Abstract
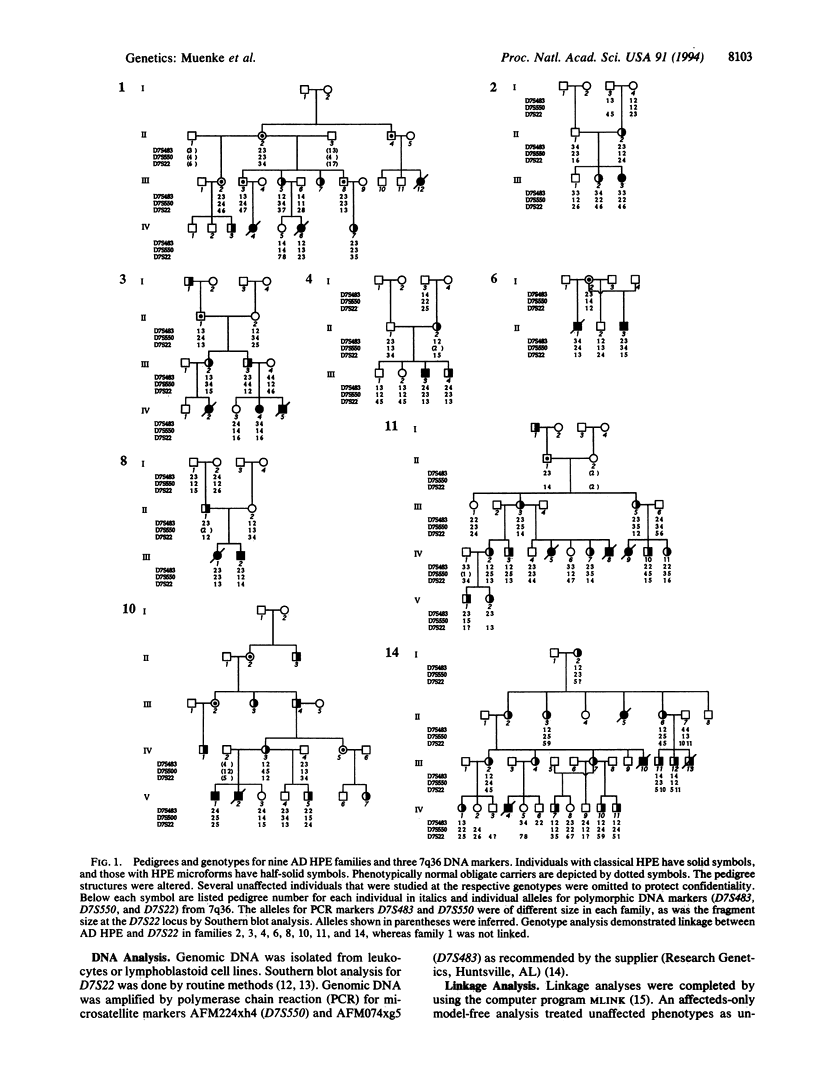
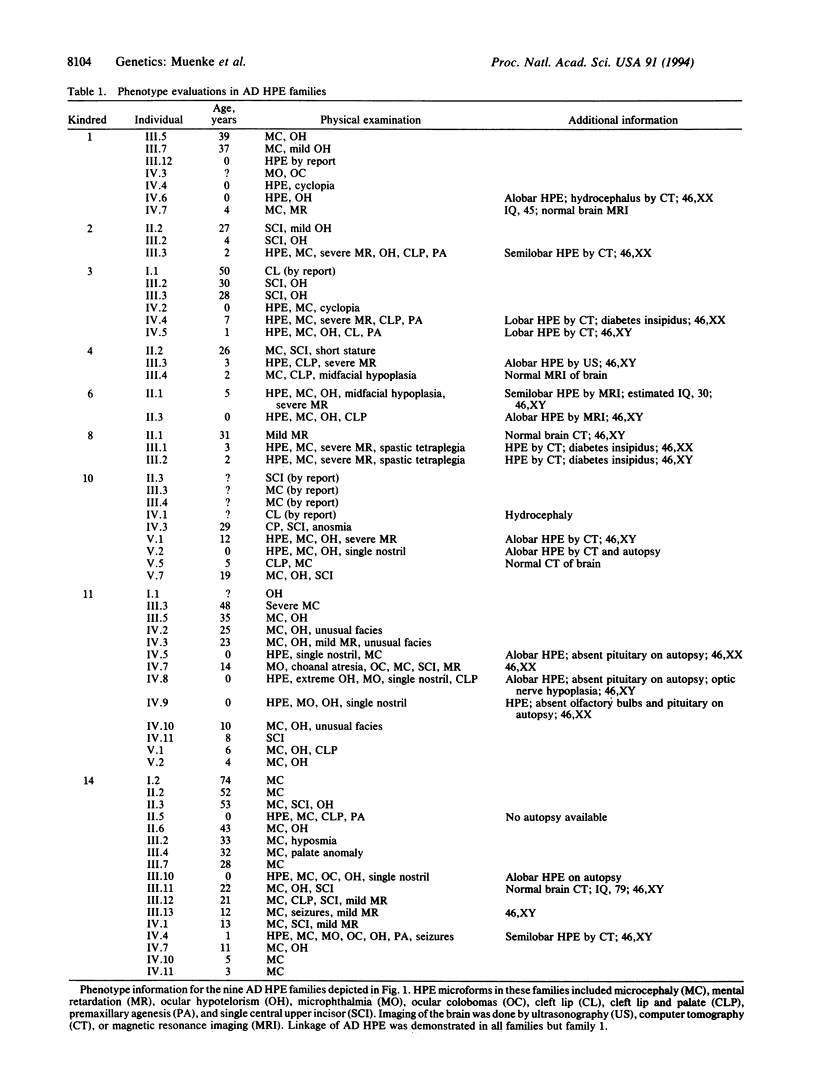
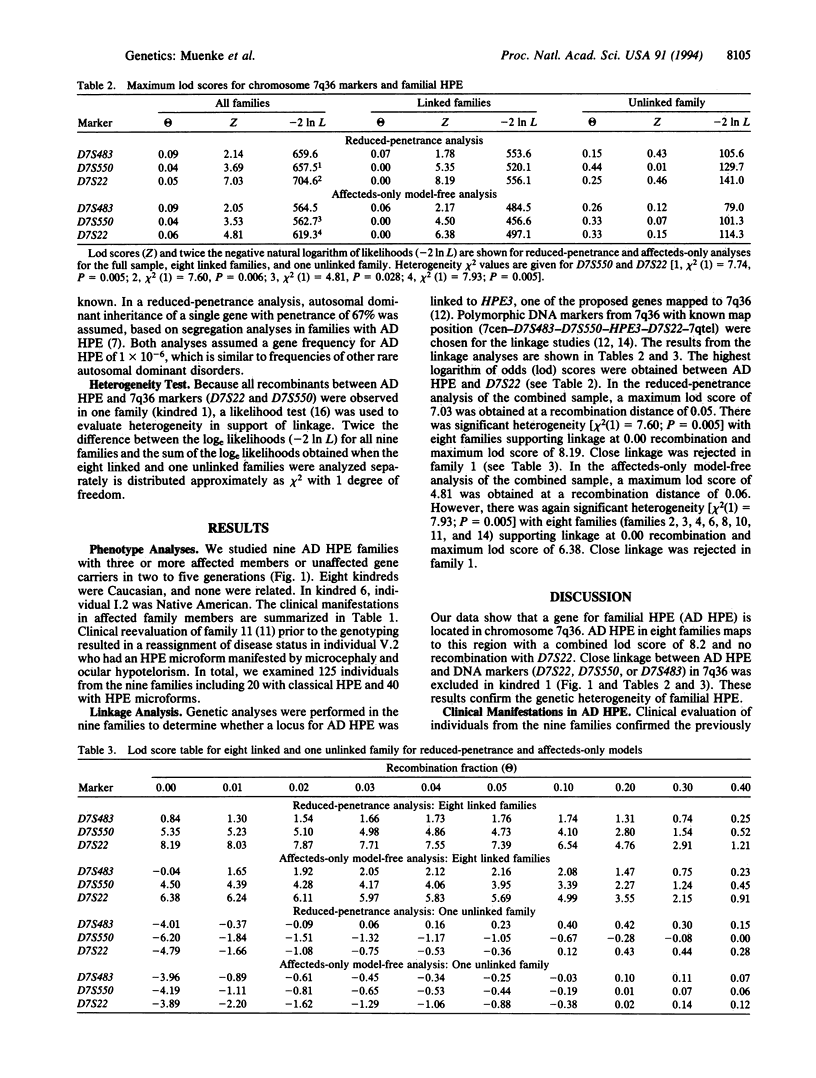
Holoprosencephaly (HPE) is a common malformation of the developing forebrain and midface characterized by incomplete penetrance and variable expressivity. Familial HPE has been reported in many families with autosomal dominant inheritance in some and apparent autosomal recessive inheritance in others. We have examined 125 individuals from nine families with autosomal dominant HPE. Expression in gene carriers varied from alobar HPE and cyclopia through microforms such as microcephaly or single central incisor to normal phenotype. We performed linkage studies by either Southern blot or polymerase chain reaction analyses with DNA markers (D7S22, D7S550, and D7S483) that are deleted from some patients with sporadic HPE and flank a translocation breakpoint in 7q36 associated with HPE. The strongest support for linkage was with D7S22, which was linked with no recombination to autosomal dominant HPE in eight of nine families with a combined logarithm of odds score of 6.4 with an affected-only model-free analysis and 8.2 with a reduced-penetrance model and all phenotypes. Close linkage to this region could be excluded in one family, and there was significant evidence of genetic heterogeneity. These results show that a gene for autosomal dominant HPE is located in a chromosomal region (7q36) known to be involved in sporadic HPE with visible cytogenetic deletions. They also demonstrate genetic heterogeneity in familial HPE. We hypothesize that mutations of a gene in 7q36, designated HPE3, are responsible for both sporadic HPE and a majority of families with autosomal dominant HPE.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ardinger H. H., Bartley J. A. Microcephaly in familial holoprosencephaly. J Craniofac Genet Dev Biol. 1988;8(1):53–61. [PubMed] [Google Scholar]
- Berry S. A., Pierpont M. E., Gorlin R. J. Single central incisor in familial holoprosencephaly. J Pediatr. 1984 Jun;104(6):877–880. doi: 10.1016/s0022-3476(84)80485-0. [DOI] [PubMed] [Google Scholar]
- Cohen M. M., Jr Perspectives on holoprosencephaly: Part I. Epidemiology, genetics, and syndromology. Teratology. 1989 Sep;40(3):211–235. doi: 10.1002/tera.1420400304. [DOI] [PubMed] [Google Scholar]
- Cohen M. M., Jr Perspectives on holoprosencephaly: Part III. Spectra, distinctions, continuities, and discontinuities. Am J Med Genet. 1989 Oct;34(2):271–288. doi: 10.1002/ajmg.1320340232. [DOI] [PubMed] [Google Scholar]
- Cohen M. M., Jr, Sulik K. K. Perspectives on holoprosencephaly: Part II. Central nervous system, craniofacial anatomy, syndrome commentary, diagnostic approach, and experimental studies. J Craniofac Genet Dev Biol. 1992 Oct-Dec;12(4):196–244. [PubMed] [Google Scholar]
- Collins A. L., Lunt P. W., Garrett C., Dennis N. R. Holoprosencephaly: a family showing dominant inheritance and variable expression. J Med Genet. 1993 Jan;30(1):36–40. doi: 10.1136/jmg.30.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurrieri F., Trask B. J., van den Engh G., Krauss C. M., Schinzel A., Pettenati M. J., Schindler D., Dietz-Band J., Vergnaud G., Scherer S. W. Physical mapping of the holoprosencephaly critical region on chromosome 7q36. Nat Genet. 1993 Mar;3(3):247–251. doi: 10.1038/ng0393-247. [DOI] [PubMed] [Google Scholar]
- Hatziioannou A. G., Krauss C. M., Lewis M. B., Halazonetis T. D. Familial holoprosencephaly associated with a translocation breakpoint at chromosomal position 7q36. Am J Med Genet. 1991 Aug 1;40(2):201–205. doi: 10.1002/ajmg.1320400216. [DOI] [PubMed] [Google Scholar]
- Johnson V. P. Holoprosencephaly: a developmental field defect. Am J Med Genet. 1989 Oct;34(2):258–264. doi: 10.1002/ajmg.1320340228. [DOI] [PubMed] [Google Scholar]
- Lathrop G. M., Lalouel J. M., Julier C., Ott J. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3443–3446. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münke M. Clinical, cytogenetic, and molecular approaches to the genetic heterogeneity of holoprosencephaly. Am J Med Genet. 1989 Oct;34(2):237–245. doi: 10.1002/ajmg.1320340222. [DOI] [PubMed] [Google Scholar]
- Weissenbach J., Gyapay G., Dib C., Vignal A., Morissette J., Millasseau P., Vaysseix G., Lathrop M. A second-generation linkage map of the human genome. Nature. 1992 Oct 29;359(6398):794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- Wong Z., Wilson V., Jeffreys A. J., Thein S. L. Cloning a selected fragment from a human DNA 'fingerprint': isolation of an extremely polymorphic minisatellite. Nucleic Acids Res. 1986 Jun 11;14(11):4605–4616. doi: 10.1093/nar/14.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]


