± 1.5 | 1.0 ± 0.0 | 15.0 ± 1.7 | 55 | 68 | + | − | − | − | − | ++ | − | − | Light |
| P58-C8 | 5.0 ± 2.0 | 3.0 ± 0.0 | 10.0 ± 1.0 | 32 | 35 | E | + | + | + | ++ | ++ | + | + | Dark |
| P64-C3 | 18.0 ± 1.2 | 3.0 ± 0.0 | 12.0 ± 1.1 | 52 | 58 | D | − | − | − | ++ | ++ | + | + | Dark |
| P64-D5 | 10.0 ± 1.1 | 3.0 ± 0.0 | 15.0 ± 1.1 | 66 | 72 | + | − | − | − | ++ | ++ | + | + | Dark |
| Swimming motility without NaCl | ||||||||||||||
| P1-4 | 48.3 ± 6.1 | 37.7 ± 4.0 | 10.3 ± 4.0 | 38 | 46 | + | + | + | + | ++ | ++ | + | + | Dark |
| P4-9 | 42.3 ± 4.9 | 20.0 ± 3.0 | 25.3 ± 2.5 | 38 | 46 | + | + | + | + | ++ | ++ | + | + | Dark |
| P56-C3 | 42.0 ± 4.4 | 35.3 ± 4.1 | 10.8 ± 3.3 | 38 | 46 | + | + | + | + | ++ | ++ | ± | + | Dark |
| Hyperswarming | ||||||||||||||
| P2-A9 | 43.3 ± 3.5 | 9.3 ± 0.6 | 48.3 ± 5.8 | 38 | 46 | + | + | + | + | ++ | ++ | + | + | Dark |
| P5-A7 | 36.0 ± 3.6 | 6.2 ± 0.6 | 22.5 ± 3.5 | 38 | 46 | + | + | + | + | ++ | ++ | + | + | Light |
| P7-A8 | 40.3 ± 2.5 | 1.0 ± 0.0 | 20.3 ± 0.6 | 38 | 46 | + | + | + | + | ++ | ++ | + | + | Dark |
| P7-A11 | 41.3 ± 1.2 | 3.3 ± 0.6 | 74.0 ± 3.6 | 38 | 46 | + | + | + | + | ++ | ++ | + | + | Dark |
| P12-A1 | 37.3 ± 0.6 | 7.0 ± 2.6 | 79.4 ± 3.4 | 40 | 50 | E | + | + | + | ++ | + | + | ++ | Dark |
| P20-D5 | 45.0 ± 3.4 | 20.0 ± 3.4 | 27.7 ± 2.5 | 38 | 46 | + | + | + | + | ++ | ++ | ± | + | Dark |
| P38-B11 | 36.3 ± 1.5 | 2.7 ± 0.6 | 16.5 ± 4.9 | 38 | 46 | + | + | + | + | ++ | + | + | + | Dark |
| P50-A6 | 42.0 ± 1.0 | 3.0 ± 1.0 | 36.7 ± 5.8 | 45 | 55 | + | + | + | + | +++ | + | + | ++ | Dark |
| P57-G9 | 29.0 ± 6.5 | 20.7 ± 0.6 | 69.0 ± 7.9 | 38 | 46 | + | + | + | + | ++ | + | + | ++ | Dark |
| P78-C2 | 49.0 ± 0.6 | 50.0 ± 13.4 | 24.9 ± 11.8 | 38 | 46 | + | + | + | + | +++ | ++ | + | + | Dark |
| P81-G6 | 45.3 ± 2.9 | 3.7 ± 0.6 | 53.3 ± 30.5 | 38 | 46 | − | + | + | + | ++ | + | + | + | Dark |
| P81-H7 | 33.0 ± 0.6 | 8.0 ± 2.6 | 68.0 ± 10.4 | 38 | 46 | D | + | + | + | ++ | + | + | + | Dark |
| P81-H8 | 58.6 ± 3.0 | 19.0 ± 1.7 | 67.0 ± 19.0 | 38 | 46 | + | + | + | + | ++ | + | + | + | Dark |
| P82-A5 | 27.0 ± 3.4 | 3.0 ± 0.0 | 80.0 ± 0.0 | 38 | 46 | + | + | + | + | + | + | + | + | Dark |
| P83-A9 | 35.0 ± 3.0 | 10.0 ± 3.4 | 41.7 ± 5.8 | 44 | 52 | + | + | + | + | ++ | + | + | + | Dark |
| P83-B10 | 64.0 ± 2.0 | 14.7 ± 1.5 | 69.0 ± 7.9 | 32 | 40 | + | + | + | + | +++ | + | + | ++ | Dark |
| Hyperswimming | ||||||||||||||
| P5-2 | 45.0 ± 4.3 | 3.0 ± 0.0 | 12.7 ± 1.2 | 38 | 46 | + | + | + | + | ++ | ++ | + | + | Dark |
| P75-A4 | 53.3 ± 2.5 | 3.5 ± 0.7 | 33.3 ± 14.2 | 40 | 48 | + | + | + | + | ++ | + | + | + | Dark |
| P93-A4 | 47.3 ± 6.6 | 5.0 ± 1.3 | 34.0 ± 13.9 | 31 | 44 | + | + | + | + | ++ | + | + | + | Dark |
The swim migration assay was carried out after 48 h of incubation at 28°C, and swim migration medium contained 0.3% Bacto agar. Values are the average swim ring diameters, with standard deviations. Results are averages of 3 measurements per independent experiment.
Swarm migration medium contained 1.25% Bacto agar, and swarm ring diameters were measured at 48 h of incubation at 28°C. Values average swim or swarm ring diameters, with standard deviations. Results are averages of 3 measurements per independent experiment. ND, not determined.
Insect pathogenesis was determined on G. mellonella larvae as described in Materials and Methods. LT50 and LT100 are the time (in hours) required to kill 50 and 100% of larvae.
Symbiosis was determined by the nematode assay and determined by the development of mature IJs. +, capable of normal nematode development similar to the parental wild type; −, incapable of supporting nematode development; E, enhanced (nematode development occurred 2 days earlier than with the parental wild type); D, delayed (nematode development occurred 2 days later than with the parental wild type).
Dye absorption on EB and MacConkey plates was noted at 48 h. A positive result on EB plates was indicated by metallic green colonies, and a negative result was indicated by dull purple colonies. A positive result on MacConkey plates was indicated by bright red colonies and a negative result by colorless or pink colonies.
DNase and protease activities were determined by measuring the sizes (in millimeters) of the halos surrounding the bacterial colonies 24 h after inoculation. +++, hyperpositive (>4-mm halo); ++, strongly positive (2- to 4-mm halo); +, positive (1- to 2-mm halo); ±, weakly positive (<1-mm halo); −, negative (no halo). Hemolytic activity was determined by clearing zones around the bacterial colonies. +, strong annular hemolysis; ±, weakly positive hemolysis, −, no hemolysis. Antibiotic production was determined by measuring the size of the halo (in millimeters) surrounding the bacterial colonies 1 day after inoculation of the tester bacterium (Micrococcus luteus). ++, strongly positive (>5-mm halo), +, positive (2- to 5-mm halo); ±, weakly positive (<2-mm halo); −, no halo.
Several phenotypic properties of the motility mutants and their parental strains were investigated (Table 3; also, see Table S7 in the supplemental material). In general, no overall patterns were observed for each mutant classification. Approximately 25 of the 86 motility mutants had altered production of extracellular enzymes. Four mutants (P35-B12, P61-A3, P5-11, and P13-7) had profiles similar to those of wild-type secondary-phase cells showing altered pigment production and reduced levels of extracellular products. The downregulation of numerous phenotypes suggests mutations in global regulatory elements. Alternatively, these mutants could have converted to secondary-phase cells.
Antibiotic production was the most altered phenotype of the motility mutants, with 18 mutants being upregulated or downregulated. Three mutants (nonmotile mutant P72-C6 and hyperswarmers P11-A6 and P83-B10) had almost 2-fold increases in the zone of inhibition compared to the wild type. Protease activity was also upregulated or downregulated in 5 mutants, while DNase activity was altered in only one mutant (P39-E6) with a 2-fold increase in activity compared to the wild type. Several mutants had reduced hemolysin production; none had increased hemolysin activity.
Identification of pathogenesis mutants.
All 86 motility mutants were screened for altered pathogenesis properties by use of the in vivo wax moth test system. Approximately 100 cells were directly injected into the host-insect G. mellonella, and mortality over time was determined (see Fig. S11 in the supplemental material). The parental wild-type primary phase cells killed the entire larval population injected by 46 h (LT100) and required 38 h to kill 50% of the population (LT50). From the first screen with a 100-cell dose, 13 of the 86 mutants tested exhibited higher or lower LT50 and LT100 values than the parental wild-type (Table 3; also, see Table S7 in the supplemental material). These mutants included the following categories: 7 hyperswarmer mutants (P12-A1, P50A6, P83-A9, and P83-B10 plus 3 mutants originally identified as hyperswimmer mutants (P62-H6, P75-A4, and P93-A4); 3 nonmotile mutants (P4-27, P20-F12, and P86-G6); and 3 motility-defective mutants (P13-7, P58-C8, and P64-D5). Of these 13 mutants, 5 mutants (P58-C8, P20-F12, P93-A4, P83-B10, and P62-H6) had lower LT50 and LT100 values, which suggested enhanced virulence. There were 8 mutants (P13-7, P64-D5, P4-27, P83-A9, P75-A4, P12-A1, P50-A6, and P86-G6) had elevated LT50 and LT100 values, implying reduced virulence. Over time, all of the mutants killed the insects. No mutants were identified that were completely defective in pathogenesis. Since all mutants and the parental wild-type cells had the same growth rate (data not shown), these differences in pathogenesis were not correlated to growth rate. These 13 mutants were selected for further study.
Confirmation of pathogenesis mutants.
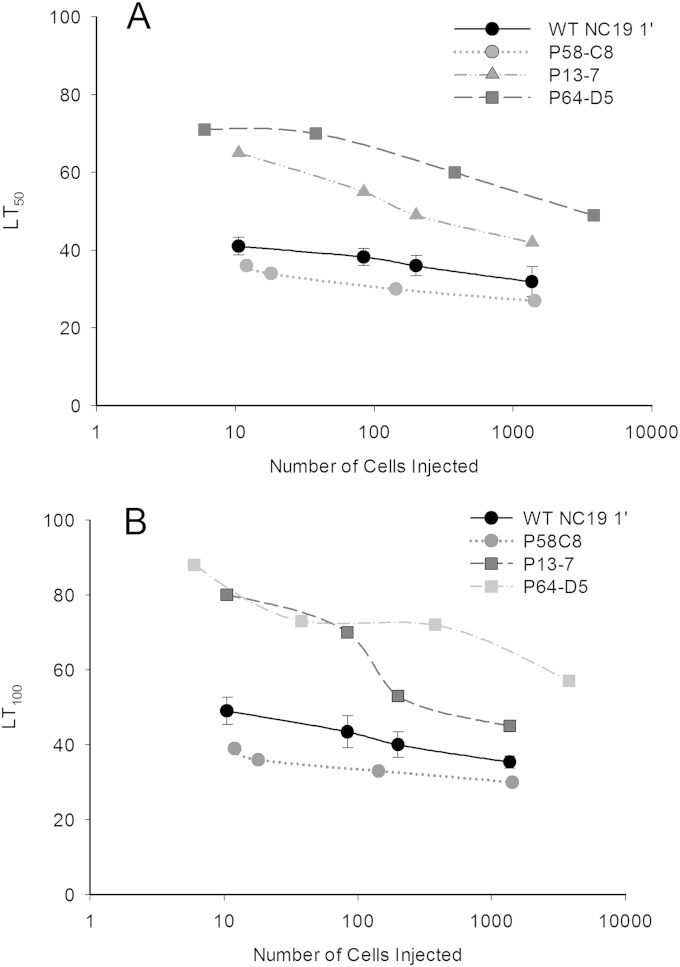
One possible explanation for the differences in virulence levels described above could be variation in viable cell numbers. To verify these changes in virulence, the effect of dosage was tested. The larvae were directly injected with dosages ranging from 10 to 10,000 cells per insect. The objective of these experiments was to identify mutants that consistently caused an altered mortality rate at each dosage. A dose-dependent response was observed for both LT50 and LT100 determinations (Fig. 6). Three motility mutants (P13-7, P58-C8, and P64-D5) maintained an altered virulence pattern over the range tested and were chosen for further study. Mutant P58-C8 exhibited enhanced virulence, causing mortality approximately 10 h sooner than the parental wild type. The other 2 mutants, P13-7 and P64-D5, exhibited a delayed pathogenesis response and killed larvae approximately 10 to 20 h later than the parental wild type. These mortality values were consistent for both LT50 (Fig. 6A) and LT100 (Fig. 6B) values.
FIG 6.
Verification of mutants exhibiting altered virulence. The effects of dosage on LT50 values (A) and LT100 values (B) are shown.
Effect on nematode symbiosis.
Besides pathogenesis, another important aspect of the Photorhabdus life cycle is the symbiosis with a nematode host. The motility mutants used as described above were also examined for their ability to support the growth and development of H. bacteriophora nematodes. The mutants were first screened for symbiosis defects or changes via a nematode plate assay described in Materials and Methods. Cultures capable of supporting nematode growth were clearly identified by a white mass of nematodes. Capabilities were compared to two positive wild-type controls, P. temperata strains NC19 primary phase and Meg1. Negative controls included P. temperata NC19 secondary phase, E. coli, and sterile lipid agar. From the initial screening (Table 3; also, see Table S7 in the supplemental material), mutant P58-C8 showed an accelerated rate of symbiosis formation that resulted in a mature mass of IJs a few days earlier than in the parental wild type. Mutant P64-D5 showed a delayed symbiosis response, forming a nematode mass a few days later than the wild type. Mutant P13-7 showed a wild-type response, developing a mature mass at the same time period. Since all mutants and the parental wild-type cells had the same growth rate (data not shown), these differences in symbiosis were not correlated to growth rate.
The symbiosis assay was repeated on individual lipid agar plates rather than a 24-well plate, and the results were confirmed. Mutant P64-D5 exhibited a delayed-symbiosis trait that resulted in nematode development occurring approximately 2 days later than in the parental wild type. Mutant P58-C8 showed an accelerated-symbiosis trait that resulted in nematode development approximately 2 days earlier than in the parental wild type. Both of these mutants also had altered insect pathogenesis responses. Mutant P58-C8 showed both accelerated nematode development and enhanced virulence. Initial pathogenesis screening indicated that P64-D5 showed both a delayed pathogenesis response and symbiosis capabilities. Mutant P13-7 showed only a delayed pathogenesis response and wild-type symbiosis. This mutant was chosen for genetic complementation.
Genetic analysis.
The mini-Tn5 insertion sites of several mutants that represented each class or had altered insect pathogenesis were identified (Table 4). Not surprisingly, two nonmotile mutants (P4-27 and P59-G8) had insertions in the structural genes flgE and flgK, whose products are part of the flagellar hook. Another nonmotile mutant P3-C8 had an insertion in flagellar biogenesis gene flhB. One of the defective motility mutants was also in flgK and was nonmotile.
TABLE 4.
Identification of transposon insertion sites
| Characteristic and mutant | Gene | Size (bp) | plu or pau homologue(s) | Site of insertion | Predicted gene function |
|---|---|---|---|---|---|
| Hyperswarming | |||||
| P2-A9 | pte_02547 | 1,026 | plu2502, pau_02024 | 1,011 | Hnr two-component system CheY-like receiver |
| P5-A7 | pte_03283 | 2,895 | plu1678, plu1719, pau_02800 | 1,308 | Hypothetical protein |
| P7-A8 | pte_03319 | 324 | None | 248 | Hypothetical protein |
| P7-A11 | pte_01279 | 972 | plu4516, pau_04019 | 626 | Multidrug resistance efflux pump |
| P12-A1 | pte_02238 | 1,077 | None | 444 | Methyltransferase domain |
| P20-D5 | pte_03319 | 324 | None | 248 | Hypothetical protein |
| P38-B11 | pte_04499 | 15,594 | plu3263, pau_03067 | 1,682 | Nonribosomal peptide synthetase modules and related proteins |
| P50-A6 | pte_03291 | 1,191 | plu1686, plu1727, plu2528, pau_02110, pau_01996 | 475 | Phage tail sheath protein FI |
| P57-G9 | pte_04499 | 15,594 | plu3263, pau_03067 | 8,343 | Nonribosomal peptide synthetase modules and related proteins |
| P78-C2 | pte_03341 | 525 | None | 376 | Transposase and inactivated derivatives |
| P81-G6 | pte_03579 | 252 | plu0884, plu0889, pau_02520, pau_00815 | 131 | Hypothetical protein |
| P81-H7 | pte_03416 | 813 | pau_01962 | 391 | Hypothetical protein (DUF1983) |
| P81-H8 | pte_03610 | 939 | plu2858, pau_01678 | 707 | Fructose-1-phosphate kinase |
| P82-A5a | |||||
| P83-A9 | pte_00950 | 1,785 | plu3457 | 794 | Integrase core domain |
| P83-B10 | pte_03283 | 2,895 | plu1678, plu1719, pau_0280, pau_02004 | 1,447 | Hypothetical protein |
| P75-A4 | pte_01774 | 900 | plu3727 | 872 | Putative transcriptional regulator AbgR |
| Swimming motility without NaCl | |||||
| P1-4 | pte_02195 | 1,419 | plu3713, plu1359, plu1143, pau_02999, pau_00954, pau_00948 | 1,094 | Site-specific recombinase XerD |
| P4-9 | pte_01778 | 675 | plu3734, pau_03032 | 34 | Unknown, probable transcriptional regulator (PAS_4 and LuxR-C); response regulator containing a CheY-like receiver domain and an HTH DNA-binding domain |
| P56-C3 | pte_01618 | 2,865 | plu0967, pau_03850 | 1,995 | TccC insecticidal toxin complex protein TccC; RHS repeat-associated core domain |
| Nonmotility | |||||
| P3-C8 | pte_00381 | 1,146 | plu1895, pau_02666 | 718 | Flagellar biogenesis protein FlhB |
| P4-27 | pte_03497 | 1,215 | plu1918, pau_02648 | 1,116 | Flagellar hook protein FlgE |
| P59-G8 | pte_03503 | 1,644 | plu1924, pau_02642 | 1,138 | Flagellar hook-associated protein (HAP1) FlgK |
| Defective motility | |||||
| P13-7 | pte_02637 | 1,944 | plu2384, pau_02113 | 1,192 | Exoribonuclease II |
| P64-D5 | pte_02122 | 726 | plu0425, pau_00335 | 330 | Uncharacterized protein conserved in bacteria (PAS and helix-turn-helix domains); yheO |
| Hyperswimming: P5-2 | pte_02486 | 927 | plu4343 | 14 | Farnesyl-diphosphate farnesyltransferase, phytoene synthase |
Intergenic region between pte_04057 and pte_004056.
Three mutants of the “swim without NaCl” class were identified. Mutant P4-9 had an insertion into pte_01778 gene that had 83% and 71% identity to P. luminescens plu3734 and P. asymbiotica pau_03032 gene products, respectively. This gene encodes a probable transcriptional regulator containing a PAS and LuxR-type DNA-binding HTH domains. This result suggests that this gene may be influenced by NaCl and potentially regulates the flagellar gene cascade in P. temperata. The second mutant, P56-C3, has an insertion in pte_01618, which encodes the insecticidal toxin complex TccC, which is a rearrangement hot spot (RHS) repeat association protein. This gene product had 71 and 80% identity to P. luminescens plu0967 and P. asymbiotica pau_03850 gene products, respectively. The third mutant, P1-4, had an insertion in pte_02195, which encodes a site-specific recombinase, XerD.
In the hyperswarmer category, mini-Tn5 insertion sites of 18 mutants were identified (Table 4). Mutants P7-A8 and P20-D5 appear to be sibling mutations in the same site (pte_03319). Two mutants had insertions at two separate sites in the pte_04499 gene, which encodes a nonribosomal peptide synthetase that was associated with biosynthetic cluster PT18 (Table 2). Mutant P2-A9 had an insertion in the pte_02547 gene, which encodes Hnr, a, two-component response regulator that has 52% identity with the Escherichia coli sigma-S factor regulatory protein RssB. Mutant P75-A5 had an insertion in pte_01774, which encodes a putative transcriptional regulator (AbgR family).
The hyperswimmer mutant P5-2 had an insertion in pte_02486, which encodes a farnesyl-diphosphate farnesyltransferase or phytoene synthase. This protein was part of biosynthetic cluster PT10, which is predicted to produce a terpene compound.
Two mutants from the defective motility category were identified (Table 4). Both of these mutants were also defective in pathogenesis. Mutant P13-7 had an insertion in the pte_02637 gene, which codes for exonuclease II. Mutant P64-D5 contained an insertion in pte_02122 gene. The product of this gene is predicted to be an uncharacterized conserved protein similar to YheO that contains a PAS and helix-turn-helix domains, suggesting a DNA-binding protein with signal capabilities.
Genetic complementation of the P13-7 mutant.
Although these transposon mutants showed these specific defects in pathogenesis and motility, the definitive link to demonstrate causality is genetic complementation. The mutant P13-7 with a disruption in pte_02637 was chosen for this experiment, and a PCR approach was taken. The wild-type pte_02637 gene was amplified using external primers, cloned into a TOPO vector and subcloned in the pBAD18-Cm expression vector as described in Materials and Methods. The resulting construct, pHR1, was introduced into P13-7 and the parental wild-type (NC19 primary form). The control plasmid pBAD18-Cm was also introduced into the parental wild-type and the mutant strain. P13-7 cells complemented with pHR1 (HNR1307) were immediately recognized and had different appearance from the P13-7 with the control vector (HNR1318). HNR1307 recovered pigment production activity, while control HNR1318 did not. Endpoint RT-PCR verified the expression of pte_02637 gene in the complemented mutant, HNR1307 (see Fig. S12 in the supplemental material).
The physiological properties of the P13-7 mutant and complemented mutant were investigated (Table 5). Genetic complementation (HNR1307) restored P13-7 to the primary-phase wild-type pattern for all of the traits tested. It was immediately noticed that the complemented mutant, HNR1307, returned to yellow pigmentation on LB agar plates. Several other physiological traits, including dye binding, protease, and hemolytic activities, were restored to the wild-type levels. Overexpression of pte_02637 (HNR0606) caused more dye binding on MacConkey plates and created more annular hemolysis on blood agar than was observed with the parental wild type.
TABLE 5.
Genetic complementation of mutant P13-7 restores wild-type phenotypic traits
| Strain | Insect pathogenesisa |
Swim motilityb | Symbiosisc | Dye bindingd |
Extracellular productione |
Pigment | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LT50 | LT100 | EB | MacConkey | DNase | Protease | Hemolysin | Antibiotic | ||||
| NC19 primary (parental) | 36 | 48 | 48.0 ± 3.3 | + | + | + | ++ | ++ | + | + | + |
| HNR0606 (NC19+ pHR1) | 33 | 43 | 62.2 ± 5.3 | + | + | + | ++ | ++ | ++ | + | + |
| HNR0018 (NC19 + pBAD32) | 36 | 48 | 52.2 ± 6.3 | + | + | + | ++ | ++ | + | + | + |
| P13-7 | 60 | 72 | 22.0 ± 1.1 | + | − | − | ++ | − | − | − | − |
| HNR1307 (P13-7 + HR1) | 39 | 49 | 49.5 ± 5.0 | + | + | + | ++ | ++ | + | + | + |
| HNR1318 (P13-7+ pBAD32) | 61 | 70 | 19.0 ± 1.1 | + | − | − | ++ | − | − | − | − |
Insect pathogenesis was determined on G. mellonella larvae as described in Materials and Methods. LT50 and LT100 are the time (in hours) required to kill 50 and 100% of larvae.
The swim migration assay was carried out after 48 h of incubation at 28°C. Values are average swim ring diameters, with standard deviations. Results are averages of 3 measurements per independent experiment.
Symbiosis was determined by the nematode assay and determined by the development of mature IJs. +, capable of normal nematode development, similar to the parental wild type.
Dye absorption on EB and MacConkey plates were noted at 48 h. A positive result on EB plates was indicated by metallic green colonies, and a negative result was indicated by dull purple colonies. A positive result on MacConkey plates was indicated by bright red colonies and a negative result by colorless or pink colonies.
DNase and protease activities were determined by measuring the sizes (in millimeters) of the halos surrounding the bacterial colonies 24 h after inoculation. ++, strongly positive (2- to 4-mm halo); +, positive (1- to 2-mm halo); −, negative (no halo). Hemolytic activity was determined by clearing zones around the bacterial colonies. ++, enhanced annular hemolysis; +, strong annular hemolysis; −, negative (no hemolysis). Antibiotic production was determined by measuring the sizes (in millimeters) of the halos surrounding the bacterial colonies 1 day after inoculation of the tester bacterium (Micrococcus luteus). +, positive (2- to 5-mm halo); −, negative (no halo).
The P13-7 mutant was originally identified as aberrant motility mutant, and genetic complementation of the mutation restored wild-type swimming behavior (Table 5; also, see Fig. S13 in the supplemental material). The control plasmid (HNR1318) did not complement the mutation. Both the control plasmid and the pte_02637 construct caused a slight increase in the swim ring diameter for the wild-type cells (HNR0018 and HNR0606). These data show causality for these physiological traits.
The key property for this study was insect pathogenesis. The P13-7 mutant showed a delayed insect pathogenesis. Genetic complementation (HNR1307) restored P13-7 to wild-type virulence levels (Table 5). In this assay, cells were pregrown with arabinose to induce expression of pte_02637. Wild-type cells overexpressing this gene were more virulent, showing lower LT50 and LT100 values than the wild-type control. These in vitro insect pathogenesis assays were followed up by the use of an in vivo assay with nematodes serving as a vector. Axenic nematodes were reared on either wild-type, P13-7, or HNR1307 (the genetically complemented P13-7 mutant) cells. Nematodes containing the P13-7 mutant was also defective for in vivo insect pathogenesis (LT50 and LT100 values of 82 h and 140 h, respectively), while nematodes containing wild-type cells had LT50 and LT100 values of 71 h and 121 h, respectively. Nematodes containing HNR1307 (the genetically complemented P13-7 mutant) exhibited LT50 and LT100 values that returned to the wild-type level (71 h and 123 h, respectively). Nematodes reared on the P64-D5 mutant were also defective for in vivo insect pathogenesis (LT50 and LT100 values of 80 h and 144 h, respectively). P13-7 was also defective in antibiotic activity, and complementation also restored antibiotic activity.
Mutant P13-7 had a disruption in the pte_02637 gene encoding RNase II, a 3′ to 5′ exoribonuclease II. As part of an essential RNA-degrading multiprotein complex, RNase II is involved in the maturation, turnover, and quality control of RNA (77, 78). For many pathogens, the activity of RNase II has been correlated with the regulation of virulence and motility (79, 80). With Escherichia coli, the results from next generation sequencing show that RNase II mutants inhibited motility and increased biofilm production. In our system, RNase II globally regulates many factors, including virulence, motility, cell surface properties, production of antimicrobial agents, and hemolytic activity.
Summary.
The P. temperata NC19 genome was sequenced and analyzed through comparative genomic approaches. A large number of biosynthetic gene clusters were identified within the P. temperata NC19 genome, and the presence of four complete T6SS and a single T3SS was revealed. The presence of a contiguous 104-kb region that was orthologous with a X. nematophila plasmid XNC1_p was also found only in the P. temperata NC19 genome. A genetic approach was started to identify genes involved in symbiosis and/or pathogenesis and mutants with altered motility. Five classes of motility mutants were identified and characterized physiologically and genetically. These mutants were screened for altered insect pathogenesis and reduced to a small pool of candidates. Only three mutants from the motility-defective category showed consistent results in altered pathogenesis. One mutant with a defective in RNase II showed decreased motility, virulence and production of extracellular factors. Genetic complementation of the mutant restored wild-type activity, showing causality. These results demonstrate a role for RNA turnover in insect pathogenesis and other physiological functions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by USDA NIFA grant 2009-35302-05257, by Hatch grant NH496, and by the College of Life Science and Agriculture, The University of New Hampshire, Durham, NH. Summer Undergraduate Research Fellowships (SURF) from the University of New Hampshire-Durham supported H.B., R.J., J.G., and U.D.
We thank Jessica MacLure, Spiros Kapolis, Katie Corso, Stephanie Huack, Ben Coffey, Ben Bailey, and Bryan Ballard for contributions in the initial stages of this project and Helge Bode for the stilbene analysis.
Footnotes
This is scientific contribution number 2563 from the NH Agricultural Experimental Station.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00197-15.
Citation Hurst S, IV, Rowedder H, Michaels B, Bullock H, Jackobeck R, Abebe-Akele F, Durakovic U, Gately J, Janicki E, Tisa LS. 2015. Elucidation of the Photorhabdus temperata genome and generation of a transposon mutant library to identify motility mutants altered in pathogenesis. J Bacteriol 197:2201–2216. doi:10.1128/JB.00197-15.
REFERENCES
- 1.Boemare N, Givaudan A, Brehelin M, Laumond C. 1997. Symbiosis and pathogenicity of nematode-bacterium complexes. Symbiosis 22:21–45. [Google Scholar]
- 2.Forst S, Dowds B, Boemare N, Stackebrandt E. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu Rev Microbiol 51:47–72. doi: 10.1146/annurev.micro.51.1.47. [DOI] [PubMed] [Google Scholar]
- 3.Forst S, Nealson K. 1996. Molecular biology of the symbiotic pathogenic bacteria Xenorhabdus spp and Photorhabdus spp. Microbiol Rev 60:21–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodrich-Blair H, Clarke DJ. 2007. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol Microbiol 64:260–268. doi: 10.1111/j.1365-2958.2007.05671.x. [DOI] [PubMed] [Google Scholar]
- 5.Clarke DJ. 2008. Photorhabdus: a model for the analysis of pathogenicity and mutualism. Cell Microbiol 10:2159–2167. doi: 10.1111/j.1462-5822.2008.01209.x. [DOI] [PubMed] [Google Scholar]
- 6.Waterfield NR, Ciche T, Clarke D. 2009. Photorhabdus and a host of hosts. Annu Rev Microbiol 63:557–574. doi: 10.1146/annurev.micro.091208.073507. [DOI] [PubMed] [Google Scholar]
- 7.Burnell AM, Stock SP. 2000. Heterorhabditis, Steinernema and their bacterial symbionts—lethal pathogens of insects. Nematology 2:31–42. doi: 10.1163/156854100508872. [DOI] [Google Scholar]
- 8.Fischer-Le Saux M, Viallard V, Brunel B, Normand P, Boemare NE. 1999. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. temperata subsp. nov. and P. asymbiotica sp. nov. Int J Syst Bacteriol 49:1645–1656. doi: 10.1099/00207713-49-4-1645. [DOI] [PubMed] [Google Scholar]
- 9.Boemare N. 2002. Interactions between the partners of the entomopathogenic bacterium nematode complexes, Steinernema-Xenorhabdus and Heterorhabditis-Photorhabdus. Nematology 4:601–603. doi: 10.1163/15685410260438863. [DOI] [Google Scholar]
- 10.Ciche TA, Ensign JC. 2003. For the insect pathogen Photorhabdus luminescens, which end of a nematode is out? Appl Environ Microbiol 69:1890–1897. doi: 10.1128/AEM.69.4.1890-1897.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endo BY, Nickle WR. 1991. Ultrastructure of the intestinal epithelium, lumen, and associated bacteria in Heterorhabditis-Bacteriophora. J Helminthol Soc Wash 58:202–212. [Google Scholar]
- 12.Milstead JE. 1979. Heterorhabditis-Bacteriophora as a vector for introducing its associated bacterium into the hemocele of Galleria-Mellonella larvae. J Invertebr Pathol 33:324–327. doi: 10.1016/0022-2011(79)90033-8. [DOI] [Google Scholar]
- 13.Poinar GOJ. 1975. Description and biology of a new insect parasite rhabditol, Heterorhabditis bactiophora n. gen, n sp (Rhabditida: Heterorhabditidae n fam). Nematologica 21:463–470. doi: 10.1163/187529275X00239. [DOI] [Google Scholar]
- 14.Grewal PS, Koppenhofer AM, Choo HY. 2005. Lawn, turfgrass, and pasture applications, p 115–146. In Grewal PS, Ehlers R-U, Shapiro-Ilan DI (ed), Nematodes as biocontrol agents. CABI Publishing, Wallingford, United Kingdom,. [Google Scholar]
- 15.Blackburn M, Golubeva E, Bowen D, Ffrench-Constant RH. 1998. A novel insecticidal toxin from photorhabdus luminescens, Toxin complex a (Tca), and its histopathological effects on the midgut of manduca sexta. Appl Environ Microbiol 64:3036–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowen DJ, Ensign JC. 1998. Purification and characterization of a high-molecular-weight insecticidal protein complex produced by the entomopathogenic bacterium photorhabdus luminescens. Appl Environ Microbiol 64:3029–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ffrench-Constant R, Bowen D. 1999. Photorhabdus toxins: novel biological insecticides. Curr Opin Microbiol 2:284–288. doi: 10.1016/S1369-5274(99)80049-6. [DOI] [PubMed] [Google Scholar]
- 18.ffrench-Constant RH, Bowen DJ. 2000. Novel insecticidal toxins from nematode-symbiotic bacteria. Cell Mol Life Sci 57:828–833. doi: 10.1007/s000180050044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt TM, Bleakley B, Nealson KH. 1988. Characterization of an extracellular protease from the insect pathogen Xenorhabdus luminescens. Appl Environ Microbiol 54:2793–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Dowds BC. 1993. Phase variation in Xenorhabdus luminescens: cloning and sequencing of the lipase gene and analysis of its expression in primary and secondary phases of the bacterium. J Bacteriol 175:1665–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brillard J, Ribeiro C, Boemare N, Brehelin M, Givaudan A. 2001. Two distinct hemolytic activities in Xenorhabdus nematophila are active against immunocompetent insect cells. Appl Environ Microbiol 67:2515–2525. doi: 10.1128/AEM.67.6.2515-2525.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas GM, Poinar GO. 1979. Xenorhabdus gen. nov., a genus of entomopathogenic, nematophilic bacteria of the family Enterobacteriaceae. Int J Syst Bacteriol 29:352–360. doi: 10.1099/00207713-29-4-352. [DOI] [Google Scholar]
- 23.Akhurst RJ. 1982. Antibiotic activity of Xenorhabdus spp, bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J Gen Microbiol 128:3061–3065. [DOI] [PubMed] [Google Scholar]
- 24.Richardson WH, Schmidt TM, Nealson KH. 1988. Identification of an anthraquinone pigment and a hydroxystilbene antibiotic from Xenorhabdus luminescens. Appl Environ Microbiol 54:1602–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derzelle S, Ngo S, Turlin E, Duchaud E, Namane A, Kunst F, Danchin A, Bertin P, Charles JF. 2004. AstR-AstS, a new two-component signal transduction system, mediates swarming, adaptation to stationary phase and phenotypic variation in Photorhabdus luminescens. Microbiology 150:897–910. doi: 10.1099/mic.0.26563-0. [DOI] [PubMed] [Google Scholar]
- 26.Akhurst RJ. 1980. Morphological and functional dimorphism in Xenorhabdus spp bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis. J Gen Microbiol 121:303–309. [DOI] [PubMed] [Google Scholar]
- 27.Boemare N, Thaler JO, Lanois A. 1997. Simple bacteriological tests for phenotypic characterization of Xenorhabdus and Photorhabdus phase variants. Symbiosis 22:167–175. [Google Scholar]
- 28.Rosner BM, Ensign JC, Schink B. 1996. Anaerobic metabolism of primary and secondary forms of Photorhabdus luminescens. FEMS Microbiol Lett 140:227–232. doi: 10.1111/j.1574-6968.1996.tb08341.x. [DOI] [Google Scholar]
- 29.Smigielski AJ, Akhurst RJ, Boemare NE. 1994. Phase variation in Xenorhabdus nematophilus and Photorhabdus luminescens—differences in respiratory activity and membrane energization. Appl Environ Microbiol 60:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaudriault S, Pages S, Lanois A, Laroui C, Teyssier C, Jumas-Bilak E, Givaudan A. 2008. Plastic architecture of bacterial genome revealed by comparative genomics of Photorhabdus variants. Genome Biol 9:R117. doi: 10.1186/gb-2008-9-7-r117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodgson MM, Day B, White DJ, Tisa LS. 2003. Effect of growth conditions on the motility of Photorhabdus temperata. Arch Microbiol 180:17–24. doi: 10.1007/s00203-003-0558-z. [DOI] [PubMed] [Google Scholar]
- 32.Michaels B, Tisa LS. 2011. Swarming motility by Photorhabdus temperata is influenced by environmental conditions and uses the same flagella as that used in swimming motility. Can J Microbiol 57:196–203. doi: 10.1139/W10-119. [DOI] [PubMed] [Google Scholar]
- 33.Ehlers R-U, Stoessel S, Wyss U. 1990. The influence of phase variants of Xenorhabdus spp. and Escherichia coli (Enterobacteriaceae) on the propagation of entomopathogenic nematodes of the genera Steinernema and Heterorhabditis. Rev Nematol 13:417–424. [Google Scholar]
- 34.Han RC, Ehlers RU. 2001. Effect of Photorhabdus luminescens phase variants on the in vivo and in vitro development and reproduction of the entomopathogenic nematodes Heterorhabditis bacteriophora and Steinernema carpocapsae. FEMS Microbiol Ecol 35:239–247. doi: 10.1111/j.1574-6941.2001.tb00809.x. [DOI] [PubMed] [Google Scholar]
- 35.Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles JF, Dassa E, Derose R, Derzelle S, Freyssinet G, Gaudriault S, Medigue C, Lanois A, Powell K, Siguier P, Vincent R, Wingate V, Zouine M, Glaser P, Boemare N, Danchin A, Kunst F. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol 21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- 36.Wilkinson P, Waterfield NR, Crossman L, Corton C, Sanchez-Contreras M, Vlisidou I, Barron A, Bignell A, Clark L, Ormond D, Mayho M, Bason N, Smith F, Simmonds M, Churcher C, Harris D, Thompson NR, Quail M, Parkhill J, Ffrench-Constant RH. 2009. Comparative genomics of the emerging human pathogen Photorhabdus asymbiotica with the insect pathogen Photorhabdus luminescens. BMC Genomics 10:302. doi: 10.1186/1471-2164-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.ffrench-Constant R, Waterfield Daborn N, Joyce P, Bennett S, Au H, Dowling C, Boundy A, Reynolds S, Clarke SD. 2003. Photorhabdus: towards a functional genomic analysis of a symbiont and pathogen. FEMS Microbiol Rev 26:433–456. doi: 10.1111/j.1574-6976.2003.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 38.Easom CA, Clarke DJ. 2008. Motility is required for the competitive fitness of entomopathogenic Photorhabdus luminescens during insect infection. BMC Microbiol 8:168. doi: 10.1186/1471-2180-8-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boemare NE, Akhurst RJ, Mourant RG. 1993. DNA relatedness between Xenorhabdus spp (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen nov. Int J Syst Bacteriol 43:249–255. doi: 10.1099/00207713-43-2-249. [DOI] [Google Scholar]
- 40.Poinar GOJ, Jackson T, Klein M. 1987. Heterorhabdus megidis sp. n. (Heterorhabditidae: Rhabdittida), parasitic on Japanese beetle, Popilla japonica (Scaraarabaeidedae: Coleoptera) in Ohio. Proc Helminthol Soc Wash 54:53–59. [Google Scholar]
- 41.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, Zhao XL, Dubchak I, Hugenholtz P, Anderson I, Lykidis A, Mavromatis K, Ivanova N, Kyrpides NC. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res 34:D344–D348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lerat E, Daubin V, Ochman H, Moran NA. 2005. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol 3:807–814. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 50.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. 2013. antiSMASH 2.0–a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. 2011. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ciche TA, Bintrim SB, Horswill AR, Ensign JC. 2001. A Phosphopantetheinyl transferase homolog is essential for Photorhabdus luminescens to support growth and reproduction of the entomopathogenic nematode Heterorhabditis bacteriophora. J Bacteriol 183:3117–3126. doi: 10.1128/JB.183.10.3117-3126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan JWYF, Goodwin PH. 1994. Extraction of genomic DNA from extracellular polysaccharide-synthesizing Gram-negative bacteria. Biotechniques 18:419–422. [PubMed] [Google Scholar]
- 54.Rawnsley T, Tisa LS. 2007. Development of a physical map for three Frankia strains and a partial genetic map for Frankia EuI1c. Physiol Plantarum 130:427–439. doi: 10.1111/j.1399-3054.2007.00896.x. [DOI] [Google Scholar]
- 55.Han RC, Ehlers RU. 2000. Pathogenicity, development, and reproduction of Heterorhabditis bacteriophora and Steinernema carpocapsae under axenic in vivo conditions. J Invertebr Pathol 75:55–58. doi: 10.1006/jipa.1999.4900. [DOI] [PubMed] [Google Scholar]
- 56.Clarke DJ, Dowds BCA. 1995. Virulence mechanisms of Photorhabdus sp strain K122 toward wax moth larvae. J Invertebr Pathol 66:149–155. doi: 10.1006/jipa.1995.1078. [DOI] [Google Scholar]
- 57.Bilgrami AL, Gaugler R, Shapiro-Ilan DI, Adams BJ. 2006. Source of trait deterioration in entomopathogenic nematodes Heterorhabditis bacteriophora and Steinernema carpocapsae during in vivo culture. Nematology 8:397–409. doi: 10.1163/156854106778493394. [DOI] [Google Scholar]
- 58.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tailliez P, Pages S, Edgington S, Tymo LM, Buddie AG. 2012. Description of Xenorhabdus magdalenensis sp. nov., the symbiotic bacterium associated with Steinernema australe. Int J Syst Evol Microbiol 62:1761–1765. doi: 10.1099/ijs.0.034322-0. [DOI] [PubMed] [Google Scholar]
- 60.Tailliez P, Laroui C, Ginibre N, Paule A, Pages S, Boemare N. 2010. Phylogeny of Photorhabdus and Xenorhabdus based on universally conserved protein-coding sequences and implications for the taxonomy of these two genera. Proposal of new taxa: X. vietnamensis sp. nov., P. luminescens subsp. caribbeanensis subsp. nov., P. luminescens subsp. hainanensis subsp. nov., P. temperata subsp. khanii subsp. nov., P. temperata subsp. tasmaniensis subsp. nov., and the reclassification of P. luminescens subsp. thracensis as P. temperata subsp. thracensis comb. nov. Int J Syst Evol Microbiol 60:1921–1937. doi: 10.1099/ijs.0.014308-0. [DOI] [PubMed] [Google Scholar]
- 61.Ciche TA, Blackburn M, Carney JR, Ensign JC. 2003. Photobactin: a catechol siderophore produced by Photorhabdus luminescens, an entomopathogen mutually associated with Heterorhabditis bacteriophora NC1 nematodes. Appl Environ Microbiol 69:4706–4713. doi: 10.1128/AEM.69.8.4706-4713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brachmann AO, Joyce SA, Jenke-Kodama H, Schwar G, Clarke DJ, Bode HB. 2007. A type II polyketide synthase is responsible for anthraquinone biosynthesis in Photorhabdus luminescens. Chembiochem 8:1721–1728. doi: 10.1002/cbic.200700300. [DOI] [PubMed] [Google Scholar]
- 63.Joyce SA, Lango L, Clarke DJ. 2011. The regulation of secondary metabolism and mutualism in the insect pathogenic bacterium Photorhabdus luminescens. Adv Appl Microbiol 76:1–25. doi: 10.1016/B978-0-12-387048-3.00001-5. [DOI] [PubMed] [Google Scholar]
- 64.Li JX, Chen GH, Wu HM, Webster JM. 1995. Identification of two pigments and a hydroxystilbene antibiotic from Photorhabdus luminescens. Appl Environ Microbiol 61:4329–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bode HB, Reimer D, Fuchs SW, Kirchner F, Dauth C, Kegler C, Lorenzen W, Brachmann AO, Grun P. 2012. Determination of the absolute configuration of peptide natural products by using stable isotope labeling and mass spectrometry. Chemistry 18:2342–2348. doi: 10.1002/chem.201103479. [DOI] [PubMed] [Google Scholar]
- 66.Brachmann AO, Brameyer S, Kresovic D, Hitkova I, Kopp Y, Manske C, Schubert K, Bode HB, Heermann R. 2013. Pyrones as bacterial signaling molecules. Nat Chem Biol 9:573–578. doi: 10.1038/nchembio.1295. [DOI] [PubMed] [Google Scholar]
- 67.Theodore CM, King JB, You JL, Cichewicz RH. 2012. Production of cytotoxic glidobactins/luminmycins by Photorhabdus asymbiotica in liquid media and live crickets. J Nat Prod 75:2007–2011. doi: 10.1021/np300623x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derzelle S, Duchaud E, Kunst F, Danchin A, Bertin P. 2002. Identification, characterization, and regulation of a cluster of genes involved in carbapenem biosynthesis in Photorhabdus luminescens. Appl Environ Microbiol 68:3780–3789. doi: 10.1128/AEM.68.8.3780-3789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joyce SA, Brachmann AO, Glazer I, Lango L, Schwar G, Clarke DJ, Bode HB. 2008. Bacterial biosynthesis of a multipotent stilbene. Angew Chem Int Ed 47:1942–1945. doi: 10.1002/anie.200705148. [DOI] [PubMed] [Google Scholar]
- 70.Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chaston JM, Suen G, Tucker SL, Andersen AW, Bhasin A, Bode E, Bode HB, Brachmann AO, Cowles CE, Cowles KN, Darby C, de Leon L, Drace K, Du Z, Givaudan A, Herbert Tran EE, Jewell KA, Knack JJ, Krasomil-Osterfeld KC, Kukor R, Lanois A, Latreille P, Leimgruber NK, Lipke CM, Liu R, Lu X, Martens EC, Marri PR, Medigue C, Menard ML, Miller NM, Morales-Soto N, Norton S, Ogier JC, Orchard SS, Park D, Park Y, Qurollo BA, Sugar DR, Richards GR, Rouy Z, Slominski B, Slominski K, Snyder H, Tjaden BC, van der Hoeven R, Welch RD, Wheeler C, Xiang B, Barbazuk B, Gaudriault S, Goodner B, Slater SC, Forst S, Goldman BS, Goodrich-Blair H. 2011. The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes. PLoS One 6:e27909. doi: 10.1371/journal.pone.0027909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shrivastava S, Mande SS. 2008. Identification and functional characterization of gene components of type VI secretion system in bacterial genomes. PLoS One 3:e2955. doi: 10.1371/journal.pone.0002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chow J, Mazmanian SK. 2010. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waterfield NR, Daborn PJ, ffrench-Constant RH. 2002. Genomic islands in Photorhabdus. Trends Microbiol 10:541–545. doi: 10.1016/S0966-842X(02)02463-0. [DOI] [PubMed] [Google Scholar]
- 75.Brugirard-Ricaud K, Givaudan A, Parkhill J, Boemare N, Kunst F, Zumbihl R, Duchaud E. 2004. Variation in the effectors of the type III secretion system among Photorhabdus species as revealed by genomic analysis. J Bacteriol 186:4376–4381. doi: 10.1128/JB.186.13.4376-4381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brugirard-Ricaud K, Duchaud E, Givaudan A, Girard PA, Kunst F, Boemare N, Brehelin M, Zumbihl R. 2005. Site-specific antiphagocytic function of the Photorhabdus luminescens type III secretion system during insect colonization. Cell Microbiol 7:363–371. doi: 10.1111/j.1462-5822.2004.00466.x. [DOI] [PubMed] [Google Scholar]
- 77.Frazao C, Mcvey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. 2006. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature 443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 78.Lu F, Taghbalout A. 2014. The Escherichia coli major exoribonuclease RNase II is a component of the RNA degradosome. Biosci Rep 34:e00166. doi: 10.1042/BSR20140113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsao MY, Lin TL, Hsieh PF, Wang JT. 2009. The 3′-to-5′ exoribonuclease (encoded by HP1248) of Helicobacter pylori regulates motility and apoptosis-inducing genes. J Bacteriol 191:2691–2702. doi: 10.1128/JB.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matos RG, Barria C, Moreira RN, Barahona S, Domingues S, Arraiano CM. 2014. The importance of proteins of the RNase II/RNB-family in pathogenic bacteria. Front Cell Infect Microbiol 4:68. doi: 10.3389/fcimb.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 82.Felsenstein J. 1985. Confidence limits on phylogenies—an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.