Abstract
Unusually low genetic diversity can be a warning of an urgent need to mitigate causative anthropogenic activities. However, current low levels of genetic diversity in a population could also be due to natural historical events, including recent evolutionary divergence, or long-term persistence at a small population size. Here, we determine whether the relatively low genetic diversity of pygmy blue whales (Balaenoptera musculus brevicauda) in Australia is due to natural causes or overexploitation. We apply recently developed analytical approaches in the largest genetic dataset ever compiled to study blue whales (297 samples collected after whaling and representing lineages from Australia, Antarctica and Chile). We find that low levels of genetic diversity in Australia are due to a natural founder event from Antarctic blue whales (Balaenoptera musculus intermedia) that occurred around the Last Glacial Maximum, followed by evolutionary divergence. Historical climate change has therefore driven the evolution of blue whales into genetically, phenotypically and behaviourally distinct lineages that will likely be influenced by future climate change.
Keywords: Australia, Balaenoptera musculus, climate change, phylogeography, speciation, endangered species
1. Introduction
The loss of genetic diversity in species and populations is often regarded as a key threatening process. Low levels of genetic diversity can act as a warning of increased extinction risk through reduced individual fitness and reduced evolutionary potential, and highlight an urgent need to mitigate causative anthropogenic activities [1]. However, low levels of genetic diversity could also be a product of natural historical events or natural long-term persistence at a small population size. One of the most notable natural events during recent history is climatic oscillation between glacial and interglacial periods. During glacial periods, the temperate and tropical regions contracted towards the equator, sea levels lowered and polar ice expanded, with the Last Glacial Maximum (LGM) marking when ice sheets were last at their maximum extension. These climatic oscillations resulted in evolutionary and demographic changes and thereby changes in genetic diversity [2].
Blue whales (Balaenoptera musculus) were severely hunted in the twentieth century and are currently endangered [3]. They typically feed at higher latitudes during summer, and then migrate to breed and feed at lower latitudes during winter. In the Northern Hemisphere, there is one subspecies (B. m. musculus), and in the Southern Hemisphere, there are two subspecies: the pygmy blue whale (B. m. brevicauda), which feeds in temperate waters, and the Antarctic blue whale (B. m. intermedia), which feeds in Antarctic waters. The lowest recorded genetic diversity in populations of blue whales is found in pygmy blue whales that feed off Australia [4–7] (table 1). Traditional genetic analyses have indicated that Australian pygmy blue whales have undergone a genetic bottleneck at an unknown time [4]. Australian pygmy blue whales are thought to have had abundances of a few or several thousand individuals before exploitation to several hundred or a few thousand immediately after exploitation [8], but this and their current abundance is unverified.
Table 1.
Genetic variation of blue whales (Balaenoptera musculus) in this study and previous studies.
| microsatellites |
mtDNA control region |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| no. samples, n | no. loci | mean no. alleles (±s.d.) | mean allelic richness | mean observed heterozygosity, HO (±s.d.) | mean expected heterozygosity, HE (±s.d.) | no. samples, n | fragment size (bp) | no. haplotypes | haplotype diversity, h (±s.d.) | nucleotide diversity, π (±s.d.) | source | |
| Australia (B. m. brevicauda) | 109 | 20 | 6.00 (3.06) | 6.00 | 0.602 (0.194) | 0.600 (0.188) | 89 | 414 | 14 | 0.680 (0.053) | 0.003 (0.002) | this study |
| 47 | 10 | 6.70 (2.79) | — | 0.659 (0.022) | 0.655 (0.042) | 67 | 394 | 14 | 0.683 (0.062) | 0.003 (0.002) | [4]a | |
| 25 | 10 | 5.80 (2.25) | — | 0.590 (0.031) | 0.625 (0.043) | 32 | 394 | 9 | 0.758 (0.070) | 0.004 (0.003) | [4]a | |
| Antarctica (B. m. intermedia) | 142 | 20 | 11.65 (5.46) | 11.31 | 0.758 (0.130) | 0.763 (0.133) | 140 | 414 | 46 | 0.968 (0.005) | 0.014 (0.008) | this study |
| — | — | — | — | — | — | 183 | 410 | 52 | 0.968 (0.004) | 0.014 (0.007) | [6] | |
| 46 | 7 | 10.43 (2.70) | — | 0.752 (0.145) | — | 47 | 414 | 26 | 0.969 (0.010) | — | [10] | |
| Chile (B. m. brevicauda; putatively novel subspecies [14]) | 52 | 7 | 7.71 (3.04) | — | 0.692 (0.160) | 0.730 (0.147) | 46 | 360 | 12 | 0.890 (0.019) | 0.011 (0.001) | [7] |
| Mexico (B. m. musculus) | 187 | 9 | 9.6 (2.4) | — | 0.74 (0.03) | 0.74 (0.09) | — | — | — | — | — | [5] |
aSeparate feeding grounds.
Here, we determine whether the low genetic diversity of Australian pygmy blue whales is due to long-term persistence at a small population size, a natural historical event, or a recent anthropogenic event. We explored different scenarios of population demography and diversification by applying recently developed, coalescent-based analytical approaches to the largest genetic dataset ever compiled to study blue whales. Long-term persistence at a small population size would show no changes in population size and a smaller population size than other lineages of blue whales. A natural historical event would show a reduction in population size linked to climate change, with subsequent population expansion. Overexploitation during the twentieth century would show a recent genetic bottleneck with no recovery yet. This study aims therefore to elucidate fundamental forces—whether natural or anthropogenic—operating on the evolution of the world's largest animal species.
2. Material and methods
The methods are described in detail in the electronic supplementary material. In brief, genetic samples were collected from 1990 to 2010 from Australia (n = 109), Antarctica (n = 142) and Chile (n = 46). For blue whales sampled off Australia and Antarctica, genetic data were compiled from 20 microsatellites genotyped by Attard et al. [9] and from a 414 bp fragment of the mtDNA control region sequenced by LeDuc et al. [10] and sequenced during this study. For blue whales sampled off Chile, genetic data were compiled from the same fragment of the mtDNA control region and sequenced by Torres-Florez et al. [7]. Genetic variation was estimated for each locality. Genetic relationships among blue whale lineages were reconstructed using a haplotype network based on the mtDNA sequence data pooled from the current and previous studies, and the evolutionary distinctness of lineages was confirmed using the Genealogical Sorting Index [11]. The same data were used to trace population size changes by means of extended Bayesian skyline plots and to determine the ancestral population using BEAST v. 1.7.5 [12]. A combined dataset comprising microsatellite and mtDNA sequence data was used to assess the relative support for competing models of long-term stability, a natural population reduction, and recent overexploitation in blue whales sampled off Australia by means of approximate Bayesian computation (ABC) in DIYABC v. 2.0.4 [13].
3. Results
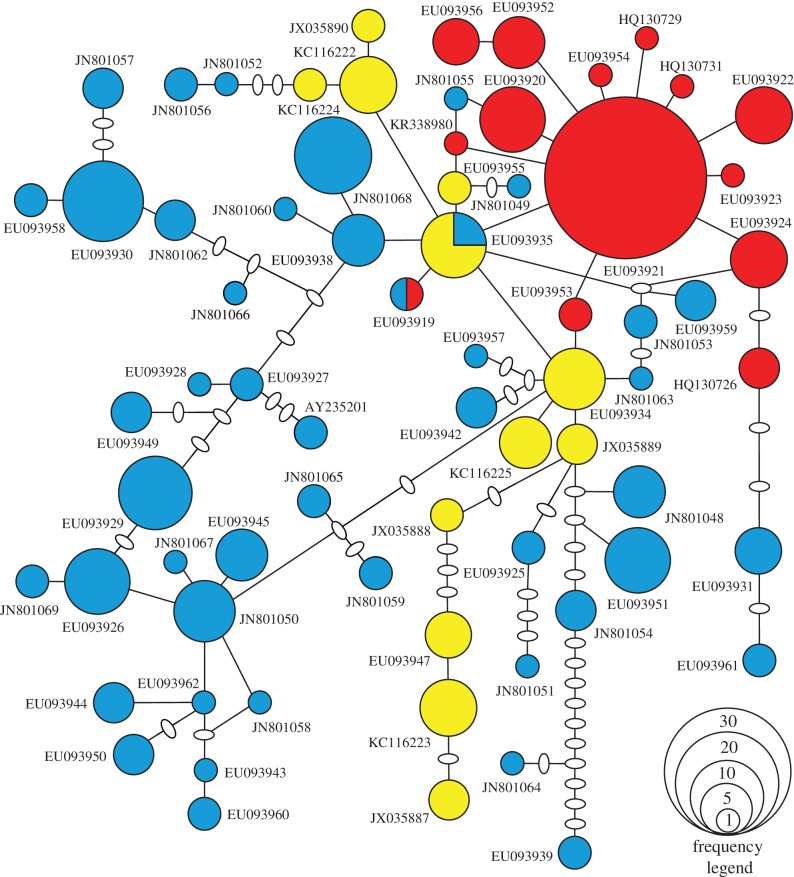
Our new dataset provides conclusive evidence that Australian pygmy blue whales have the lowest levels of genetic variation reported in blue whales (table 1). There was strong evidence that the low genetic variation is due to Australian pygmy blue whales being founded from Antarctic blue whales around the LGM. This is supported by the combination of the star phylogeny of Australian pygmy blue whales among the diverse Antarctic blue whale lineage (figure 1), extended Bayesian skyline plots that include an increase in population size for Australian blue whales starting around the LGM (figure 2), a posterior probability of 0.980 that Antarctic blue whales represent the ancestors of the Australian and Chilean blue whales, and the ABC analysis showing less than 0.1% probability for models that included a decrease in population size due to anthropogenic impacts (electronic supplementary material). Population size changes also occurred in Antarctic blue whales, but not in Chilean blue whales (figure 2).
Figure 1.
Haplotype network of mtDNA control region for blue whales off Australia (red online or dark grey in print), Antarctica (blue online or medium grey in print) and Chile (yellow online or light grey in print). Each circle represents a haplotype with size corresponding to its observed frequency. Each line represents a single nucleotide difference and ovals represent unsampled or extinct haplotypes. GenBank accession numbers are shown. (Online version in colour.)
Figure 2.
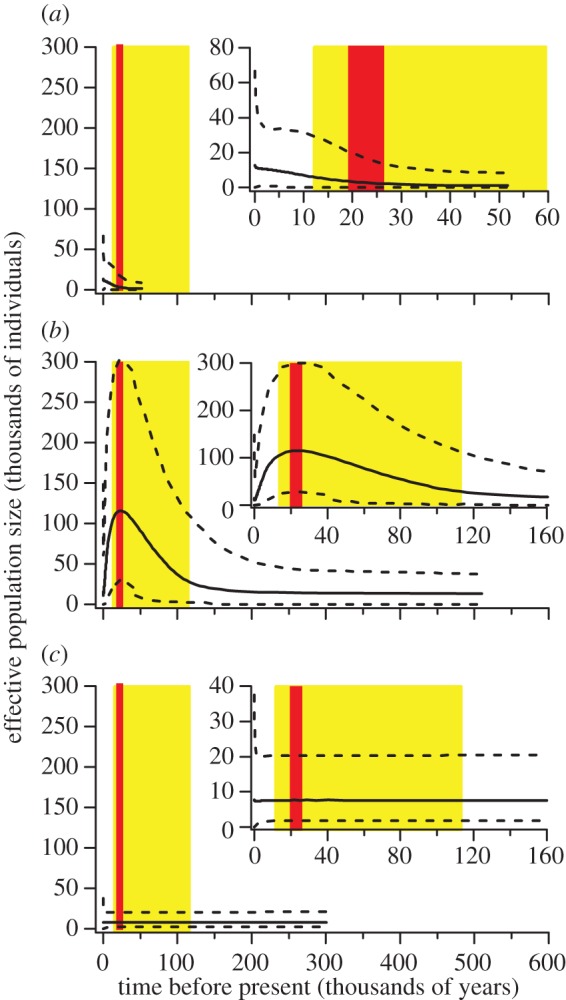
Extended Bayesian skyline plots depicting changes in effective population size over time for (a) Australian pygmy blue whales, (b) Antarctic blue whales and (c) Chilean blue whales. Shown are the median (solid lines) and the 95% highest posterior density interval (dashed lines) of the analyses, and the time of the last glacial period (yellow online or light grey in print) and the LGM (red online or dark grey in print). Plot inserts are a zoomed-in part of the associated plot. (Online version in colour.)
4. Discussion
The low level of genetic diversity in Australian pygmy blue whales is likely the result of a natural founder event followed by evolutionary diversification, rather than a recent anthropogenic event or a naturally small but constant population size. Our results indicate that Antarctic blue whales founded Australian pygmy blue whales around the time of the LGM. These findings provide the first insight into the divergence of any blue whale subspecies or population. The naturally low genetic diversity of Australian pygmy blue whales means they likely have a lower ability to respond to today's changing environment compared with other blue whale populations [1]. After being founded, the Australian pygmy blue whales became phenotypically and behaviourally distinct from Antarctic blue whales. Changes included body length [14] and song type [15]. This suggests that Australian pygmy blue whales not only became genetically different through the stochastic processes of mutation and genetic drift, but adaptively different through natural selection.
The peak population size of Antarctic blue whales was at the LGM, as shown in their Bayesian skyline plot, which may have increased the likelihood of individuals dispersing and founding the Australian population. The population size changes of the Antarctic blue whales match the gradual increase in sea ice extent during the last glacial period followed by the rapid decrease after the LGM [16]. The distribution of Antarctic blue whales likely shifted with the expansion and recession of the ice edge, as suggested for other Antarctic fauna [17]. This suggests that the carrying capacity of Antarctic blue whale habitat increased as sea ice extended, either due to increased circumference and therefore potentially increased spatial extent of Antarctic blue whale habitat, greater biological productivity, or a combination of both. Chilean blue whales did not change in population size, consistent with the results of a separate study [7].
Biological productivity is likely a key driver in the evolution and demography of blue whales. Blue whale populations require biologically productive habitats, such as upwelling and frontal regions, with high densities of krill [18]. Antarctic krill (Euphausia superba) is the primary prey of blue whales in Antarctic waters and, as expected if biological productivity is a driver of demographic change, have undergone a recent population expansion like that of Antarctic blue whales [19]. The founding during the LGM and subsequent evolutionary divergence of Australian pygmy blue whales would have relied on the presence of biological productivity in their habitat.
We have shown that climate change has shaped the evolutionary diversification of the largest extant animal into genetically, phenotypically and behaviourally distinct lineages. The young age of the Australian pygmy blue whale population accounts for their low genetic diversity. This also indicates that blue whale ecology and evolution will likely be influenced by future climate change.
Supplementary Material
Acknowledgements
We thank Kelly M. Robertson and Paula A. Olson for their input. Acknowledgements are detailed elsewhere for sampling [7,9].
Ethics statement
Samples were collected under the ethics requirements of the country, as detailed elsewhere [7,9].
Data accessibility
Microsatellite genotypes are in DRYAD under Attard et al. [9] (DRYAD entry doi:10.5061/dryad.8m0t6). Sequences are in GenBank under the accession numbers in figure 1.
Funding statement
Australian Marine Mammal Centre within the Australian Antarctic Division, Flinders University, Macquarie University, the Royal Zoological Society of NSW and an Australian Postgraduate Award to C.R.M.A.
Authors' contributions
C.R.M.A. contributed to the conception and design of the study, the acquisition, analysis and interpretation of data, and drafted the article. L.M.M. and L.B.B. contributed to the conception and design of the study, the acquisition, analysis and interpretation of data, and critically revised the article. K.C.S.J., P.C.G., M.-N.M.J. and M.G.M. contributed to the acquisition of data and critically revised the article. P.R.T. contributed to the analysis and interpretation of data and critically revised the article. All authors approved the published version.
Competing interests
We have no competing interests.
References
- 1.Frankham R, Ballou JD, Briscoe DA. 2002. Introduction to conservation genetics. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 2.Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405, 907–913. ( 10.1038/35016000) [DOI] [PubMed] [Google Scholar]
- 3.Branch TA, et al. 2007. Past and present distribution, densities and movements of blue whales Balaenoptera musculus in the Southern Hemisphere and northern Indian Ocean. Mammal. Rev. 37, 116–175. ( 10.1111/j.1365-2907.2007.00106.x) [DOI] [Google Scholar]
- 4.Attard CRM, Beheregaray LB, Jenner C, Gill P, Jenner M, Morrice M, Bannister J, LeDuc R, Möller L. 2010. Genetic diversity and structure of blue whales (Balaenoptera musculus) in Australian feeding aggregations. Conserv. Genet. 11, 2437–2441. ( 10.1007/s10592-010-0121-9) [DOI] [Google Scholar]
- 5.Costa-Urrutia P, Sanvito S, Victoria-Cota N, Enríquez-Paredes L, Gendron D. 2013. Fine-scale population structure of blue whale wintering aggregations in the Gulf of California. PLoS ONE 8, e58315 ( 10.1371/journal.pone.0058315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sremba AL, Hancock-Hanser B, Branch TA, LeDuc RL, Baker CS. 2012. Circumpolar diversity and geographic differentiation of mtDNA in the critically endangered Antarctic blue whale (Balaenoptera musculus intermedia). PLoS ONE 7, e32579 ( 10.1371/journal.pone.0032579) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres-Florez JP, Hucke-Gaete R, Rosenbaum H, Figueroa CC. 2014. High genetic diversity in a small population: the case of Chilean blue whales. Ecol. Evol. 4, 1398–1412. ( 10.1002/ece3.998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zemsky VA, Sazhinov EG. 1982. Distribution and current abundance of pygmy blue whales. In Marine mammals (ed. Arsen'ev VA.), pp. 53–70. Moscow, Russia: All-Union Research Institute of Marine Fisheries and Oceanography (in Russian) [Transl. by Gurevich VS in 1994, translation edited by Donahue MA, Brownell Jr RL as Southwest Fisheries Science Center Administrative Report LJ-94-02.] [Google Scholar]
- 9.Attard CRM, Beheregaray LB, Jenner KCS, Gill PC, Jenner M-N, Morrice MG, Robertson KM, Möller LM. 2012. Hybridization of Southern Hemisphere blue whale subspecies and a sympatric area off Antarctica: impacts of whaling or climate change? Mol. Ecol. 21, 5715–5727. ( 10.1111/mec.12025) [DOI] [PubMed] [Google Scholar]
- 10.LeDuc RG, Dizon AE, Goto M, Pastene LA, Kato H, Nishiwaki S, LeDuc CA, Brownell RL. 2007. Patterns of genetic variation in Southern Hemisphere blue whales and the use of assignment test to detect mixing on the feeding grounds. J. Cetacean Res. Manage. 9, 73–80. [Google Scholar]
- 11.Cummings MP, Neel MC, Shaw KL. 2008. A genealogical approach to quantifying lineage divergence. Evolution 62, 2411–2422. ( 10.1111/j.1558-5646.2008.00442.x) [DOI] [PubMed] [Google Scholar]
- 12.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornuet J-M, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin J-M, Estoup A. 2014. DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30, 1187–1189. ( 10.1093/bioinformatics/btt763) [DOI] [PubMed] [Google Scholar]
- 14.Branch TA, Abubaker EMN, Mkango S, Butterworth DS. 2007. Separating southern blue whale subspecies based on length frequencies of sexually mature females. Mar. Mam. Sci. 23, 803–833. ( 10.1111/j.1748-7692.2007.00137.x) [DOI] [Google Scholar]
- 15.McDonald MA, Mesnick SL, Hildebrand JA. 2006. Biogeographic characterisation of blue whale song worldwide: using song to identify populations. J. Cetacean Res. Manage. 8, 55–65. [Google Scholar]
- 16.Crosta X, Sturm A, Armand L, Pichon J-J. 2004. Late Quaternary sea ice history in the Indian sector of the Southern Ocean as recorded by diatom assemblages. Mar. Micropaleontol. 50, 209–223. ( 10.1016/S0377-8398(03)00072-0) [DOI] [Google Scholar]
- 17.Thatje S, Hillenbrand C-D, Mackensen A, Larter R. 2008. Life hung by a thread: endurance of Antarctic fauna in glacial periods. Ecology 89, 682–692. ( 10.1890/07-0498.1) [DOI] [PubMed] [Google Scholar]
- 18.Goldbogen JA, Calambokidis J, Oleson E, Potvin J, Pyenson ND, Schorr G, Shadwick RE. 2011. Mechanics, hydrodynamics and energetics of blue whale lunge feeding: efficiency dependence on krill density. J. Exp. Biol. 214, 131–148. ( 10.1242/jeb.048157) [DOI] [PubMed] [Google Scholar]
- 19.Bortolotto E, Bucklin A, Mezzavilla M, Zane L, Patarnello T. 2011. Gone with the currents: lack of genetic differentiation at the circum-continental scale in the Antarctic krill Euphausia superba. BMC Genet. 12, 32 ( 10.1186/1471-2156-12-32) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microsatellite genotypes are in DRYAD under Attard et al. [9] (DRYAD entry doi:10.5061/dryad.8m0t6). Sequences are in GenBank under the accession numbers in figure 1.