Abstract
Recent studies provided evidence that the macroalga Cladopohora in lakes hosts associated Escherichia coli, with consequences on the environmental and human health. We expanded these investigations to other macroalgae (Ulva spp., Sargassum muticum and Undaria pinnatifida) widespread in the lagoon of Venice (Italy). Attached E. coli were abundant, accounting up to 3,250 CFU gram−1 of alga. Macroalgal-associated isolates belonged to all E. coli phylogroups, including pathogenic ones, and to Escherichia cryptic clades. Attached E. coli showed potential to grow even at in situ temperature on macroalgal extracts as only source of carbon and nutrients, and ability to produce biofilm in vitro. The genotypic diversity of the attached isolates was high, with significant differences between algae and the overlying water. Our evidences suggest that attached populations consist of both resident and transient strains, likely resulting from the heterogeneous input of fecal bacteria from the city. We report that cosmopolitan and invasive macroalgae may serve as source of E. coli, including pathogenic genotypes, and that this habitat can potentially support their growth. Considering the global diffusion of the macroalgae here studied, this phenomenon is likely occurring in other coastal cities worldwide and deserves further investigations from either the sanitary and ecological perspectives.
The fecal indicator bacterium (FIB) Escherichia coli can persist in a variety of secondary habitats, such as water, sand, sediments and others1,2,3,4,5. These studies have suggested that some strains can replicate in the environment, and be reintroduced to the primary host through water and food6. Strains isolated from the environment display different characters than commensal strains from animals or humans7,8,9. In this perspective, there is a human health concern due to potential acquisition of new properties (virulence factors or antibiotic resistances)10 suggesting the need to study the ecology of E. coli in secondary habitats.
E. coli strains can be assigned to 8 phylogroups, seven (A, B1, B2, C, D, E and F) belonging to E. coli sensu stricto11 and one corresponding to the cryptic Escherichia clade I11, which is phylogenetically the closest lineage of E. coli among the five cryptic clades12. The cryptic lineages (II to V) have been identified in the Escherichia genus, which are genetically but not phenotypically distinct from E. coli12,13. Strains belonging to different phylogroups can occupy separate ecological niches and display diverse properties or ability to cause infections11,14. Many commensal strains belong to groups A and B1, whereas E. coli associated with human extraintestinal infections frequently belong to groups B2 and D15,16,17. Phylogroup A and B1 are typically over-represented in waters2,18 and B1 strains are more persistent in soil19. The primary niche of the clades III, IV and V may be outside of the host gut, and they are believed to represent environmentally-adapted Escherichia isolates13,20. Cryptic lineages have been suggested to unlikely represent a significant human risk12,20, but other authors reported that clade V strains isolated from marine sediments show gene repertoires and adhesion properties similar to those of pathogenic strains, suggesting potential virulence9. The existence of “naturalized” populations21, whose distribution in the aquatic environment is understudied, poses questions about the reliability of E. coli as a fecal indicator12.
Marine algae are colonized by abundant microbes22, with a role in the development, defence and metabolism of the plant23. Algal exudates are a nutrient source and contain substrates able to sustain bacterial growth24, such as polysaccharides and glycoproteins25, and algae can provide survival advantages to associated pathogenic microbes26. Studies in lakes showed that Cladophora and wracks can harbour FIB24,27,28,29,30. Macroalgae are a potentially favourable environment for FIB, by providing sites for adhesion, protection against UV radiations and predation, and nutrients. However, information available is limited to the freshwater macroalga Cladophora, while studies have not addressed widely spread macroalgal genera, such as the cosmopolitan Ulva31 or the invasive Undaria and Sargassum32,33.
The Venice lagoon is the largest in Italy (ca. 550 km2) and one of the largest in the Mediterranean Sea34. It is a highly–polluted environment, especially in the area surrounding the city, due to a variety of industrial and agricultural waste, and the presence of domestic wastes deriving from the lack of adequate sewage treatment infrastructures35,36,37. The lagoon has experienced several macroalgal blooms in response to eutrophication34,38. About 300 macroalgal species have been recorded in the area, among which Chlorophyceae, Rhodophyceae, Phaeophyceae and Chrysophyceae39, and there are continuous records of newly–introduced species. The invasive species Sargassum muticum (Yendo) Fensholt and Undaria pinnatifida (Harvey) Suringar, introduced into the lagoon in the early 1990s, are the most abundant invaders colonizing the hard substrata of the historical centre of Venice37. U. pinnatifida is a cold-temperate species originating from Asia, while S. muticum is a temperate species distributed along a large latitudinal range37. Conversely, Ulva species have a nearly ubiquitous distribution in a wide range of environments40. Ulva spp., S. muticum and U. pinnatifida colonize the hard substrates of the lagoon and, especially during spring, the edges of the city canals are largely covered by the algal thalli. S. muticum and U. pinnatifida reach high densities (up to 15 kg f wt m−2 in April–May) and considerable length (up to 7 meters)34,41, creating concern and problems to navigation. Investigating whether they can accumulate FIB appears important, considering the potential evolution of pathogenic strains on their surface, and the spread of contaminated thalli in areas of the lagoon hosting aquaculture farms34 or in bathing beaches located immediately outside the lagoon inlets.
We studied the association between E. coli and the dominant macroalgae in the city of Venice (Fig. 1), with the aim of i) quantifying the abundance of E. coli attached to the macroalgae, to test the hypothesis that seaweeds are sources or reservoirs for E. coli; ii) describing the population structure, by assigning isolates to the phylogenetic group or the cryptic clades; iii) performing experiments to investigate whether macroalgae support the growth of enteric bacteria, and to test the isolates’ ability to produce biofilm in vitro, and iv) describing the genotypic diversity of attached E. coli, to verify whether seaweeds host genetically-distinct types. To our knowledge, this is the first study performed so far to describe the association of E. coli with three common macroalgal genera (Ulva, Sargassum and Undaria) by adopting a temporal and spatial sampling strategy.
Figure 1. Study area and sampling sites.
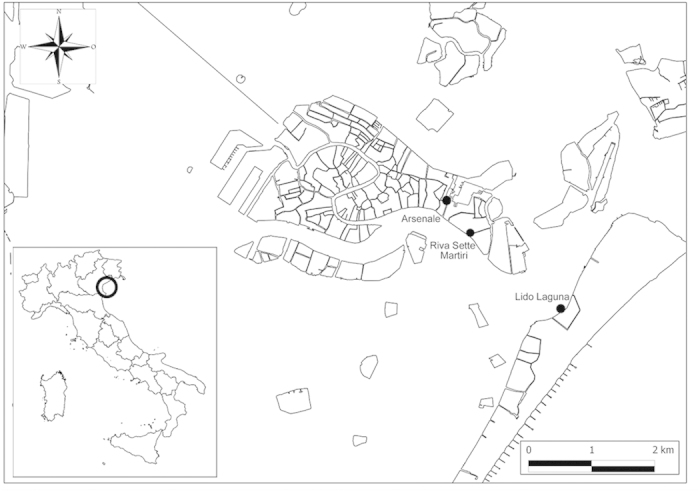
The study area and the sampling sites located around the city of Venice (Italy). The sites were selected on the basis of the presumptive level of fecal contamination. The sites SM and A were located closer to the historical centre of Venice, thus under closer proximity to the sources of fecal bacteria. The LL site was located in the Lido island, at higher distance from the city centre and closer to the Lido inlet, which exchanges water with the sea, thus representing a potentially less contaminated site. The latitude and longitude of the three sites were: 45°25’48.77”N, 12°21’16.56”E (site SM), 45°26’2.24”N, 12°21’0.32”E (site A), 45°25’10.21”N, 12°22’26.38” E (site LL). The map was created with QGIS v2.2.0 software.
Results
E. coli abundance and distribution
E. coli was typically detected in all the investigated macroalgae, and the abundance varied according to the macroalga, sampling site and time (Fig. 2). During the low tide regime, abundance ranged from 5 ± 4 to 605 ± 83 CFU g−1 in Ulva, from 25 ± 21 to 3250 ± 636 CFU g−1 in Sargassum and from 6 ± 3 to 100 ± 71 CFU g−1 in Undaria, while ranged from 2 ± 1 to 416 ± 80 CFU mL−1 in the overlying water. The three-way ANOVA showed a significant interaction between macroalgae, sampling time and site on the E. coli abundance (Table S1). The SM site showed the highest abundance, with higher values observed on February 19th and March 7th. The A site showed lower E. coli contamination in either macroalgae and water. The LL site, the farthest from the city center, showed the lowest concentration of attached E. coli, and a very low abundance in the water (as low as 1 CFU mL−1).
Figure 2. Abundance of E. coli in the three sampling sites during the low tide regime.
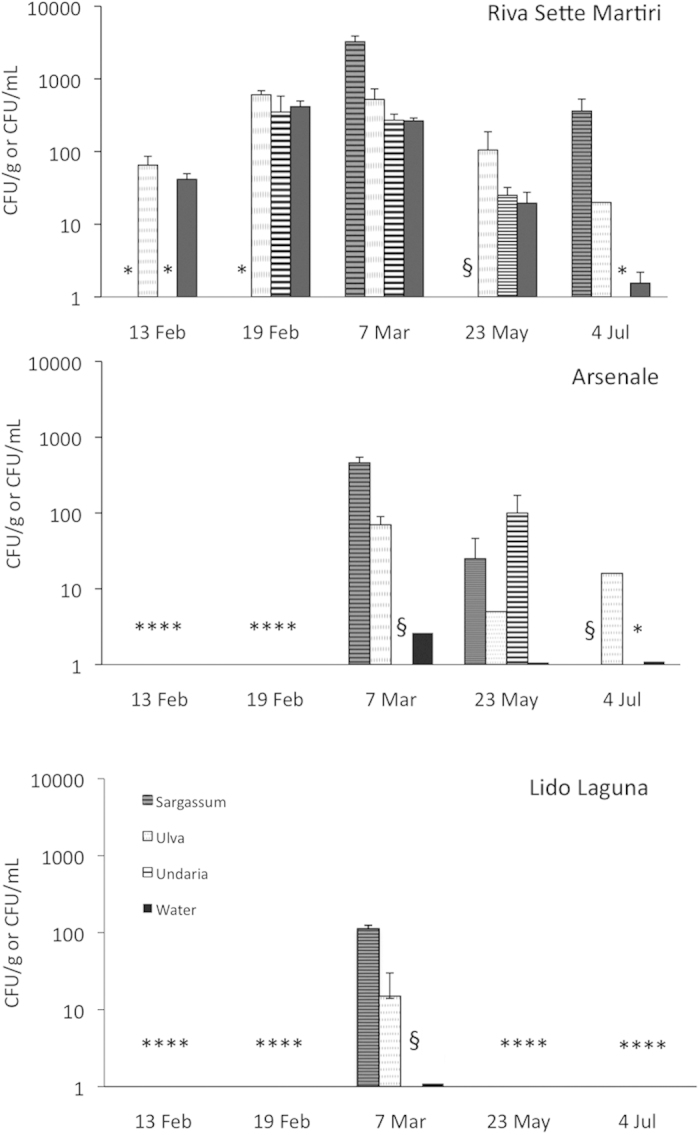
Data represent average ± standard error and are expressed as CFU g−1 (for macroalgae) or mL−1 (for water). * = data not available due to the absence of the macroalga in the sampling site; § = no E. coli growth was observed.
In the SM site, Sargassum generally hosted the highest abundances of E. coli, with values up to 6 times higher than the other macroalgae. Conversely, Undaria showed the lowest abundance. Similarly, in the A site, Sargassum showed the highest abundance of E. coli, while ranged from undetectable to 100 ± 71 CFU g−1 in Undaria. In both SM and A sampling sites, significant differences in E. coli abundances were observed according to sampling times in both water and macroalgae (ANOVA, p < 0.01). In the LL site, representing the less microbiologically polluted site, the higher E. coli concentrations were found in Sargassum.
The abundance of macroalgae-associated E. coli varied according to the tidal regime of the lagoon (Fig. S1), with higher values typically observed during low tide. The same temporal pattern was observed for E. coli abundance in water. Similar patterns were observed in the A site (data not shown). A significant, positive relationship between the E. coli abundance in macroalgae and water was observed (n = 101, R = 0.4169, p < 0.01).
Phylotyping of E. coli isolates
Among the 378 isolates, 287 (76%) were identified as E. coli by the uidA gene amplification. The further characterization of confirmed isolates for their phylogroup of origin showed that the relative abundance of the phylogenetic groups changed among macroalgae and the overlying water (Fig. 3).
Figure 3. Distribution of the E. coli phylogroups in the macroalgae and water samples.
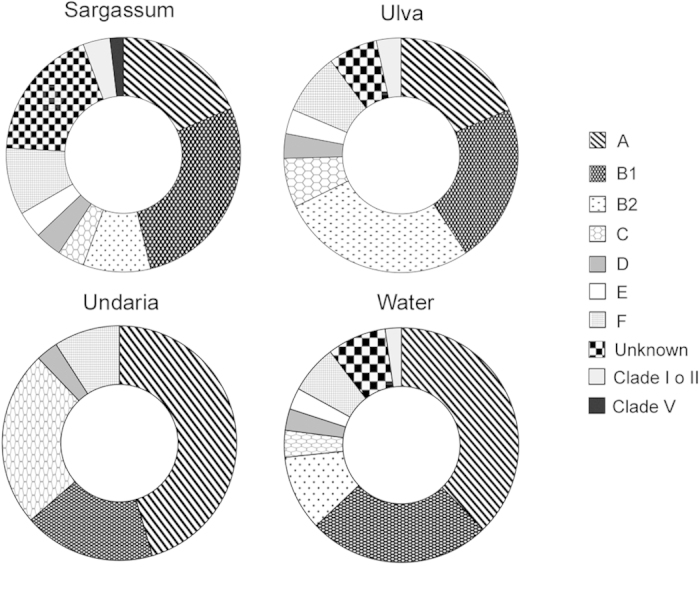
Data are expressed as percentage (%).
In the macroalgal samples, Ulva showed the highest percentage of isolates belonging to the B2 phylogroup (27%). Sargassum showed the highest fraction of isolates (18.5%) which could not be assigned to any phylogroup (defined “unknown”, according to the Clermont et al.11 method and requiring further MLST characterization), while Undaria showed the dominance of the A (46%) and C (24%) and the absence of B2 and E phylogroups. Sargassum hosted prevalently B1 isolates (27.8%), and comparable values of A and “unknown” (18.5% in both cases) isolates. The F phylogroup was observed in all the seaweed types. A low fraction (3.7 and 3.4%, respectively) of isolates from Sargassum and Ulva belonged to the cryptic clades I or II, and one isolate from Sargassum was assigned to the cryptic clade V.
As opposed to macroalgae, the phylogroups A and B1 were more frequently isolated in water (38 and 25%, respectively), followed by B2 (10.4%), F (6.7%) and all the other known groups (11.9%), including a small percentage of clade I or II (2.2%).
When all the isolates were grouped together (Fig. S2), independently from the isolation source (macroalga or water, hereafter defined as “habitat”), the largest fraction belonged to A (31%) and B1 (24.2%), followed by B2 (12.5%), F (7.8%), C (6.8%), D (3.2%) and E (2.9%) phylogroups. A percentage of isolates was “unknown” (8.9%), while a low fraction (ca. 3%) belonged to cryptic clades (I or II, and V).
E. coli ability to grow on macroalgal extracts and produce biofilm
A subset of E. coli isolates, selected on the basis of their habitat of isolation or the sampling site, was used to perform two types of laboratory experiments (growth and biofilm production).
To test the potential of the isolates to multiply on macroalgae, we set-up a medium containing macroalgal extracts as the only source of carbon and nutrients. We tested 6 strains, two of which previously isolated from Sargassum (strain M340 and M349), two from Ulva (M223 and M360), one from Undaria (strain M306) and one from seawater (M378). Each strain was tested on all three macroalgal-based media and at three different temperatures, i.e., 10 °C, 20 °C and 37 °C. All the macroalgal extracts supported the growth of the tested strains under the different temperatures (Fig. 4A–C, Fig. S3A-F), suggesting that E. coli can grow on macroalgae as the only source of carbon and nutrients. However, the growth was not dependent upon the habitat of isolation. At 10 °C and 20 °C, the tested strains showed an overall higher and faster growth on Sargassum extract and, only in some instances, isolates grew better on the extract prepared with the macroalga of isolation (Fig. S3B-C). At 37 °C, E. coli strains grew faster when compared with 10 °C and 20 °C, by reaching the stationary phase within 4–6 hours, and generally showed higher OD600 values in the Ulva extracts. However, these patterns were not consistently observed, and other isolates showed comparable growth on media prepared with two or even three different macroalgae (Fig. 4A–C, Fig. S3A,C). This was especially true at 20 °C while at 10 °C, for most of the tested strains, a higher variability in OD600 values was observed among algal extracts (Fig. S3A-C). The strain isolated from seawater demonstrated ability to grow on all macroalgal media at all tested temperatures (data not shown).
Figure 4. Growth curves on macroalgal extracts at 20 °C for representative E. coli isolates.
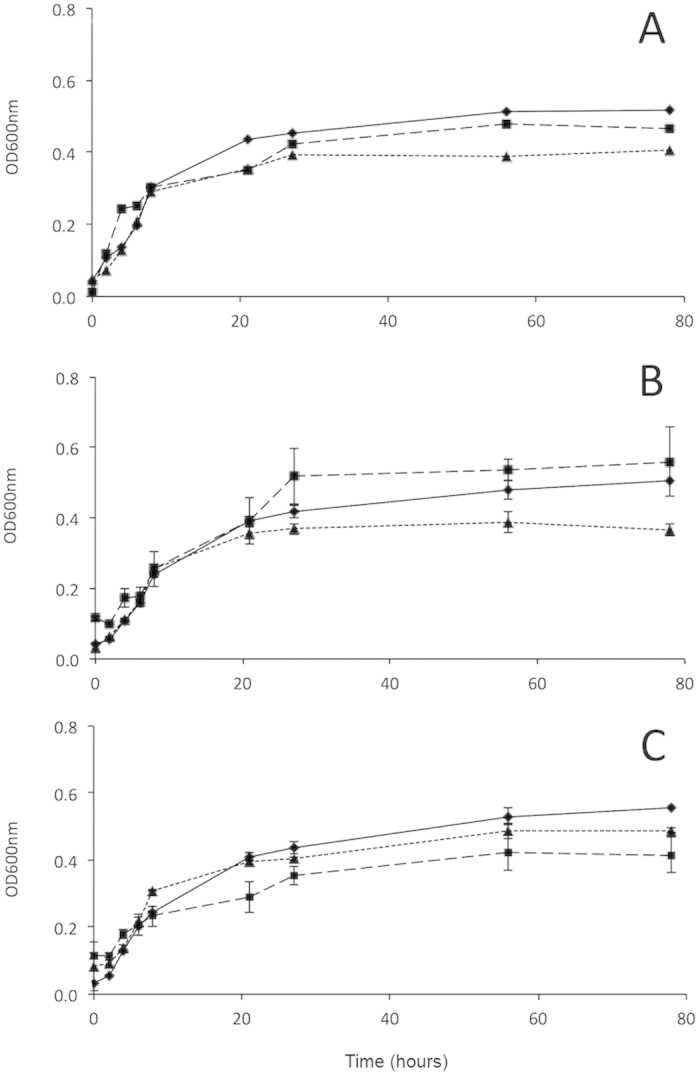
A = strain #223 isolated from Ulva; B = strain #349 isolated from Sargassum; C = strain #306 isolated from Undaria. Triangles refer to the growth on the Undaria medium, while rhombuses to the growth on Sargassum and squares on Ulva. The data are averages ± standard errors of the averages. OD600nm = optical density at 600 nm.
A second subset of isolates, including those previously used for growth experiments, was tested for the ability to form biofilm at 20 °C and 37 °C. We tested 21 isolates from all three seaweed types (1 from Undaria, 7 from Sargassum, 8 from Ulva) and the overlying seawater (5 isolates). The isolates were variably able to form biofilm, ranging from no production to strong production (Fig. 5). At 20 °C, several E. coli isolates showed capability to produce biofilm, evident from the large percentage of strong (12.5%), moderate (6.25%) and weak (68.75%) producers, while only 12.5% of isolates did not form biofilm. At 37 °C, a high but lower percentage of macroalgal isolates was able to produce biofilm (6% strong, 13% moderate and 31% weak producers), while the remaining 50% of isolates did not form biofilm. Most isolates from water did not produce biofilm (60%), and the remaining isolates were weak or moderate producers (20% in both cases). Conversely, no strong biofilm producers E. coli were isolated from water. Similarly, for the water-isolated strains, all non producers isolates observed at 37 °C became weak biofilm producers (for a total of 80%) at 20 °C.
Figure 5. Biofilm producer E. coli strains.
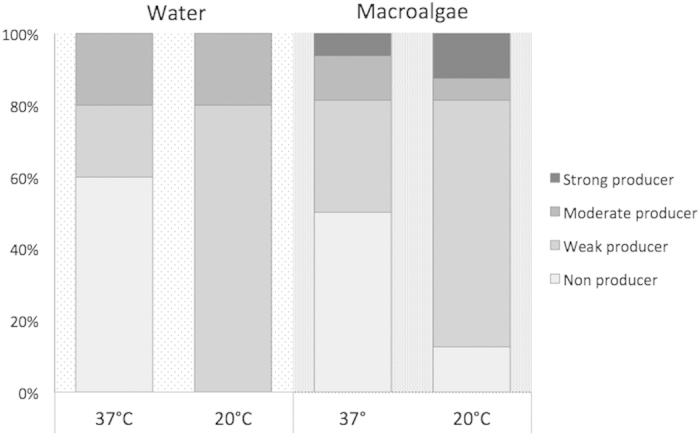
Percentages of biofilm producers in vitro among the E. coli isolated from macroalgae and the overlying water at 37 °C and 20 °C.
Genetic diversity of E. coli isolates
Ninety-nine isolates, selected on the basis of their habitat of isolation and the sampling event, were typed by an improved high-resolution RAPD (Random[ly] Amplified Polymorphic DNA) protocol. This protocol allowed discriminating differences as small as 3–5 base pairs (for fragments ≤ 500 bp), a value much higher than obtained on conventional RAPD protocols on agarose gel. Replicated RAPD analyses performed on the same isolates yielded similarity percentages from 98 to 100% (data not shown). We subsequently used the most conservative value (98%) as value above which two isolates are considered as clones.
The dendrograms divided per habitat and the overall dendrogram are reported in the Supplemental Material (Fig. S4-S5 respectively). The results highlighted a wide genetic diversity of E. coli attached to macroalgae, with very few clonal isolates (only observed in Ulva, e.g. M17–M56, and M212–M223) and a large number of unique genotypes. Similarity values ranged from ca. 6 to 88% in Sargassum, from ca. 14 to 100% in Ulva and 8 to 88% in Undaria. The diversity of E. coli isolated from water samples was similarly very high but, as opposed to seaweeds, showed higher frequency of clonal clusters (e.g. M210–M282–M332).
When all isolates were grouped together (Fig. S5), the ANOSIM analysis showed significant differences in the genetic diversity according to the habitat (macroalgae vs. water; Global R = 0.148, significance level < 0.001), with significant differences observed between Undaria and water (Global R = 0.308, significance level < 0.001), Sargassum and water (Global R = 0.192, significance level < 0.001) and Ulva and water (Global R = 0.102, significance level < 0.001) and no differences among macroalgae (for all comparisons, ns). Conversely, the ANOSIM did not reveal differences in the distribution of E. coli genotypes according to the sampling time or site (ANOSIM, ns). Typically, isolates from the same macroalgae did not group with each other, showing a high genetic diversity even within the same sample. However, identical clones were sometimes found associated with different macroalgae from the same site and sampling event (M237–M223, and M162–M193). Similarly, the same clones were occasionally shared by macroalgae and the water collected in a different site (e.g. M234–M32 and M304–M282–M332). Some genotypically identical isolates were observed in the water collected in different sites (M210–M282–M332).
Discussion
A large body of evidence demonstrated that E. coli can persist outside the hosts, likely due to its high versatility and genetic diversity42. Studying the fate of E. coli in non-enteric habitats, and identifying environmental reservoirs, is crucial to evaluate the potential acquisition of virulence or antibiotic-resistance properties, leading to infections by pathogenic E. coli (such as the O157:H7 serotype) suspected to evolve in the environment43,44. Recent studies identified reservoirs such as beach sands2,45, marine sediments and wetlands3,4,46. Freshwater macroalgae, such as Cladophora, can harbor FIB in lakes24,27,29,47. However, similar studies have not been performed in coastal marine and transitional environments, nor have investigated the role of common macroalgae, such as those belonging to the genera Ulva, Sargassum and Undaria, as E. coli sources.
We demonstrated that E. coli can be associated with live macroalgae other than Cladophora. The abundance was highly variable on both spatial and temporal scales. The brown algae Sargassum muticum and Undaria pinnatifida are invasive species, typically found in temperate to cold-temperate habitats, while green Ulva species have a nearly ubiquitous distribution31. These algal genera display different features potentially influencing bacterial adhesion and survival, including life cycle, light requirement, biochemical composition48 and the surface-to-volume (S/V) ratio. Due to its branching morphology, Sargassum offer higher S/V ratio than the other seaweeds, characterized by a laminar morphology, which may explain the higher concentrations observed in Sargassum. Because of the wide metabolic and genetic versatility and ability to colonize diverse macroalgae, it is likely that also other macroalgal genera may serve as unidentified sources or reservoirs for E. coli. The only studies available for comparison refer to the macroalga Cladophora, and have been performed across the Lake Michigan (USA). The authors report a similar range of abundance, and comparable spatial and temporal variability in E. coli abundance in live and floating macroalgae21,27. Olapade et al.49 reported up to 60,000 CFU 100 g−1 in Cladophora mats in lake shores, while Imamura et al.30 reported that dead macroalgae (algal wracks) stranded in California beaches hosted > 4 log CFU dry g−1.
The E. coli abundance in water showed values comparable to previous findings in the area50. The presence of large populations of E. coli is clearly linked to the lack, in the historical centre of Venice, of modern depuration plants and the input of almost untreated wastes51, despite the efforts in place to provide the city with more efficient sewage treatment plants. The abundance in water changed according to the tidal regime, with higher abundances during low tide. This pattern is possibly driven by the water exchanges between the lagoon and the sea during the high tide, favouring inputs of less contaminated water from the sea. The water circulation in the lagoon is mainly driven by tides (up to 1-m excursion) and wind, and the amount of salt water flowing in and out at each tidal cycle amounts to one third of the lagoon volume52. This pattern was reflected in the macroalgae, which showed higher bacterial abundance during low tide. Moreover, the abundance of algal-attached E. coli was positively related with the abundance in the overlying water. Similar studies reported that E. coli concentrations in Cladophora and lake water were correlated27,53. Englebert et al.54 reported that E. coli abundance was higher in the water above Cladophora mats than in water far away, suggesting release from mats due to wind and wave action. Taken together, these results suggest that, while E. coli may not be a typical member of the epiphytic bacterial flora in non-polluted environments22, it can colonize the macroalgal surface in chronically-polluted areas.
Phylogenetic analysis highlighted the presence of all known phylogroups of E. coli sensu stricto, including the commensal A and B1, the potentially pathogenic B2 and D (causing extra-intestinal infections)55 and the recently described C (distinct but closely related to B1)56,57, E (formerly a set of unassigned strains, of which the O157:H7 is the best known member)11 and F57,58. These results are the first on the occurrence, using the recent method by Clermont et al.11, of C, E and F phylogroups in association with macroalgae and, more broadly, in the aquatic environment. Studies carried to investigate the prevalence of phylogroups have mostly addressed estuarine and freshwater environments59,60 rather than marine ones3. We also highlighted potential phylogroup-macroalgal associations, as in the case of Sargassum being the only seaweed hosting all phylogroups and the cryptic clade V. Conversely, neither cryptic clades nor B2, E and “unknown” isolates were observed in Undaria. This non-random distribution of phylogroups among seaweeds let hypothesize that the different characteristics of the algae may select for certain phylogroups. However, this hypothesis needs to be tested on a larger number of isolates. Phylogrouping analyses highlighted presence of a low percentage of cryptic clades. Limited information is available about cryptic lineages in aquatic environments. They are likely representing environmentally adapted Escherichia lineages12,13 and seem to be more abundant in certain environments, such as sediments9,12. We suggest that the macroalgal habitat may favor the evolution of some Escherichia cryptic clades, which deserves further investigations.
Laboratory experiments performed at in situ and 37 °C temperatures showed that the three macroalgae can, under laboratory conditions, support the growth of E. coli as only source of carbon and nutrients, and that several strains can form biofilms. A similar in vitro ability was demonstrated for fecal Cladophora isolates24. These findings emphasize the role of seaweeds as potential E. coli sources in coastal areas. The ability of different isolates to grow on different algae, and similarly the ability of E. coli isolated from water to grow on macroalgal extracts, suggests that E. coli can adapt to grow on a variety of organic substrates, and pass from water to the macroalgal habitat and viceversa. Finally, the ability of several macroalgal isolates to produce biofilm at different temperatures suggests that a biofilm lifestyle may be common among attached E. coli and raises questions about the potential virulence of environmental E. coli. In the environment, the biofilm lifestyle may confer several ecological advantages, such as higher resistance to predators or UV radiations, and increased resource availability20. Similarly to our findings, Moreira et al.61 reported superior ability to form biofilm by E. coli isolated from freshwater periphyton in comparison with human strains, suggesting that the biofilm lifestyle increased persistence in the environment.
RAPD analyses highlighted a high genotypic diversity of macroalgae-associated E. coli, consistently to previous results47. Our similarity values were in a range similar to those reported, using the REP-PCR fingerprinting technique, in Cladophora mats28,47. Despite the ANOSIM test revealed significant differences in the distribution of E. coli genotypes among macroalgae and water, we didn’t find clear clustering of isolates according with time, site or macroalga. This high genetic heterogeneity suggests that macroalgae are not selecting for specific genotypes, and that the majority of attached genotypes are likely transient members, whose large genotypic diversity reflects the wide variety of sources of fecal contamination from the city, which is visited yearly by more than 10 millions of tourists from worldwide. This agrees with Byappanahalli et al.24, who suggested that the association between E. coli and macroalgae may be not algal-specific, and is supported by our experimental findings that different isolates grow on all the macroalgal types. However, at the same time, we found that some genotypically identical isolates were associated to diverse macroalgae collected from different sites. This would suggest that at least some genotypes can be consistently associated with macroalgae, as later reported by Byappanahalli et al.47 in Cladophora on a larger dataset, and that the association may be advantageous for survival of those genotypes. Further genotypic analyses on a larger dataset will allow to clarify this issue.
This study is of relevance for the city of Venice, where high macroalgal biomasses are present in certain periods of the year. The waters around the city receive large loads of FIB, and are not suitable for bathing. The presence of FIB determines human health hazards through unusual routes of exposure, such as aerosolization of polluted waters by the frequent boats traffic (which may lead to inhalation of pathogens) or the recurrent floods (“acqua alta”) inundating streets and houses and determining waterborne disease exposure scenarios36. This large macroalgal source of FIB represents an additional, so far unrecognized, health hazard and poses extra questions about the spread of waterborne infectious diseases in the area. Macroalgae could serve as a continuous source of fecal and potentially pathogenic bacteria, likely able to alter the water quality of surrounding coastal areas.
We conclude that cosmopolitan and invasive macroalgae can serve as environmental sources or reservoirs of E. coli, including potentially pathogenic strains. Given the wide geographic diffusion of the studied macroalgae and the E. coli ability to colonize different seaweeds, this phenomenon is likely to occur in other coastal cities worldwide and can potentially alter the water quality of shorelines, deserving investigations from either the human health and ecological perspectives.
Methods
Study area and sampling activities
Sargassum muticum (Yendo) Fensholt, Undaria pinnatifida (Harvey) Suringar and Ulva species and overlying water were collected aseptically from three sites (Sette Martiri – SM, Arsenale – A, Lido Laguna – LL; Fig. 1 and S6). Samples were collected typically once per month, with the exception of February when samples were collected twice to firstly evaluate the differences under a shorter temporal scale (February 13th and 19th, March 7th, May 23th and July 4th 2013). Samples were collected twice during both low and high tide, except on February. Live macroalgal samples (attached to substrates such as canal embankments or docks) were collected and analysed (two replicates) using sterilized tweezers to avoid contamination. Seaweeds were put into sterile containers, maintained at 4 °C and analysed within 2–6 hours from sampling. One liter of water overlying the macroalgae was put into sterile bottles and stored at 4 °C in the dark until processing as above.
Abundance of E. coli in macroalgae and water
Aliquots of 1 gram of macroalgae were transferred into sterile tubes and suspended into 9 mL of sterile 0.8% NaCl. For bacterial detachment, similarly to previous studies on macroalgae24,27,47, samples were vigorously shaken and sonicated (3 times, 1 minute each cycle with 30 sec intervals within cycles). Serial ten-fold dilutions (1:10 and sometimes 1:100) were performed. Aliquots of 1 mL of the supernatant and the obtained dilutions were filtered and analysed using the Membrane Filtration technique as described previously3. Filters were plated onto mFC agar plates (Biolife) and incubated for 24 hours at 44.5 °C. For water samples, 1, 10 and 100 mL aliquots were filtered (three replicates) and the filters plated and incubated as above. Blue colonies were considered as presumptive E. coli and isolated for further analyses. The abundance of presumptive E. coli was reported as CFU per gram of wet macroalgae or mL of water.
Identification of E. coli isolates
Well-separated presumptive E. coli colonies were streaked on agar for isolation and cultured overnight in Tryptone Soy Broth (Biolife) with 0.3% Yeast Extract (Biolife). Lysates of overnight cultures were used as a DNA template in all PCRs protocols in this study. Identification of presumptive E. coli colonies (n = 378) was performed by amplifying the uidA gene62. PCR products were separated by agarose gel electrophoresis (1%) and visualized with GelRed (Biotium).
Determination of phylogenetic groups and cryptic clades
The 287 Escherichia isolates were analyzed using the method by Clermont et al.11, which allows to assign isolates to the seven phylogroups (A, B1, B2, C, D, E and F) or to the cryptic Escherichia clades. Isolates belonging to cryptic clades I or II and III, IV or V were confirmed by the Clermont et al.57 protocol. PCR products were separated by agarose gel electrophoresis (2%) and visualized with GelRed (Biotium).
Growth of E. coli on macroalgal extracts
Macroalgal extracts were used as growth medium to investigate the ability of E. coli to multiply using algae as only carbon and nutrient source. Ulva spp., Sargassum muticum and Undaria pinnatifida samples were collected and put at −80 °C for 24 hours to kill the epiphytic microbes. This procedure was preferred to other procedures (e.g. autoclaving), which would have altered the algal chemical composition and destroyed important macromolecules for bacterial growth. E. coli strains were inoculated in algal extracts at 10 °C, 20 °C and 37 °C. The first two temperatures were chosen as representative for the late winter (February to April) and spring-summer (May to July) periods on the basis of periodical measurements performed by the local municipality (data not shown). The growth curve at 37 °C was performed as representative of the optimal growth temperature for E. coli. The efficiency of the freezing procedure was tested on uninoculated macroalgal extracts, incubated at the three different temperatures. The tests revealed that extracts did not show growth over a 80 hours period (for the tests carried out at 10 °C and 20 °C), while contamination was detected after about 8 hours when testing the growth at 37 °C. This suggested to perform shorter incubations (e.g. until 6 hours) for experiments at 37 °C to ensure measuring the growth of only the studied strains. To prepare the algal-made medium, each algae was put into a sterile tube and added with sterile 0.8% NaCl (1:10 w/v). This mixture was whisked aseptically with an electric blender for 2 minutes. The supernatant was filtered through a sterile Whatman #3 membrane (to eliminate large particles) and the crude algal extract collected into a sterile bottle. The growth curves for the isolates (six E. coli tested in triplicate) were obtained by measuring spectrophotometrically the optical density (λ = 600 nm) of extracts inoculated with 1% (v/v) of a single strain overnight broth culture (OD600 = 0.9 - 1) and incubated aerobically at 37 °C up to 6 hours.
Biofilm production in vitro
The biofilm production test was performed as described by Vignaroli et al.4 and references therein. Bacterial cells were grown overnight in Luria-Bertani broth (LB) (Oxoid) supplemented with 1% glucose (LBG) at 37 °C and diluted to an OD625 of 0.1. These suspensions (0.2 mL) were incubated overnight at 20 °C and 37 °C in 96-well polystyrene microtiter plates (Falcon, Becton Dickinson Labware). The broth cultures were aspirated and the wells washed three times with 0.2 mL of phosphate-buffered saline (PBS). Microtiters were dried at 60 °C for 1 hour, and stained with 0.1 mL of Hucker’s Crystal Violet (CV) solution (10% crystal violet in 20% ethanol containing 1% ammonium oxalate) for 10 min. CV was aspirated, and wells were washed with sterile water. CV was extracted from adhering bacterial cells by ethyl alcohol/acetone (80:20 v/v), and the OD690 was measured using a microplate reader (Thermo Electron Corporation, Madison, WI). Each assay was performed in triplicate. Strains were classified as non producer (OD ≤ ODc), weak producer (ODc < OD ≤ 2 × ODc), moderate producer (2 × ODc < OD < 4 × ODc) or strong producer (OD > 4 × ODc). The OD cutoff (ODc) was defined as 3 standard deviations above the mean OD of the negative control represented by uninoculated wells containing LBG. The strong biofilm producer S. epidermidis ATCC 35984 was used as the positive control.
High-resolution genotyping
Ninety-nine E. coli strains were genotyped by using RAPD (Random[ly] Amplified Polymorphic DNA) PCR, and the amplicons analyzed using the high-resolution Qiaxcel Advanced capillary electrophoresis System (Qiagen). We selected the RAPD technique which, despite certain limitations, is a useful tool to provide initial insights on the genotypic diversity of large numbers of isolates63,64,65. PCR were performed as described by Regua-Mangia et al.66 and Luna et al.3, using 1 μL of bacterial lysate. Amplification products were purified and analysed using Qiaxcel. A dendrogram was obtained by analyzing electrophoretic peaks on Bionumerics v7.0 software (Applied Maths), using the Dice similarity coefficient and UPGMA (Unweighted Pair Group Method of Averages) as clustering method.
Statistical analyses
Differences in E. coli abundance between macroalgae were assessed using a three-way analysis of variance (ANOVA) carried out on the low-tide abundance dataset. The analysis treated the factor site (S, 2 levels: A and SM sites) as fixed, time (T, 5 levels: February 13th, February 19th, March 7th, May 23th and July 4th) as fixed and crossed with S, and habitat type (H, 4 levels: Ulva, Undaria, Sargassum and water) as random and orthogonal. The LL site was excluded due to the low number of samples. When significant differences (p < 0.05) were observed for fixed factors, a post-hoc Student-Newman-Kuels’ test (SNK) was performed. Prior to the analysis, the homogeneity of variance was checked using the Cochran’s test on appropriately transformed data, whenever necessary. If the transformation did not allow to obtain an homogeneous variance, a more conservative level of significance was considered. ANOVA and SNK tests were carried out using GMAV (University of Sydney). Differences in the genotypic diversity of E. coli isolates from different habitats were assessed on the RAPD dataset using the analysis of similarity (ANOSIM) tool based on a Bray-Curtis similarity matrix. The presence of statistical differences between samples is indicated by a significance level at least p < 0.05. The ANOSIM analysis was performed using the PRIMER 6 + software (http://www.primer-e.com/).
Additional Information
How to cite this article: Quero, G. M. et al. Understanding the association of Escherichia coli with diverse macroalgae in the lagoon of Venice. Sci. Rep. 5, 10969; doi: 10.1038/srep10969 (2015).
Supplementary Material
Acknowledgments
This work was possible thanks to funds to G.M.L. by the programme RITMARE (SP3-WP2-A2 “Strumenti innovativi per la valutazione degli effetti di contaminanti emergenti sulle comunità biologiche”) and the IPA Project “BALMAS” (code 1° STR/0005) funded by EU. We thank Dr. Laura Perini and Dr. Arianna Mazzariol for their precious support at all stages, and Dr. Alessandro Buosi for his grateful assistance during macroalgal sampling and identification.
Footnotes
Author Contributions G.M.L. and L.F. designed the research. G.M.Q. conducted the fieldwork, performed all the experiments and analyzed the data. C.V. performed laboratory experiments on biofilm. G.M.Q. and G.M.L. wrote the manuscript, C.V. and L.F. reviewed and edited the manuscript.
References
- Byappanahalli M. N., Whitman R. L., Shively D. A., Sadowsky M. J. & Ishii S. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8, 504–513 (2006). [DOI] [PubMed] [Google Scholar]
- Walk S. T., Alm E. W., Calhoun L. M., Mladonicky J. M. & Whittam T. S. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9, 2274–2288 (2007). [DOI] [PubMed] [Google Scholar]
- Luna G. M. et al. Extraintestinal Escherichia coli carrying virulence genes in coastal marine sediments. Appl. Environ. Microbiol. 76, 5659–5668 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignaroli C. et al. Epidemic Escherichia coli ST131 and Enterococcus faecium ST17 in coastal marine sediments from an italian beach. Environ. Sci. Technol. 47, 13772–13780 (2013). [DOI] [PubMed] [Google Scholar]
- Sidhu J. P., Hanna J. & Toze S. G. Survival of enteric microorganisms on grass surfaces irrigated with treated effluent. J. Water Health 6, 255–262 (2008). [DOI] [PubMed] [Google Scholar]
- Ishii S. & Sadowsky M. J. Escherichia coli in the environment: implications for water quality and human health. Microb. Environ. 23, 101–108 (2007). [DOI] [PubMed] [Google Scholar]
- Gordon D. M., Bauer S. & Johnson J. R. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148, 1513–1522 (2002). [DOI] [PubMed] [Google Scholar]
- Cohan F. M. & Kopac S. M. Microbial genomics: E. coli relatives out of doors and out of body. Curr. Biol. 21, R587–R589 (2011). [DOI] [PubMed] [Google Scholar]
- Vignaroli C. et al. Adhesion of marine cryptic Escherichia isolates to human intestinal epithelial cells. ISME J. 9, 508–515 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martìnez J. L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902 (2009). [DOI] [PubMed] [Google Scholar]
- Clermont O., Christenson J. K., Denamur E. & Gordon D. M. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65 (2013). [DOI] [PubMed] [Google Scholar]
- Luo C. et al. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc. Natl. Acad. Sci. 108, 7200–7205 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walk S. T. et al. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 75, 6534–6544 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. M., Clermont O., Tolley H. & Denamur E. Assigning Escherichia coli strains to phylogenetic groups: multilocus sequence typing versus the PCR triplex method. Environ. Microbiol. 10, 2484–2496 (2008). [DOI] [PubMed] [Google Scholar]
- Ahmed W. et al. Occurrence of intestinal and extraintestinal virulence genes in Escherichia coli isolates from rainwater tanks in Southeast Queensland, Australia. Appl. Environ. Microbiol. 77, 7394–7400 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrouzian F. L., Adlerberth I. & Wold A. E. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. 8, 834–840 (2006). [DOI] [PubMed] [Google Scholar]
- Gordon D. M., Stern S. E. & Collignon P. J. Influence of the age and sex of human hosts on the distribution of Escherichia coli ECOR groups and virulence traits. Microbiology 151, 15–23 (2005). [DOI] [PubMed] [Google Scholar]
- Power M. L., Littlefield-Wyer J., Gordon D. M., Veal D. A. & Slade M. B. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ. Microbiol. 7, 631–640 (2005). [DOI] [PubMed] [Google Scholar]
- Bergholz P. W., Noar J. D. & Buckley D. H. Environmental patterns are imposed on the population structure of Escherichia coli after fecal deposition. Appl. Environ. Microbiol. 77, 211–219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle D. J. et al. Biofilm formation by and thermal niche and virulence characteristics of Escherichia spp. Appl. Environ. Microbiol. 77, 2695–2700 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S. et al. Cladophora (Chlorophyta) spp. harbor human bacterial pathogens in nearshore water of Lake Michigan. Appl. Environ. Microbiol. 72, 4545–4553 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S. et al. The seaweed holobiont: understanding seaweed–bacteria interactions. FEMS Microbiol. Rev. 37, 462–476 (2013) [DOI] [PubMed] [Google Scholar]
- Tujula N. A. et al. Variability and abundance of the epiphytic bacterial community associated with a green marine Ulvacean alga. ISME J. 4, 301–311 (2010). [DOI] [PubMed] [Google Scholar]
- Byappanahalli M. N. & Shively D. A., Nevers, M. B., Sadowsky, M. J. & Whitman, R. L. Growth and survival of Escherichia coli and enterococci populations in the macro‐alga Cladophora (Chlorophyta). FEMS Microbiol. Ecol. 46, 203–211 (2003). [DOI] [PubMed] [Google Scholar]
- Rahman M. A. & Halfar J. First evidence of chitin in calcified coralline algae: new insights into the calcification process of Clathromorphum compactum. Sci. Rep. 4, 6162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli L., Pruzzo C., Huq A. & Colwell R. R. Environmental reservoirs of Vibrio cholerae and their role in cholera. Environ. Microbiol. Rep. 2, 27–33 (2010). [DOI] [PubMed] [Google Scholar]
- Whitman R. L., Shively D. A. & Pawlik H., Nevers, M. B. & Byappanahalli, M. N. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl. Environ. Microbiol. 69, 4714–4719 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badgley D., Nayak B. S. & Harwood V. J. The importance of sediment and submerged aquatic vegetation as potential habitats for persistent strains of enterococci in a subtropical watershed. Water Res. 44, 5857–5866 (2010). [DOI] [PubMed] [Google Scholar]
- Badgley B. D. et al. 2011. Multi-scale temporal and spatial variation in genotypic composition of Cladophora-borne Escherichia coli populations in Lake Michigan. Water Res. 45:721–731 (2011). [DOI] [PubMed] [Google Scholar]
- Imamura G. J., Thompson R. S., Boehm A. B. & Jay J. A. Wrack promotes the persistence of fecal indicator bacteria in marine sands and seawater. FEMS Microbiol. Ecol. 77, 40–49. (2011) [DOI] [PubMed] [Google Scholar]
- Hayden H. S. et al. Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. Eur. J. Phycol. 38, 277–294 (2003). [Google Scholar]
- Hay C. H. The dispersal of sporophytes of Undaria pinnatifida by coastal shipping in New Zealand, and implications for further dispersal of Undaria in France. Brit. Phycol. J. 25, 301–313 (1990). [Google Scholar]
- Deysher L. & Norton T. A. Dispersal and colonization in Sargassum muticum (Yendo) Fensholt. J. Exp. Mar. Biol. Ecol. 56, 179–195 (1981). [Google Scholar]
- Solidoro C. et al. [Response of Venice Lagoon ecosystem to natural and anthropogenic pressures over the last 50 years] Coastal lagoons: critical habitats of environmental change [Kennish M. H. K., Paerl H. (ed)] [483–512] (CRC Press, Boca Raton, Florida) 2010. [Google Scholar]
- Pavoni B., Calvo C., Sfriso A. & Orio A. A. Time trend of PCB concentrations in surface sediments from a hypertrophic, macroalgae populated area of the lagoon of Venice. Sci. Total Environ. 91, 13–21 (1990). [DOI] [PubMed] [Google Scholar]
- Rose M. A., Dhar A. K., Brooks H. A., Zecchini F. & Gersberg R. M. Quantitation of hepatitis A virus and enterovirus levels in the lagoon canals and Lido beach of Venice, Italy, using real-time RT-PCR. Water Res. 40, 2387–2396 (2006). [DOI] [PubMed] [Google Scholar]
- Sfriso A. & Facca C. Annual growth and environmental relationships of the invasive species Sargassum muticum and Undaria pinnatifida in the lagoon of Venice. Est. Coast. Shelf Sci. 129, 162–172 (2013). [Google Scholar]
- Sfriso A., Facca C. & Ghetti P. F. Temporal and spatial changes of macroalgae and phytoplankton in a Mediterranean coastal area: the Venice Lagoon as case study. Mar. Environ. Res. 56, 617–636 (2003). [DOI] [PubMed] [Google Scholar]
- Sfriso A. & Curiel D. Check-list of seaweeds recorded in the last 20 years in Venice lagoon, and a comparison with the previous records. Bot. Mar. 50, 22–58 (2007). [Google Scholar]
- Wolf M. A., Sciuto K., Andreoli C. & Moro I. Ulva (Chlorophyta, Ulvales) biodiversity in the North Adriatic Sea (Mediterranean, Italy): cryptic species and new introductions. J. Phycol. 48, 1510–1521 (2012). [DOI] [PubMed] [Google Scholar]
- Sfriso A., Facca C. & Ghetti P. F. Validation of the macrophyte quality index (MaQI) set up to assess the ecological status of Italian marine transitional environments. Hydrobiologia 617, 117–141 (2009). [Google Scholar]
- Touchon M. et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5, e1000344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elsas J. D., Semenov A. V., Costa R. & Trevors J. T. Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 5, 173–183 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekabab S. M., Paquin-Veillette J., Dozois C. M. & Harel J. The ecological habitat and transmission of Escherichia coli O157:H7. FEMS Microbiol. Lett. 341, 1–12 (2013). [DOI] [PubMed] [Google Scholar]
- Sabino R. et al. Routine screening of harmful microorganisms in beach sands: implications to public health. Sci. Total Environ. 472, 1062–1069 (2014). [DOI] [PubMed] [Google Scholar]
- Perchec-Merien A. M. & Lewis G. D. Naturalized Escherichia coli from New Zealand wetland and stream environments. FEMS Microbiol. Ecol. 83, 494–503 (2013). [DOI] [PubMed] [Google Scholar]
- Byappanahalli M. N. et al. Population structure of Cladophora-borne Escherichia coli in nearshore water of Lake Michigan. Water Res. 41, 3649–3654 (2007). [DOI] [PubMed] [Google Scholar]
- Jard G. et al. French Brittany macroalgae screening: composition and methane potential for potential alternative sources of energy and products. Bioresource technol. 144, 492–498 (2013). [DOI] [PubMed] [Google Scholar]
- Olapade O. A., Depas M. M., Jensen E. T. & McLellan S. L. Microbial communities and fecal indicator bacteria associated with Cladophora mats on beach sites along Lake Michigan shores. Appl. Environ. Microbiol. 72, 1932–1938 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari G. et al. in Rapporto sullo stato ambientale delle acque dei rii di Venezia e delle aree lagunari limitrofe. Campagna Di Monitoraggio 2008 – 2009. Ministero delle Infrastrutture e dei Trasporti Magistrato Alle Acque, Ufficio Tecnico per l’Antinquinamento della laguna di Venezia del Magistrato alle Acque. p. 73 (2010). [Google Scholar]
- Sherwin M. R., Van Vleet E. S., Fossato V. U. & Dolci F. Coprostanol (5β-cholestan-3β-ol) in lagoonal sediments and mussels of Venice, Italy. Mar. Poll. Bull. 26, 501–507 (1993). [Google Scholar]
- Ferrarin C., Cucco A., Umgiesser G., Bellafiore D. & Amos C. L. Modelling fluxes of water and sediment between Venice Lagoon and the sea. Contin. Shelf Res. 30, 904–914 (2010). [Google Scholar]
- Vanden Heuvel A. et al. The green alga, Cladophora, promotes Escherichia coli growth and contamination of recreational waters in Lake Michigan. J. Environ. Qual. 39, 333–344 (2010). [DOI] [PubMed] [Google Scholar]
- Englebert E. T., McDermott C. & Kleinheinz G. T. Effects of the nuisance algae, Cladophora, on Escherichia coli at recreational beaches in Wisconsin. Sci. Tot. Environ. 404, 10–17 (2008). [DOI] [PubMed] [Google Scholar]
- Johnson J. R. & Russo T. A. Extraintestinal pathogenic Escherichia coli: “the other bad E. coli”. J. Lab. Clin. Med. 139, 155–162 (2002). [DOI] [PubMed] [Google Scholar]
- Moissenet D. et al. Meningitis caused by Escherichia coli producing TEM-52 extended-spectrum beta-lactamase within an extensive outbreak in a neonatal ward: epidemiological investigation and characterization of the strain. J. Clin. Microbiol. 48, 2459–2463 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O., Gordon D. M., Brisse S., Walk S. T. & Denamur E. Characterization of the cryptic Escherichia lineages: rapid identification and prevalence. Environ. Microbiol. 13, 2468–2477 (2011). [DOI] [PubMed] [Google Scholar]
- Tenaillon O., Skurnik D., Picard B. & Denamur E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8, 207–217 (2010). [DOI] [PubMed] [Google Scholar]
- Berthe T., Ratajczak M., Clermont O., Denamur E. & Petit F. Evidence for coexistence of distinct Escherichia coli populations in various aquatic environments and their survival in estuary water. Appl. Environ. Microbiol. 79, 4684–4693 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak M. et al. Influence of hydrological conditions on the Escherichia coli population structure in the water of a creek on a rural watershed. BMC Microbiol. 10, 222 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira S. et al. Persistence of Escherichia coli in freshwater periphyton: biofilm‐forming capacity as a selective advantage. FEMS Microbiol. Ecol. 79, 608–618 (2012). [DOI] [PubMed] [Google Scholar]
- McDaniels A. E. et al. Confirmational identification of Escherichia coli, a comparison of genotypic and phenotypic assays for glutamate decarboxylase and beta-D-glucuronidase. Appl. Environ. Microbiol. 62, 3350–3354 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J. et al. Stability, genotypic and phenotypic diversity of Shewanella baltica in the redox transition zone of the Baltic Sea. Environ. Microbiol. 16, 1854–1866 (2014). [DOI] [PubMed] [Google Scholar]
- Jost T., Lacroix C., Braegger C. P., Rochat F. & Chassard C. Vertical mother–neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 16, 2891–2904 (2013). [DOI] [PubMed] [Google Scholar]
- França L., Lopéz-Lopéz A., Rosselló-Móra R. & da Costa M. S. Microbial diversity and dynamics of a groundwater and a still bottled natural mineral water. Environ. Microbiol. 10.1111/1462-2920.12430 (2014). [DOI] [PubMed] [Google Scholar]
- Regua-Mangia A. H. et al. Genetic relatedness of a non-motile variant O157 enteropathogenic Escherichia coli (EPEC) strain and E. coli strains belonging to pathogenic related groups. Microbiol. Res. 163, 225–233 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


