Abstract
Malpighian tubules play an essential role in excretion, osmoregulation and immunity of most insects. Exceptionally, aphids lack Malpighian tubules, providing the opportunity to investigate the fate of genes expressed in an organ that has undergone evolutionary reduction and loss. Making use of the sequenced genomes of Drosophila melanogaster and the pea aphid Acyrthosiphon pisum, we demonstrated that more than 50% of Drosophila genes expressed specifically in the Malpighian tubules had orthologues in the pea aphid genome and that most of the pea aphid orthologues with detectable expression were identified in the gut transcriptome. Relative to the whole genome, genes functioning in amino acid metabolism are significantly over-represented among the pea aphid orthologues of Malpighian tubule genes, likely reflecting the central importance of amino acid acquisition and metabolism in aphids. This study demonstrates that the evolutionary loss of a key insect organ, the Malpighian tubules, has not been associated with the coupled loss of molecular functions.
Keywords: Acyrthosiphon pisum, Drosophila melanogaster, gene orthologue, Malpighian tubules, organ loss
1. Background
Generally, the size of different organs in an animal body is tightly regulated, such that the various organs scale allometrically with overall body size, both within and across species [1]. Deviations from these allometric patterns provide the basis to investigate the selection pressures and molecular mechanisms that determine organ size and function [2–4]. In particular, vestigialization and loss of structures are increasingly being used to study the relationship between reductive evolution of an organ and patterns of gene expression [5–7].
This study concerns the molecular correlates of the evolutionary loss of an insect organ, the Malpighian tubules. The Malpighian tubules are paired outpocketings of the gut that arise at the junction between the midgut and hindgut (figure 1a). They function in osmoregulation, nitrogen excretion, detoxification and immunity [8]. Malpighian tubules are near-universal in insects. Exceptionally, they are absent from the aphids (Aphidoidea), in which the gut comprises a tube without any discernible evaginations (figure 1a) [9,10]. Aphids are plant phloem sap-feeding insects of the order Hemiptera. Phloem feeding through the life cycle has evolved multiple times in hemipteran insects but apparently no other animals. Other phloem-feeding hemipterans either possess Malpighian tubules (e.g. scale insects, planthoppers and heteropteran bugs) or have outpocketings from the gut that have been described as either ceca or Malpighian tubules (e.g. whiteflies and psyllids) [9–11]. These data indicate that the absence of Malpighian tubules is compatible with, but not necessary for, the phloem-feeding habit.
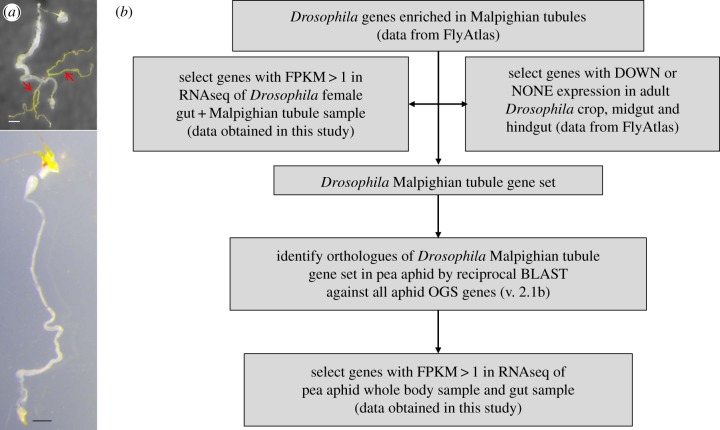
Figure 1.
Insect Malpighian tubules and Malpighian tubule genes. (a) The dissected gut of an adult female D. melanogaster (top: arrows, Malpighian tubules) and adult female pea aphid (bottom). Scale bar, 0.5 mm. (b) Flow chart for identification of orthologues of Drosophila Malpighian tubule genes in the pea aphid alimentary tracts. See §2 for descriptions of BLAST, FlyAtlas and RNAseq. (Online version in colour.)
We hypothesized that the evolutionary loss of Malpighian tubules in aphids is associated with the allocation of certain functions to different organ(s), especially the gut, which, like the Malpighian tubules, plays an important role in water relations and osmoregulation of insects [12]. To investigate this hypothesis we adopted a molecular approach: specifically, to identify a panel of genes preferentially expressed in Malpighian tubules of one insect, and then determine the incidence of the orthologues of these genes in an aphid. We used Drosophila melanogaster and the pea aphid Acyrthosiphon pisum because the genome sequences of both insects have excellent annotations (flybase.org, aphidbase.com) and the molecular physiology of Malpighian tubule function of Drosophila has been studied extensively [8].
2. Material and methods
The experimental insects were: 6-day-old adult female D. melanogaster Canton-S and 3-day-old adult parthenogenetic female pea aphid A. pisum Harris (CWR09/18) from laboratory cultures (see electronic supplementary material).
RNA was extracted from whole insects and dissected guts using the RNeasy Mini kit (Qiagen) and reverse-transcribed into cDNA. Multiplexed Illumina TruSeq libraries (barcode information in electronic supplementary material, table S1) were sequenced using an Illumina Hi-Seq2000 platform (100 bp single-end reads). After quality filtering, the reads were aligned to reference genomes of D. melanogaster (BDGP 5.25) and A. pisum (v. 2.1), using TopHat for Illumina (v. 1.5.0) [13,14] (see the electronic supplementary material for details). In total, 24 192 960 and 21 859 046 reads were assigned to Drosophila genes for the whole body and gut-and-Malpighian tubules samples, respectively, and 21 866 904 and 24 845 384 reads were assigned to the pea aphid whole body and gut samples. FPKMs (fragments per kilobase of transcript per million mapped reads) were obtained using Cufflink (v. 2.1.1).
The Drosophila Malpighian tubule gene set and pea aphid orthologues were identified as in figure 1b (see the electronic supplementary material for details). Briefly, the Drosophila genes enriched at least two-fold in tubules according to FlyAtlas microarray data were, first, filtered to remove genes that were enriched in other gut tissues (crop, midgut and hindgut) in FlyAtlas. We also excluded genes with low transcript abundance (FPKM < 1) in the Drosophila RNAseq analysis of this study. This filtered gene list comprised our Drosophila Malpighian tubule gene set. Pea aphid orthologues were obtained by reciprocal BLAST between pea aphid proteins (AphidBase:ACYPI proteins v. 2.1b) and all Drosophila genes (Flybase).
3. Results
Our initial set of Drosophila Malpighian tubule genes obtained from the FlyAtlas microarray database of gene expression data comprised 269 genes (electronic supplementary material, table S2a). Recognizing that microarray data represent relative expression and not absolute expression levels, we additionally excluded genes with very low expression levels in Drosophila. Specifically, we quantified the expression of the initial gene set in RNAseq datasets obtained for the whole body and dissected Malpighian tubule-and-gut preparations of adult Drosophila; the samples were exclusively female Drosophila for comparison with the pea aphid, which comprises parthenogenetic females. In total, 191 (71% of the 269 Malpighian tubule genes) yielded FPKM > 1 in the Malpighian tubule-and-gut sample of adult female Drosophila, and these were used as our Drosophila Malpighian tubule gene set for subsequent analysis (electronic supplementary material, table S2a).
Of the 191 Drosophila Malpighian tubule genes, 99 (52%) had orthologues in the pea aphid genome, as determined by reciprocal best hits in BLAST (electronic supplementary material, table S2b). Of these, 14 genes were orthologous to two or more pea aphid genes and two genes were orthologous to one pea aphid gene (FBgn0039049 and FBgn0039050 versus ACYPI009740), giving 123 pea aphid orthologues. RNAseq analysis of pea aphid whole bodies and dissected guts (electronic supplementary material, table S2b) with validation of selected genes by qPCR (electronic supplementary material, table S3) detected 110 transcripts with FPKM > 1 in the whole body samples, 95 (86%) of which were detected in the gut transcriptome (electronic supplementary material, table S2b). These results are consistent with our prediction (see §1) that the evolutionary loss of Malpighian tubules in aphids may be associated with the allocation of certain functions to the gut.
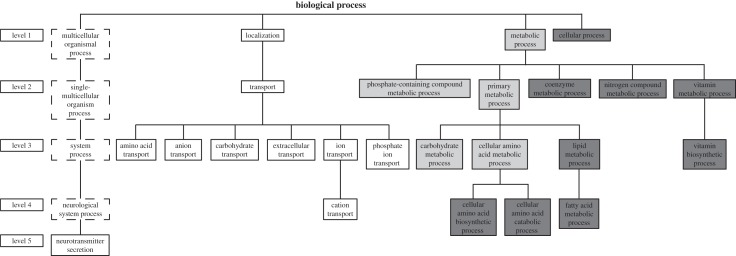
The functions represented by the Drosophila Malpighian tubule genes and their pea aphid orthologues were investigated by mapping the Drosophila and pea aphid gene sets to PANTHER for GO analysis [15]. (Multiple pea aphid genes corresponding to a single Drosophila gene were scored as a single gene, to avoid artefactual inflation of certain functional categories.) Annotated functions were assigned to 181 Drosophila genes and 93 pea aphid genes (electronic supplementary material, table S4). These genes were used to identify biological functions that are over-represented in the Drosophila Malpighian tubule gene set and their pea aphid orthologues, relative to the total annotated gene set in the respective insect genomes. The genes involved in various transport functions and neurotransmitter secretion are over-represented in the Drosophila Malpighian tubule gene set; these included many of the 92 Drosophila Malpighian tubules genes without orthologues in the pea aphid (electronic supplementary material, table S5). Genes in amino acid and fatty acid metabolism (including coenzyme and vitamin metabolism related to amino acid and fatty acid metabolism) were over-represented in the pea aphid orthologues (figure 2; electronic supplementary material, table S6). Furthermore, pea aphid orthologues of Malpighian tubule genes were significantly more likely than other genes of the same functional group to be expressed in the pea aphid gut (table 1).
Figure 2.
Significantly over-represented functions in the Drosophila Malpighian tubule gene set (white) and pea aphid orthologues (dark grey), relative to the annotated gene set in the genomes. Significantly over-represented functions in both species are highlighted in light grey; non-significant functions are in dotted-line boxes.
Table 1.
Number of Malpighian tubule orthologues and non-orthologues expressed in the pea aphid gut. Data are presented for functional groups in figure 2.
| functional group (no. of genes in pea aphid genome) | no. genes expressed in gut/total number of genes (proportion) |
|
|---|---|---|
| Malpighian tubule orthologues | non-orthologues | |
| neurotransmitter secretion (67)a | 1/3 (0.33) | 22/64 (0.34) |
| amino acid transport (131)a | 2/4 (0.50) | 40/127 (0.32) |
| anion transport (132)a | 0/1 (0) | 61/131 (0.47) |
| carbohydrate transport (217)a | 2/4 (0.50) | 84/213 (0.39) |
| extracellular transport (180)a | 2/5 (0.40) | 96/175 (0.55) |
| cation transport (436)a | 5/7 (0.71) | 180/429 (0.42) |
| phosphate ion transport (158)a | 4/6 (0.67) | 59/152 (0.39) |
| phosphate-containing compound metabolic process (401)b | 7/8 (0.88) | 218/393 (0.55) |
| carbohydrate metabolic process (846)b | 11/15 (0.73) | 345/831 (0.42) |
| cellular amino acid biosynthesis process (145)c | 7/7 (1) | 68/138 (0.49) |
| cellular amino acid catabolic process (66)c | 6/6 (1) | 32/60 (0.53) |
| coenzyme metabolic process (99)c | 6/6 (1) | 63/93 (0.68) |
| fatty acid metabolic process (157)c | 7/7 (1) | 80/150 (0.53) |
| nitrogen compound metabolic process (206)c | 8/9 (0.89) | 122/197 (0.62) |
| vitamin biosynthetic process (41)c | 2/3 (0.67) | 18/38 (0.47) |
| t = 2.66, p = 0.009d | ||
aOver-represented in Malpighian tubule gene set of Drosophila, relative to Drosophila genome.
bOver-represented in Malpighian tubule gene set of Drosophila and Malpighian tubule orthologue set of pea aphid, relative to respective genomes.
cOver-represented in Malpighian tubule orthologue gene set of pea aphid, relative to pea aphid genome.
dPaired t-test comparison of proportion of Malpighian tubule orthologues and non-orthologues, after asin-square-root transformation to obtain normal distributions (Anderson Darling test).
4. Discussion
The pea aphid genome codes for orthologues of over half the Drosophila Malpighian tubule gene set, even though the pea aphid lacks Malpighian tubules. This result is indicative of evolutionary changes in the expression patterns of the lineage(s) giving rise to one or both of these insects. As genome sequences with high-quality annotations become available for many insect species, especially hemipterans with Malpighian tubules, it will be increasingly feasible to discriminate between the Malpighian tubule-associated genes gained/lost in the lineages giving rise to the Diptera (including Drosophila) and Hemiptera (including aphids) and the genes lost specifically from insects that lack Malpighian tubules.
Our analysis additionally provides insight into the biological correlates of the differences in gene functions represented by the Drosophila Malpighian tubule gene set and the pea aphid orthologues. In particular, the significant over-representation of genes associated with amino acid metabolism in the pea aphid is congruent with the central role of amino acid metabolism in these insects, linked to their metabolic integration of amino acid inputs from their diet of plant phloem sap and endosymbiotic bacterial symbionts [16]. The under-representation of transport functions in the pea aphid gene set may reflect the relatively uniform ionic composition and low diversity of nutrients in phloem sap, dominated by organic solutes of low molecular weight (sugars, amino acids, organic acids, etc.). These results, notwithstanding, gene families coding for certain amino acid and sugar transporters have undergone dramatic evolutionary expansion in the aphids [17,18], possibly linked to the phloem-feeding habit, but these are not represented in our analysis where ‘one-to-many’ Drosophila to pea aphid orthologues are treated as a single orthologue.
Our research illustrates that the evolutionary loss of structures is not necessarily tightly coupled to loss of molecular function. Some genes may be retained because they have pleiotropic functions (i.e. different functions in different organs), as reported for opsin eye pigments in cave-dwelling amphipods with vestigial eyes [5]. The retention of other genes in the pea aphid genome may, however, be a consequence of the evolutionary recruitment of Malpighian tubule gene expression to other organs, especially the gut (table 1). Organ loss may be evolutionarily less ‘difficult’ in lineages where the expression of relatively few genes is restricted to the organ in question, or in which gene expression patterns generally are evolutionarily labile, so facilitating the recruitment of molecular functions to alternative organs.
Supplementary Material
Supplementary Material
Data accessibility
RNAseq data are deposited in NCBI_SRA (accession no. SRP053295). All other data underlying the findings described in this manuscript are provided in the electronic supplementary material.
Funding statement
This work was supported by NIFA grant no. NYW-2011–04650.
Authors' contributions
X.J. and A.E.D. conceived the study. X.J. and X.Y. conducted the experimental work. X.J. and T.A.W. did the computational analysis. X.J. and A.E.D. wrote the manuscript. T.A.W. and X.Y. commented on manuscript drafts. All authors approved the final manuscript.
Conflict of interests
The authors declare no competing financial interests.
References
- 1.Schmidt Nielsen K. 1984. Scaling: why is animal size so important? Cambridge, UK: Cambridge University Press. [Google Scholar]
- 2.Emlen DJ, Warren IA, Johns A, Dworkin I, Lavine LC. 2012. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science 337, 860–864. ( 10.1126/science.1224286) [DOI] [PubMed] [Google Scholar]
- 3.Kaiser A, Klok CJ, Socha JJ, Lee WK, Quinlan MC, Harrison JF. 2007. Increase in tracheal investment with beetle size supports hypothesis of oxygen limitation on insect gigantism. Proc. Natl Acad. Sci. USA 104, 13 198–13 203. ( 10.1073/pnas.0611544104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rilling JK. 2006. Human and nonhuman primate brains: are they allometrically scaled versions of the same design? Evol. Anthropol. 15, 65–77. [Google Scholar]
- 5.Carlini DB, Satish S, Fong DW. 2013. Parallel reduction in expression, but no loss of functional constraint, in two opsin paralogs within cave populations of Gammarus minus (Crustacea: Amphipoda). BMC Evol. Biol. 13, 89 ( 10.1186/1471-2148-13-89) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klaus S, Mendoza JC, Liew JH, Plath M, Meier R, Yeo DC. 2013. Rapid evolution of troglomorphic characters suggests selection rather than neutral mutation as a driver of eye reduction in cave crabs. Biol. Lett. 9, 20121098 ( 10.1098/rsbl.2012.1098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohner N, et al. 2013. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science 342, 1372–1375. ( 10.1126/science.1240276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beyenbach KW, Skaer H, Dow JA. 2010. The developmental, molecular, and transport biology of Malpighian tubules. Annu. Rev. Entomol. 55, 351–374. ( 10.1146/annurev-ento-112408-085512) [DOI] [PubMed] [Google Scholar]
- 9.Goodchild AJ. 1966. Evolution of the alimentary canal in Hemiptera. Biol. Revs. 41, 97–120. ( 10.1111/j.1469-185X.1966.tb01540.x) [DOI] [Google Scholar]
- 10.Cicero JM, Hiebert E, Webb SE. 1995. The alimentary canal of Bemisia tabaci and Trialeurodes abutilonea (Homoptera, Sternorrhynchii): histology, ultrastructure and correlations to function. Zoomorphology 115, 31–39. ( 10.1007/BF00397932) [DOI] [Google Scholar]
- 11.Cicero JM, Brown JK, Roberts PD, Stansly PA. 2009. The digestive system of Diaphorina citri and Bactericera cockerelli (Hemiptera: Psyllidae). Ann. Entomol. Soc. Am. 102, 650–665. ( 10.1603/008.102.0410) [DOI] [Google Scholar]
- 12.Dow JAT. 2013. Excretion and salt and water regulation. In The insects: structure and function (eds Simpson SJ, Douglas AE.), pp. 546–587, 5th edn Cambridge, UK: Cambridge University Press. [Google Scholar]
- 13.Kim D, Pertea F, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 ( 10.1186/gb-2013-14-4-r36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trapnell C. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. ( 10.1038/nprot.2012.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi H, Muruganujan A, Thomas PD. 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386. ( 10.1093/nar/gks1118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douglas AE. 2015. Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34. ( 10.1146/annurev-ento-010814-020822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncan RP, Husnik F, Van Leuven JT, Gilbert DG, Dávalos LM, McCutcheon JP, Wilson ACC. 2014. Dynamic recruitment of amino acid transporters to the insect/symbiont interface. Mol. Ecol. 23, 1608–1623. ( 10.1111/mec.12627) [DOI] [PubMed] [Google Scholar]
- 18.Price DR, Gatehouse JA. 2014. Genome-wide annotation and functional identification of aphid GLUT-like sugar transporters. BMC Genomics 15, 647 ( 10.1186/1471-2164-15-647) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data are deposited in NCBI_SRA (accession no. SRP053295). All other data underlying the findings described in this manuscript are provided in the electronic supplementary material.