SUMMARY
It has long been thought that clonal deletion efficiently removes almost all self-specific T cells from the peripheral repertoire. But here we found that self peptide-MHC specific CD8+ T cells in the blood of healthy humans were present in frequencies similar to those specific for non-self antigens. For the Y chromosome encoded SMCY antigen, self-specific T cells exhibited only a three-fold lower average frequency in males versus females and were anergic with respect to peptide activation, although this inhibition could be overcome by a stronger stimulus. We conclude that clonal deletion prunes but does not eliminate self-specific T cells and suggest that to do so would create holes in the repertoire that pathogens could readily exploit. In support of this hypothesis, we detected T cells specific for all 20 amino acid variants at the p5 position of a hepatitis C virus epitope in a random group of blood donors.
INTRODUCTION
To create a diverse repertoire of antigen receptors, maturing B and T lymphocytes bring together V, J, and, in some loci, D gene segments to form functional genes to express a very large number of immunoglobulin or T cell receptors (TCR), respectively (Tonegawa, 1983; Davis and Bjorkman, 1988). The semi-random process of V(D)J recombination not only generates antigen receptors with the ability to recognize foreign epitopes, but also endogenously expressed self epitopes as well. The potential to mount an immune response against self must therefore be controlled in order to avoid autoimmune disease, an issue raised over 100 years ago by Paul Ehrlich (Silverstein, 2001).
The clonal selection theory, associated most closely with the work of F. Macfarlane Burnet, provides a model for immunologic tolerance to self: lymphocytes only express antigen receptors of one specificity and those lymphocytes specific for self are clonally deleted (Burnet, 1959). With respect to the control of self-specific helper and cytotoxic αβ T cells, mice have been the main experimental animal model used in support of this theory. Classic experiments by Kappler and Marrack showed that specific Vβ expressing thymocytes were efficiently deleted in mouse strains which expressed particular endogenous superantigens (Kappler et al., 1987; Herman et al., 1991). This was followed by a series of TCR transgenic studies in which it was shown that the presence of the relevant peptide-major histocompatibility complex (MHC) ligand of the TCR in the thymus led to massive thymocyte death by apoptosis at the double positive stage (Kisielow et al., 1988; Sha et al., 1988; Hogquist et al., 2005). Similar results were obtained in studies of TCR transgenics by other laboratories, including ours, where we found extensive thymic deletion of TCR β- expressing transgenic thymocytes in a CD4+ system (Berg et al., 1989). More recently, identification of the Aire gene has demonstrated how otherwise tissue-specific genes may be expressed in the thymus to precipitate the deletion of self-specific thymocytes (Anderson et al., 2002).
As a result of these studies in mice, it became generally accepted that the deletion of self-specific αβ T cells is a very efficient mechanism for reducing the threat of autoimmunity (von Boehmer, 1990; Herman et al., 1991; Hogquist et al., 2005). This paradigm implies that peripheral tolerance regulates only a small number of escaping T lymphocytes that bind to self-antigen with low affinity. A further implication is that the efficient deletion of self-specific T cells will result in gaps in the universe of ligands recognizable by the TCR repertoire (Vidovic and Matzinger, 1988). As a consequence, pathogens could make use of these immunologic blind spots to escape detection.
Because of their relatedness in evolution and as components of the immune system, it is of interest to compare the escape of self-specific αβ T cells to other lymphocyte lineages. Up to 20% of human mature circulating B cells are self-reactive and may contribute to natural antibody production (Wardemann et al., 2003). In the case of mouse γδ T cells, Jensen et al. find that γδ T cells specific for the non-classical class I molecule T10 and the closely related T22, are not appreciably deleted in the thymi of non-transgenic mice expressing these antigens, despite previous results showing the extensive deletion of γδ TCR transgenic T cells having that specificity (Jensen et al., 2008).
In the case of human αβ T cells, assessing the effect of clonal deletion has been more difficult, although there are sporadic reports mentioning the peripheral survival of self-specific T cells (Delluc et al., 2010; Velthuis et al., 2010; Su et al., 2013). In this study, we further explore the fate of self-specific CD8+ αβ T cells using the unique resource of healthy blood donors. We used specific peptide HLA-A*0201 tetramers and a modification of the enrichment scheme of Jenkins and colleagues (Moon et al., 2007) to directly measure the frequency of particular CD8+ T cells, and found that the frequency of CD8+ T cells recognizing endogenous peptides was roughly equivalent to that of naïve CD8+ T cells that recognize foreign epitopes. This is also consistent with what we have found previously with CD4+ T cells in healthy human volunteers (Su et al., 2013).
These results strongly suggest that clonal deletion does not play an absolute role in shaping the peripheral repertoire, but because they do not directly compare the same specificity in both self and non-self situations, we surveyed the frequency of SMCY specific T cells in males (who express this Y chromosome encoded antigen) and females (who do not), and found only a 3-fold reduction in males. A parallel experiment in mice produced a similar result. We then derived male versus female human T clones specific for SMCY and found that they have overlapping functional avidities. To explore whether a genetic program within self-specific CD8+ T cells might contribute to their apparent tolerance, we performed microfluidics-based single cell quantitative PCR (qPCR) of SMCY specific T cells from women and men and found distinct patterns of gene expression that suggested impaired expansion in self-specific (male) cells. We then confirmed this observation functionally by showing that the activation of primary, self-specific CD8+ T cells from blood is impaired after in vitro stimulation.
These results indicate that clonal deletion is a factor, but not the most critical one, in mechanisms that establish tolerance. Instead, it may be that self-specific CD8+ T cells are imprinted with a less harmful genetic program, either in the thymus or in the periphery, and that this, together other peripheral tolerance mechanisms such as CD4+ regulatory T cells or the expression of Aire in peripheral lymphoid organs (Gardner et al., 2008; Wing and Sakaguchi, 2010), is the principal bulwark against autoimmunity. We also suggest that the wholesale removal of T cell specificities is avoided because infectious pathogens are a much greater threat to evolutionary fitness than autoimmunity and that it is therefore imperative that T cells cover every possible peptide-MHC variant. In support of this model, we show that there are CD8+ T cells specific for every natural amino acid at position 5 of a hepatitis C virus (HCV) epitope presented by HLA-A*0201, including one peptide variant reported to represent a blind spot in the TCR repertoire caused by clonal deletion (Wolfl et al., 2008).
RESULTS
The Frequencies of CD8+ T Cells Specific for Self and Non-Self are Similar in Healthy Adults
In order to establish a benchmark comparison, we first determined the frequency of naïve antigen specific CD8+ T cells in healthy adults. We performed peptide-MHC tetramer enrichment using HLA-A*0201 tetramers containing peptides derived from cytomegalovirus (CMV), human immunodecificiency virus (HIV), hepatitis C virus, and the avian influenza A (H5N1) virus on peripheral blood mononuclear cells (PBMCs) obtained from HLA-A*0201+ blood bank donors (seronegative for antibodies against HIV, HCV, and CMV and very unlikely to have been infected by the H5N1 avian influenza virus; Figure 1A and 1B, Table 1) (Jenkins et al., 2010; Newell et al., 2009; Toebes et al., 2006; Wolfl et al., 2008). The frequency of the CD8+ T cells recognizing the foreign antigens ranged from 1:105 to 1:106 in comparison to total CD8+ T cells (Figure 2A). This agrees roughly with the naïve T cell frequencies measured in mice (1:104 – 1:106) and correlates well with the values reported recently in humans (Moon et al., 2007; Kotturi et al., 2008; Obar et al., 2008; Alanio et al., 2010; Schmidt et al., 2011; Su et al., 2013).
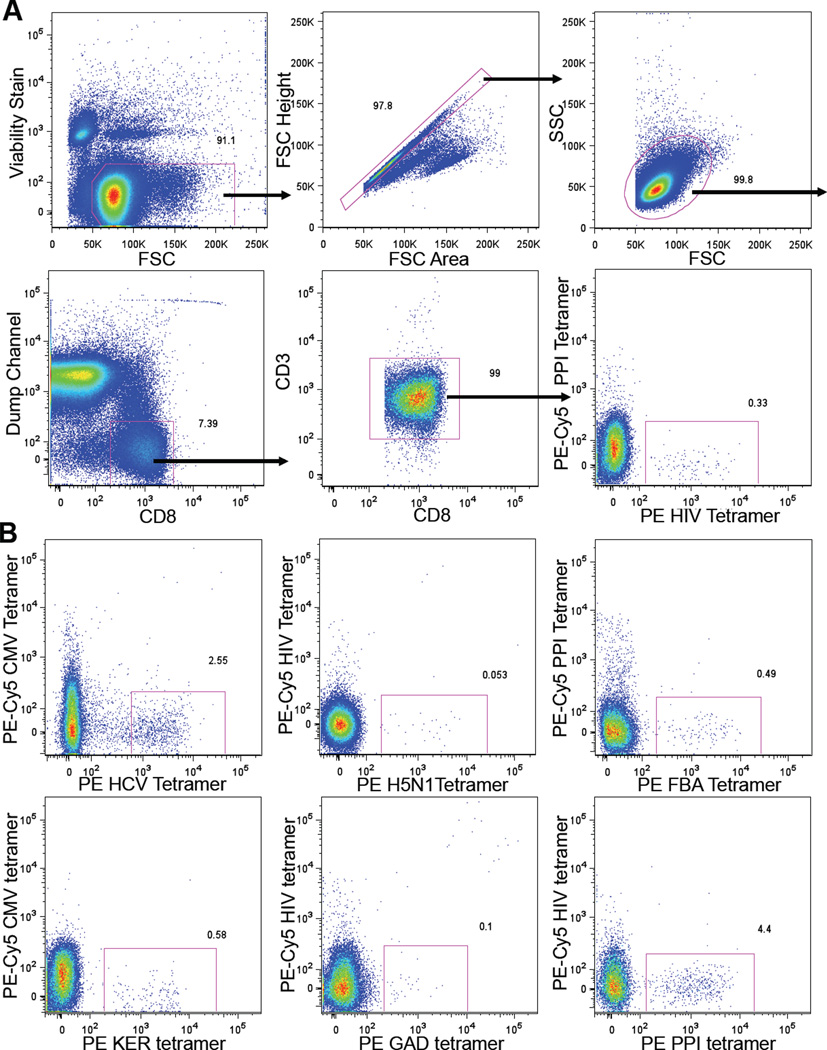
Figure 1. Flow Cytometry of Peptide HLA-A*0201 Tetramer Enriched CD8+ T Cells.
(A) Flow cytometry gating scheme. PBMCs from a HLA-A*0201+ blood donor were concentrated for CD8+ T cells by depletion, followed by HIV:HLA-A*0201 tetramer enrichment over a magnetized column before flow cytometric analysis. Dump channel includes cells labeled with antibodies against CD4, CD14, CD16, CD19, and γδ TCR. In this case, the PE-Cy5 peptide HLA-A*0201 tetramer was only used as control for peptide MHC specific binding.
(B) Representative flow cytometric plots of different peptide HLA-A*0201 tetramer enriched CD8+ T cells. Panels shown are gated on CD8+ T cells. See also Figure S1.
Table 1.
Peptides Loaded onto HLA-A*0201 Tetramers
| Foreign peptides to which donors have not been exposed: | |
| Human immunodeficiency virus (HIV) | SLYNTVATL |
| Avian influenza virus (H5N1) | AMDSNTLEL |
| Hepatitis C virus (HCV) | KLVALGINAV |
| Cytomegalovirus (CMV) | NLVPMVATV |
| Endogenous peptides: | |
| Fructose bisphosphate aldolase (FBA) | ALSDHHIYL |
| Keratin (KER) | ALLNIKVKL |
| SMCY (male specific) | FIDSYICQV |
| Endogenous peptides associated with autoimmunity: | |
| Preproinsulin (PPI) | ALWMRLLPL |
| Glutamic acid decarboxylase 65 (GAD) | VMNILLQYVV |
Epitopes used for peptide-MHC tetramer enrichment of CD8+ T cells from human blood bank donor PBMCs in Figure 2. See text for references.
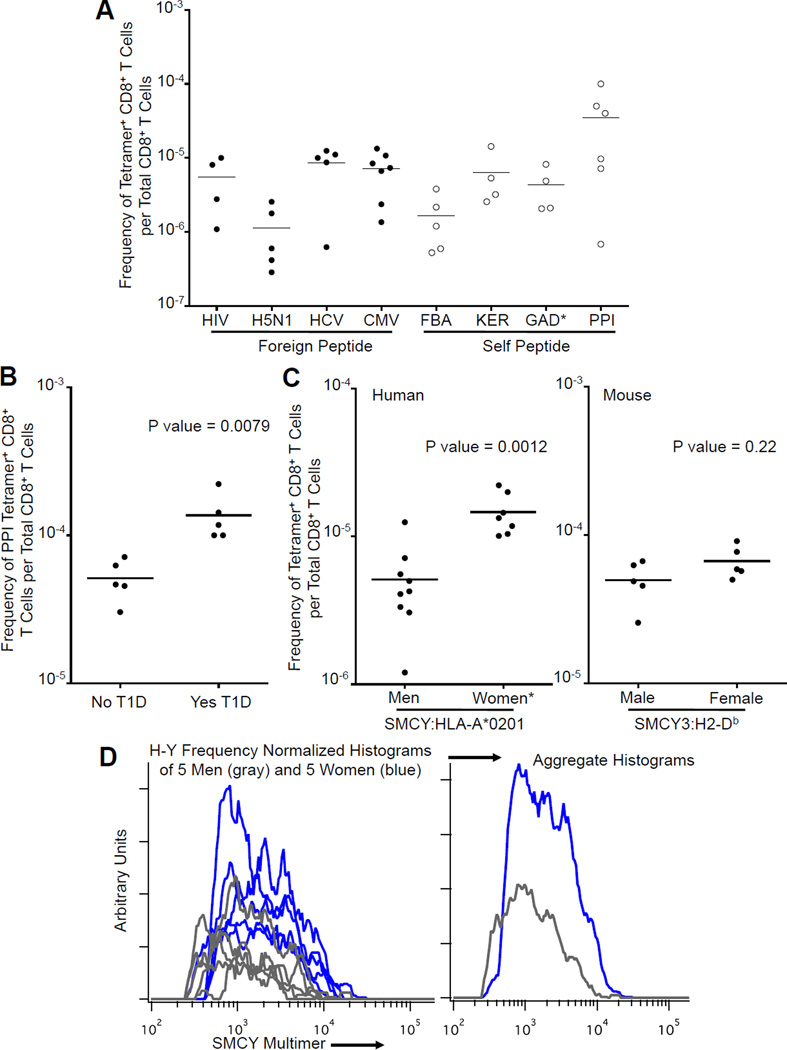
Figure 2. Frequency of Antigen Specific CD8+ T Cells.
(A) Frequency of CD8+ T cells binding foreign versus self peptide HLA-A*0201 tetramers (human blood). The frequency of tetramer+ CD8+ T cells per total CD8+ T cells was determined using tetramer enrichment. Each point represents one sample from a separate individual. Bar indicates mean. (*GAD tetramer+ T cells not detected in two samples.) See also Table 1.
(B) Frequency of PPI peptide specific CD8+ T cells in HLA-A*0201+ individuals with type 1 diabetes mellitus (T1D) versus controls. HLA-A*0201 tetramer enrichment was used to calculate the frequency of tetramer+ CD8+ T cells per total CD8+ T cells in whole blood. Each point represents one human blood sample. Bar indicates mean. P value calculated using Mann-Whitney test. See also Table S2.
(C) Frequency of SMCY peptide specific CD8+ T cells in males versus females. Tetramer enrichment was used to calculate the frequency of tetramer+ CD8+ T cells per total CD8+ T cells. Each point represents one human blood sample or one mouse. Bar indicates mean. P value calculated using Mann-Whitney test. Left: Frequency of SMCY:HLA-A*0201 binding CD8+ T cells in men versus women. (*One blood sample from a female with a frequency of 1 in 3.5×102 was not included (see main text).) Right: Frequency of SMCY3:H2-Db binding CD8+ T cells in male mice versus female mice.
(D) Histograms showing the intensity distribution of SMCY:HLA-A*0201 tetramer fluorescence on CD8+ T cells in men (gray) and women (blue). Histogram areas are normalized for the relative frequency of H-Y+ CD8+ T cells per total CD8+ T cells for each blood donor.
Several controls were performed to confirm the accuracy of the tetramer enrichment method: 1) Minimal double labeling was observed when PBMCs were incubated with two HLA-A*0201 tetramers loaded with distinct peptide epitopes (Figure 1). 2) Spiking experiments demonstrated that when present at a starting frequency of 1 in 5,400,000 total CD8+ T cells, 80% of antigen specific cells could be recovered by tetramer enrichment (Figure S1A). 3) Tetramer enriched T cells were also sorted as single cells by fluorescence activated cell sorter (FACS) and expanded in vitro. 46% to 60% of the clones that grew bound tetramer upon reanalysis (Figure S1B).
Next we examined the frequency of CD8+ T cells recognizing self, as opposed to foreign, peptides bound to HLA-A*0201 tetramers. We chose two endogenous epitopes derived from fructose bisphosphate aldolase (FBA) and keratin (KER) (Barnea et al., 2002; Weinschenk et al., 2002) as well as two autoimmune disease-associated epitopes derived from preproinsulin (PPI) and glutamic acid decarboxylase (GAD) 65 (Table 1 and Figure 1B) (Mallone et al., 2007). In a few cases, self-specific CD8+ T cells were not detectable. Unexpectedly, however, in the majority of cases the frequency of CD8+ T cells recognizing these four endogenously expressed epitopes was 1:104 to 1:106 – in the same range as CD8+ T cells recognizing the foreign antigens (Figure 2A). In addition, the intensity of tetramer staining for the self epitopes was robust and comparable to that for foreign epitopes (Figure 1B), and therefore most consistent with agonist level TCR affinities (Savage et al., 1999).
To explore the differentiation status of these antigen specific cells, we assessed the surface expression of the molecules CCR7, CD27, CD28 and CD45RA, which are reported to be expressed by naïve CD8+ T cells (Appay et al., 2002). Some degree of lowered expression in at least one of these four surface molecules was seen in 57% of the antigen specific CD8+ T cell populations recognizing one of the four foreign epitopes and 47% of those recognizing endogenous antigens (Table S1 and Figure S1C). This result suggests that, similarly to recent work on human CD4+ T cells, there may be a significant degree of TCR crossreactivity to some other antigen or antigens that the subjects have been exposed to (Su et al., 2013).
Of all epitopes analyzed, preproinsulin was recognized by CD8+ T cells at the highest frequency, up to 1:104 in healthy blood bank donors. Preproinsulin is of particular interest as it is associated with type 1 diabetes mellitus (T1D) (Mallone et al., 2007; Todd, 2010). To determine whether there is expansion of PPI specific CD8+ T cells in T1D, we measured their frequency in the peripheral blood of five HLA-A*0201+ individuals with T1D and five age matched controls. We found a 2.66 fold increase in CD8+ T cells specific for the PPI epitope in T1D individuals (p= 0.0079), consistent with results seen by other groups (Figure 2B and Table S2) (Velthuis et al., 2010; Kronenberg et al., 2012). It is tempting to speculate that in those susceptible to diabetes, even a partial failure of peripheral tolerance might permit the activation and expansion of some cells in this pool, making this a possible risk factor in this disease.
T Cells Specific for the SMCY/H-Y antigen in Males and Females
To directly test the effects of deletional tolerance in humans, the frequency of CD8+ T cells recognizing a Y chromosome specific SMCY (the H-Y equivalent) epitope was measured in male and female HLA-A*0201+ blood donors (Table 1 and Figure 2C) (Meadows et al., 1997). In nine men, the frequency of SMCY specific CD8+ T cells ranged from ~1:80k to ~1:800k with a mean frequency of 1 in 2×105 CD8+ T cells (Figure 2C). This falls well within the range of frequencies seen for both self and foreign antigens (Figure 2A) and represents a substantial number of T cells with this specificity in an average adult male (~106). Among eight women the frequency of SMCY specific T cells ranged from 1 in 5×104 to 1×105 with the exception of one individual who had recently given birth to a son and had a frequency of 1 in 3.5×102 (Figure 2C and data not shown). In this latter case, 80% of these cells were CD45RAlo, suggesting that the exceptionally high frequency of these cells was due to the expansion of these T cells after exposure to SMCY antigen from the male fetus. In the remaining seven women, for whom we do not have a complete reproductive history, the average frequency of CD8+ T cells recognizing the SMCY epitope was approximately 1 in 6.8×104, approximately three-fold greater than in men. This indicates that a large fraction (1/3) of SMCY-specific T cells escape clonal deletion in males.
To determine whether this is also true in non-transgenic mice, we analyzed H-Y specific CD8+ T cells, in which the peptide KCSRNRQYL, derived from the Y chromosome encoded protein SMCY3, is presented by H2-Db (Uematsu et al., 1988). The frequency of these CD8+ T cells was determined using the same tetramer enrichment method for C57BL/6 male and female mice (5 each). There was a trend toward a higher frequency of SMCY3 specific CD8+ T cells in females in comparison to males, but this difference was not statistically significant (Figure 2C). This result corroborates the human results in that a significant fraction of self-specific T cells escape clonal deletion. Thus this is not a species-specific difference.
To test whether CD8+ T cells from men express TCR of sufficient affinity for SMCY to be functionally competent, we used single cell sorting and in vitro expansion with anti- CD3/CD28 or phytohemagglutinin (PHA) stimulation - both strong stimuli - to generate SMCY-specific T clones from five women and five men, and then compared their functional avidity using the CD107 mobilization assay (Table S3) (Rubio et al., 2003; Stuge et al., 2004).
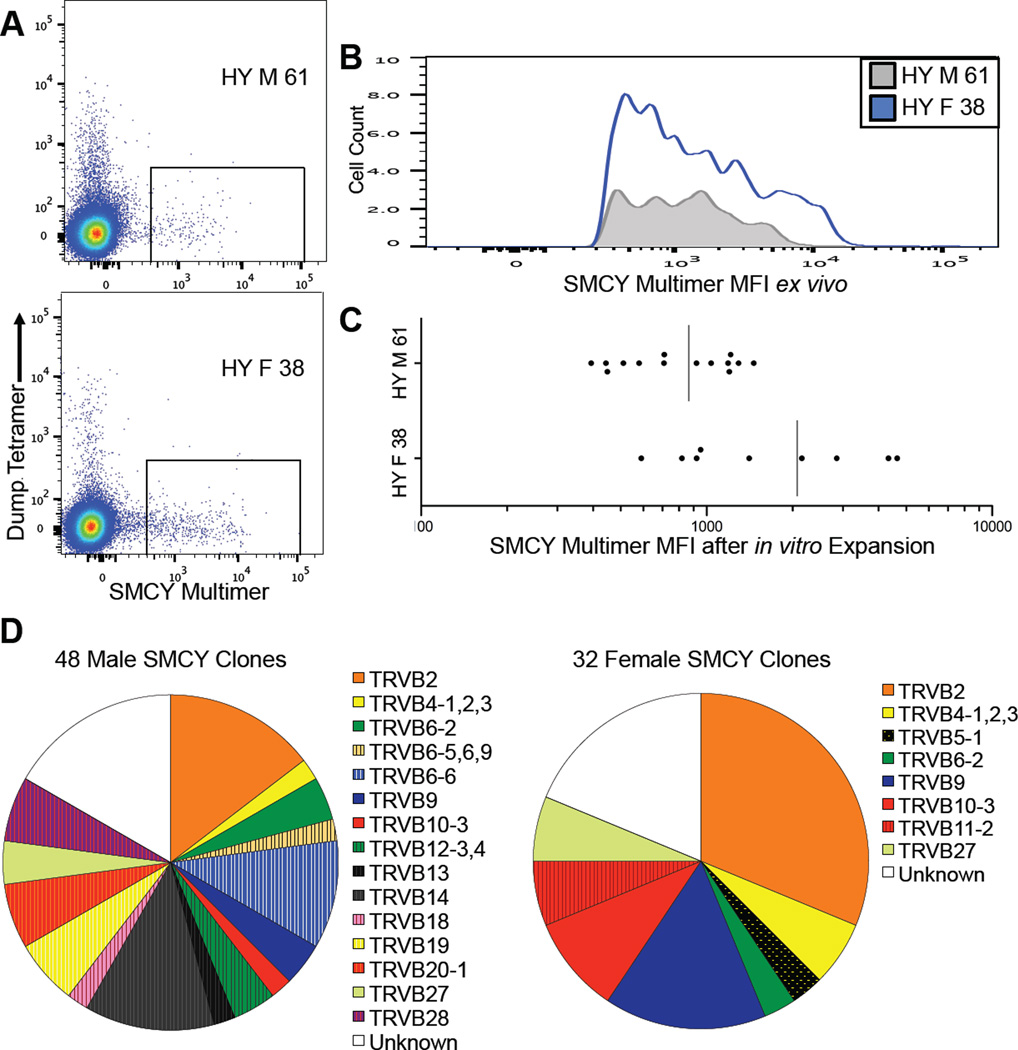
We found that, ex vivo, CD8+ T cells from men tended to have an intensity of SMCY tetramer staining that overlapped extensively with that of women, but this intensity distribution was depleted of cells in males, consistent with clonal deletion (Figures 2D, 3A and 3B). After in vitro expansion, we reassayed T cell clones for SMCY tetramer staining and found a similar difference in the distribution of intensities between women and men, indicating that we had recovered a representative sample of the original population (Figure 3C). We also measured the distribution of TCR Vβ gene segment families in SMCY specific T cell clones by antibody staining and found a comparable diversity of Vβ usage in women and men, indicating that the high frequency of self-specific CD8+ T cells in humans is not due to the peripheral expansion of a few escaping clones (Figure 3D). In addition, the Vβ usage of SMCY T cells was skewed in comparison with bulk peripheral blood T cells, consistent with the former being a subpopulation of antigen-specific T cells (van den Beemd et al., 2000).
Figure 3. Single Cell FACS and in vitro Expansion of SMCY:HLA-A*0201 Specific CD8+ T Cells.
(A) FACS plots gated for CD8+ T cells from one woman with no history of pregnancy (bottom, ID 38) and one man (top, ID 61) after tetramer enrichment. Gates shown for single cell FACS of SMCY:HLA-A*0201 binding CD8+ T cells used for in vitro expansion of T cells clones.
(B) Plot of mean fluorescence intensity (MFI) of SMCY:HLA-A*0201 tetramer binding, primary CD8+ T cells from Figure 3A.
(C) MFI of SMCY CD8+ T cell clones indicates that they are representative of the original population. Plot of MFI for SMCY:HLA-A*0201 multimer binding CD8+ T cell clones after in vitro expansion from one woman with no history of pregnancy (bottom, ID 38) and one man (top, ID 61). Each point represents one distinct T cell clone. Compare with relative fluorescence intensity of primary male or female derived CD8+ T cells in Figure 3A and 3B).
(D) Pie chart showing TCR Vβ family expression of in vitro expanded SMCY:HLA-A*0201 binding CD8+ T cell clones from four men (IDs 61, 69, 74, 390) and four women (IDs 38, 67, 84, 86). Vβ antibody panel from Beckman Coulter. See also Table S3.
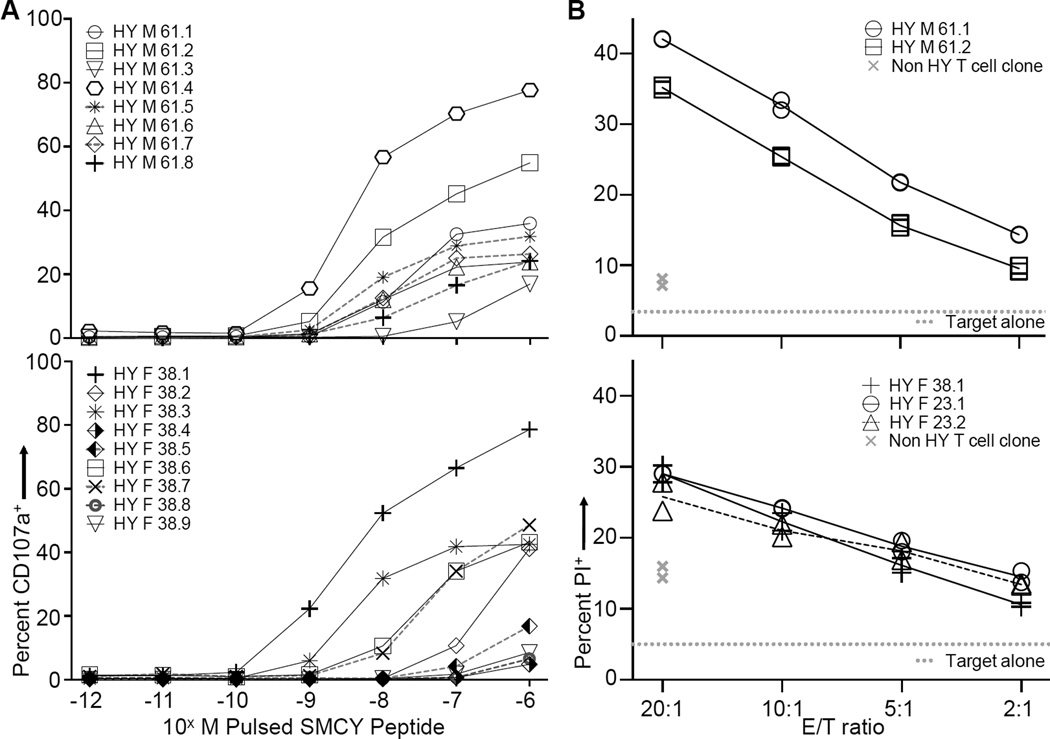
We then performed a CD107 mobilization assay on SMCY specific CD8+ T cell clones from five women and five men. CD107a, or lysosomal associated membrane protein-1, is transiently expressed on the cell surface of CD8+ T cells during the release of cytotoxic granules (Rubio et al., 2003) and thus represents a functional response. The sensitivity to SMCY antigen ranged from 10−7M to 10−9M in both men and women (Figures 4A, S2A and S2B,Table S3). To determine whether the SMCY tetramer binding clones are able to react to endogenously processed and presented antigen, we incubated male and female derived T cell clones with either male (JY) or female (OH) HLA-A 0201+, EBV-transformed B lymphoblastoid target cells. SMCY specific T cells clones from both genders preferentially killed male target cells with similar efficiency in two different assay types (Figures 4B, S3A–E). We conclude that SMCY tetramer binding CD8+ T cells from men express TCRs with a ligand sensitivity that overlaps with females and are able to recognize endogenously expressed antigen.
Figure 4. Broadly Overlapping Functional Avidity of SMCY:HLA-A*0201 Specific CD8+ T Cell Clones Derived from Men and Women.
(A) CD107 mobilization assay performed on SMCY specific CD8+ T cell clones pulsed with 10−6M SMCY peptide followed by tenfold dilutions. Each line represents one distinct T cell clone. None of the clones responded to 10−6M negative control peptide (PPI) by this assay. T cell clones derived from female ID 38 (bottom) and male ID 61 (top). See also Figure S2A and S2B, and Table S3 for more clones.
(B) Propidium iodide (PI) cytotoxicity assay. Graphs show the percentage of PI+ JY (male) target cells after incubation with SMCY specific CD8+ T cell clones at the indicated effector to target (E/T) ratios (symbols with black lines). Representative clones are from females ID 38 and 23 (bottom), and male ID 61 (top). A non specific CD8+ T cell clone derived from the same individuals is shown in each panel (gray “x” symbols). The gray dotted line indicates the background level of JY cell death in the absence of T cell clones. Performed in duplicate and representative of three to six experiments. The bottom panel combines data from two experiments; for that panel, values for the non HY T cell clone control and background target cell death were averaged. See also Figure S3C–E for additional clone and controls. An alternate cytotoxicity assay based on relative target cell survival is shown in Figure S3A and S3B.
The CD8 coreceptor is known to contribute to peptide MHC-TCR binding (Legoux et al., 2010). To determine whether CD8 differentially contributes to SMCY binding by T cells in males versus females, we tested the ability of nine female (donor ID 38) and fourteen male T cell clones (donor ID 61) to bind SMCY HLA-A*0201 wildtype tetramer versus a D227K/T228A CD8 binding mutant of the same MHC molecule. Tetramer binding was dependent on CD8 in both females and males (Figure S4A). In addition, a large and similar degree of CD8 dependence between self and nonself was observed when performing peptide-MHC tetramer enrichment with pools of eleven or more self or foreign peptides (Table S4 and Figure S4B and S4C).
Foreign-Specific CD8+ T Cells Express a Distinct Set of Gene Transcripts and Expand After Stimulation, Whereas Self-Specific CD8+ T Cells Do Not
Given their similarities in ligand sensitivity, we hypothesized that self-specific CD8+ T cells might be kept in check by tolerance mechanisms characterized by distinct patterns of gene expression. A precedent for this has been found in the case in γδ T cells, where self- and non-self specific cells express different cytokines (Jensen et al., 2008). Therefore, we examined the single cell gene expression of 96 genes in male and female SMCY-specific T cells using a microfluidics-based quantitative polymerase chain reaction (qPCR) (Table S5) (Diehn et al., 2009). We primarily chose cytokines, cytokine receptors, and genes involved with cell survival as we reasoned that these might be affected in a tolerant cell. After a 14 hour ex vivo stimulation with peptide and anti-CD28 antibody, 152 CD8+ T cells from four men and 154 cells from four women (with no history of pregnancy) were examined (example qPCR run, Figure S5A). Among those genes for which for transcript was detectable, the cycle threshold value was similar between cells (not shown). Therefore, genes were considered expressed or not in a given cell depending on whether detectable transcript levels were measured, and only cells expressing detectable levels of the housekeeping gene GAPDH were considered for analysis. ICOS, an activation inducible gene, was expressed in 52% to 62% of cells; a similar heterogeneity in ICOS expression (~60%) was seen in CMV:HLA-A*0201 tetramer positive memory CD8+ T cells stimulated similarly (data not shown).
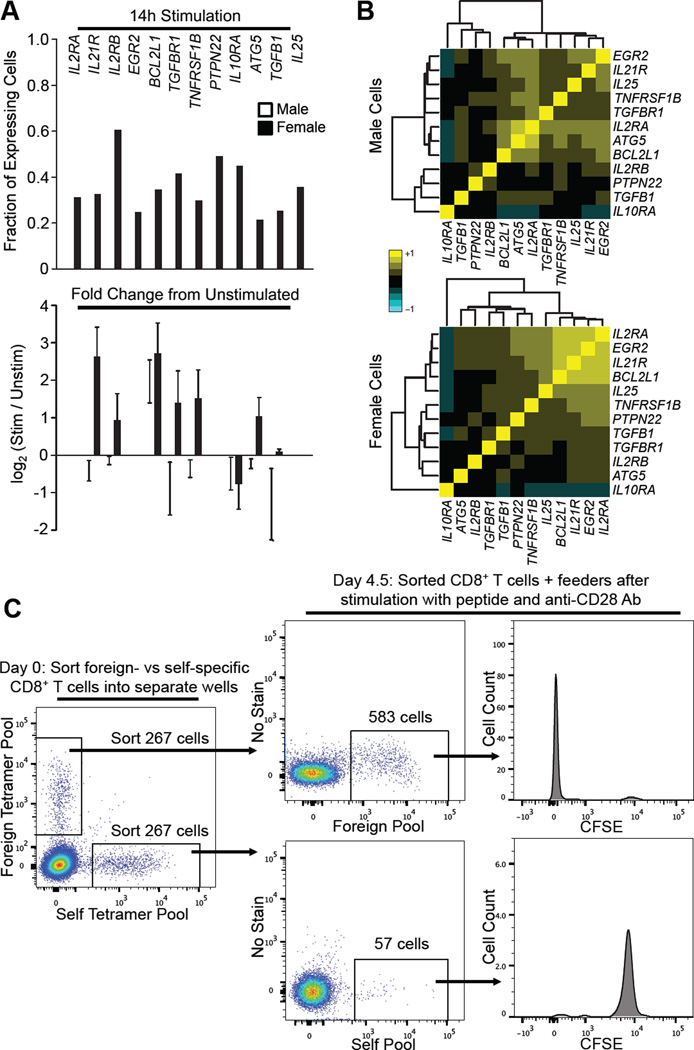
Twelve genes showed a difference in the likelihood that they would be expressed between women and men with p values <0.05 (Figure 5A). With the exception of IL10RA, each gene was detected in a greater proportion of naïve CD8+ T cells versus self-specific cells. We then clustered these genes based on correlated coexpression at the single cell level. In women, but not in men, a statistically significant group of four genes was found to be expressed together (p = 0.025, Figures 5B, S5B and S5C). Three of these genes, IL2RA, IL21R, and BCL2L1 (also known as BCLXL), are associated with T cell proliferation and survival (Choi and Schwartz, 2007; Todd, 2010). IL2 transcripts were not detected at the single cell level, most likely due to the inefficiency of the PCR primers, as transcripts were only reliably detected in stimulated CMV-specific memory CD8+ T cells after increasing the initial amount of cell lysate (not shown). To control for the influence of gender, we compared CD8+ T cells from men and women stimulated with either a self peptide (PPI) or foreign peptide (HIV) and observed no consistent statistically significant gender based differences (Figure S5D). Details of these experiments are in the supplemental methods.
Figure 5. Foreign-Specific CD8+ T Cells Expand in Response to Peptide + anti-CD28 Stimulation, while Self-Specific CD8+ T Cells Do Not.
(A) Microfluidics based qPCR was performed individually on single CD8+ T cells binding the SMCY:HLA-A*0201 tetramer. Top: 152 cells from four males and 154 cells from four females (no history of pregnancy) were analyzed. Cells were stimulated 14 hours with SMCY peptide and anti-CD28 antibody. Only genes differentially expressed between women and men with p <0.05 and q < 0.15 are shown. Bottom: Fold change in gene expression after stimulation. qPCR was performed on unstimulated single CD8+ T cells binding the SMCY:HLA-A*0201 tetramer. 41 cells from one man and 38 cells from one woman were used in comparison to calculate fold difference in gene expression after 14 hours stimulation. Fold change could not be calculated for genes that were not expressed in unstimulated cells.
(B) Coordinated mRNA expression of genes associated with proliferation and survival in female SMCY specific CD8+ T cells but not in males. Two-way clustered heatmaps of the correlation matrices for differentially expressed genes (p < 0.05 and q < 0.15) between male (top) and female (bottom) cells stimulated 14 hours with SMCY antigen and anti-CD28 antibody. Approximate value of correlation coefficient (R) indicated by color; 1.0 (yellow) indicates perfect correlation in the expression of two different genes on a per cell basis.
(C) Cell expansion after peptide + anti-CD28 antibody stimulation of foreign-, but not self-specific CD8+ T cells. Equal numbers of tetramer+ CD8+ T cells, labeled by pooled tetramers loaded with either self or foreign peptides (Table S6, top), were sorted from a single blood sample into separate wells. Approximately 267 sorted cells for each group (self or foreign) were then stimulated in the presence of autologous feeder PBMCs with the same peptides with which they were tetramer selected at 1.5ug/ml and with anti-CD28 antibody at 5ug/ml. After 4.5 days, each sample was analyzed by flow cytometry. Panels gated on live CD8+ T cells. Sample in this figure is representative of five experiments, analyzed at 4.5 and/or 7.5 days; three of the five experiments included CFSE staining. See also Figure S5E and S5F, and Table S6.
We then compared the response of self- versus foreign-specific primary human CD8+ T cells to a longer period of in vitro stimulation with peptide antigen and anti-CD28 antibody. PBMCs from five human blood donors were each stained with two pools of HLA-A*0201 tetramers: one pool loaded with six self peptides, the other with six foreign peptides to which the blood donors should be naïve (Table S6, top). We used tetramer pools so that there would be a sufficient number of antigen specific cells for a robust readout. In addition, we examined the protein expression of IL2RA and IL21R, as these molecules are easily detected on the cell surface by fluorescent antibody. An equal number of self- versus foreign-specific CD8+ T cells from each donor was sorted into separate wells containing autologous PBMCs. The sorted cells were then stimulated with the same pool of peptides used for tetramer sorting as well as anti-CD28 antibody, and analyzed by flow cytometry after either 4.5 or 7.5 days.
In each case, the foreign-specific pool of CD8+ T cells showed increased numbers of tetramer+ cells and upregulation of IL2RA and IL21R (Figures 5C and S5E). In contrast, the self-specific CD8+ T cells decreased in number after stimulation, and showed no increase in IL2RA or IL21R staining. Samples from three of the five blood donors were also stained with carboxyfluorescein succinimidyl ester (CFSE): for all three, the foreign-specific CD8+ T cells lost CFSE staining, indicating that they had undergone proliferation, whereas the self-specific T cells retained high intensity CFSE staining (Figure 5C). This experiment was repeated using a nonoverlapping set of self and foreign peptides (Table S6, bottom). The same result was seen in three more blood samples (Figure S5F). To determine whether the expanding cells were representative of the input mixture, we used combinatorial tetramer staining (Newell et al., 2009) of an eight peptide-MHC pool post stimulation to determine that at least five of eight foreign specificities were represented in the proliferating T cells in two individuals (data not shown). Thus self-specific human CD8+ T cells are anergic with respect to peptide stimulation, but foreign-specific CD8+ T cells are not.
Comprehensive Coverage of HCV Peptide Variants by the TCR Repertoire
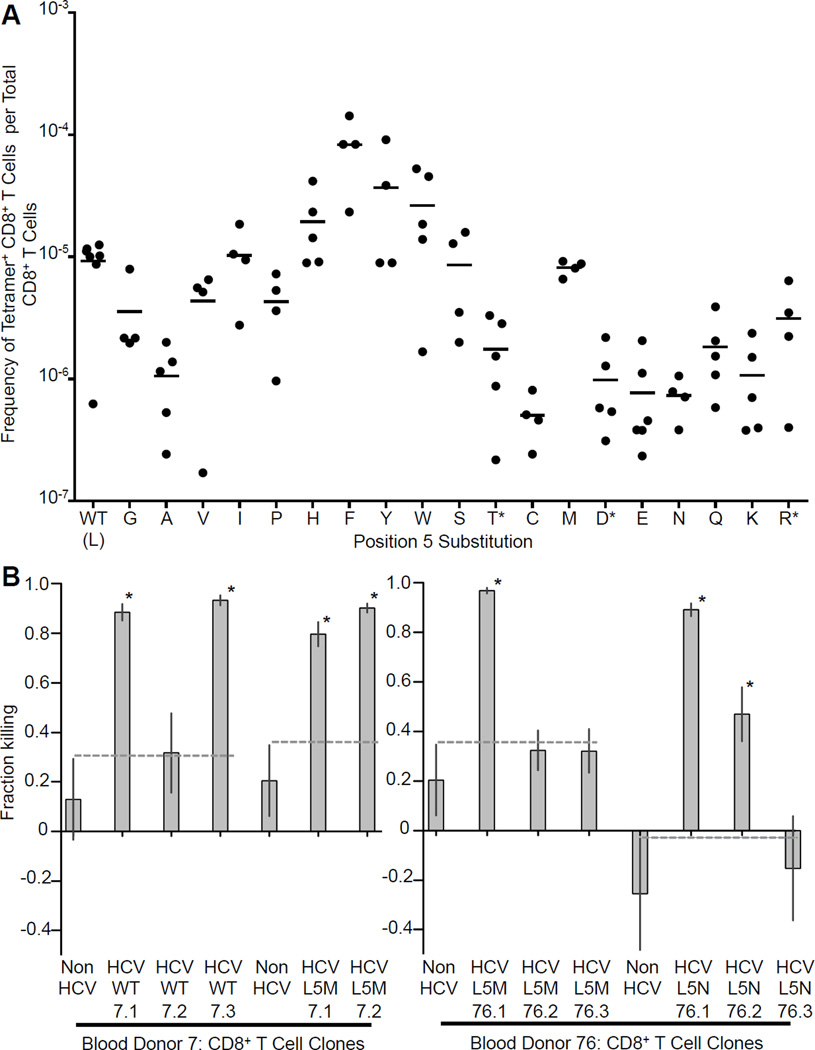
Our finding of an extensive pool of self-specific CD8+ T cells led us to reevaluate the effect of clonal deletion on ligand coverage by the TCR population as a whole. Gaps in the repertoire of epitopes recognized by T cells have been long been proposed to exist due to the effect of clonal deletion during thymic maturation but it has not been possible previously to determine whether T cells of a particular specificity were actually deleted or just less able to respond to stimuli (Vidovic and Matzinger, 1988; Wolfl et al., 2008). To investigate this further, we chose to focus on the HLA-A*0201 restricted epitope KLVALGINAV from the HCV protein NS3. A leucine to methionine escape mutation (L5M) at position 5 seen in chronic HCV infection has been proposed to represent a hole in the T cell ligand repertoire (Wolfl et al., 2008). We therefore made HLA-A*0201 tetramers for this epitope and also for all possible amino acid variants at position 5, in order to systematically examine T cell epitope coverage. Because the aliphatic anchor residues for HLA-A*0201 are at the ends of the peptide, these mutations are not expected to affect peptide binding to the MHC.
Tetramer enrichment of HLA-A*0201+ PBMCs from blood bank donors demonstrated that CD8+ T cells recognizing the L5M variant are present at a comparable frequency to those recognizing the wildtype epitope, showing that previous difficulties in detecting T cells specific for this variant are likely due to a defect in clonal expansion during in vitro priming rather than a physical hole in the repertoire (Figure 6A). Furthermore, in the majority of blood samples tested, all the peptide variants are detectable, suggesting that the T cell repertoire can cover all or almost all possible peptide MHC combinations.
Figure 6. Broad Recognition of All Position 5 Amino Acid Substitutions of the HLA-A* 0201 Restricted Peptide KLVALGINAV.
(A) Frequency of CD8+ T cells recognizing position 5 amino acid substitutions of the HLA-A* 0201 restricted peptide KLVALGINAV. Frequency of tetramer+ CD8+ T cells per total CD8+ T cells calculated by tetramer enrichment. Each point represents one sample. Bar indicates mean. (*Tetramer+ T cells not detected in one sample.)
(B) Cytotoxicity assay (based on relative target cell survival): CD8+ T cell clones recognize endogenously processed and presented HCV WT or variant antigen. Preferential killing of JY target cells expressing HCV NS3 protein containing the target HCV variant peptide versus a negative control HCV variant peptide after 18 hours incubation with a twenty fold excess of CD8+ T cell clone. 95% confidence intervals shown for experiments performed in triplicate. WT indicates wildtype HCV epitope. The dotted line indicates upper 95% confidence interval of a negative control T cell clone (label: non HCV) that does not bind any of the HCV variant tetramers. (*p<0.025: indicates significant difference in killing from negative control.) See also Figure S6 for CD107 mobilization assay with HCV peptide titration curves, and Table S7.
Interestingly, at position five, amino acid residues with ring structures tend to be recognized at a higher frequency than residues with charged or polar side chains. Since amino acids which have aromatic or ring containing side chains are well known from structural studies to be able to form a wide variety of bonds in protein-protein interactions, this result suggests that they also have a greater propensity for TCR cross-reactivity, resulting in a higher frequency of T cells that recognize them (Villar and Kauvar, 1994). In addition, a consistent pattern of frequency variation depending on the type of amino acid side chain substitution is an additional validation of the accuracy of the tetramer enrichment method.
To address whether the CD8+ T cells specific for HCV variants detected by tetramer staining are able to mount a functional response, we generated T cell clones specific for the wild type and L5M HCV epitopes, and also the L5N epitope, which was detected at lower frequency in comparison to other variants (Figure 6A and Table S7). When comparing within the same blood donor, we observed a similar sensitivity to a dose titration of the variant peptides by the CD107 mobilization assay (Figure S6A and S6B). We then generated JY target cells expressing the entire HCV NS3 protein containing the wild type, L5M, or L5N epitope. We were again able to generate CD8+ T cell clones that preferentially killed the appropriate target cell for all three cases (Figure 6B).
DISCUSSION
Perhaps the most surprising result in our study is that self-specific CD8+ T cells are so abundant in the peripheral blood of healthy adult people. We make this conclusion after making over 40 separate measurements from blood bank donors to determine the frequency of CD8+ T cells recognizing four self and four foreign epitopes (not including SMCY). These self-specific T cells stained robustly with tetramers at an intensity generally consistent with agonist level affinities (Kd ~100 to 1 uM) (Savage et al., 1999). We surveyed nine additional self epitopes and eleven foreign epitopes with comparable results. Using the H-Y system, we showed that clones derived from male SMCY (and therefore self-) specific CD8+ T cells overlap broadly with female derived SMCY specific clones in their functional avidity to both pulsed and endogenously presented antigen. This is in addition to published data showing that even CD8+ T cells with lower affinity for endogenous antigens can be drawn into immune responses in the setting of inflammation (Zehn et al., 2009; Enouz et al., 2012). We wish to emphasize that our results are not consistent with immunologic ignorance. Specifically we showed that male derived SMCY specific CD8+ T cells are found at a lower frequency and have a pattern of gene transcript expression that is distinct in comparison to the equivalent cells in females. In addition, we found that self-specific, primary CD8+ T cells are resistant to activation ex vivo in comparison to foreign-specific cells.
It is relevant to note two reports by Bousso and Bevan and their colleagues, in which they analyzed two different TCR β chain transgenic mouse systems which bias T cells towards the recognition of an endogenous antigen (SMCY3 and ovalbumin, respectively) (Bouneaud et al., 2000; Zehn and Bevan, 2006). In both systems, T cells specific for the endogenous antigen persist, yet have an approximately 100-fold reduction in functional avidity. Our experimental system differs in that we directly observed multiple self-antigen specific CD8+ T cells, from unmanipulated humans, that stain with tetramers at an intensity consistent with TCR ligand agonists (Savage et al., 1999). In addition, in the case of the SMCY epitope, CD8+ T cell clones from both men and women exhibited very similar functional responsiveness to antigen.
One explanation for why so many of the early mouse studies on negative selection in the thymus showed massive deletion could be that TCR transgenes almost always originated from T cell clones that were the best responders to a given antigen, which may have predisposed them to clonal deletion. In addition, in comparison to unmanipulated mice, the introduction of α and β TCR transgenes is known to increase the surface density of TCRs and also results in the early expression of mature TCR at the double negative (DN) thymocyte stage (von Boehmer and Kisielow, 2006). This shift in timing can precipitate lineage diversion and impair proliferation between the DN and double positive thymocyte stages. Such differences may impact negative selection, although in a H-Y TCR transgenic mouse model with appropriately timed expression of the TCR α chain, comparable clonal deletion was seen (McCaughtry et al., 2008). A final consideration is the effect of repertoire skewing in transgenic mice. A reduced efficiency in positive selection or diversion into the CD4+ regulatory T cell lineage has been reported for particular TCR transgenes when their proportion in the thymus exceeds 5% or 1%, respectively (Huesmann et al., 1991; Bautista et al., 2009). A limiting number of environmental niches has been proposed as a basis for this phenomenon. The survival and fate of self-specific CD8+ T cells might similarly depend on a finite number of niches.
Our results parallel the developmental observations made in other lymphocyte lineages. In the B cell lineage, 20% of mature B cells are reported to recognize self antigen in the periphery in humans (Wardemann et al., 2003). Jensen et al. demonstrate that self-specific γδ T cells are not deleted in the thymus of normal non-transgenic mice, in contrast to earlier work using TCR transgenic γδ T cells (Jensen et al., 2008). Our results are also consistent with the work of Nepom and colleagues in CD4+ T cells (Mallone et al., 2005), as well as the work of Jenkins and colleagues, who show that the clonal deletion of CD4+ T cells that recognize an antigen expressed endogenously by a transgene in the mouse (Moon et al., 2011) is incomplete, with approximately one third of the specific T cells migrating to the periphery. Recently, Sakaguchi and colleagues have reported the presence of anergic MART-1 specific CD8+ T cells in healthy people, similar to a population previously reported by Romero and colleagues (Pittet et al., 1999; Maeda et al., 2014).
Given that we detect such a large pool of self-specific CD8+ T cells, how are they kept in check? Our results showed that self-specific CD8+ T cells are significantly anergic compared to foreign-specific cells, although this can be overcome in vitro by anti-CD3 and anti-CD28 crosslinking combined with IL2. The resistance of self-specific CD8+ T cells to activation was indicated on the transcript level with single cell qPCR in the H-Y system, in which we found the preferential and correlated expression of IL2RA, IL21R, and BCL2L1 (i.e., BCLXL) - genes associated with survival and proliferation - in ex vivo stimulated T cells from women as compared to men. This phenotype was then confirmed functionally by following stimulated, antigen-specific CD8+ T cells that had been sorted using pooled tetramers over several days, and observing proliferation and the protein expression of IL2RA and IL21R in foreign-specific T cells, but not in self-specific cells. Interestingly, IL-2 is required for T cell proliferation, and lack of signaling through the IL-2 receptor has been associated with T cell tolerance, while the expression of BCLXL is associated with increased cell survival (Smith, 1988; Boise et al., 1995; Choi and Schwartz, 2007). The IL21 pathway in turn contributes to cytotoxic T cell expansion and has been linked to the pathogenesis of diabetes mellitus in the NOD mouse (Zeng et al., 2005; McGuire et al., 2009). It is tempting to speculate that the inertia against activation in self-specific CD8+ T cells may be maintained by a protective gene program induced through interactions with any number of cell types in the periphery, including regulatory T cells. Alternatively, such a gene program might be imprinted centrally in the thymus, as both γδ T cells and regulatory CD4+ T cells seem subject to thymic imprinting as well (Jensen et al., 2008; Wing and Sakaguchi, 2010).
The presence of an abundant pool of self-specific peripheral T cells – as opposed to their elimination by clonal deletion - is important because it further shifts burden of maintaining tolerance to other mechanisms that must function for the life of the individual. For this reason, we find it significant that preproinsulin reactive CD8+ T cells can be found at a relatively high frequency in healthy individuals and especially so in type 1 diabetics. In mice, proinsulin, the proteolytic product of preproinsulin, is proposed as an early target in the epitope spreading hierarchy of NOD diabetes (Nakayama et al., 2005). In humans, insulin and preproinsulin is proposed to be an immunologic target associated with T1D patients (Kent et al., 2005; Martinuzzi et al., 2008; Skowera et al., 2008). It is possible that the presence of a substantial pool of peripheral insulin reactive T cells contributes to an increased risk of developing T1D when other mechanisms of tolerance break down.
So why aren’t more of these self-specific cells removed in the thymus? We suggest that the reason is that they may still be needed to defend against pathogens, which historically are a much greater threat than autoimmunity to children and young adults, the main drivers of a population’s survival. If T cells specific for self epitopes were efficiently deleted it would leave many “holes in the T cell repertoire” which pathogens would almost certainly exploit. In contrast, if the immune system retained self-specific T cells in an anergic state, they could become activated with the strong stimulus of infection (Ohashi et al., 1991). The fact that we were able to stimulate and expand self-specific T cells with anti-CD3/CD28 antibodies to make clones shows that this is possible and suggests that such a mechanism exists in vivo. Consistent with this hypothesis is a recent report by Jameson and colleagues showing in mice that CD5hi naïve CD8+ T cells, which are thought to interact more strongly with endogenous peptide MHC, respond better to infection than CD5lo CD8+ naïve T cells (Fulton et al., 2015). In this context, it is interesting to note that within medicine there are numerous examples of autoimmune phenomena, cellular and humoral, following an earlier infection (Blank et al., 2007; Chen et al., 2012). Over the years, various holes in the TCR ligand repertoire have been postulated, but these have largely depended on antigen dependent proliferation assays, and thus could easily miss anergic cells (Vidovic and Matzinger, 1988; Wolfl et al., 2008). We also found that for at least one viral epitope, there is an unbroken “wall” of T cell specificities for all possible variants at a key position with respect to T cell recognition. Thus we propose that the immune system prunes away only the most self-reactive T cells while retaining a sizable pool of TCR specificities against self such that every possible peptide bound to a given MHC can be recognized.
EXPERIMENTAL PROCEDURES
Human PBMCs
Human PBMCs were obtained from platelet apheresis donors through the Stanford Blood Bank according to IRB protocol. All donors were HLA-A*0201+, and all were Caucasian except for one Asian donor. A separate IRB protocol was used to collect 50 to 100ml of whole blood from individuals with T1D as well as age matched controls.
Tetramer Enrichment
For human samples, LRS chambers containing PBMCs from platelet donors were processed within 24 hours. CD8+ T cells were concentrated by negative depletion with RosetteSep (Stemcell Technologies). Red blood cells (RBCs) were then lysed with ACK buffer, filtered through 70um mesh, and resuspended in flow cytometry buffer (Ca2+/Mg2+ free PBS with 2% heat inactivated FCS, 0.5mM EDTA. 0.1% sodium azide was added except when cells were sorted for cloning or qPCR). For mice, 4–6 week old C57BL/6 were sacrificed and their lymph nodes and spleens were resuspended in RBC lysis buffer prior to resuspension in flow cytometry buffer.
Cells resuspended in flow cytometry buffer at ~ 50×106/100ul were incubated 1hr at room temperature with peptide MHC tetramer(s) (~20nM each, labeled with PE or PE-Cy conjugate), fluorophore labeled anti-CD8 antibody, purified anti-CD32, APC anti-CD16, and 50uM biotin. After washing, cells were incubated with anti-PE Microbeads (Miltenyi) and tetramer enriched on magnetized Miltenyi LS columns. After enrichment, the column fraction and an aliquot of the flow through were stained for 30 minutes on ice with a viability stain (either propidium iodide or Aqua Live/Dead Stain (Invitrogen)) and an additional antibody mixture before flow cytometric analysis on a BD LSRII or Aria. The antibodies used are listed in the supplementary section.
CD107 Mobilization Assay
Performed similarly as previously described (Rubio et al., 2003) with modifications: Initially, the assay was performed with peptide pulsed T2 cells and T cell clone in a 1:1 ratio. Depending on the amount of T cell clone available, 50,000 or 100,000 of each cell type were incubated together in 50ul media the presence of anti-CD107a antibody and 10nM monensin for 4.5 hours. We obtained a similar to more sensitive readout by pulsing the T cell clones directly with peptide in the absence of T2 cells (data not shown). Consequently this latter method of performing the CD107 assay was used from then on.
in vitro Stimulation of CD8+ T cells Enriched with Pooled Peptide MHC Tetramer
We generated HLA-A*0201 tetramers loaded with six different self peptides, all using the same fluorophore (e.g, PE), and also generated a second pool of six foreign peptide HLA-A* 0201 tetramers with a different fluorophore (e.g., PE-Cy7) in order to be able to distinguish between the two groups (Table S6). For this series of experiments, the HLA-A* 0201 monomer used incorporated a myc tag (Newell et al., 2012), and so tetramer enrichment was performed as above, but with anti-myc Microbeads (Miltenyi) instead of anti-PE Microbeads. Cell sorting and stimulation were performed as for the single cell qPCR experiments with the following exceptions: the incubation times were either 4.5 or 7.4 days, no Alexa Fluor tracer was used, and where indicated, CFSE staining was performed on the mixture of sorted cells and autologous feeder cells before peptide and anti-CD28 antibody were added.
Supplementary Material
HIGHLIGHTS.
Similar numbers of human blood CD8+ T cells recognize self vs. novel foreign antigens.
H-Y T cells in men are 1/3 as frequent as in women but have similar functional avidity.
Self-specific CD8+ T cells are resistant to activation and/or expansion.
Inefficient self-specific T cell deletion may allow better protection against infection.
ACKNOWLEDGEMENTS
We thank Peter Lee, Hongxiang Yu, Tor Stuge, Alexandre Johannsen, Alasdair Leslie, Tao Dong, and Carol Clayberger for help and advice with cell culture; Jeffrey Glenn and Menashe Elazar for the HCV NS3 plasmid; Karin Kealoha, Linda Enomoto, and Cynthia Evora from the Stanford Blood Bank for their assistance in obtaining blood samples; Christina Meyer and Xun Zeng from the Yueh-Hsiu Chien laboratory for help generating JY cells expressing HCV NS3; Olivia M. Martinez and Olivia Hatton for the OH cell line; Marty Bigos, Catherine Crumpton, Ometa Herman, Tim Knaak, David Parks, and Jonathan van Dyke for assistance at the flow cytometry facility; Tandy Aye, Leo Hansmann, and Ileana Rau for helpful suggestions; Barbara Whyte and Rick Cuevas for administrative assistance. This work was supported by the Damon Runyon Cancer Research Foundation (W.Y.: Postdoctoral Fellowship; N.J.: Damon Runyon-Rachleff Innovator, DRR-32-15), the NIH (M.M.D.: U19-AI090019 and U19-AI57229; W.Y.: 1K08DK093709-01; N.J.: R00AG040149), the Cancer Prevention and Research Institute of Texas (N.J: R1120), the Fondation ARC pour la Recherche sur le Cancer (S.V.: EML2012090493), the Institut National du Cancer (S.V.: INCa 2012-054), the Agence Nationale de la Recherche (Laboratoire d'Excellence Toulouse Cancer, S.V.), and the Howard Hughes Medical Institute to W.Y. and M.M.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alanio C, Lemaitre F, Law HK, Hasan M, Albert ML. Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood. 2010;115:3718–3725. doi: 10.1182/blood-2009-10-251124. [DOI] [PubMed] [Google Scholar]
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- Appay V, Dunbar PR, Callan M, Klenerman P, Gillespie GM, Papagno L, Ogg GS, King A, Lechner F, Spina CA, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8:379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- Barnea E, Beer I, Patoka R, Ziv T, Kessler O, Tzehoval E, Eisenbach L, Zavazava N, Admon A. Analysis of endogenous peptides bound by soluble MHC class I molecules: a novel approach for identifying tumor-specific antigens. Eur J Immunol. 2002;32:213–222. doi: 10.1002/1521-4141(200201)32:1<213::AID-IMMU213>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Bautista JL, Lio CW, Lathrop SK, Forbush K, Liang Y, Luo J, Rudensky AY, Hsieh CS. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol. 2009;10:610–617. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg LJ, Fazekas de St Groth B, Pullen AM, Davis MM. Phenotypic differences between alpha beta versus beta T-cell receptor transgenic mice undergoing negative selection. Nature. 1989;340:559–562. doi: 10.1038/340559a0. [DOI] [PubMed] [Google Scholar]
- Blank M, Barzilai O, Shoenfeld Y. Molecular mimicry and auto-immunity. Clin Rev Allergy Immunol. 2007;32:111–118. doi: 10.1007/BF02686087. [DOI] [PubMed] [Google Scholar]
- Boise LH, Minn AJ, Noel PJ, June CH, Accavitti MA, Lindsten T, Thompson CB. CD28 costimulation can promote T cell survival by enhancing the expression of Bcl-XL. Immunity. 1995;3:87–98. doi: 10.1016/1074-7613(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Bouneaud C, Kourilsky P, Bousso P. Impact of negative selection on the T cell repertoire reactive to a self-peptide: a large fraction of T cell clones escapes clonal deletion. Immunity. 2000;13:829–840. doi: 10.1016/s1074-7613(00)00080-7. [DOI] [PubMed] [Google Scholar]
- Burnet FM. The Clonal Selection Theory of Acquired Immunity. Nashville: Vanderbilt University Press; 1959. [Google Scholar]
- Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, Cheng Y, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Schwartz RH. Molecular mechanisms for adaptive tolerance and other T cell anergy models. Semin Immunol. 2007;19:140–152. doi: 10.1016/j.smim.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Delluc S, Ravot G, Maillere B. Quantification of the preexisting CD4 T-cell repertoire specific for human erythropoietin reveals its immunogenicity potential. Blood. 2010;116:4542–4545. doi: 10.1182/blood-2010-04-280875. [DOI] [PubMed] [Google Scholar]
- Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enouz S, Carrie L, Merkler D, Bevan MJ, Zehn D. Autoreactive T cells bypass negative selection and respond to self-antigen stimulation during infection. J Exp Med. 2012;209:1769–1779. doi: 10.1084/jem.20120905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton RB, Hamilton SE, Xing Y, Best JA, Goldrath AW, Hogquist KA, Jameson SC. The TCR's sensitivity to self peptide-MHC dictates the ability of naive CD8(+) T cells to respond to foreign antigens. Nat Immunol. 2015;16:107–117. doi: 10.1038/ni.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JM, Devoss JJ, Friedman RS, Wong DJ, Tan YX, Zhou X, Johannes KP, Su MA, Chang HY, Krummel MF, Anderson MS. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science. 2008;321:843–847. doi: 10.1126/science.1159407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman A, Kappler JW, Marrack P, Pullen AM. Superantigens: mechanism of Tcell stimulation and role in immune responses. Annu Rev Immunol. 1991;9:745–772. doi: 10.1146/annurev.iy.09.040191.003525. [DOI] [PubMed] [Google Scholar]
- Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- Huesmann M, Scott B, Kisielow P, von Boehmer H. Kinetics and efficacy of positive selection in the thymus of normal and T cell receptor transgenic mice. Cell. 1991;66:533–540. doi: 10.1016/0092-8674(81)90016-7. [DOI] [PubMed] [Google Scholar]
- Jenkins MK, Chu HH, McLachlan JB, Moon JJ. On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu Rev Immunol. 2010;28:275–294. doi: 10.1146/annurev-immunol-030409-101253. [DOI] [PubMed] [Google Scholar]
- Jensen KD, Su X, Shin S, Li L, Youssef S, Yamasaki S, Steinman L, Saito T, Locksley RM, Davis MM, et al. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immunity. 2008;29:90–100. doi: 10.1016/j.immuni.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler JW, Roehm N, Marrack P. T cell tolerance by clonal elimination in the thymus. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- Kent SC, Chen Y, Bregoli L, Clemmings SM, Kenyon NS, Ricordi C, Hering BJ, Hafler DA. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature. 2005;435:224–228. doi: 10.1038/nature03625. [DOI] [PubMed] [Google Scholar]
- Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- Kotturi MF, Scott I, Wolfe T, Peters B, Sidney J, Cheroutre H, von Herrath MG, Buchmeier MJ, Grey H, Sette A. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J Immunol. 2008;181:2124–2133. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg D, Knight RR, Estorninho M, Ellis RJ, Kester MG, de Ru A, Eichmann M, Huang GC, Powrie J, Dayan CM, et al. Circulating preproinsulin signal peptide-specific CD8 T cells restricted by the susceptibility molecule HLA-A24 are expanded at onset of type 1 diabetes and kill beta-cells. Diabetes. 2012;61:1752–1759. doi: 10.2337/db11-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legoux F, Debeaupuis E, Echasserieau K, De La Salle H, Saulquin X, Bonneville M. Impact of TCR reactivity and HLA phenotype on naive CD8 T cell frequency in humans. J Immunol. 2010;184:6731–6738. doi: 10.4049/jimmunol.1000295. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Nishikawa H, Sugiyama D, Ha D, Hamaguchi M, Saito T, Nishioka M, Wing JB, Adeegbe D, Katayama I, Sakaguchi S. Detection of self-reactive CD8(+) T cells with an anergic phenotype in healthy individuals. Science. 2014;346:1536–1540. doi: 10.1126/science.aaa1292. [DOI] [PubMed] [Google Scholar]
- Mallone R, Kochik SA, Reijonen H, Carson B, Ziegler SF, Kwok WW, Nepom GT. Functional avidity directs T-cell fate in autoreactive CD4+ T cells. Blood. 2005;106:2798–2805. doi: 10.1182/blood-2004-12-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallone R, Martinuzzi E, Blancou P, Novelli G, Afonso G, Dolz M, Bruno G, Chaillous L, Chatenoud L, Bach JM, van Endert P. CD8+ T-cell responses identify beta-cell autoimmunity in human type 1 diabetes. Diabetes. 2007;56:613–621. doi: 10.2337/db06-1419. [DOI] [PubMed] [Google Scholar]
- Martinuzzi E, Novelli G, Scotto M, Blancou P, Bach JM, Chaillous L, Bruno G, Chatenoud L, van Endert P, Mallone R. The frequency and immunodominance of islet-specific CD8+ T-cell responses change after type 1 diabetes diagnosis and treatment. Diabetes. 2008;57:1312–1320. doi: 10.2337/db07-1594. [DOI] [PubMed] [Google Scholar]
- McCaughtry TM, Baldwin TA, Wilken MS, Hogquist KA. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J Exp Med. 2008;205:2575–2584. doi: 10.1084/jem.20080866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire HM, Vogelzang A, Hill N, Flodstrom-Tullberg M, Sprent J, King C. Loss of parity between IL-2 and IL-21 in the NOD Idd3 locus. Proc Natl Acad Sci U S A. 2009;106:19438–19443. doi: 10.1073/pnas.0903561106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows L, Wang W, den Haan JM, Blokland E, Reinhardus C, Drijfhout JW, Shabanowitz J, Pierce R, Agulnik AI, Bishop CE, et al. The HLA-A*0201-restricted H-Y antigen contains a posttranslationally modified cysteine that significantly affects T cell recognition. Immunity. 1997;6:273–281. doi: 10.1016/s1074-7613(00)80330-1. [DOI] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Dash P, Oguin TH, 3rd, McClaren JL, Chu HH, Thomas PG, Jenkins MK. Quantitative impact of thymic selection on Foxp3+ and Foxp3- subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc Natl Acad Sci U S A. 2011;108:14602–14607. doi: 10.1073/pnas.1109806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, Yu L, Wegmann DR, Hutton JC, Elliott JF, Eisenbarth GS. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005;435:220–223. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell EW, Klein LO, Yu W, Davis MM. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat Methods. 2009;6:497–499. doi: 10.1038/nmeth.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obar JJ, Khanna KM, Lefrancois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi PS, Oehen S, Buerki K, Pircher H, Ohashi CT, Odermatt B, Malissen B, Zinkernagel RM, Hengartner H. Ablation of "tolerance" and induction of diabetes by virus infection in viral antigen transgenic mice. Cell. 1991;65:305–317. doi: 10.1016/0092-8674(91)90164-t. [DOI] [PubMed] [Google Scholar]
- Pittet MJ, Valmori D, Dunbar PR, Speiser DE, Lienard D, Lejeune F, Fleischhauer K, Cerundolo V, Cerottini JC, Romero P. High frequencies of naive Melan-A/MART-1-specific CD8(+) T cells in a large proportion of human histocompatibility leukocyte antigen (HLA)-A2 individuals. J Exp Med. 1999;190:705–715. doi: 10.1084/jem.190.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Stuge TB, Singh N, Betts MR, Weber JS, Roederer M, Lee PP. Ex vivo identification, isolation and analysis of tumor-cytolytic T cells. Nat Med. 2003;9:1377–1382. doi: 10.1038/nm942. [DOI] [PubMed] [Google Scholar]
- Savage PA, Boniface JJ, Davis MM. A kinetic basis for T cell receptor repertoire selection during an immune response. Immunity. 1999;10:485–492. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- Schmidt J, Neumann-Haefelin C, Altay T, Gostick E, Price DA, Lohmann V, Blum HE, Thimme R. Immunodominance of HLA-A2-restricted hepatitis C virus-specific CD8+ T cell responses is linked to naive-precursor frequency. J Virol. 2011;85:5232–5236. doi: 10.1128/JVI.00093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha WC, Nelson CA, Newberry RD, Kranz DM, Russell JH, Loh DY. Positive and negative selection of an antigen receptor on T cells in transgenic mice. Nature. 1988;336:73–76. doi: 10.1038/336073a0. [DOI] [PubMed] [Google Scholar]
- Silverstein AM. Autoimmunity versus horror autotoxicus: the struggle for recognition. Nat Immunol. 2001;2:279–281. doi: 10.1038/86280. [DOI] [PubMed] [Google Scholar]
- Skowera A, Ellis RJ, Varela-Calvino R, Arif S, Huang GC, Van-Krinks C, Zaremba A, Rackham C, Allen JS, Tree TI, et al. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J Clin Invest. 2008;118:3390–3402. doi: 10.1172/JCI35449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KA. Interleukin-2: inception, impact, and implications. Science. 1988;240:1169–1176. doi: 10.1126/science.3131876. [DOI] [PubMed] [Google Scholar]
- Stuge TB, Holmes SP, Saharan S, Tuettenberg A, Roederer M, Weber JS, Lee PP. Diversity and recognition efficiency of T cell responses to cancer. PLoS Med. 2004;1:e28. doi: 10.1371/journal.pmed.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Toebes M, Coccoris M, Bins A, Rodenko B, Gomez R, Nieuwkoop NJ, van de Kasteele W, Rimmelzwaan GF, Haanen JB, Ovaa H, Schumacher TN. Design and use of conditional MHC class I ligands. Nat Med. 2006;12:246–251. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Uematsu Y, Ryser S, Dembic Z, Borgulya P, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. In transgenic mice the introduced functional T cell receptor beta gene prevents expression of endogenous beta genes. Cell. 1988;52:831–841. doi: 10.1016/0092-8674(88)90425-4. [DOI] [PubMed] [Google Scholar]
- van den Beemd R, Boor PP, van Lochem EG, Hop WC, Langerak AW, Wolvers-Tettero IL, Hooijkaas H, van Dongen JJ. Flow cytometric analysis of the Vbeta repertoire in healthy controls. Cytometry. 2000;40:336–345. doi: 10.1002/1097-0320(20000801)40:4<336::aid-cyto9>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Velthuis JH, Unger WW, Abreu JR, Duinkerken G, Franken K, Peakman M, Bakker AH, Reker-Hadrup S, Keymeulen B, Drijfhout JW, et al. Simultaneous detection of circulating autoreactive CD8+ T-cells specific for different islet cell-associated epitopes using combinatorial MHC multimers. Diabetes. 2010;59:1721–1730. doi: 10.2337/db09-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic D, Matzinger P. Unresponsiveness to a foreign antigen can be caused by selftolerance. Nature. 1988;336:222–225. doi: 10.1038/336222a0. [DOI] [PubMed] [Google Scholar]
- Villar HO, Kauvar LM. Amino acid preferences at protein binding sites. FEBS Lett. 1994;349:125–130. doi: 10.1016/0014-5793(94)00648-2. [DOI] [PubMed] [Google Scholar]
- von Boehmer H. Developmental biology of T cells in T cell-receptor transgenic mice. Annu Rev Immunol. 1990;8:531–556. doi: 10.1146/annurev.iy.08.040190.002531. [DOI] [PubMed] [Google Scholar]
- von Boehmer H, Kisielow P. Negative selection of the T-cell repertoire: where and when does it occur? Immunol Rev. 2006;209:284–289. doi: 10.1111/j.0105-2896.2006.00346.x. [DOI] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Weinschenk T, Gouttefangeas C, Schirle M, Obermayr F, Walter S, Schoor O, Kurek R, Loeser W, Bichler KH, Wernet D, et al. Integrated functional genomics approach for the design of patient-individual antitumor vaccines. Cancer Res. 2002;62:5818–5827. [PubMed] [Google Scholar]
- Wing K, Sakaguchi S. Regulatory T cells exert checks and balances on self tolerance and autoimmunity. Nat Immunol. 2010;11:7–13. doi: 10.1038/ni.1818. [DOI] [PubMed] [Google Scholar]
- Wolfl M, Rutebemberwa A, Mosbruger T, Mao Q, Li HM, Netski D, Ray SC, Pardoll D, Sidney J, Sette A, et al. Hepatitis C virus immune escape via exploitation of a hole in the T cell repertoire. J Immunol. 2008;181:6435–6446. doi: 10.4049/jimmunol.181.9.6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehn D, Bevan MJ. T cells with low avidity for a tissue-restricted antigen routinely evade central and peripheral tolerance and cause autoimmunity. Immunity. 2006;25:261–270. doi: 10.1016/j.immuni.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R, Spolski R, Finkelstein SE, Oh S, Kovanen PE, Hinrichs CS, Pise-Masison CA, Radonovich MF, Brady JN, Restifo NP, et al. Synergy of IL-21 and IL-15 in regulating CD8+ T cell expansion and function. J Exp Med. 2005;201:139–148. doi: 10.1084/jem.20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.