ABSTRACT
Integrons are bacterial genetic elements able to capture and express genes contained within mobile gene cassettes. Gene cassettes are expressed via a Pc promoter and can be excised from or integrated into the integron by integrase IntI. Although the mechanisms of gene cassette integration and excision are well known, the kinetics and modes of gene cassette shuffling leading to new gene cassette arrays remain puzzling. It has been proposed that under antibiotic selective pressure, IntI-mediated rearrangements can generate integron variants in which a weakly expressed gene cassette moves closer to Pc, thus leading to higher-level resistance. To test this hypothesis, we used an integron with four gene cassettes, intI1-aac(6′)-Ib-dfrA15-aadA1-catB9, and applied selective pressure with chloramphenicol, resistance to which is encoded by catB9. Experiments were performed with three different Pc variants corresponding to three IntI1 variants. All three integrases, even when not overexpressed, were able to bring catB9 closer to Pc via excision of the dfrA15 and aadA1 gene cassettes, allowing their host bacteria to adapt to antibiotic pressure and to grow at high chloramphenicol concentrations. Integrase IntI1R32_H39, reported to have the highest recombination activity, was able, when overexpressed, to trigger multiple gene cassette rearrangements. Although we observed a wide variety of rearrangements with catB9 moving closer to Pc and leading to higher chloramphenicol resistance, “cut-and-paste” relocalization of catB9 to the first position was not detected. Our results suggest that gene cassette rearrangements via excision are probably less cost-effective than excision and integration of a distal gene cassette closer to Pc.
IMPORTANCE Integrons are bacterial genetic elements able to capture and express gene cassettes. Gene cassettes are expressed via a Pc promoter; the closer they are to Pc, the more strongly they are expressed. Gene cassettes can be excised from or integrated into the integron by integrase IntI. The kinetics and modes of gene cassette shuffling, leading to new gene cassette arrays remain puzzling. We used an integron with 4 antibiotic resistance gene cassettes and applied selective pressure with the antibiotic for which resistance was encoded by cassette 4. All IntI variants were able to bring cassette 4 closer to Pc. Rearrangements occur via excision of the previous gene cassettes instead of cut-and-paste relocalization of the fourth gene cassette.
INTRODUCTION
Integrons are bacterial genetic elements able to capture and express genes contained within mobile gene cassettes (1). Integrons are composed of three key elements: (i) an intI gene encoding an integrase that mediates gene cassette integration and excision through site-specific RecA-independent recombination, (ii) recombination site attI, and (iii) a promoter, Pc (2). Integrons containing antibiotic resistance gene cassettes are located on transposons or plasmids and are widely involved in the spread of antibiotic resistance among Gram-negative bacteria. More than 130 gene cassettes, encoding resistance to nearly all antibiotic families, have been described (3). Several classes of integron have been described on the basis of the amino acid sequence of the IntI integrase (4), class 1 being the most frequently described for multidrug-resistant Gram-negative bacteria (5–7).
Gene cassettes are mobilizable units composed of an open reading frame followed by a recombination site, attC. Gene cassettes are usually promoterless and are transcribed under the control of Pc (8). Expression of gene cassettes is also influenced by their position within the cassette array: the closer they are to Pc, the more strongly they are expressed (9). Among the 13 Pc variants that have been described for class 1 integrons, 4 predominate, namely, PcS, PcWTGN-10, PcH1, and PcW (listed from strongest to weakest, with a 25-fold difference in strength between PcS and PcW) (8). In class 1 integrons, Pc is located within the intI1 gene and Pc polymorphism affects the IntI1 amino acid sequence at positions 32 and 39; three IntI1 variants, IntI1R32_N39, IntI1P32_H39, and IntI1R32_H39, correspond to the four main Pc variants (8). We have previously shown that the weaker the Pc variant, the more active the encoded IntI1: the IntI1R32_H39 variant (corresponding to the weak promoter sequences PcW and PcH1) has the most efficient recombination activity (8). Moreover, intI1 transcription is regulated via the SOS response (10). This global regulatory network is controlled by the transcriptional repressor LexA (11) and is induced by stresses such as exposure to some antibiotics (quinolones, β-lactams, and trimethoprim). PintI1 lies face to face with Pc. Pc interferes with the level of intI1 transcription, but this effect depends on the Pc variant: the strong Pc variant PcS prevents intI1 expression, contrary to the other variants (12). Drug resistance driven by class 1 integrons thus results from (i) the level of gene cassette expression, (ii) the level of integrase expression, (iii) the recombinogenic efficiency of the integrase variant, and (iv) the shuffling capacity of the gene cassettes, driven by their attC site folding (13).
The integrase is able to catalyze gene cassette excision by attC × attC recombination events and gene cassette integration by attC × attI recombination events. The mechanisms of gene cassette integration and excision are well known (14, 15), notably the attC sites, for which recombination occurs in single-stranded form, only the folded bottom strand being active (16, 17). However, there are very few data on the dynamics of gene cassette shuffling leading to new gene cassette arrays. It has been proposed that under antibiotic selective pressure, integrase-mediated gene cassette rearrangements could create integron variants in which a weakly expressed gene cassette moves closer to Pc, leading to higher-level resistance (9). For this to occur, the integrase has to recombine two attC sites, leading to excision of a circular gene cassette that can subsequently be integrated at the attI site. To verify this hypothesis experimentally, we constructed a class 1 integron [aac(6′)-Ib-dfrA15-aadA1-catB9] and applied selective pressure with chloramphenicol, the antibiotic to which resistance is encoded by the last gene cassette (catB9). Experiments were performed with three different Pc variants and the three different IntI1 variants, under different conditions of intI1 expression. Although we observed a wide variety of rearrangements with catB9 closer to Pc and leading to higher chloramphenicol resistance, “cut-and-paste” relocalization of catB9 to the first position was not detected. The most frequent rearrangement was obtained by excision of the two gene cassettes upstream of catB9.
MATERIALS AND METHODS
Bacteria and growth conditions.
Bacteria (Table 1) were grown at 37°C in brain heart infusion broth (BHI; AES Chemunex, Bruz, France) supplemented when necessary with kanamycin (Km; 25 mg/liter), ampicillin (Amp; 100 mg/liter), chloramphenicol (Cm; at concentrations depending on exerted selection pressure), and arabinose (0.2%). Antibiotics and arabinose were obtained from Sigma-Aldrich, Lyon, France.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| Enterobacter aerogenes BM2688 | Contains class 1 RI In40 with gene cassette array aac(6′)-Ib, qacF, cmlA2, and oxa-9 and the strong Pc variant (PcS). Resistant to tobramycin. | 13 |
| Shigella dysenteriae 3Sh | Contains class 1 RI with gene cassette array dfrA15-aadA1. Resistant to streptomycin, spectinomycin, and trimethoprim. | Laboratory collection |
| Escherichia coli 1314 | pSU38::catB9 | 14 |
| Escherichia coli DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ- thi-1 gyrA96 relA1. Resistant to nalidixic acid. Used as recipient strain | Laboratory collection |
| Plasmids | ||
| pSU38ΔtotlacZ | Vector carrying the lacZ coding sequence with no translation initiation region or promoter | 8 |
| p1S | Integron 1S intI1-aac(6′)-Ib-dfrA15-aadA1-catB9 cloned into pSU38ΔtotlacZ with PcS promoter | This study |
| p1W | p1S mutated with primer PcWmut to create p1W with PcW promoter | This study |
| p1WTGN-10 | p1S mutated with primer PcWTGN-10mut to create p1WTGN-10 with PcWTGN-10 promoter | This study |
| p1SL | p1S mutated with primer LexAmut2L to create p1SL (constitutive expression of intI1R32_N39) | This study |
| p1WL | p1W mutated with primer LexAmut2L to create p1WL (constitutive expression of intI1R32_H39) | This study |
| p1WTGN-10L | p1WTGN-10 mutated with primer LexAmut2L to create p1WTGN-10L (constitutive expression of intI1P32_H39) | This study |
| pBad-intI1*R32_H39 | intI1 containing the PcW variant cloned into pBAD18 (arabinose-inducible expression vector) in which PcW is inactivated | 8 |
| pBad-intI1*R32_N39 | intI1 containing the PcS variant cloned into pBAD18 in which PcS is inactivated | 8 |
| pBad-intI1*P32_H39 | intI1 containing the PcWTGN-10 variant cloned into pBAD18 in which PcWTGN-10 is inactivated | 8 |
| p2S | Integron 2S intI1-catB9 cloned into pSU38ΔtotlacZ with PcS promoter | This study |
| p2W | p2S mutated with primer PcWmut to create p2W with PcW promoter | This study |
| p2WTGN-10 | p2S mutated with primer PcWTGN-10mut to create p2WTGN-10 with PcWTGN-10 promoter | This study |
| p2SL | p2S mutated with primer LexAmut2L to create p2SL (constitutive expression of intI1R32_N39) | This study |
| p2WL | p2W mutated with primer LexAmut2L to create p2WL (constitutive expression of intI1R32_H39) | This study |
| p2WTGN-10L | p2WTGN-10 mutated with primer LexAmut2L to create p2WTGN-10L (constitutive expression of intI1P32_H39) | This study |
| p3Sa | intI1-aac(6′)-Ib-catB9 cloned into pSU38ΔtotlacZ | This study |
| p4Sa | intI1-aac(6′)-Ib-aac(6′)-Ib-catB9 cloned into pSU38ΔtotlacZ | This study |
| p5Sa | intI1-dfrA15-aadA1-catB9 cloned into pSU38ΔtotlacZ | This study |
| p6Sa | intI1-aac(6′)-Ib cloned into pSU38ΔtotlacZ | This study |
| p7Sa | intI1-aac(6′)-Ib-dfrA15-catB9 cloned into pSU38ΔtotlacZ | This study |
| p8Sa | intI1-aadA1-catB9 cloned into pSU38ΔtotlacZ | This study |
| p9Sa | intI1-aac(6′)-Ib-dfrA15-aadA1- dfrA15-aadA1-catB9 cloned into pSU38ΔtotlacZ | This study |
| p10Sa | intI1 cloned into pSU38ΔtotlacZ | This study |
| p11Sa | intI1-aac(6′)-Ib-dfrA15-aac(6′)-Ib-dfrA15-aadA1-catB9 cloned into pSU38ΔtotlacZ | This study |
| p12Sa | intI1-aac(6′)-Ib-aac(6′)-Ib-dfrA15-aadA1-catB9 cloned into pSU38ΔtotlacZ | This study |
| p13Sa | intI1-aac(6′)-Ib-catB9-aac(6′)-Ib-catB9 cloned into pSU38ΔtotlacZ | This study |
| p14Sa | intI1-aac(6′)-Ib-dfrA15-aadA1-aac(6′)-Ib-dfrA15-aadA1-dfrA15-aadA1-catB9 cloned into pSU38ΔtotlacZ | This study |
| p15Sa | intI1-aac(6′)-Ib-catB9-catB9 cloned into pSU38ΔtotlacZ | This study |
| p16Sa | intI1- aac(6′)-Ib-dfrA15-aadA1-catB9- aac(6′)-Ib-dfrA15-aadA1-catB9 cloned into pSU38ΔtotlacZ | This study |
| p8741 | pBad::intI1Y312F | 20 |
| p1SintI1Y312F | p1S with the inactive integrase IntI1Y312F | This study |
Obtained after Cm pressure assays and cloned into pSU38ΔtotlacZ.
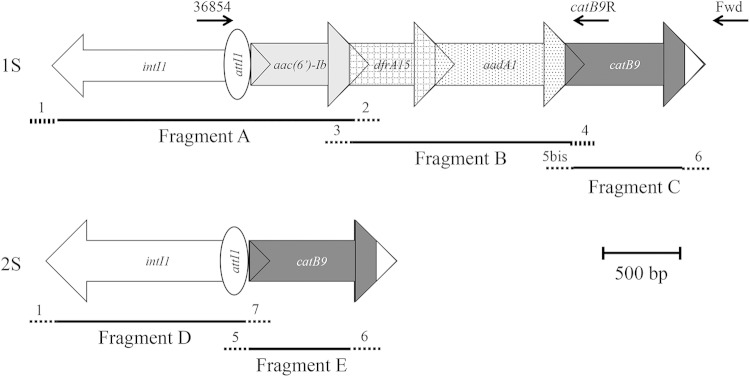
Construction of synthetic class 1 integrons (Fig. 1).
FIG 1.
Construction of integrons 1S and 2S. Integrons 1S and 2S were obtained by assembly PCR. To construct integron 1S, fragment B+C (2.2 kb) was first obtained and then coupled with fragment A. To construct integron 2S, fragments D and E were fused. Integron 1S was 3.9 kb, and integron 2S 1.9 kb. The primers used for assembly are represented by dotted lines and indicated by numbers (see Table 1S in the supplemental material), and the primers used for catB9 detection are represented by black arrows (Fwd is located in pSU38ΔtotlacZ).
PCR assembly was used to generate two synthetic class 1 integrons: integron 1S, intI1-aac(6′)-Ib-dfrA15-aadA1-catB9 (see Text S1 in the supplemental material) (3.9 kb) and integron 2S, intI1-catB9 (1.9 kb). Integron 1S contains four gene cassettes, encoding resistance to amikacin, tobramycin, and netilmicin [aac(6′)-Ib], trimethoprim (dfrA15), spectinomycin and streptomycin (aadA1), and chloramphenicol (catB9). Fragments A (1.8 kb) and D (1.2 kb) were amplified from Enterobacter aerogenes BM2688 genomic DNA (18) using, respectively, primers 1 and 2 and primers 1 and 7 (Fig. 1; see also Table S1 in the supplemental material). Fragment B (1.5 kb) was obtained from Shigella dysenteriae 3Sh genomic DNA (laboratory collection) using primers 3 and 4. Fragments C (0.8 kb) and E (0.7 kb) were amplified from Escherichia coli 1314 genomic DNA (19) using, respectively, primers 5bis and 6 and primers 5 and 6. To construct integron 1S, fragment B+C was prepared first and then coupled to fragment A. To construct integron 2S, fragments D and E were fused. The final PCR products (3.9 kb for 1S and 1.9 kb for 2S) were then cloned into the unique restriction sites EcoRI and BamHI of pSU38ΔtotlacZ to obtain plasmids p1S and p2S (Table 1). The different Pc variants and LexA binding-site mutants were obtained from integrons 1S and 2S by using the GeneEditor Site-directed Mutagenesis system (Promega, Charbonnieres, France) and specific primers (see Table S1).
We also constructed plasmid p1SintI1Y312F containing the inactive integrase IntI1Y312F (20). Using plasmids p1S and p8741 digested with BsaI and SpeI enzymes, we replaced the functional integrase of p1S by the inactive integrase IntI1Y312F, the catalytic site of which is mutated.
Chloramphenicol selection pressure assays.
E. coli DH5α containing p1S and its derivatives were cultured in BHI broth and subjected at successive 24-h intervals to increasing chloramphenicol concentrations. Each day, the broth containing the highest Cm concentration permitting visible bacterial growth (turbidity) was diluted 1/100 in fresh BHI broth containing a higher concentration of Cm and was also seeded on antibiotic-free BHI plates. The protocol was continued until no visible growth was observed. Experiments were performed at least three times, with integrons 1S, 1W, and 1WTGN-10, with LexA binding-site mutants 1SL, 1WL, and 1WTGN-10L (Table 1), and with or without overexpression of intI1 in trans, using the arabinose-inducible plasmid pBad-intI1 (arabinose was added every day). For intI1 overexpression, we used the three plasmids corresponding to three different IntI1 variants (pBad-intI1*R32_N39, pBad-intI1*P32_H39, and pBad-intI1*R32_H39) (8).
To locate the position of the catB9 gene cassette within the integron, 100 colonies were analyzed each day by PCR with primers 36854 located in the attI1 site, catB9R located in the catB9 gene, and Fwd located in the vector downstream of the integron (Fig. 1; see also Table S1 in the supplemental material). When PCR products of unexpected sizes were obtained, plasmid DNA was extracted and digested with BamHI and EcoRI and/or sequenced. When two or three fragments were obtained, each fragment was cloned into the unique restriction sites EcoRI and BamHI of plasmid pSU38ΔtotlacZ, and the sequence of each fragment was analyzed.
Chloramphenicol susceptibility testing and quantification of catB9 transcripts.
Chloramphenicol MICs were determined at least three times for each integron-containing E. coli strain according to CLSI recommendations (http://www.clsi.org).
Total RNA was extracted with the FastRNA Pro Blue kit (Q-BIOgene, Illkirch, France) by following the manufacturer's recommendations. Contaminating DNA was removed from RNA samples by using the Turbo DNA-free kit (Ambion, Courtaboeuf, France). cDNAs were synthesized from 1 μg of DNase-treated total RNA by using degenerate primers and Superscript III reverse transcriptase (Invitrogen, Cergy-Pontoise, France). cDNA was quantified in an Mx3005P qPCR system (Agilent Technologies) using the LightCycler FastStart DNA Master Hybridization Probes mix (Roche) according to the supplier's instructions, with appropriate oligonucleotides catB9LC1, catB9LC2, dxsLC1, and dxsLC2 and dxs and catB9 probes (see Table S1 in the supplemental material). Three independent experiments were performed, each in triplicate. Expression of the catB9 gene was estimated by normalizing absolute transcript values to those of the housekeeping gene dxs and by comparing catB9 expression to that in the native integron (DH5α plus p1S), used as calibrator.
RESULTS
Construction of the integron and chloramphenicol susceptibility.
Up to 10 gene cassettes have been described for class 1 integrons (GenBank accession number DQ112222). Each gene cassette can move independently from the others through site-specific recombination catalyzed by the integrase. To investigate the different modes of gene cassette rearrangement under antibiotic selective pressure, we used a chimeric integron in order to obtain an array of at least four gene cassettes. The gene cassettes encoded different mechanisms of resistance to different antibiotic families, thereby avoiding bias during antibiotic selective pressure. We chose four gene cassettes amplified from clinical strains harboring a class 1 integron. This integron possesses all the features of wild-type integrons; the attC sites required for recombination are conserved, as are the open reading frames encoding antimicrobial resistance.
We determined, as controls, the level of catB9-encoded Cm resistance in E. coli cells containing p1S or p2S and their derivatives, by determining MICs and by quantifying catB9 transcripts (see Table S2 in the supplemental material). As expected, higher MICs were obtained when catB9 was in the first position and the level of resistance differed according to the Pc variant as previously described. No difference with the LexA-binding site mutants was observed, confirming the lack of influence of intI1 transcription on gene cassette expression (12).
Chloramphenicol selection pressure assays.
Three different experiments were performed: one under wild-type conditions where integrase expression was repressed by LexA and two in which the integrase was expressed either under the control of its own promoter containing a mutation of the LexA-binding site (derepression) or by overexpression in trans by an inducible vector. Durations of the experiments and Cm concentrations reached each day of the assays are shown in Table 2. At the end of the protocol, bacteria were able to grow at Cm concentrations representing 10 times the initial MIC.
TABLE 2.
Chloramphenicol selection pressure assaysa
| Integron | Cm concn (mg/liter) on day: |
Level of gene cassette rearrangementsd | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| Integrons 1S (initial Cm MIC: 32 mg/liter) | ||||||
| p1S (PcS, IntI1R32_N39) | 50 | 75 | 100 | 300 | 400 | + |
| p1S + pBad-intI1*P32_H39 | 100 | 250 | 400 | + | ||
| p1S + pBad-intI1*R32_H39 | 100 | 300 | 400 | +++ | ||
| p1S + pBad-intI1*R32_N39b | 100 | 300 | 400 | + | ||
| p1SLc | 100 | 150 | 400 | + | ||
| p1SintI1Y312F | 50 | 75 | 100 | 150 | − | |
| Integrons 1W (initial Cm MIC: 8 mg/liter) | ||||||
| p1W (PcW, IntI1R32_H39) | 5 | 5 | 15 | 75 | + | |
| p1W + pBad-intI1*P32_H39 | ND | ND | ND | ND | ND | ND |
| p1W + pBad-intI1*R32_H39b | 5 | 10 | 75 | +++ | ||
| p1W + pBad-intI1*R32_N39 | ND | ND | ND | ND | ND | ND |
| p1WLc | 5 | 15 | 60 | + | ||
| Integrons 1WTGN-10 (initial Cm MIC: 16 mg/liter) | ||||||
| p1WTGN-10 (PcWTGN-10, IntI1P32_H39) | 25 | 25 | 100 | 250 | 350 | + |
| p1WTGN-10 + pBad-intI1*P32_H39b | 25 | 100 | 250 | 350 | + | |
| p1WTGN-10 + pBad-intI1*R32_H39 | 50 | 150 | 350 | +++ | ||
| p1WTGN-10 + pBad-intI1*R32_N39 | ND | ND | ND | ND | ND | ND |
| p1WTGN-10Lc | 25 | 50 | 300 | + | ||
Values indicate Cm concentrations reached in BHI broth each day of the protocol using DH5α strains. The native integron organization, without intI1 overexpression, and its corresponding values are presented in bold. Each experiment was performed at least 3 times. ND, not determined.
intI1 overexpression was obtained with the same integrase as those present in the synthetic integron.
“L” indicates a mutated LexA-binding site.
To locate the position of the catB9 gene cassette within the integron, 100 colonies were analyzed each day by PCR and sequencing. Symbols: +, single rearrangement with integron intI1-aac(6′)-Ib-catB9; +++, multiple rearrangements; −, no rearrangement.
Surprisingly, under wild-type conditions, when the integrase was repressed by LexA, we obtained a rearrangement at day 3 of Cm selection pressure, creating an integron with two gene cassettes, intI1-aac(6′)-Ib-catB9, regardless of the Pc variant and the IntI1 variant. The only difference between the integrons used was the Cm concentration reached according to the Pc variant, from 15 mg/liter for the weak variant PcW to 100 mg/liter for stronger variants (PcS and PcWTGN-10) on day 3 (Table 2). However, the initial integron with the four gene cassettes was still present in all studied colonies, and we failed to separate this integron from the aac(6′)-Ib-catB9-containing integron, suggesting the presence of cointegrates.
When the integrase was expressed either under the control of its own promoter with a mutation in the LexA-binding site or by overexpression in trans, the maximal Cm concentrations reached were identical to those obtained under wild-type conditions but occurred 1 or 2 days earlier, whatever the IntI1 variant (Table 2). When constitutively expressed from their own promoter, the three IntI1 variants gave the same rearrangement as that observed without forced intI1 expression in trans, i.e., intI1-aac(6′)-Ib-catB9. The dynamics of rearrangement differed when the promoter was constitutively expressed or repressed by LexA: for example, with DH5α plusp1SL, colonies harboring the gene cassette rearrangement represented 1% on day 1, up to 95% on day 2, and 100% on day 3, whereas with p1S the rearrangement occurred only on day 3, representing 66% of colonies on day 3 and 100% on days 4 and 5.
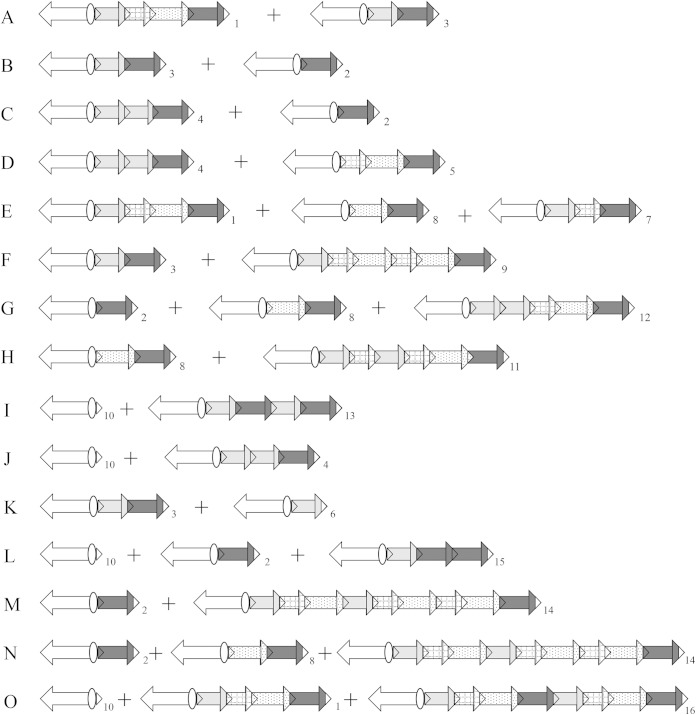
When overexpressed from the strong inducible promoter pBad, the same unique rearrangement was obtained, with IntI1*R32_N39 and IntI1*P32_H39 with comparable dynamics (5% of gene cassette rearrangement on day 1 and up to 95% on day 2 in all experiments), whereas IntI1*R32_H39 overexpression led to multiple gene cassette rearrangements from the first day (95% of colonies), whatever the initial integron (1S, 1W, or 1WTGN-10) (Table 2). These multiple rearrangements corresponded to gene cassette excisions, always moving catB9 closer to Pc, and/or to catB9 duplication (Fig. 2). The observed rearrangement in 60% of colonies on day 1 was the same as that obtained without intI1 overexpression, i.e., aac(6′)-Ib-catB9, but we detected up to 15 different gene cassette rearrangements (Fig. 2). However, most of the DH5α strains contained 2 or 3 different integrons, probably due to gene cassette recombination between two plasmid copies, leading to cointegrates, as previously shown (21, 22).
FIG 2.
Example of multiple gene cassette rearrangements observed under Cm selection pressure, starting with DH5α plus p1S plus pBad-intI1*R32_H39. Integron 1 represents the initial integron intI1-aac(6′)-Ib-dfrA15-aadA1-catB9; the intI1 gene is represented by a white arrow, the aac(6′)-Ib gene cassette by a gray arrow, the dfrA15 gene cassette by a cross-hatched arrow, the aadA1 gene cassette by a stippled arrow, and catB9 gene cassette by a black arrow. For example, integron 2 harbors the gene cassette catB9 (black arrow), and integron 3 harbors the two gene cassettes aac(6′)-Ib and catB9 (gray arrow followed by a black arrow). In this assay, after gene cassette rearrangement, all colonies harbored at least 2 different integrons. The different rearrangements are numbered arbitrarily from 3 to 16. The letters (A to O) correspond to integron combinations observed within a given strain.
Expression of the catB9 gene.
We then individually cloned into pSU38ΔtotlacZ the rearranged integrons obtained under Cm selective pressure from the initial integron 1S with intI1 overexpression, yielding p3S to p16S (Table 1), and analyzed catB9 gene expression relative to that observed with the native integron in p1S (Table 3). Cm MICs ranged from 16 to 256 mg/liter and showed no clear relation to the position of catB9, whereas transcript numbers gradually increased (from 0.18 to 26.4 copies per cell), and a good correlation was found with the proximity of catB9 to Pc and/or with the number of catB9 gene cassettes (Table 3).
TABLE 3.
Comparison of Cm MICs and catB9 transcripts in E. coli DH5α strains according to the position of catB9 in the integrona
| Integron | Relative quantity of catB9 transcripts | Cm MIC (mg/liter) |
|---|---|---|
| p2S (intI1-catB9) | 26.40 ± 3.54 | 256 |
| p15S (intI1-catB9-catB9) | 14.95 ± 1.48 | 256 |
| p13S [intI1-aac(6′)-Ib-catB9-aac(6′)-Ib-catB9] | 8.18 ± 0.81 | 256 |
| p3S [intI1-aac(6′)-Ib-catB9] | 6.78 ± 0.21 | 256 |
| p4S [intI1- aac(6′)-Ib -aac(6′)-Ib-catB9] | 3.78 ± 0.24 | 256 |
| p5S (intI1-dfrA15-aadA1-catB9) | 3.54 ± 0.54 | 128 |
| p8S (intI1aadA1-catB9) | 3.40 ± 0.46 | 128 |
| p7S [intI1-aac(6′)-Ib-dfrA15-catB9] | 2.07 ± 0.28 | 128 |
| p1S [intI1-aac(6′)-Ib-dfrA15-aadA1-catB9] | 1 | 32 |
| p16S [intI1-aac(6′)-Ib-dfrA15-aadA1-catB9-aac(6′)-Ib-dfrA15-aadA1-catB9] | 0.95 ± 0.08 | 32 |
| p9S [intI1-aac(6′)-Ib-dfrA15-aadA1-dfrA15-aadA1-catB9] | 0.49 ± 0.11 | 32 |
| p11S [intI1-aac(6′)-Ib-dfrA15-aac(6′)-Ib-dfrA15-aadA1-catB9] | 0.32 ± 0.02 | 32 |
| p12S [intI1-aac(6′)-Ib-aac(6′)-Ib-dfrA15-aadA1-catB9] | 0.27 ± 0.01 | 32 |
| p14S [intI1-aac(6′)-Ib-dfrA15-aadA1-aac(6′)-Ib-dfrA15-aadA1-dfrA15-aadA1-catB9] | 0.18 ± 0.05 | 16 |
| p6S [intI1-aac(6′)-Ib] | 0 | 4 |
| p10S (intI1) | 0 | 4 |
Strains are classified according to their quantities of catB9 transcripts relative to DH5α plusp1S (underlined) considered as the calibrator strain.
DISCUSSION
The aim of this study was to determine if the class 1 integrase is able to catalyze gene cassette rearrangements within an integron under antibiotic selective pressure and under different conditions of gene cassette and intI1 expression. In our model, whatever the conditions of Cm selection pressure, the catB9 gene cassette always succeeded in being closer to Pc, even when the integrase was not overexpressed. The latter results were somewhat unexpected, considering that intI1 expression is repressed by LexA and that, in previous studies, integrase overexpression was systematically required to obtain gene cassette excision or integration at detectable or measurable frequencies (21–23). It might be argued that under our wild-type conditions, intI1 expression could have resulted from Cm induction of the SOS response, but while Cm indeed induces the SOS response in Vibrio cholerae, this is not the case in E. coli (24). Moreover, we used E. coli strain DH5α, which is DrecA, thus preventing SOS response induction. We thus showed that the integrase was able to catalyze gene cassette rearrangement at a very low frequency even when the SOS response was not induced. A possible integrase-independent mechanism, such as slippage (20), could not be ruled out. We thus repeated the experiment with the inactive integrase IntI1Y312F (p1SintI1Y312F) and found that (i) the Cm concentrations reached on day 4 were lower than with p1S and (ii) no rearrangement occurred (Table 2). Our results thus suggest that the very weak expression which has been observed previously from the promoter of intI1, despite LexA repression (12), might be sufficient for gene cassette rearrangement. On the other hand, with strains carrying the LexA binding-site mutation or the pBad-inducing intI1 overexpression vector, the time required to reach the highest Cm concentrations and gene cassette rearrangements was shortened by 1 or 2 days, whatever the integrase variant (Table 2). Few studies have analyzed the dynamics of gene cassette rearrangement, and these have been carried out mainly under conditions of integrase overexpression (21, 23). We provide comparative data on the ability of the integrase to trigger rearrangements when IntI1 is repressed by LexA and when IntI1 is derepressed or overexpressed. We found that the dynamics of gene cassette rearrangement increased when IntI1 was expressed at a higher level. Even though this might seem logical, it had never previously been shown experimentally.
All our assay conditions, except those in which IntI1*R32_H39 was overexpressed from pBad, led to the same unique rearrangement, intI1-aac(6′)-Ib-catB9, resulting from excision of both the dfrA15 and aadA1 gene cassettes. This is in keeping with our previous data on recombination efficiency showing that IntI1R32_N39 andIntI1P32_H39 were poorly efficient (8, 12). On the other hand, IntI1R32_H39 is known to be the most active integrase (8), and its overexpression led to many gene cassette rearrangements (up to 15 identified) (Fig. 2), whatever the Pc variant present in the initial integron (Table 2).
Most of the rearrangements observed in this study resulted from multiple reordering events, including gene cassette duplication. The main rearrangements likely occurred through the formation of cointegrates. Previous studies have shown that plasmid cointegrates and duplicated gene cassettes are the main IntI1-mediated recombination products (21, 22). These duplications could also been explained by the probability that the attC × attC recombination process is replicative, leading to replication of the original substrate (15). We never observed that catB9 excised and reintegrated in first position, in front of the aac(6′)-Ib, dfrA15, and aadA1 gene cassettes. This could be explained by the fact that gene cassette excision and cointegrate formation, leading to the intI1-aac(6′)-Ib-catB9 rearrangement, require only one recombination event. This recombination event could be either an intramolecular event (excision event on the same plasmid) between the two attC sites of the aac(6′)-Ib and aadA1 gene cassettes or an intermolecular event (between two copies of the plasmid, generating cointegrates) between the attC site of the aadA1 gene cassette carried by one plasmid copy and the attC site of the aac6′-Ib gene cassette carried by another plasmid copy. In contrast, the cut-and-paste mechanism leading to catB9 excision and reintegration in the first position would require two recombination events. Previous observations show that single reordering events are more frequent than multiple ones (25).
The intI1-aac(6′)-Ib-catB9 rearrangement yielded high-level resistance to Cm in DH5α as strains carrying an integron with catB9 in the first position (Table 3). In most of the observed rearrangements, the aac(6′)-Ib gene cassette remained in first position (Fig. 2). This could be explained by a lower efficiency of the integrase to excise a gene cassette located in first position; indeed, attI × attC recombinations occur at lower frequencies than attC × attC recombinations (21). Another explanation lies in the structure of the folded bottom strand of the attC site, with the presence of extrahelical bases (17). IntI1 is more efficient for gene cassette excision using T-N6-G or T-N6-C motifs (22, 26), and this was the case in our integron for both dfrA15 attC (T-N6-C) and aadA1 attC (T-N6-G) sites, whereas aac(6′)-Ib attC and catB9 attC sites contain C-N6-G motifs for which no excision activity has been observed with IntI1 (22, 26). Furthermore, in our model, the large size of the catB9 attC site (125 bp) might also contribute to the relatively low recombination efficiency of catB9. Indeed, in their model with blaVEB-1 and aadB gene cassettes, Aubert et al. demonstrated that both an optimal motif (like T-N6-G) and a short variable terminal structure (VTS) of the attC site (close to 60 bp) are needed to obtain detectable gene cassette rearrangements (22). Moreover, the fact that the aadA1 and dfrA15 gene cassettes were rarely excised separately might be due to the imperfect complementarity between the 1R (GTTAAAC) and 1L (GTCTAAC) sites of aadA1, as previously shown for other gene cassettes (22). The dynamics of gene cassette shuffling is likely largely dependent on the number of gene cassettes within the gene cassette array and on the attC site efficiency of the gene cassettes. Our data were obtained in a model of four gene cassettes with attC sites of different recombination efficiencies. Even if these conditions likely exist in natural integrons, the observed dynamics of gene cassette rearrangement cannot be generalized to all integrons, and further experiments with other gene cassettes and gene cassette arrays will be needed.
Analysis of catB9 expression showed, for all rearrangements, that relocation of catB9 closer to Pc always enhanced Cm resistance (Table 3). As shown in other studies, the closer the gene cassette was to Pc, the more strongly the gene was expressed (9).
In vivo gene cassette rearrangement was recently described for a class 1 integron in Pseudomonas aeruginosa after induction of integrase expression by metronidazole, an SOS-inducing antibiotic, leading to the excision of a gene cassette and allowing the expression of a downstream β-lactamase-encoding gene cassette (27). However, this work did not explore the dynamics of gene cassette shuffling. Our work reinforced this in vivo finding, as this was the first study to analyze, under antibiotic selective pressure, the modes and dynamics of the movement of an unexpressed or weakly expressed gene cassette located in a position distal to Pc. In our experimental model, we showed that (i) the class 1 integrase was able to catalyze gene cassette rearrangement even when its promoter was repressed by LexA, (ii) rearrangement of the catB9 gene cassette closer to Pc was mainly due to movements of the upstream gene cassettes through excision and not to the cut-and-paste movement of the catB9 gene cassette itself, and (iii) only the IntI1*R32_H39 integrase was able to promote multiple rearrangements.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from Ministère de l'Enseignement Supérieur et de la Recherche and Institut National de la Santé et de la Recherche Médicale (INSERM).
We thank Didier Mazel, Thomas Jové, and Sandra Da Re for helpful discussions and Didier Mazel for the kind gift of the catB9-containing strain and p8741 plasmid.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02455-14.
REFERENCES
- 1.Stokes HW, Hall RM. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol 3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 2.Cambray G, Guerout AM, Mazel D. 2010. Integrons. Annu Rev Genet 44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 3.Partridge SR, Tsafnat G, Coiera E, Iredell JR. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev 33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 4.Mazel D. 2006. Integrons: agents of bacterial evolution. Nat Rev Microbiol 4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 5.Daikos GL, Kosmidis C, Tassios PT, Petrikkos G, Vasilakopoulou A, Psychogiou M, Stefanou I, Avlami A, Katsilambros N. 2007. Enterobacteriaceae bloodstream infections: presence of integrons, risk factors, and outcome. Antimicrob Agents Chemother 51:2366–2372. doi: 10.1128/AAC.00044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leverstein-van Hall MA, Blok MH, Donders TA, Paauw A, Fluit AC, Verhoef J. 2003. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J Infect Dis 187:251–259. doi: 10.1086/345880. [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Freijo P, Fluit AC, Schmitz FJ, Grek VS, Verhoef J, Jones ME. 1998. Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother 42:689–696. doi: 10.1093/jac/42.6.689. [DOI] [PubMed] [Google Scholar]
- 8.Jové T, Da Re S, Denis F, Mazel D, Ploy MC. 2010. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet 6:e1000793. doi: 10.1371/journal.pgen.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collis CM, Hall RM. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother 39:155–162. doi: 10.1128/AAC.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, Gonzalez-Zorn B, Barbe J, Ploy MC, Mazel D. 2009. The SOS response controls integron recombination. Science 324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 11.Erill I, Campoy S, Barbe J. 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev 31:637–656. doi: 10.1111/j.1574-6976.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- 12.Guérin E, Jove T, Tabesse A, Mazel D, Ploy MC. 2011. High-level gene cassette transcription prevents integrase expression in class 1 integrons. J Bacteriol 193:5675–5682. doi: 10.1128/JB.05246-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loot C, Bikard D, Rachlin A, Mazel D. 2010. Cellular pathways controlling integron cassette site folding. EMBO J 29:2623–2634. doi: 10.1038/emboj.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacDonald D, Demarre G, Bouvier M, Mazel D, Gopaul DN. 2006. Structural basis for broad DNA-specificity in integron recombination. Nature 440:1157–1162. doi: 10.1038/nature04643. [DOI] [PubMed] [Google Scholar]
- 15.Loot C, Ducos-Galand M, Escudero JA, Bouvier M, Mazel D. 2012. Replicative resolution of integron cassette insertion. Nucleic Acids Res 40:8361–8370. doi: 10.1093/nar/gks620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouvier M, Demarre G, Mazel D. 2005. Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J 24:4356–4367. doi: 10.1038/sj.emboj.7600898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouvier M, Ducos-Galand M, Loot C, Bikard D, Mazel D. 2009. Structural features of single-stranded integron cassette attC sites and their role in strand selection. PLoS Genet 5:e1000632. doi: 10.1371/journal.pgen.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ploy MC, Courvalin P, Lambert T. 1998. Characterization of In40 of Enterobacter aerogenes BM2688, a class 1 integron with two new gene cassettes, cmlA2 and qacF. Antimicrob Agents Chemother 42:2557–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demarre G, Frumerie C, Gopaul DN, Mazel D. 2007. Identification of key structural determinants of the IntI1 integron integrase that influence attC × attI1 recombination efficiency. Nucleic Acids Res 35:6475–6489. doi: 10.1093/nar/gkm709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loot C, Parissi V, Escudero JA, Amarir-Bouhram J, Bikard D, Mazel D. 2014. The integron integrase efficiently prevents the melting effect of Escherichia coli single-stranded DNA-binding protein on folded attC sites. J Bacteriol 196:762–771. doi: 10.1128/JB.01109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collis CM, Hall RM. 1992. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol 174:1574–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aubert D, Naas T, Nordmann P. 2012. Integrase-mediated recombination of the veb1 gene cassette encoding an extended-spectrum beta-lactamase. PLoS One 7:e51602. doi: 10.1371/journal.pone.0051602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collis CM, Grammaticopoulos G, Briton J, Stokes HW, Hall RM. 1993. Site-specific insertion of gene cassettes into integrons. Mol Microbiol 9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 24.Baharoglu Z, Mazel D. 2011. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrob Agents Chemother 55:2438–2441. doi: 10.1128/AAC.01549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bikard D, Julie-Galau S, Cambray G, Mazel D. 2010. The synthetic integron: an in vivo genetic shuffling device. Nucleic Acids Res 38:e153. doi: 10.1093/nar/gkq511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larouche A, Roy PH. 2011. Effect of attC structure on cassette excision by integron integrases. Mob DNA 2:3. doi: 10.1186/1759-8753-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hocquet D, Llanes C, Thouverez M, Kulasekara HD, Bertrand X, Plesiat P, Mazel D, Miller SI. 2012. Evidence for induction of integron-based antibiotic resistance by the SOS response in a clinical setting. PLoS Pathog 8:e1002778. doi: 10.1371/journal.ppat.1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.