Abstract
Human cardiac electrophysiology (EP) is a unique system for computational modelling at multiple scales. Due to the complexity of the cardiac excitation sequence, coordinated activity must occur from the single channel to the entire myocardial syncytium. Thus, sophisticated computational algorithms have been developed to investigate cardiac EP at the level of ion channels, cardiomyocytes, multicellular tissues, and the whole heart. Although understanding of each functional level will ultimately be important to thoroughly understand mechanisms of physiology and disease, cardiac arrhythmias are expressly the product of cardiac tissue—containing enough cardiomyocytes to sustain a reentrant loop of activation. In addition, several properties of cardiac cellular EP, that are critical for arrhythmogenesis, are significantly altered by cell-to-cell coupling. However, relevant human cardiac EP data, upon which to develop or validate models at all scales, has been lacking. Thus, over several years, we have developed a paradigm for multiscale human heart physiology investigation and have recovered and studied over 300 human hearts. We have generated a rich experimental dataset, from which we better understand mechanisms of arrhythmia in human and can improve models of human cardiac EP. In addition, in collaboration with computational physiologists, we are developing a database for the deposition of human heart experimental data, including thorough experimental documentation. We anticipate that accessibility to this human heart dataset will further human EP computational investigations, as well as encourage greater data transparency within the field of cardiac EP.
Keywords: Human cardiac electrophysiology
Introduction
Since the start of the genomic research era, substantial research efforts have led to mapping the human genome,1,2 considering genes as drivers of system interactions.3–6 Then, investigators have isolated many of these genes, one-by-one, to detail the proteins they encode and their anatomical and functional signatures. However, the totality of genetic sequence data does not inform us about physiological function,5,7 nor does a gene or protein's role in a reductionist context necessarily pertain in the greater environment of tissues, organs, and systems.8 Emergent properties arise from non-linear responses when components interact within profoundly complex multidimensional and multiscale networks. As was elegantly stated by O'Malley and Dupre,3 ‘it is crucial … to analyze systems as systems, and not as mere collections of parts in order to understand the emergent properties of component interactions’. Thus, computational tools that enable us to understand systems-level networks will enhance biomedical research and discovery.
While human cardiac cell electrophysiology (EP) models have become very sophisticated and highly complex, extensions of these models to the tissue and whole-organ level are needed to elucidate pro-arrhythmic mechanisms. However, more tissue-level experimental data must be incorporated to generate improved models with cardiac cell-to-cell coupling. While data on human cardiac tissues are still emerging, we have developed a paradigm for multiscale investigation of the human heart, from molecules and cells to intact tissue and organ preparations. To date, we have procured over 300 live donor and failing human hearts, which we have studied for arrhythmogenic properties. In addition, using criteria established by the Physiome project,9 we are making these data available through an open-access, searchable database. We aim to review the existing human ventricular cell EP models, and their incorporation into tissue-level simulations. We will also discuss our programme for multiscale investigation of human heart physiology in health and disease, and standards for data sharing and transparency for cardiac EP.
Modelling human cardiac cellular electrophysiology
Computational modelling of cardiac myocyte EP is well established and arose from the pioneering work of Hodgkin and Huxley (HH) on cellular excitability.10,11 The HH current equations have served as the foundation for computational EP due to similarities between neuronal and cardiac excitable properties. As a modification of the HH current equations, FitzHugh and Nagumo (FHN) developed simplified equations, which could simulate excitation and propagation.12–14 Though the FHN equations do not describe individual ionic currents, reducing excitation and propagation to variables for voltage, stimulus, and recovery, they enable simulation of two-dimensional (2D) and 3D cardiac tissues with less computational requirement.15,16 In contrast, for more recent cardiac cellular models, more sophisticated Markov chain equations have largely supplanted HH formalism, enabling independent ion channel representations and improved model fidelity. This section aims to briefly trace the development of EP models for the human ventricular myocyte and specialized conduction system. We will discuss foundational non-human models, which served as the basis for existing human simulations, and the currently established human ventricular EP models. For greater detail on other cardiomyocyte models, we refer to other reviews on the subject.11,17,18
Purkinje cell modelling
Purkinje cells were the first simulated cardiac myocytes, in the foundational work of Denis Noble.19,20 This initial model of the cardiac Purkinje cell used modified HH formalism, accounting for Na, K, and background Cl currents. Later, McAllister, Noble, and Tsien extended this general model to include the newly discovered inward Ca current, using HH formalism.21,22 While these early models are limited due to challenges with experimental voltage-clamp studies,23 they provided key conceptual innovations for computational cardiac EP. The DiFrancesco–Noble (DN)24 model of the Purkinje cell was later created through elaboration of early ventricular and Purkinje cell models. The DN model incorporated updated ionic currents, automaticity, and dynamic Ca handling.23 This was the first model to depart from HH formalism to account for variations in K and Na during the AP.17 Due to the highly innovative nature of this model, it has served as the foundation for numerous future models of various cardiac cell types.
Ventricular cell models
Following the Purkinje cell model, the foundational ventricular cell model was the HH formulated Beeler–Reuter (BR) model.25 This model was notable as the first to include variations in intracellular Na concentration, intracellular and extracellular K concentrations, and sodium-potassium (Na-K) pump and sodium-calcium exchanger (NCX) activities.22 Subsequently, due to the similarity between guinea pig and human ventricular action potentials (APs), sophisticated computational models were developed based guinea pig experimental data, most notably that of Luo and Rudy (LR).26 This model was based on BR, with reformulation of key depolarizing and repolarizing currents from improved experimental data. Luo and Rudy then updated their original model by incorporating dynamic Na, K, and Ca concentrations, which is referred to as the LR dynamic model (LRd).17,27,28
The first cellular EP model of the human ventricular myocyte was published by Priebe and Beuckelmann (PB)29 and was formulated largely based upon the LRd model. This model incorporated INa, INCX, INa-K, and background currents from the LRd model, while reformulating Ca and K currents based on human data. The PB model had the benefit that Beuckelmann and colleagues generated much of the human experimental data, and thus had a deep understanding of the collection and analysis conditions. This model is also notable for including data from both non-failing and failing human hearts. Unfortunately, the PB model failed to attract attention from the modelling community, because it was less freely distributed than other models.22
Until relatively recently, two human ventricular models were predominantly used: the Ten Tusscher, Noble, Noble, and Panfilov model (TNNP) and the Iyer, Mazhari, and Winslow (IMW) model.30–32 Both models use Markov chain equations for relevant ionic currents and are based on the LRd model, with updated experimental data for certain channels. As such, guinea pig data make up a big portion of the experimental foundation for these human models (Figure 1). Also, there were significant gaps in available human data when these models were developed. However, the more recently improved TNNP model includes updated Ca dynamics, IKs, and ICa,L.34
Figure 1.

Tree map representations of the (A) IMW and (B) TNNP human cellular models, showing relative proportions of experimental data from specific species. Due to lack of EP data from non-failing human hearts, both models were generated based on a large fraction of non-human work. Experimental data proportions for each of these models were obtained from Niederer et al.33
More recently, several additional human ventricular models have been developed to improve fidelity with human data, including those of Grandi, Pasqualini, and Bers (GPB); O'Hara, Virag, Varro, and Rudy (OVVR); and Carro, Rodriguez, Laguna, and Pueyo (CRLP)34–36 The GPB model of human ventricular AP includes different subcellular regions, including subsarcolemmal and junctional compartments, along with several ionic currents and intricate Ca handling from its rabbit model predecessor.36,37 Consequently, it also mimics the rabbit model AP restitution properties. Due to the inability of the GPB model to replicate human AP restitution properties and heart rate adaptation, CRLP successfully updated GPB model to have more consistency with current human experimental data. Carro, Rodriguez, Laguna, and Pueyo introduced fast and slow ICa,L inactivation gates and reformulated NCX flux and K currents.34 Importantly, only human data were used to update and refine the CRLP model. For the OVVR model, O'Hara and colleagues pull extensively from human ventricular experimental data. Thus, the OVVR model is the most up-to-date model of the human ventricular AP, and it includes parameterization for ICa,L, Ito, INCX, IK1, IKr, IKs, INa, and INa-K.35 In addition, the authors are transparent about model limitations, especially with regard to deficiencies in human data.
Multiscale systems physiology approach to cardiac modelling
The goal of systems physiology is to integrate multiscale physiological and anatomical data to understand complex behaviours from cell to organ system.38 However, to truly understand physiology, the functional unit of a system must be of utmost consideration. For disorders such as cancer, one might argue that the functional unit of disease is a single cell, as cancer is initiated by abnormal signalling within a single rogue cell. Thus, modelling properties of a cell may be scalable to help understand more complex tumour behaviours, such as immune system avoidance and metastasis. Though this approach may be suitable for some organs and/or diseases, most physiological function cannot be reduced as simply. Cardiac EP is a keen example of the inability to reduce function to the single-cell physiological scale. Though aberrant electrical behaviour can be demonstrated in single isolated cardiac myocytes, most types of arrhythmia do not exist in isolated cells. Generally, a ring of cardiomyocytes large enough to sustain a reentrant loop of activation is the minimum requirement.
Thus, traditional approaches to modelling, including ‘bottom-up’, which starts with genes and proteins and works upward to higher-level function, or ‘top-down’, which begins with higher-level function and then sorts out the details, should be augmented by ‘middle-out’ methodologies as well.3,39,40 In the field of cardiac EP, one might consider the bottom-up approach as modelling of specific ionic currents and cells, and the top-down strategy as working down from the whole heart or circulatory system. A middle-out approach might instead involve starting with a functional middle ground, such as multicellular tissue preparations, that are able to encapsulate the functional properties and architecture of the entire relevant system, i.e. the ‘functional unit’ of cardiac arrhythmias. From this systems level, one could understand the behaviour of arrhythmias, and then work towards the ‘bottom’ to understand the underlying molecular/cellular changes and towards the ‘top’ to understand the ultimate effects on human health and well-being.
The importance of tissue-level modelling to comprehend human heart EP is demonstrated by several electrical properties, fundamental to arrhythmia development, that do not exist in individual isolated myocytes, but emerge with cell-to-cell coupling.41 For example, such basic properties as conduction velocity (CV), conduction safety (CS), and wavefront curvature (WC) are critical determinants of reentrant wavefront maintenance, and all three are properties of tissues, not cells. The primary determinant of CV is the degree of cellular coupling, not the cellular-level correlate of maximum AP upstroke velocity. Conduction safety and WC also depend on cell-to-cell coupling and cannot be conceptualized in a single-cell model. In addition, other properties of cardiac AP morphology, such as amplitude, restitution, and alternans, are significantly altered in tissues when compared with single cells.
Modelling electrophysiology of human cardiac tissue
Due to computational feasibility of simulating interconnected cardiac myocytes at the tissue or whole-heart levels, propagation of electrical activity is often conceptualized as a well-coupled syncytium and modelled by the bidomain or monodomain equations.42–44 In the more comprehensive, but computationally costly, bidomain model, the myocardium is approximated as a continuum of interconnected, anisotropic resistive components, described by two separate (3D) intracellular and extracellular conductivity tensors.45 The monodomain equations are a greater simplification of the bidomain model, in which the myocardium is treated as a single compartment.41 However, as computational power continues to increase, individual cell representations can be incorporated into tissue-level models. While model detail must still be balanced by computational efficiency, using more comprehensive cellular models will hopefully enable greater fidelity to the properties of the myocardial syncytium.
Human tissue models incorporating cellular units
Several human tissue-level computational studies have now been published with varying degrees of cellular model simplification. By incorporating a modified version of their human TNNP cellular model into a 2D sheet model, Ten Tusscher and Panfilov46 were able to investigate the role of restitution slope in the generation of alternans and electrical instability. Then, by again reformulating their human ventricular cell model, they developed a model that is computationally efficient enough for simulations in cardiac tissue, while maintaining detail to represent individual ionic current parameters.31 Thus, they could reliably simulate the effects of ionic current abnormalities that occur in human disorders, such as long QT syndrome. In addition, they demonstrated consistent behaviour of the simplified cell model with four times greater computational efficiency.
Another benefit of simulating human cardiac EP properties within tissue is that additional parameters can be included to represent various myocardial heterogeneities. These include fibre orientation and anatomical differences in ionic currents or conduction properties. In addition, cardiac extracellular matrix is very dense and can become even more pronounced in disease, and fibroblasts make up approximately half of the cells within the heart.47 Thus, Ten Tusscher and Panfilov48 again adjusted their TNNP tissue model to account for effects of fibrosis by incorporating inexcitable, no-flux boundaries into 2D and 3D geometries. These boundaries lead to increased pro-arrhythmic behaviour, including CV slowing and spiral wave formation.
Furthering this work, the group of Dr Rahul Pandit examined the effects of inhomogeneities in the TNNP cardiac tissue model in a series of consecutive studies. First, Majumder et al.49 incorporated fibre orientation and anatomical differences in cardiomyocyte EP properties, resulting in altered scroll-wave dynamics, with anchoring occurring at regions of heterogeneity. In the next model iteration, they incorporate randomly inserted fibroblasts, which were non-excitable but able to couple to a neighbouring cardiomyocyte.50 Importantly, conduction properties in the model were altered by the degree of coupling between fibroblasts and myocytes. Nayak et al.51 then expanded the previous models to allow for more complex coupling between myocytes and fibroblasts. With this model, they demonstrated more complex, non-linear relationships of CV to fibroblast coupling parameters. In addition, local fibroblast inhomogeneities could serve as anchoring centres for spiral waves.
Comparisons of human cardiomyocyte models within tissue simulations
Given that there are several human cardiac cell models, with differing parameters and resulting behaviour, tissue-level models can also vary based on which cellular model is incorporated. Bueno-Orovio, Cherry, and Fenton (BCF)52 developed a 2D tissue model based on the minimal number of differential equations needed to reproduce experimentally derived EP properties from human epicardial, mid-myocardial, and endocardial cells within tissue. They also compared the results of their model to 2D simulations based on the PB, TNNP, and IMW models. In their simulations of reentrant spiral waves, the models showed different results for wave stability and dynamics. The TNNP and BCF minimal ventricular models demonstrate stable reentrant waves with similar dominant frequencies. In contrast, the PB and IMW models both develop wave breakup, with the IMW model producing a wide range of dominant frequencies that are inconsistent with the range of ventricular tachycardia frequencies in patients. Similarly, following the publication of the OVVR and GPB human ventricular cell models, Elshrif and Cherry53 compared the behaviour of these models in tissue. Both models agreed reasonably well with experimentally measured AP duration, but other tissue-level properties were less consistently reproduced. Such direct comparisons of cellular models within tissue will hopefully enable other modellers to choose the appropriate computational method to answer their question of interest.
Challenges of simulating individual cardiac myocytes within tissue
In addition to differences among cellular models, several other challenges arise when incorporating cellular units into larger scale models. Existing cellular models, that are freely available in such repositories as cell markup language (CellML), systems biology markup language (SBML), and FieldML, are not formulated as tissue model components.54–57 Thus, relevant parameters must be extracted from the cell model for incorporation into a tissue model. Support for this process has been developed with the Chaste (‘Cancer, heart and soft-tissue environment’) software library, via the PyCml software, such that appropriately annotated cell models can be used within a tissue simulation.58–60 However, simply inheriting cell models and their parameters into tissue-scale models does not guarantee that the output will behave as tissue would behave.61
Incorporating single-cell models and data into multicellular tissue models, without consideration of data reflecting tissue-specific behaviour, will lead to oversimplification and, potentially, inconsistent model results. To extend cardiac cell models to 2D- and 3D-level simulations of cardiac EP and arrhythmogenesis, tissue characteristics must be reproduced, which impact reentrant wave initiation, dynamics, and stability.52 Important considerations to assess cardiac EP function and arrhythmogenesis at the tissue level include anatomical differences in EP properties, dependence of CV and repolarization on rate, tissue resistance anisotropy, and WC. In addition, AP morphologies are known to vary significantly between isolated cardiac myocytes and tissue, presumably due to electrotonic effects or consequences of the isolation procedure.62
Bridging middle-out with top-down cardiac models
Although the tissue-level approach to cardiac EP modelling enables arrhythmia simulation with some inhomogeneity, whole-heart architecture is much more complex than a block of tissue. The heart has many constituent parts that behave differently, including atria, ventricles, and the conduction system, and the interplay of these different components can significantly impact cardiac electrical behaviour. Also, regionally specific myocardial disease can promote arrhythmias, which might not be observed in a uniform, isolated tissue. Thus, studying EP behaviour in tissue has the limitation that one must extrapolate properties to other heart regions. Tissue-level models must ultimately converge with whole-heart simulations, which are built from in vivo patient EP and imaging data and can replicate global arrhythmia behaviour.63
Coupling model development with experimental data from the human heart
Computational modelling and wet-lab experimentation are like an iterative loop, continually feeding back, with each set of data informing the other. ‘Without data, there is nothing to model; and without models, there is no source of deep predictive understanding’.64 Models rely on experimental data for their formalism and parameters. Few research groups have successfully coupled experimental and computational studies of cardiac EP, and reviews on this powerful research paradigm have been published.43,65 However, most computational modellers rely upon published literature from other investigators, making the original data less easily accessible and restricting the understanding of the conditions under which the experimental data were derived. In addition, once a model has been developed, validation using independent experimental data must occur.42 An important limitation of model development and validation for human cardiac EP has been the supply of relevant data collected from human hearts.
Multiscale human cardiac physiology data
Experimental basis for current models
Though there are several models specifically designed to reproduce the behaviour of human cardiomyocytes, the data upon which these models rely are inconsistent. Even for ‘human models’, experimental data have been collected from different species or cell types and under differing experimental conditions. Previously developed models from various species are often inherited into newer models, including human models, for efficient model generation. Thus, models continue to propagate older datasets, from assorted species, frequently with little reference to the primary data.33,66
From the standpoint of choosing reliable and consistent experimental data for incorporation into models, Fink et al.67 argue that establishing species and experimental conditions consistency are main factors that will enable models to accurately represent physiology. In addition, they argue that the cardiac modelling community needs to reconcile the availability of current experimental data with the complexity of the models being developed, and databases should be constructed for deposition of experimental data for use in modelling studies. Such a repository of original data and information regarding experimental context in which it was acquired will enable improved validation of model components.
Multiscale experimental physiology in the human heart
To this end, we present an experimental paradigm for multiscale human cardiac structure and function investigation. Until recently, in vitro investigation of human heart physiology was a daunting task, achieved primarily at the cellular level only in a limited number of research centres, associated with robust organ recovery organizations. Recent development of the first standalone organ recovery centre in St Louis created a unique opportunity for human organ transplantation and research.68 The logistics of the human heart tissue recovery have been fully detailed elsewhere.69 Briefly, donor human hearts rejected for transplantation or end-stage failing hearts from transplant recipients are recovered at the time of explantation in the operating rooms of the Mid-America Transplant Services (MTS) or Barnes-Jewish Hospitals, respectively. Hearts are then treated with similar measures to hearts taken for transplantation—they are perfused and maintained in ice-cold cardioplegic solution during transport to the research laboratory. Heart tissues can then be dissected for various functional experiments or collected and preserved for later molecular and histological analyses. Since the initiation of this programme, our laboratory has acquired over 300 human hearts, with a historical average recovery frequency of ∼1 heart per week. Since programme inception, heart recovery numbers have increased annually, presumably in part due to improved coordination with procurement teams over time (Figure 2C).
Figure 2.
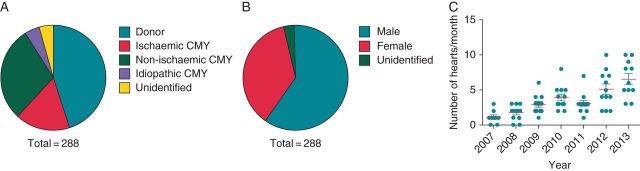
Multiscale human heart physiology programme distributions. Pie charts representing the distribution of recovered human hearts by (A) disease and (B) gender. (C) Chart representing the average number of hearts collected per month for the years spanning from 2007 to 2013. Points represent collection numbers for each individual month. Since initiation of the programme, number of hearts recovered has been increasing annually.
These human heart data serve as a superb platform from which computational models of the human heart in health and disease can be generated, refined, and/or validated. Data types acquired from this invaluable human heart tissue resource range from gene and protein expression levels to functional measurements conducted in living human heart tissues. In many of our experiments, we have used optical imaging of transmembrane potential and intracellular calcium to examine conduction, repolarization, and Ca handling properties in donor vs. failing hearts.70–74 Structures of the specialized conduction system, including the sinoatrial node and atrioventricular junction have also been characterized.75,76 In addition, we have begun conducting studies of structural and/or metabolic remodelling in heart failure in relationship to EP changes. Importantly, because we recover both donor and failing hearts, we have data for EP properties in non-failing human hearts, as well as on remodelling in disease.
Comparison with other human experimental data
An advantage of our experimental approach for generating modelling data is that our functional EP studies are conducted on intact tissue preparations, typically tens of cm3 in volume. We have conducted studies from perfused left ventricular wedge, right ventricular, atrial, and conduction system preparations. In contrast, most currently published human heart functional EP data are either from isolated cells or from in vivo patient measurements during EP study.77–80 Isolated cells enable recordings of transmembrane potential to obtain AP morphology, and they also allow individual ion currents to be isolated with voltage-clamp protocols. However, properties of isolated cells differ significantly from the properties of cardiomyocytes in tissue. Electrotonic effects of cell-to-cell coupling are lost, and there are other potentially detrimental effects on ionic current function due to disruption of normal cell connections to the extracellular matrix scaffold.62 In addition, we can couple functional EP data with molecular and histological analyses, in a way that is difficult to achieve with in vivo patient studies, to help examine the mechanistic underpinnings of functional alterations in disease. In other work, we have taken advantage of the large and diverse number of samples to answer more specific questions, such as the role of gender in EP remodelling at the gene expression level in the atria and ventricles.81
While in vivo EP measures have the important benefit of being collected under conditions with normal autonomic input and neurohormonal signals, these recordings are typically limited in type or number and/or by anatomical region. Improvements in catheter mapping have enabled increases in the number of electrogram recordings; however, these still lack the detail provided by transmembrane potential recordings.82 Instead, in vivo recordings of membrane voltage can only be collected through monophasic AP recordings, which are typically limited in number.78,83,84 In addition, concerns of patient safety limit the regions from which in vivo recordings can be collected. Electrophysiology study recordings are less frequently obtained from the left ventricle due to concerns of embolus formation and potential stroke as a result of procedure.
Other non-invasive methods have been developed to examine in vivo human cardiac EP. Electrocardiographic imaging is a method by which electrogram morphologies can be calculated from a large number of body surface potential recordings combined with anatomical data collected via computed tomography scan.85 Currently, though, this technology is only able to resolve electrograms on the epicardial surface, as the inverse problem using body surface electrograms to determine cardiac electrogram morphologies is ill-posed.
Modelling of donor and failing human heart properties from human experimental data
Already, data from our human donor and failing heart gene expression analysis have been utilized for computational study.81,86 mRNA expression levels were used to determine parameter values in the OVVR human cardiac cell model for ionic currents, which were up- or downregulated in failure. Simulations revealed that alterations in gene expression could determine EP remodelling, as cellular EP properties were consistent with EP changes in human heart failure, including AP prolongation and increased Ca transient duration. In addition, this computational study predicts the relative importance of the different ionic currents for driving EP remodelling in heart failure. This work highlights the important interplay of wet-lab experiments and computational models, as experimental data lead to parameter optimization, and then the model produced results that were not plausibly obtained by experiment.
Implementation of a multiscale human experimental data repository
In addition to generating a large amount of data from human hearts in health and disease, we also aim to make this information accessible to the modelling community based on standards that have been developed for the Physiome project87–89 and the Minimum Information for Cardiac Electrophysiology Experiments (MICEE).90 In collaboration with the laboratory of Dr Alexander Panfilov from University of Ghent, we are developing a database for deposition of human heart experimental data from each of our hearts. We continue to populate and update this database as new data are available for distribution. This includes analysed data, as well as raw data measurements, and the methods with which these data were acquired (Figure 3).
Figure 3.
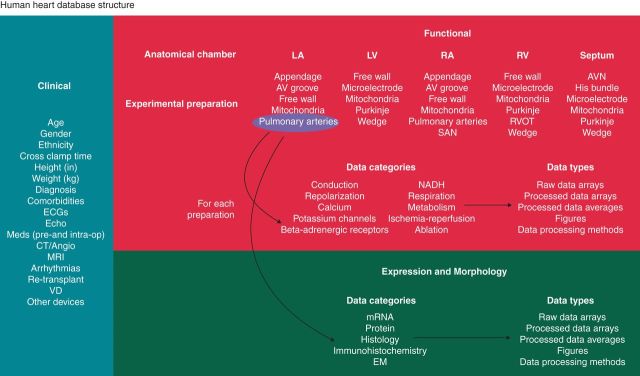
Structural representation of our multiscale human physiology database. Human heart data are identified based on heart number, and three overarching categories encapsulate the available data for each heart: (i) clinical, (ii) functional, and (iii) expression and morphology. For ease of locating data from specific heart regions, functional and expression and morphology data are indexed by anatomical chamber and experimental preparations for each chamber. For each of the experimental preparations, as highlighted by the pulmonary arteries example, several categories of data will be available, and the data will be accessible in formats ranging from processed data and to raw data arrays, with methodological details.
Transparency of models and experimental data
The International Union of Physiological Sciences (IUPS) initiated the Physiome project in 1993, with the goal to ultimately develop a quantitative model for the intact organism.9,91,92 To achieve this, computational biologists must develop standards for defining models, such that they can communicate with one another. These standards span from seemingly simple details, such as units employed in models, to establishing a language that can encapsulate many of these models. Markup languages, such as CellML and SBML, ensure that models are in consistent form and can be imported into simulation packages in standard format.55,58 In addition, many conventions for experimental data must be established, including ontologies for structure and function from the micro to macro scales.
Challenges with model development, inheritance, and transparency
Because cardiac modelling is the most advanced field of organ system modelling, many key issues have surfaced which need to be addressed to ensure model accuracy. Several important problems were raised in the meta-analysis by Niederer et al.33 In particular, they demonstrate the pervasive re-use of model components within the modelling community.33 This pattern is seen in single-cell modelling as well as whole-organ models.39,42,63 In addition, due to the historical lack of non-failing human data, experimental results have often been borrowed from non-human species, even in the construction of ‘human’ cell models. In their network evaluation of the TNNP and IMW human ventricular models, Niederer and colleagues found that only 50% of experimental data came from humans, 25% came from guinea pigs, and the last 25% came from a variety of other species. In addition, only 60% of the data originated from ventricular cell experiments.93 We have represented the relative contributions of data from various species for the TNNP and IMW models in Figure 1A and B.
In a review on Physiome project standards, Smith et al.66 outline how experimental data for cardiac myocyte contraction have been inherited into successive generations of computational models. The same datasets, generated 25–30 years ago, on binding affinity of the Ca ion to troponin C have continued to propagate into current models for cardiac excitation and contraction. However, since the development of the original models, much more reliable experimental data have been acquired, while few models have updated their parameters with these current data. The same issue is prevalent in human cardiomyocyte EP modelling, with the TNNP and IMW models both relying heavily on data from the 1990s, generated using dated methods.33
In contrast to this inheritance paradigm for model development, Niederer et al.94 developed a model for cardiac relaxation, in which all parameters were determined from many experimental sources after extensive literature review. In addition, they provide original references for their formulation of each parameter, addressing the challenge of tracing experimental data sources used for model development. This particular model serves as an exemplary standard for computational model development and documentation.
Challenges for cardiac electrophysiology experimentalists: data transparency and model usage
Accurate model development not only depends on computational biology, but also on access to relevant experimental data. The primary goal set out in the MICEE publication is that experimental EP data should be made available in online repositories, which are referenced in the corresponding publication.90 Quinn and colleagues highlight five key components that represent the most important aspects of any cardiac EP experiment and publication, and these include (i) material, (ii) environment, (iii) protocols, (iv) recordings, and (v) analysis. Also, standards for the treatment of data are likely going to become more common, as funding agencies and journals will continue to develop more rigorous data-sharing requirements, which will likely result in improved tools for data sharing and annotation. Currently, some foundational data-sharing resources have been implemented, including the database of Genotypes and Phenotypes (dbGAP; http://www.ncbi.nlm.nih.gov/gap/) and the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/).
In addition, experimentalists often fail to appreciate the significance of computational models, underutilizing their predictive power to help determine important experimental questions to address. Models can provide key insights into cardiac EP function, providing direction for experiments, or even completely substituting for wet-lab studies in some cases. Importantly, while much experimental research must take a reductionist approach to tease out physiological mechanisms, computational models have the ability to integrate information from many experiments and scales; thus, models can address more systems-level questions that are difficult to address by experiment.
Conclusions
In tracing the development of cardiomyocyte EP models, several important themes arise including (i) model re-use, resulting in overarching models that are assembled from many different species of data and models; (ii) significant differences in model behaviour; (iii) failure to incorporate improved experimental data as it arises; (iv) the lack of sufficient data, especially from non-failing human hearts; and (v) the scarcity of appropriately documented cardiac EP experimental results. To address many of these issues, formalized reporting standards should be developed and adopted, for both computational and experimental cardiac EP studies. In addition, for the lack of relevant data from the human heart, we have presented a powerful research paradigm for human cardiac physiology investigation. The growing body of research resulting from our multiscale studies, and the open availability of data, will enable improvements in human heart EP modelling. In turn, we hope for greater progress in transparency for the greater cardiac EP community.
Conflict of interest: none declared.
Funding
This work was supported by the National Institutes of Health grant R01 HL114395 and by the Agence Nationale de la Recherche grant number ANR-10-IAHU04-LIRYC.
References
- 1.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. doi:10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi:10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.O'Malley MA, Dupre J. Fundamental issues in systems biology. Bioessays. 2005;27:1270–6. doi: 10.1002/bies.20323. doi:10.1002/bies.20323. [DOI] [PubMed] [Google Scholar]
- 4.Hood L, Galas D. The digital code of DNA. Nature. 2003;421:444–8. doi: 10.1038/nature01410. doi:10.1038/nature01410. [DOI] [PubMed] [Google Scholar]
- 5.Ehrenberg M, Elf J, Aurell E, Sandberg R, Tegnér J. Systems biology is taking off. Genome Res. 2003;13:2377–80. doi: 10.1101/gr.1763203. doi:10.1101/gr.1763203. [DOI] [PubMed] [Google Scholar]
- 6.Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Ann Rev Genomics Human Genet. 2001;2:343–72. doi: 10.1146/annurev.genom.2.1.343. doi:10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 7.Mesarovic M, Sreenath S, Keene J. Search for organising principles: understanding in systems biology. Syst Biol. 2004;1:19–27. doi: 10.1049/sb:20045010. doi:10.1049/sb:20045010. [DOI] [PubMed] [Google Scholar]
- 8.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–4. doi: 10.1126/science.1069492. doi:10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 9.Bassingthwaighte JB. Strategies for the physiome project. Ann Biomed Eng. 2000;28:1043–58. doi: 10.1114/1.1313771. doi:10.1114/1.1313771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noble D, Rudy Y. Models of cardiac ventricular action potentials: iterative interaction between experiment and simulation. Philos Trans R Soc Lond A Math Phys Eng Sci. 2001;359:1127–42. doi:10.1098/rsta.2001.0820. [Google Scholar]
- 12.FitzHugh R. Impulses and physiological states in theoretical models of nerve membrane. Biophys J. 1961;1:445–66. doi: 10.1016/s0006-3495(61)86902-6. doi:10.1016/S0006-3495(61)86902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izhikevich EM, FitzHugh R. Fitzhugh-nagumo model. Scholarpedia. 2006;1:1349. doi:10.4249/scholarpedia.1349. [Google Scholar]
- 14.Nagumo J, Arimoto S, Yoshizawa S. An active pulse transmission line simulating nerve axon. Proc IRE. 1962;50:2061–70. doi:10.1109/JRPROC.1962.288235. [Google Scholar]
- 15.Pertsov AM, Davidenko JM, Salomonsz R, Baxter WT, Jalife J. Spiral waves of excitation underlie reentrant activity in isolated cardiac muscle. Circ Res. 1993;72:631–50. doi: 10.1161/01.res.72.3.631. doi:10.1161/01.RES.72.3.631. [DOI] [PubMed] [Google Scholar]
- 16.Aliev RR, Panfilov AV. A simple two-variable model of cardiac excitation. Chaos Solitons Fractals. 1996;7:293–301. doi:10.1016/0960-0779(95)00089-5. [Google Scholar]
- 17.Rudy Y, Silva JR. Computational biology in the study of cardiac ion channels and cell electrophysiology. Q Rev Biophys. 2006;39:57–116. doi: 10.1017/S0033583506004227. doi:10.1017/S0033583506004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nygren A, Leon L, Giles W. Simulations of the human atrial action potential. Philos Trans R Soc Lond A Math Phys Eng Sci. 2001;359:1111–25. doi:10.1098/rsta.2001.0819. [Google Scholar]
- 19.Noble D. A modification of the Hodgkin–Huxley equations applicable to Purkinje fibre action and pacemaker potentials. J Physiol. 1962;160:317–52. doi: 10.1113/jphysiol.1962.sp006849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noble D. Applications of Hodgkin-Huxley equations to excitable tissues. Physiol Rev. 1966;46:1–50. doi: 10.1152/physrev.1966.46.1.1. [DOI] [PubMed] [Google Scholar]
- 21.McAllister RE, Noble D, Tsien R. Reconstruction of the electrical activity of cardiac Purkinje fibres. J Physiol. 1975;251:1–59. doi: 10.1113/jphysiol.1975.sp011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenton FH, Cherry EM. Models of cardiac cell. Scholarpedia. 2008;3:1868. doi:10.4249/scholarpedia.1868. [Google Scholar]
- 23.Winslow RL, Cortassa S, O'Rourke B, Hashambhoy YL, Rice JJ, Greenstein JL. Integrative modeling of the cardiac ventricular myocyte. Wiley Interdiscip Rev Syst Biol Med. 2011;3:392–413. doi: 10.1002/wsbm.122. doi:10.1002/wsbm.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiFrancesco D, Noble D. A model of cardiac electrical activity incorporating ionic pumps and concentration changes. Philos Trans R Soc Lond B Biol Sci. 1985;307:353–98. doi: 10.1098/rstb.1985.0001. doi:10.1098/rstb.1985.0001. [DOI] [PubMed] [Google Scholar]
- 25.Beeler GW, Reuter H. Reconstruction of the action potential of ventricular myocardial fibres. J Physiol. 1977;268:177–210. doi: 10.1113/jphysiol.1977.sp011853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo C-H, Rudy Y. A model of the ventricular cardiac action potential: depolarization, repolarization, and their interaction. Circ Res. 1991;68:1501–26. doi: 10.1161/01.res.68.6.1501. doi:10.1161/01.RES.68.6.1501. [DOI] [PubMed] [Google Scholar]
- 27.Luo C-H, Rudy Y. A dynamic model of the cardiac ventricular action potential. I. Simulations of ionic currents and concentration changes. Circ Res. 1994;74:1071–96. doi: 10.1161/01.res.74.6.1071. doi:10.1161/01.RES.74.6.1071. [DOI] [PubMed] [Google Scholar]
- 28.Luo C-H, Rudy Y. A dynamic model of the cardiac ventricular action potential. II. Afterdepolarizations, triggered activity, and potentiation. Circ Res. 1994;74:1097–113. doi: 10.1161/01.res.74.6.1097. doi:10.1161/01.RES.74.6.1097. [DOI] [PubMed] [Google Scholar]
- 29.Priebe L, Beuckelmann DJ. Simulation study of cellular electric properties in heart failure. Circ Res. 1998;82:1206–23. doi: 10.1161/01.res.82.11.1206. doi:10.1161/01.RES.82.11.1206. [DOI] [PubMed] [Google Scholar]
- 30.Ten Tusscher K, Noble D, Noble P, Panfilov A. A model for human ventricular tissue. Am J Physiol Heart Circ Physiol. 2004;286:H1573–89. doi: 10.1152/ajpheart.00794.2003. doi:10.1152/ajpheart.00794.2003. [DOI] [PubMed] [Google Scholar]
- 31.Ten Tusscher K, Panfilov A. Cell model for efficient simulation of wave propagation in human ventricular tissue under normal and pathological conditions. Phys Med Biol. 2006;51:6141. doi: 10.1088/0031-9155/51/23/014. doi:10.1088/0031-9155/51/23/014. [DOI] [PubMed] [Google Scholar]
- 32.Iyer V, Mazhari R, Winslow RL. A computational model of the human left-ventricular epicardial myocyte. Biophys J. 2004;87:1507–25. doi: 10.1529/biophysj.104.043299. doi:10.1529/biophysj.104.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niederer S, Fink M, Noble D, Smith N. A meta-analysis of cardiac electrophysiology computational models. Exp Physiol. 2009;94:486–95. doi: 10.1113/expphysiol.2008.044610. doi:10.1113/expphysiol.2008.044610. [DOI] [PubMed] [Google Scholar]
- 34.Carro J, Rodriguez JF, Laguna P, Pueyo E. A human ventricular cell model for investigation of cardiac arrhythmias under hyperkalaemic conditions. Philos Trans R Soc A-Math Phys Eng Sci. 2011;369:4205–32. doi: 10.1098/rsta.2011.0127. doi:10.1098/rsta.2011.0127. [DOI] [PubMed] [Google Scholar]
- 35.O'Hara T, Virag L, Varro A, Rudy Y. Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLoS Comput Biol. 2011;7:e1002061. doi: 10.1371/journal.pcbi.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grandi E, Pasqualini FS, Bers DM. A novel computational model of the human ventricular action potential and Ca transient. J Mol Cell Cardiol. 2010;48:112–21. doi: 10.1016/j.yjmcc.2009.09.019. doi:10.1016/j.yjmcc.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shannon TR, Wang F, Puglisi J, Weber C, Bers DM. A mathematical treatment of integrated Ca dynamics within the ventricular myocyte. Biophys J. 2004;87:3351–71. doi: 10.1529/biophysj.104.047449. doi:10.1529/biophysj.104.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popel AS, Hunter PJ. Systems biology and physiome projects. Wiley Interdiscip Rev Syst Biol Med. 2009;1:153–8. doi: 10.1002/wsbm.67. doi:10.1002/wsbm.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noble D. Modeling the heart—from genes to cells to the whole organ. Science. 2002;295:1678–82. doi: 10.1126/science.1069881. doi:10.1126/science.1069881. [DOI] [PubMed] [Google Scholar]
- 40.Bray D. Molecular networks: the top-down view. Science. 2003;301:1864–5. doi: 10.1126/science.1089118. doi:10.1126/science.1089118. [DOI] [PubMed] [Google Scholar]
- 41.Clayton RH, Bernus O, Cherry EM, Dierckx H, Fenton FH, Mirabella L, et al. Models of cardiac tissue electrophysiology: progress, challenges and open questions. Prog Biophys Mol Biol. 2011;104:22–48. doi: 10.1016/j.pbiomolbio.2010.05.008. doi:10.1016/j.pbiomolbio.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 42.Carusi A, Burrage K, Rodríguez B. Bridging experiments, models and simulations: an integrative approach to validation in computational cardiac electrophysiology. Am J Physiol Heart Circ Physiol. 2012;303:H144–55. doi: 10.1152/ajpheart.01151.2011. doi:10.1152/ajpheart.01151.2011. [DOI] [PubMed] [Google Scholar]
- 43.Trew ML, Caldwell BJ, Sands GB, Hooks DA, Tai DC-S, Austin TM, et al. Cardiac electrophysiology and tissue structure: bridging the scale gap with a joint measurement and modelling paradigm. Exp Physiol. 2006;91:355–70. doi: 10.1113/expphysiol.2005.031054. doi:10.1113/expphysiol.2005.031054. [DOI] [PubMed] [Google Scholar]
- 44.Henriquez CS. Simulating the electrical behavior of cardiac tissue using the bidomain model. Crit Rev Biomed Eng. 1992;21:1–77. [PubMed] [Google Scholar]
- 45.Sobie EA, Susil RC, Tung L. A generalized activating function for predicting virtual electrodes in cardiac tissue. Biophys J. 1997;73:1410–23. doi: 10.1016/S0006-3495(97)78173-6. doi:10.1016/S0006-3495(97)78173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ten Tusscher K, Panfilov A. Alternans and spiral breakup in a human ventricular. Am J Physiol Heart Circ Physiol. 2006;291:H1088–100. doi: 10.1152/ajpheart.00109.2006. doi:10.1152/ajpheart.00109.2006. [DOI] [PubMed] [Google Scholar]
- 47.Rossi MA. Connective tissue skeleton in the normal left ventricle and in hypertensive left ventricular hypertrophy and chronic chagasic myocarditis. Med Sci Monit. 2000;7:820–32. [PubMed] [Google Scholar]
- 48.Ten Tusscher KH, Panfilov AV. Influence of diffuse fibrosis on wave propagation in human ventricular tissue. Europace. 2007;9:vi38–45. doi: 10.1093/europace/eum206. doi:10.1093/europace/eum206. [DOI] [PubMed] [Google Scholar]
- 49.Majumder R, Nayak AR, Pandit R, Majumder R, Nayak A, Pandit R. Scroll-wave dynamics in human cardiac tissue: lessons from a mathematical model with inhomogeneities and fiber architecture. PLoS ONE. 2011;6:e18052. doi: 10.1371/journal.pone.0018052. doi:10.1371/journal.pone.0018052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Majumder R, Nayak AR, Pandit R. Nonequilibrium arrhythmic states and transitions in a mathematical model for diffuse fibrosis in human cardiac tissue. PLoS ONE. 2012;7:e45040. doi: 10.1371/journal.pone.0045040. doi:10.1371/journal.pone.0045040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nayak AR, Shajahan T, Panfilov A, Pandit R. Spiral-wave dynamics in a mathematical model of human ventricular tissue with myocytes and fibroblasts. PLoS ONE. 2013;8:e72950. doi: 10.1371/journal.pone.0072950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bueno-Orovio A, Cherry EM, Fenton FH. Minimal model for human ventricular action potentials in tissue. J Theor Biol. 2008;253:544–60. doi: 10.1016/j.jtbi.2008.03.029. doi:10.1016/j.jtbi.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 53.Elshrif MM, Cherry EM. A quantitative comparison of the behavior of human ventricular cardiac electrophysiology models in tissue. PLoS ONE. 2014;9:e84401. doi: 10.1371/journal.pone.0084401. doi:10.1371/journal.pone.0084401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cooper J, Corrias A, Gavaghan D, Noble D. Considerations for the use of cellular electrophysiology models within cardiac tissue simulations. Prog Biophys Mol Biol. 2011;107:74–80. doi: 10.1016/j.pbiomolbio.2011.06.002. doi:10.1016/j.pbiomolbio.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 55.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–31. doi: 10.1093/bioinformatics/btg015. doi:10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 56.Hunter P, Robbins P, Noble D. The IUPS human physiome project. Pflügers Archiv. 2002;445:1–9. doi: 10.1007/s00424-002-0890-1. doi:10.1007/s00424-002-0890-1. [DOI] [PubMed] [Google Scholar]
- 57.Christie GR, Nielsen PM, Blackett SA, Bradley CP, Hunter PJ. FieldML: concepts and implementation. Philos Trans R Soc A Math Phys Eng Sci. 2009;367:1869–84. doi: 10.1098/rsta.2009.0025. doi:10.1098/rsta.2009.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garny A, Nickerson DP, Cooper J, dos Santos RW, Miller AK, McKeever S, et al. CellML and associated tools and techniques. Philos Trans R Soc A Math Phys Eng Sci. 2008;366:3017–43. doi: 10.1098/rsta.2008.0094. doi:10.1098/rsta.2008.0094. [DOI] [PubMed] [Google Scholar]
- 59.Cooper J, Mirams GR, Niederer SA. High-throughput functional curation of cellular electrophysiology models. Prog Biophys Mol Biol. 2011;107:11–20. doi: 10.1016/j.pbiomolbio.2011.06.003. doi:10.1016/j.pbiomolbio.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Mirams GR, Arthurs CJ, Bernabeu MO, Bordas R, Cooper J, Corrias A, et al. Chaste: an open source C++ library for computational physiology and biology. PLoS Comput Biol. 2013;9:e1002970. doi: 10.1371/journal.pcbi.1002970. doi:10.1371/journal.pcbi.1002970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pathmanathan P, Mirams GR, Southern J, Whiteley JP. The significant effect of the choice of ionic current integration method in cardiac electro-physiological simulations. Int J Numer Methods Biomed Eng. 2011;27:1751–70. doi:10.1002/cnm.1438. [Google Scholar]
- 62.Yue L, Feng J, Li G, Nattel S. Transient outward and delayed rectifier currents in canine atrium: properties and role of isolation methods. Am J Physiol Heart Circ Physiol. 1996;270:H2157–68. doi: 10.1152/ajpheart.1996.270.6.H2157. [DOI] [PubMed] [Google Scholar]
- 63.Trayanova NA, Rice JJ. Cardiac electromechanical models: from cell to organ. Front Physiol. 2011;2:43. doi: 10.3389/fphys.2011.00043. doi:10.3389/fphys.2011.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bassingthwaighte JB. The Physiome project: the macroethics of engineering toward health. Bridge. 2002;32:24–9. [Google Scholar]
- 65.Quinn TA, Kohl P. Combining wet and dry research: experience with model development for cardiac mechano-electric structure-function studies. Cardiovasc Res. 2013;97:601–11. doi: 10.1093/cvr/cvt003. doi:10.1093/cvr/cvt003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith NP, Crampin EJ, Niederer SA, Bassingthwaighte JB, Beard DA. Computational biology of cardiac myocytes: proposed standards for the physiome. J Exp Biol. 2007;210:1576–83. doi: 10.1242/jeb.000133. doi:10.1242/jeb.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fink M, Niederer SA, Cherry EM, Fenton FH, Koivumäki JT, Seemann G, et al. Cardiac cell modelling: observations from the heart of the cardiac physiome project. Prog Biophys Mol Biol. 2011;104:2–21. doi: 10.1016/j.pbiomolbio.2010.03.002. doi:10.1016/j.pbiomolbio.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 68.Doyle M, Vachharajani N, Wellen J, Lowell J, Shenoy S, Ridolfi G, et al. A Novel organ donor facility: a decade of experience with liver donors. Am J Transplant. 2014;14:615–20. doi: 10.1111/ajt.12607. doi:10.1111/ajt.12607. [DOI] [PubMed] [Google Scholar]
- 69.Efimov I, Fedorov VV, Glukhov A, Lou Q, Ambrosi C, Janks D, et al. Multiscale imaging of the human heart: building the foundation for human systems physiology and translational medicine. 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC),; 2010. pp. 5177–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lou Q, Fedorov VV, Glukhov AV, Moazami N, Fast VG, Efimov IR. Transmural heterogeneity and remodeling of ventricular excitation-contraction coupling in human heart failure. Circulation. 2011;123:1881–90. doi: 10.1161/CIRCULATIONAHA.110.989707. doi:10.1161/CIRCULATIONAHA.110.989707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lou Q, Janks DL, Holzem KM, Lang D, Onal B, Ambrosi CM, et al. Right ventricular arrhythmogenesis in failing human heart: the role of conduction and repolarization remodeling. Am J Physiol Heart Circ Physiol. 2012;303:H1426–34. doi: 10.1152/ajpheart.00457.2012. doi:10.1152/ajpheart.00457.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glukhov AV, Fedorov VV, Kalish PW, Ravikumar VK, Lou Q, Janks D, et al. Conduction remodeling in human end-stage non-ischemic left ventricular cardiomyopathy. Circulation. 2012;125:1835–47. doi: 10.1161/CIRCULATIONAHA.111.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Glukhov AV, Fedorov VV, Lou Q, Ravikumar VK, Kalish PW, Schuessler RB, et al. Transmural dispersion of repolarization in failing and nonfailing human ventricle. Circ Res. 2010;106:981–91. doi: 10.1161/CIRCRESAHA.109.204891. doi:10.1161/CIRCRESAHA.109.204891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holzem KM, Glukhov AV, Efimov IR. The role of IKr in transmural repolarization abnormalities in human heart failure [Abstract] Circulation. 2011;124:A16014. [Google Scholar]
- 75.Hucker WJ, Fedorov VV, Foyil KV, Moazami N, Efimov IR. Optical mapping of the human atrioventricular junction. Circulation. 2008;117:1474–7. doi: 10.1161/CIRCULATIONAHA.107.733147. doi:10.1161/CIRCULATIONAHA.107.733147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fedorov VV, Glukhov AV, Chang R, Kostecki G, Aferol H, Hucker WJ, et al. Optical mapping of the isolated coronary-perfused human sinus node. J Am Coll Cardiol. 2010;56:1386–94. doi: 10.1016/j.jacc.2010.03.098. doi:10.1016/j.jacc.2010.03.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holzem KM, Efimov IR. Arrhythmogenic remodelling of activation and repolarization in the failing human heart. Europace. 2012;14(Suppl 5):v50–7. doi: 10.1093/europace/eus275. doi:10.1093/europace/eus275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Franz MR, Swerdlow CD, Liem LB, Schaefer J. Cycle length dependence of human action potential duration in vivo. Effects of single extrastimuli, sudden sustained rate acceleration and deceleration, and different steady-state frequencies. J Clin Invest. 1988;82:972–9. doi: 10.1172/JCI113706. doi:10.1172/JCI113706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beuckelmann DJ, Nabauer M, Erdmann E. Alterations of K+ currents in isolated human ventricular myocytes from patients with terminal heart failure. Circ Res. 1993;73:379–85. doi: 10.1161/01.res.73.2.379. doi:10.1161/01.RES.73.2.379. [DOI] [PubMed] [Google Scholar]
- 80.Beuckelmann DJ, Erdmann E. Ca(2+)-currents and intracellular [Ca2+]i-transients in single ventricular myocytes isolated from terminally failing human myocardium. Basic Res Cardiol. 1992;87(Suppl 1):235–43. doi: 10.1007/978-3-642-72474-9_19. [DOI] [PubMed] [Google Scholar]
- 81.Ambrosi CM, Yamada KA, Nerbonne JM, Efimov IR. Gender differences in electrophysiological gene expression in failing and non-failing human hearts. PLoS ONE. 2013;8:e54635. doi: 10.1371/journal.pone.0054635. doi:10.1371/journal.pone.0054635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Andrikopoulos G, Tzeis S, Vardas PE. Invasive therapy for atrial fibrillation: recent developments in ablation, navigation and mapping technology. Heart. 2011;97:237–43. doi: 10.1136/hrt.2009.190017. doi:10.1136/hrt.2009.190017. [DOI] [PubMed] [Google Scholar]
- 83.Yue AM, Franz MR, Roberts PR, Morgan JM. Global endocardial electrical restitution in human right and left ventricles determined by noncontact mapping. J Am Coll Cardiol. 2005;46:1067–75. doi: 10.1016/j.jacc.2005.05.074. doi:10.1016/j.jacc.2005.05.074. [DOI] [PubMed] [Google Scholar]
- 84.Nash MP, Bradley CP, Sutton PM, Clayton RH, Kallis P, Hayward MP, et al. Whole heart action potential duration restitution properties in cardiac patients: a combined clinical and modelling study. Exp Physiol. 2006;91:339–54. doi: 10.1113/expphysiol.2005.031070. doi:10.1113/expphysiol.2005.031070. [DOI] [PubMed] [Google Scholar]
- 85.Ramanathan C, Ghanem RN, Jia P, Ryu K, Rudy Y. Noninvasive electrocardiographic imaging for cardiac electrophysiology and arrhythmia. Nat Med. 2004;10:422–8. doi: 10.1038/nm1011. doi:10.1038/nm1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Walmsley J, Rodriguez JF, Mirams GR, Burrage K, Efimov IR, Rodriguez B. mRNA expression levels in failing human hearts predict cellular electrophysiological remodeling: a population-based simulation study. PLoS ONE. 2013;8:e56359. doi: 10.1371/journal.pone.0056359. doi:10.1371/journal.pone.0056359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ribba B, Tracqui P, Boix J-L, Boissel J-P, Thomas SR. QxDB: a generic database to support mathematical modelling in biology. Philos Trans R Soc A Math Phys Eng Sci. 2006;364:1517–32. doi: 10.1098/rsta.2006.1784. doi:10.1098/rsta.2006.1784. [DOI] [PubMed] [Google Scholar]
- 88.Goldberg RN, Tewari YB, Bhat TN. Thermodynamics of enzyme-catalyzed reactions—a database for quantitative biochemistry. Bioinformatics. 2004;20:2874–7. doi: 10.1093/bioinformatics/bth314. doi:10.1093/bioinformatics/bth314. [DOI] [PubMed] [Google Scholar]
- 89.Popel A, Greene A, Ellis C, Ley K, Skalak T, Tonellato P. The microcirculation physiome project. Ann Biomed Eng. 1998;26:911–3. doi: 10.1114/1.112. doi:10.1114/1.112. [DOI] [PubMed] [Google Scholar]
- 90.Quinn T, Granite S, Allessie M, Antzelevitch C, Bollensdorff C, Bub G, et al. Minimum information about a cardiac electrophysiology experiment (MICEE): standardised reporting for model reproducibility, interoperability, and data sharing. Prog Biophys Mol Biol. 2011;107:4–10. doi: 10.1016/j.pbiomolbio.2011.07.001. doi:10.1016/j.pbiomolbio.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hunter PJ, Borg TK. Integration from proteins to organs: the Physiome Project. Nat Rev Mol Cell Biol. 2003;4:237–43. doi: 10.1038/nrm1054. doi:10.1038/nrm1054. [DOI] [PubMed] [Google Scholar]
- 92.Crampin EJ, Halstead M, Hunter P, Nielsen P, Noble D, Smith N, et al. Computational physiology and the physiome project. Exp Physiol. 2004;89:1–26. doi: 10.1113/expphysiol.2003.026740. doi:10.1113/expphysiol.2003.026740. [DOI] [PubMed] [Google Scholar]
- 93.Niederer SA, Smith NP. At the heart of computational modelling. J Physiol Lond. 2012;590:1331–8. doi: 10.1113/jphysiol.2011.225045. doi:10.1113/jphysiol.2011.225045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Niederer S, Hunter P, Smith N. A quantitative analysis of cardiac myocyte relaxation: a simulation study. Biophys J. 2006;90:1697–722. doi: 10.1529/biophysj.105.069534. doi:10.1529/biophysj.105.069534. [DOI] [PMC free article] [PubMed] [Google Scholar]


