Abstract
Genetic differentiation in natural populations is driven by geographic distance and by ecological or physical features within and between natural habitats that reduce migration. The primary population structure in wild barley differentiates populations east and west of the Zagros Mountains. Genetic differentiation between eastern and western populations is uneven across the genome and is greatest on linkage groups 2H and 5H. Genetic markers in these two regions demonstrate the largest difference in frequency between the primary populations and have the highest informativeness for assignment to each population. Previous cytological and genetic studies suggest there are chromosomal structural rearrangements (inversions or translocations) in these genomic regions. Environmental association analyses identified an association with both temperature and precipitation variables on 2H and with precipitation variables on 5H.
Keywords: environmental association, local adaptation, population structure, chromosome structural variation, wild barley
The wild progenitors of major crops have long been recognized as a valuable genetic resource (Harris 1990). Natural populations of crop wild relatives have the potential to serve as a source of alleles that contribute to valuable agronomic traits, including cold or drought tolerance (Volis et al. 2002a, 2004), improved disease resistance (Fetch et al. 2003), and yield increase (Cohen and Galinat 1984). Plant germplasm repositories have made substantial investments in the preservation of accessions of crops and their wild relatives (Schoen and Brown 2001). Modern genetic approaches have increased the value of these resources as quantitative trait locus (QTL) mapping, association studies, and molecular population genetic studies have combined to uncover specific alleles associated with traits of potential value for crop improvement (Ross-Ibarra et al. 2007; Takeda and Matsuoka 2008).
Wild barley presents an especially valuable source of potentially useful genes because of its broad geographic distribution and ecological adaptation, spanning ∼3500 km east to west from the Levant (the eastern Mediterranean) and Anatolia (present day Turkey) to Central Asia, occurring across much of Southwestern Asia. Volis et al. (2002c) identified four wild barley ecotypes that exhibit differences in their level and patterns of phenotypic plasticity. In common garden studies, water stress caused a greater plastic response in the desert ecotype than in the Mediterranean ecotype, whereas for nutrient stress, plasticity was higher in the Mediterranean ecotype than in the desert ecotype (Volis et al. 2002b). The observed differences in ecotype plasticity or local variation in phenotypic traits suggest environmentally induced local adaptation (Volis et al. 2000, 2002c). Moreover, alleles from wild barley (Hordeum vulgare ssp. spontaneum) have been used in barley breeding programs for cultivated barley improvement (Von Korff et al. 2005, 2008; Schmalenbach et al. 2009). QTL analyses in an advanced backcross double haploid population derived from a cross between a barley cultivar and a wild barley accession demonstrates that wild barley harbors valuable alleles that can improve yield (Von Korff et al. 2006; Schmalenbach et al. 2009) and malting quality traits (Von Korff et al. 2008) and strongly reduce disease symptoms (Von Korff et al. 2005). QTL analysis of a recombinant inbred line population and an advanced backcross population derived from crosses between a barley cultivar and a wild barley accession reveals that the wild barley accession contains alleles that confer resistance to multiple fungal pathogens (Yun et al. 2005, 2006). Recently, association and candidate gene resequencing studies have begun to uncover evidence that alleles contributing to agronomically important phenotypes, such as flowering time, may have been introduced from geographic regions outside the initial region of barley domestication, with locally adaptive variants contributing to successful cultivation at higher latitudes (Jones et al. 2008).
Examination of population structure in crop wild progenitors can result in a better understanding of the number and geographic region of domestication events (Morrell and Clegg 2007), which is fundamental to understanding the processes that drove human domestication of plants. The potential to fully exploit genetic variation in the wild relatives of a crop depends, in part, on determining which portions of the range of wild progenitors have and have not contributed to diversity in the domesticates (Zohary and Hopf 2000).
A number of previous studies have examined wild barley genetic diversity (Nevo et al. 1986; Morrell and Clegg 2007; Jakob et al. 2014). Analysis of 27 allozyme loci in 2125 individuals sampled from Israel, Turkey, and Iran suggested geographic differentiation in wild barley populations (Nevo et al. 1986) but did not sample the complete geographic range of wild barley. Genetic assignment analysis based on resequencing of 18 loci in a sample of 25 to 45 wild barley individuals identified a primary geographic partition of samples into regions east and west of the Zagros Mountains (Morrell and Clegg 2007). The assignment analysis was limited by sample size but suggested that other geographically distinct wild barley subpopulations might be identified in a larger sample (Morrell and Clegg 2007; Saisho and Purugganan 2007).
The natural range of wild barley includes a variety of environmental conditions, from arid regions in Central Asia and the Syrian desert to coastal regions along the Mediterranean with relatively high rainfall, and from the cold environments in the Zagros Mountains, the western reaches of the Himalayas, and the Iranian plateau to relatively warm lowland Mediterranean coastal regions. By associating the geographic distribution of sequence polymorphisms in georeferenced samples with environmental variables, genomic regions and individual polymorphisms correlated with differences in drought or cold tolerance or other environmental factors may be identified for further investigation.
The questions we seek to explore in this article are what ecological factors are most associated with observed population structure, how does the geographic distribution of populations relate to allele frequency differentiation genome-wide, and how can we use this information to improve conservation and utilization of wild barley genetic diversity? We report the examination of geographic structure and genetic differentiation using 3072 SNPs (Close et al. 2009) genotyped in a sample of 318 wild barley accessions. We detect two primary populations of wild barley separated by the Zagros Mountains and three subpopulations within each of the two primary populations. Comparison of the two primary populations reveals two pericentromeric regions on the long arms of 2H and 5H that are associated with a substantial fraction of the geographic differentiation in allele frequencies observed in wild barley. The genetic variation in these genomic regions suggests cryptic chromosomal structural rearrangements. Environmental association analyses reveal strong association between these genomic regions and precipitation or temperature.
Materials and Methods
Plant materials
The 318 sampled wild barley accessions are known as the Wild Barley Diversity Collection (WBDC) (Steffenson et al. 2007). WBDC accessions were selected to be representative of the geographic range of wild barley, accounting for multiple ecogeographic features (e.g., latitude, elevation, temperature range, and rainfall). The majority of accessions (77.4%) are from the Fertile Crescent, with the balance from Central Asia (15.7%), North Africa (3.8%), and the Caucasus region (2.8%). Individual accessions were self-fertilized for three generations to create inbred lines. Accession numbers, latitude, and longitude information for each sample are in Supporting Information, Table S1.
Genotypic data
The WBDC accessions were genotyped using the Illumina Golden Gate Genotyping Assay with two Barley Oligo Pool Assay (BOPA) chips (BOPA1 and BOPA2), each including 1536 SNPs (Close et al. 2009). The SNPs were discovered by comparison of DNA sequence from expressed sequence tags and sequenced PCR amplicons, derived principally from one wild barley accession and eight malting barley cultivars, primarily from Europe and the United States (Close et al. 2009).
The program ALCHEMY (Wright et al. 2010) was used to generate machine-scored, automated genotype calls. The program incorporates estimated inbreeding coefficients for each sample to improve accuracy of genotype estimation. Unlike programs such as GenomeStudio, ALCHEMY does not assume Hardy-Weinberg Equilibrium (HWE) genotypic frequencies at each SNP. ALCHEMY is based on a Bayesian model of the raw intensity data and can accurately call genotypes. We used three approaches to verify the accuracy of genotype calls. First, WBDC355 (OUH602) has been genotyped separately, with variants segregating in a mapping population (OUH602 by the cultivar Haruna Nijo) (Sato et al. 2009; Muñoz-Amatriaín et al. 2011). Second, genotypes from two lines (WBDC218 and WBDC228) were estimated from RNA-Seq (see below for details of RNA-Seq data processing), so these data were used for validation. Third, all SNPs on BOPA1 were also called manually in GenomeStudio. Only 5% of SNPs have posterior probability of genotype calls <0.95 from ALCHEMY. We considered these SNPs as missing data.
Before running ALCHEMY, SNPs in BOPA1 and BOPA2 with strong compression or multiple clusters were removed. Subsequent to initial SNP calling, the following quality control steps were applied to the genotyping data. First, we eliminated SNPs that were monomorphic in the wild barley sample. Second, we removed all SNPs that included ≥15% missing data based on the rationale that large amounts of missing data at a SNP could be associated with inaccurate genotypes. Finally, observed heterozygosity was used as an additional quality control measure. Wild barley is a highly self-fertilizing species (Brown et al. 1978) and the WBDC accessions have been subject to three rounds of inbreeding. SNPs with observed heterozygosity >10% were removed on the rationale that the genotypes were likely in error. SNPs with centromeric genetic map positions were identified based on the consensus genetic map of Muñoz-Amatriaín et al. (2011). In the inbred WBDC lines, observed heterozygosity was extremely low (0.2%), thus we treat the data as haploid genotypes.
SNPs were annotated using the program SNPMeta (Kono et al. 2014). Annotation for each SNP included GenBank ID, gene short name, whether the SNP occurs in coding or noncoding sequence, the SNP position within a codon, and determination of whether the SNP is silent or induces an amino acid replacement. Among the 3072 BOPA SNPs, 2508 were annotated, with 338 annotations derived from named genes.
Along with SNP data, we examined 29 microsatellite loci in all WBDC accessions (Ramsay et al. 2000; Li et al. 2003; Varshney et al. 2007). Microsatellites offer the advantage of reduced ascertainment bias, because the microsatellites are selected to be polymorphic, but the selection process is not conditional on the presence of an individual polymorphism (Haasl and Payseur 2010). Microsatellite locus names, repeat number, repeat unit length, total size, and heterozygosity for each microsatellite can be found in Table S2. This information was obtained from GrainGenes: A Database for Triticeae and Avena (Matthews et al. 2003). The program RST Calc (Goodman 1997) was used to compare allele frequency differences among partitions of the sample, assuming a stepwise mutation model (Slatkin 1995).
RNA-Seq data processing
RNA-Seq reads from Hordeum bulbosum accession Cb2920/4 (used as outgroup to infer ancestral state) and wild barley WBDC218 and WBDC228 were trimmed of adapter contamination with Scythe (https://github.com/vsbuffalo/scythe) and then aligned to the Morex draft sequence (Mayer et al. 2012) with Bowtie 2 (Langmead and Salzberg 2012). We adjusted read mapping parameters to accommodate the expected divergence between H. vulgare lines (∼1%) and between H. vulgare and H. bulbosum (∼3%) (Morrell et al. 2014). Alignments were processed with Samtools (Li et al. 2009) and the Genome Analysis Toolkit (GATK) (McKenna et al. 2010; DePristo et al. 2011) according to GATK best practices (http://www.broadinstitute.org/gatk/guide/best-practices). For realignment around indels, we used a set of high-confidence indels reported in Sanger resequencing datasets (Caldwell et al. 2006; Morrell et al. 2006; Morrell and Clegg 2007; Morrell et al. 2014). We then extracted the base calls at each BOPA SNP location using tools from the GATK. For all SNPs where it can be inferred, the ancestral state is listed in Table S3.
Geographic differentiation
We examined the geographic structure within the range of wild barley with Bayesian genetic assignment implemented in the program STRUCTURE (Pritchard et al. 2000; Falush et al. 2003). We treated individual samples as haploid and explored both admixture and no admixture models and models with correlated and uncorrelated allele frequencies with K = 2 − 10 clusters. Because a model with no admixture and uncorrelated allele frequencies resulted in higher likelihoods, it was used for the final analyses. For each value of K, we used 10 replicate runs with a burn-in length of 100,000 iterations and a run length of 100,000 iterations.
Haplotypes were defined based on five adjacent SNPs and used both for comparison of haplotype diversity and as input for genetic assignment analysis. Combining markers into haplotypes results in multi-allelic data that can improve inference of population structure (Haasl and Payseur 2010; Gattepaille and Jakobsson 2012). To infer missing data, we used the program fastPHASE (Scheet and Stephens 2006) with 20 random starts and 25 iterations of the Expectation-Maximization algorithm. The number of SNPs within each haplotype was determined based on optimal numbers from the simulation study of Gattepaille and Jakobsson (2012) and the total number of SNPs in our samples. To deal with label switching (where cluster names change between replicate runs) and with true multimodality (where individual samples switch clusters in replicate runs), we used CLUMPP (Jakobsson and Rosenberg 2007) to summarize assignment results across replicate runs. The program Infocalc was used to calculate the informativeness for assignment (In) (Rosenberg et al. 2003) for each haplotype using the clusters identified by STRUCTURE. Informativeness for assignment identifies the information content for genetic assignment for markers based on the degree to which each locus (or haplotype) contributes to discrimination among populations (Rosenberg et al. 2003).
The prcomp function in R (R Development Core Team 2012) was used to perform principal component analysis (PCA). Each WBDC accession was assigned to clusters based on significant principal components (PCs) using the Ward clustering method in the R hclust function. To compare the similarity between genetic variation on a PCA plot and geographic maps of sample locations, we used Procrustes analysis to find a rotation that maximizes the similarity (Wang et al. 2012).
Within and among inferred clusters, we estimated hierarchical F-statistics (Yang 1998) using the Hierfstat package (Goudet 2005) in R. We calculated summary statistics, including number of segregating sites, and number of private alleles in each cluster using tools from the libsequence library (Thornton 2003) and estimated average pairwise SNP diversity within each cluster using the R package ape (Paradis et al. 2004). Rarefaction was used to analyze allelic diversity across populations while correcting for sample size differences using the program ADZE (Szpiech et al. 2008).
Linkage disequilibrium (LD) as measured by r2 (Hill and Robertson 1968) was calculated for all possible pairwise comparisons on each linkage group based on SNPs with minor allele frequency (MAF) >5%. The LDheatmap package (Shin et al. 2006) was used to generate plots of LD relative to genetic distance.
Environmental–genetic correlations
To identify genomic segments potentially contributing to local adaptation, we divided samples into the two primary clusters (the Eastern and Western populations) and six clusters identified by genetic assignment in STRUCTURE and calculated FST (Weir and Cockerham 1984). The assumption is that SNPs linked to genomic regions contributing to local adaptation will show greater allele frequency divergence (higher FST) than those affected only by demography (Cavalli-Sforza 1966; Lewontin and Krakauer 1973).
Environmental variables, including altitude, monthly precipitation, monthly maximum and minimum temperatures, and 19 additional bioclimatic variables were downloaded from www.worldclim.org (Hijmans et al. 2005). DIVA-GIS (Hijmans et al. 2001) was used to extract climate data at 5 arc-min (∼10 km) resolution for each sample. We focused on measurements within the growing season for wild barley, from late autumn to spring, so we removed environmental variables related to summer temperature and precipitation. The environmental variables used are listed in Table S4A. Environmental variables were scaled to a mean of 0 and SD of 1 and summarized into principal components. The significant PCs of environmental variables were used for Bayenv (Coop et al. 2010).
We used Bayenv to identify the correlation between SNPs and environmental variables. Bayenv requires population information to account for population structure, making use of allele frequency within a set of samples representing a localized environment to correct for population structure in environmental association. We used PCA to group all samples into 17 clusters with a mean sample size of 17 accessions. The number of optimal stratifications in our data was determined using Velicer’s minimum average partial test (Shriner 2011). We used all SNPs to construct the covariance matrix. Two independent runs of 30,000 iterations were compared to control for convergence and the final covariance matrix is the mean of these two independent runs. Bayenv was used to estimate the Bayes factor for each SNP with each environmental variable using 50,000 iterations. SNPs were considered candidates contributing to local adaptation if they had an average Bayes factor above the 95th percentile genome-wide for five separate runs.
Because the wild barley samples cover a large continuous geographic range, spatial ancestry analysis (SPA) (Yang et al. 2012) was used to model allele frequency change for each SNP as a function of the location of the individual in geographic space. SNPs contributing to local adaptation potentially have larger gradients in allele frequency, reflected in a high SPA score (Yang et al. 2012). SNPs were considered candidates for local adaptation if their SPA score was above the 95th percentile genome-wide.
SNPs that are outliers (above the 95th percentile genome-wide) in the FST, environmental association, or SPA analyses can be considered candidate variants either causative or, more likely, linked to loci involved in local adaptation. Enrichment analysis of candidates among genic vs. nongenic and nonsynonymous vs. synonymous was performed by resampling the number of SNPs in each candidate list randomly from the genome 1000 times. Enrichment analyses included sets of SNPs genome-wide, centromeric regions, and the two regions that are putative structural rearrangements (inversions or translocations). This generated a distribution of ratios of the number of genic to the number of nongenic SNPs and ratios of the number of nonsynonymous to the number of synonymous SNPs in each analysis, which can be compared to the observed ratios from the data.
Results
Initial screening of relatedness among accessions identified 30 wild barley accessions either with a large genetic distance from the majority of accessions or that appear to be duplicated within the sample. The large genetic distance was also associated with a high degree of identity to barley landraces genotyped on the same platform. The increase in genetic distance appears to result from ascertainment bias due to the discovery of barley SNPs primarily among cultivated samples, which results in a larger number of segregating polymorphisms in barley landrace accessions than in wild barley (Russell et al. 2011). The 30 accessions were excluded from further analysis because they constitute duplicate accessions or because genotypic composition suggests they either could be feral barley accessions or were subject to recent introgression. Four accessions do not have known latitude and longitude of origin, so they were removed from analyses; thus, 284 wild barley accessions from the WBDC were used in this study (Table S1). After all quality control measures, 2330 SNPs were assayed in each accession (Table S5).
Microsatellite loci were extremely polymorphic among wild barley accessions with an average of 20 alleles per locus. Allele size for individual microsatellites is normally distributed and thus accords with a stepwise mutation model (Ohta and Kimura 1973; Valdes et al. 1993).
Population structure
A Mantel test identifies a positive correlation between geographic distance and genetic distance (Mantel statistic: 0.37; significance at 0.001), consistent with isolation by distance among these wild barley accessions.
STRUCTURE results based on single-SNP and 5-SNP haplotypes for K = 2 identified populations east and west of the Zagros Mountains. Samples from a broad portion of the range, including Central Asia (particularly east of the Caspian Sea), the Iranian Plateau, and most samples in Northern Mesopotamia (modern Northern Iraq and Syria) form an Eastern population, and samples west of the Zagros Mountains, including samples from the Levant, and around the Mediterranean to North Africa form a Western population (Figure 1A). There is also isolation by distance within the Eastern population (Mantel statistic: 0.31; significance at 0.001) and the Western population (Mantel statistic: 0.20; significance at 0.001). For genetic assignment based on individual SNPs, there are six accessions from Central Asia assigned to the Western population (data not shown). Thus, genetic assignment from haplotype data shows greater consistency with geographic location of origin. The broad-scale geographic patterns identified from SNP and 5-SNP haplotypes are not readily reflected in genetic assignment based on the 29 microsatellite loci (data not shown). The individual alleles are relatively rare and have low informativeness for assignment, an issue attributable to the high levels of polymorphism in these microsatellites (Table S2).
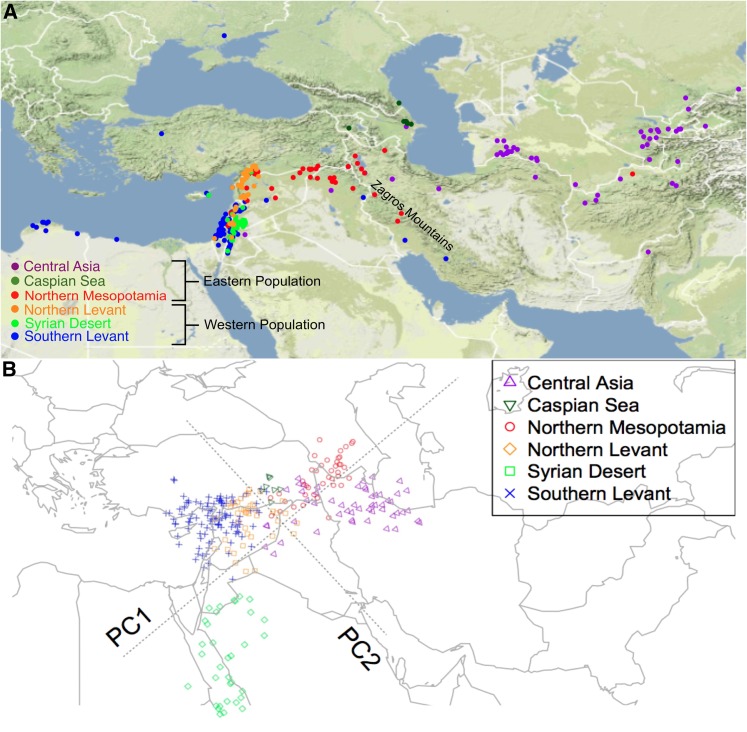
Figure 1.
(A) Population structure in wild barley. Each of the six colors represents one of six subpopulations. There are three subpopulations nested within both the Eastern and Western populations. (B) Procrustes-transformed PCA plot of genetic variation in wild barley.
For STRUCTURE analysis based on haplotype data, when K = 3, a group of accessions from the Syrian desert region becomes an independent cluster from the Western population. As K increases to 4, the samples along the east coast of the Mediterranean split into two groups, one in the north (Northern Levant) and the other in the south (Southern Levant). The Eastern population begins to differentiate as K is increased to 5. The samples in Central Asia are separated from samples from Northern Mesopotamia and those from west of the Caspian Sea. With K = 6, the seven samples from the Caspian Sea become an independent cluster (Figure 1A). For the present sample, K = 6 provides a clear distinction among populations and the genetic assignment is not constrained by very small sample size within individual clusters. The average informativeness for assignment for individual SNPs is 0.03 for K = 2 and 0.10 for K = 6, increasing to 0.14 for K = 2 and 0.47 for K = 6 when based on haplotypes (Figure S1).
Among the six clusters, three are within the Eastern population (Central Asia, Caspian Sea, and Northern Mesopotamia) and three are in the Western population (Northern Levant, Syrian Desert, and Southern Levant), constituting hierarchical population structure nested within the major Eastern and Western populations. The comparative FST analyses identify both a significant effect of individuals within subpopulations (p-value = 0.01; 100 permutations; observed likelihood ratio statistics = 94,259.55) and a significant effect of subpopulations within populations (p-value = 0.04; 100 permutations; observed likelihood ratio statistics = 33,632.57). Population structure is best explained by a six-subpopulation model (variance components = 17.7%); in contrast, the two-population model provides a poorer fit to the data (variance components = 10.6%). FST values are also higher at the subpopulation level than the population level (Table 1). Within the two higher-level populations, 20.5% of the genetic differentiation can be explained by the three subpopulations within the Eastern population, whereas 13.2% can be explained by the three subpopulations within the Western population.
Table 1. Hierarchical F-statistics comparing different levels of the hierarchical population structure.
| Population | Subpopulation | |
|---|---|---|
| Total | 0.042 | 0.191 |
| Population | 0.000 | 0.155 |
The values reported include the F statistics for two primary populations vs. total, six subpopulations vs. two primary populations, and six subpopulations vs. total.
In the PCA examination of population structure, the first PC separates all samples into the Eastern and Western populations. When adding the second PC, all six subpopulations are clearly differentiated. A PCA plot of genetic variation closely reflects geography in the population after a 42.44° counterclockwise rotation of the PCA plot using Procrustes analysis. The boundary of the Eastern and Western population approximately parallels the Zagros Mountains (Figure 1B). The Syrian Desert subpopulation is noteworthy in showing greater PC distance within and among samples, possibly because of greater genetic drift within this subpopulation. A mean pairwise FST analysis indicates a higher mean FST value for this subpopulation (0.09) than for the other two Western subpopulations (0.05 and 0.04).
Population differentiation
The average genome-wide FST between the Eastern and Western populations is 0.07. The boundary of these two populations is close to the Zagros Mountains (Figure 1A), which forms a potential barrier to migration (Lin et al. 2001; Morrell et al. 2003). The most northerly portion of the Zagros Range is at ∼48° E longitude, trending from the northwest to the southeast, so we also compared the allele frequency differentiation between the two populations east and west of 48° E. For this geographic contrast, the average FST genome-wide is 0.06 and the correlation coefficient (r) between this partition based on the Zagros Mountains and the previous partition based on genetic assignment analysis is 0.473, which supports the hypothesis that the Zagros Mountains act as a natural barrier that bisects wild barley into the Eastern and Western populations. For the 29 microsatellite loci, the average RST between the Eastern and Western populations is 0.15.
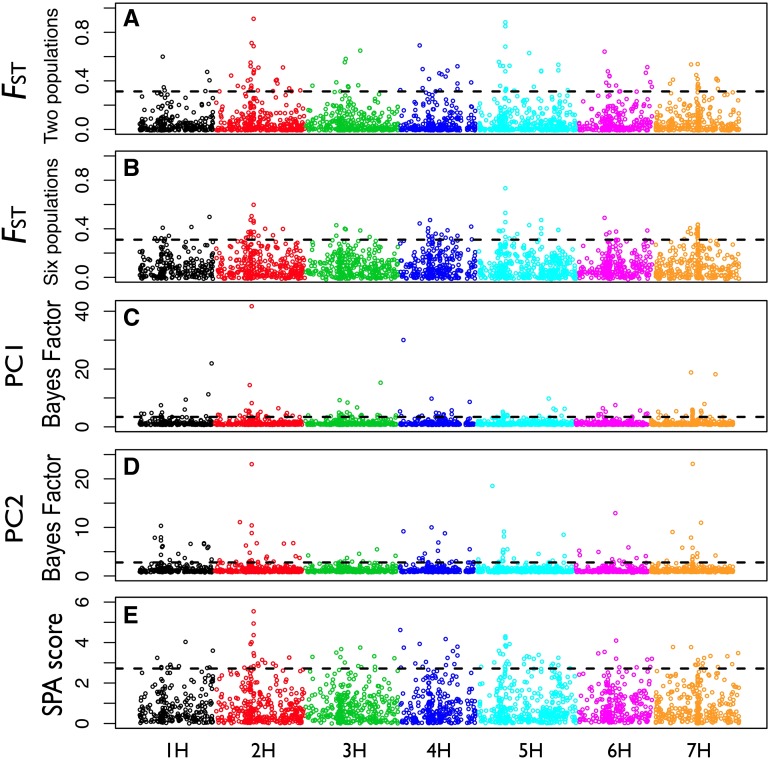
SNPs with high FST values between the Eastern and Western populations (Figure 2A) or among all the six subpopulations (Figure 2B) are most abundant in two genomic regions, one on linkage group 2H, from ∼67 to 74 cM, and the other on 5H, from ∼47 to 52 cM. For the 2H and 5H regions, the mean FSTs are 0.20 (56 SNPs) and 0.17 (32 SNPs), respectively, vs. a genome-wide FST = 0.07.
Figure 2.
(A) FST between the Eastern and Western wild barley populations. (B) Pairwise FST based on all six subpopulations. The dotted line is the 95th percentile of FST genome-wide. (C) Bayes factors for correlation between allele frequencies and PC1. (D) Bayes factors for correlation between allele frequencies and PC2. (E) SPA score genome-wide from spatial analysis. The 95th percentile of the distribution of Bayes factors or SPA scores is indicated by a horizontal dashed line.
Many SNPs or haplotypes genome-wide with high informativeness for assignment are in these two high FST regions (Figure S1). Among all SNPs above the 95th and 99th percentiles (In = 0.31 and 0.42), 33% and 52% are in these two regions (Figure S1). However, when we perform genetic assignment analysis after masking these two high FST regions, the probability of assignment for each wild barley accession into the Eastern and Western population is nearly identical for all but one accession. This result reflects the relatively high informativeness for assignment observed for SNPs within pericentromeric regions on all linkage groups (Figure S1). Therefore, population structure in these two high FST regions is similar to the genome-wide pattern and the genome-wide pattern is driven by a high degree of differentiation in pericentromeric regions.
The joint unfolded site frequency spectrum demonstrates that there are more rare variants in the Western population than in the Eastern population (Figure S2). Percent pairwise differences are lower in the Eastern population than in the Western population (Table 2). There are more private SNPs in the Western population than in the Eastern population (430 vs. 86) (Table 2). Because the sample size is different between the Eastern and Western populations, we used rarefaction to correct for sample size. Despite the correction, both the mean number of distinct alleles per locus (Figure S3A) and the mean number of private alleles per locus (Figure S3B) are higher in the Western than the Eastern population. The Southern Levant subpopulation has the highest values for the number of segregating sites and number of private SNPs, whereas the Caspian Sea subpopulation has the lowest values for these summary statistics (Table 2).
Table 2. Diversity summary statistics for the two populations and six subpopulations.
| Population | Size | # Segregating Sites | # Private SNPs | Percent Pairwise Difference (SD) | Microsatellite Expected Heterozygosity |
|---|---|---|---|---|---|
| Eastern | 101 | 2196 | 86 | 0.20 (0.04) | 0.740 |
| Caspian Sea | 7 | 1146 | 2 | 0.10 (0.02) | 0.672 |
| Central Asia | 53 | 2027 | 22 | 0.19 (0.04) | 0.734 |
| Northern Mesopotamia | 41 | 1975 | 13 | 0.17 (0.03) | 0.717 |
| Western | 183 | 2285 | 430 | 0.23 (0.03) | 0.742 |
| Northern Levant | 42 | 2033 | 19 | 0.21 (0.02) | 0.740 |
| Southern Levant | 107 | 2197 | 49 | 0.22 (0.02) | 0.736 |
| Syrian Desert | 34 | 1916 | 6 | 0.16 (0.03) | 0.722 |
Data include the sample size, number of segregating sites, number of private SNPs, percent pairwise difference with SD, and microsatellite expected heterozygosity .
Structural rearrangements
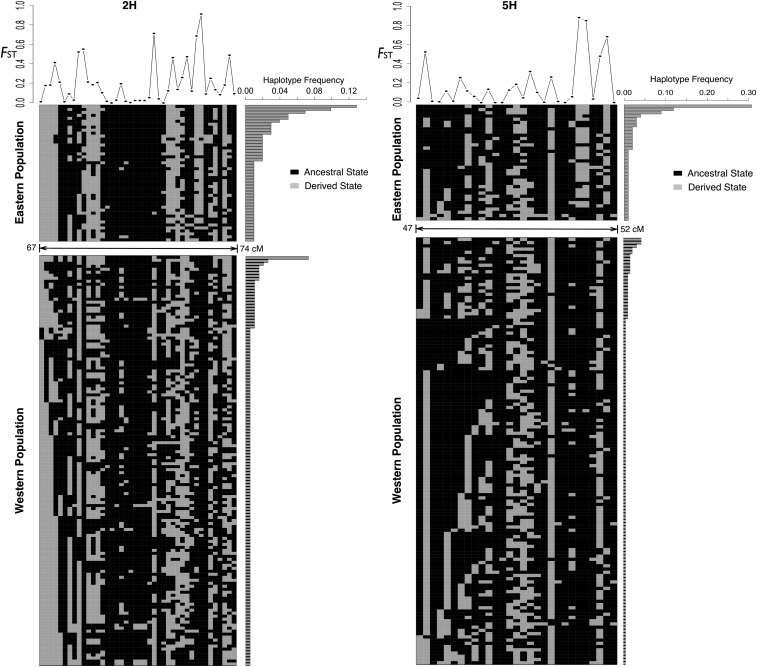
The high FST regions on 2H and 5H are potentially attributable to chromosomal structural variants. Population genetic variation, particularly patterns of LD, can suggest structural variation (Huynh et al. 2011; Long et al. 2013). The average pairwise LD in the high FST region on 2H (r2 = 0.085) is higher than other regions of 2H (r2 = 0.018) and the adjacent regions with similar size left and right of the high FST region, where r2 = 0.021 for both regions. The average pairwise LD in the high FST region on 5H (r2 = 0.097) is also higher than other regions on 5H (r2 = 0.019) and the adjacent regions with similar size to the left and right of the high FST region (r2 = 0.017 and 0.020). The 5-SNP segment with the lowest haplotype number (4) on 2H is within the high FST region (Figure S4). As on other linkage groups, the segment with the lowest haplotype number on 2H and 5H is within the centromeric region (Figure S4). The centromeric region on 2H overlaps with the high FST region, whereas the centromere is distal to the high FST region on 5H (Figure S4). The observed LD, FST, and haplotype number patterns are consistent with recent positive selection and/or chromosome structural rearrangements. There are more unique haplotypes in the Western population than in the Eastern population (Figure 3). In the Eastern population, both of the two regions on 2H and 5H are dominated by a few haplotypes (Figure 3), which is potentially consistent with selection favoring these haplotypes.
Figure 3.
Diagram of haplotype diversity in the two putative chromosome structural rearrangements on 2H and 5H. Haplotypes are divided into the two primary populations identified by genetic assignment. Each SNP is represented by either the ancestral state (black) or the derived state (gray). The frequencies of each of the haplotypes from the Eastern population (top) and from the Western population (bottom) are shown on the right. FSTs for each SNP between these two populations are shown at the top of the haplotype diagram.
Evidence for local adaptation
SNPs that occur as outliers in the FST analysis may indicate genomic regions involved in local adaptation. Annotation information for SNPs that are above the 95th percentile for FST between the Eastern and Western populations is listed in Table S6.
Environmental association analysis was used to identify genetic polymorphisms potentially involved in local adaptation. PCA reveals two major clusters of environmental variables (Figure S5). The first two PCs explain 80% of the total variance (Figure S5). The first PC includes most temperature variables, whereas most precipitation variables and altitude are in the second PC (Table S4B).
SNPs associated with both environmental PC1 and PC2 (above the 95th percentile) are distributed on all linkage groups (Figure 2, C and D). The high FST region on 2H is highly associated with both PC1 and PC2 (Figure 2 C and D). The high FST region on 5H is also associated with PC2 (Figure 2D). The annotation information for SNPs that are above the 95th percentile of association with PC1 and PC2 is listed in Table S7.
SPA analysis reveals that ∼20% of the SNPs in the high FST regions on both 2H (11 out of 56) and 5H (7 out of 32) show strong geographic gradients in allele frequencies as their SPA scores are above the 95th percentile (2.72) (Figure 2E). The SNP with the highest SPA score (5.54) is in the high FST region on 2H. There are 24 SNPs with SPA score above the 99th percentile (3.75), and nearly half of these SNPs (11) are in these two high FST regions on 2H and 5H. The annotation information for the SNPs with SPA scores above the 95th percentile is in Table S8.
Enrichment analysis reveals that the two putative chromosomal structural rearrangements are enriched for genic SNPs in the outliers of PC2 from environmental analysis and SPA analysis (Figure S6A). There is no evidence of enrichment for nonsynonymous SNPs (Figure S6B).
Discussion
Hierarchical population structure
Wild barley shows strong hierarchical population structure, with primary structure east and west of the Zagros Mountains and three subpopulations identified in both the Eastern and Western populations (Figure 1A). Previous studies of sequence diversity in wild barley have identified population structure that strongly differentiates the Eastern and Western wild barley populations (Lin et al. 2001; Morrell and Clegg 2007; Saisho and Purugganan 2007). The present study samples a much larger number of accessions and includes only 22 SNPs in common with those sampled in previous studies. Despite the limited overlap of sampled SNPs, genetic assignment with K = 2 uncovers a similar geographic pattern. Moreover, the results of this study go beyond previous work by revealing a division into six subpopulations that explains 7.1% more variance compared with the primary, two-population division.
A nearly continuous geographic range represents a particular challenge for efforts to identify population structure and geographic discontinuities. However, populations of wild barley are much more common in the western portion of the range and below 1500 m (Zohary and Hopf 2000), thus the 3000-m to 4500-m peaks of the Zagros Mountains and the high elevation regions of the Iranian plateau are disruptions of an otherwise continuous range.
The Western population is more diverse than the Eastern population (Table 2), at least in part because the SNP discovery panel was composed primarily of Western cultivars. It should be noted that the discovery panel also included OUH602 (WBDC355), which assigns to the Central Asia (Eastern) wild barley population. Estimates of diversity based on resequencing a more limited set of wild barley samples indicate moderately higher levels of diversity in the Western than in the Eastern wild barley populations (Morrell and Clegg 2007; Morrell et al. 2014). The effect of ascertainment bias on the frequency spectrum depends on the population in which SNPs were discovered (Albrechtsen et al. 2010). Therefore, we observe more rare alleles in the Western population (Figure S2). There is a small but clear effect of ascertainment bias, which leads to an increased estimate of diversity in the Western population, because the Western population is more similar to the discovery panel. The size of a discovery panel is less important than the composition, as long as it is not extremely small (<4 chromosomes) (Albrechtsen et al. 2010). Based on coalescent simulations, a discovery panel of eight samples with a minimum allele count of three best-reflects the design parameters of the BOPA SNPs (Fang et al. 2013).
Two putative chromosome rearrangements
Resequencing studies identified a large degree of heterogeneity in wild barley, in terms of both degree of population structure and levels of nucleotide sequence diversity (Lin et al. 2001; Morrell et al. 2003). Using simple coalescent simulations, Morrell et al. (2003) argued that stochastic variation alone was insufficient to explain intralocus heterogeneity and that selection, either through selective sweeps at some loci or through local adaptation at others, was necessary to explain observed patterns of diversity (Wright et al. 2005). Strong genetic differentiation between the Eastern and Western populations for the two regions on 2H and 5H suggest that some of the heterogeneity may result not from selection acting on individual loci, but rather on structural variants (Figure 2). Structural variants are quickly lost due to drift and purifying selection unless they confer a locally adaptive advantage. A structural variant that captures two or more alleles adapted to the local environment has a selective advantage that can cause it to spread (Kirkpatrick and Barton 2006).
The high FST region on 2H occurs in the same approximate chromosomal location as a chromosomal rearrangement identified in an eastern wild barley accession based on meiotic pairing studies (Konishi and Linde-Laursen 1988). Konishi and Linde-Laursen (1988) report a reciprocal translocation with 4H in a sample from Turkmenistan with the breakpoints of the translocation near the centromere on 2H. Using a three-point linkage test, Ramage and Suneson (1961) identify evidence of both an inversion and translocations on the long arm of 5H.
The two genomic regions with high FST also have above average levels of LD and low haplotype number (Figure S4). Environmental association analysis identifies multiple SNPs in these regions associated with both temperature and precipitation (Figure 2). SPA analysis identifies half of the SNPs in these two regions as outliers with dramatic change in allele frequency gradients, as identified by SPA scores above the 99th percentile. The SPA method is particularly sensitive to SNPs that have steep geographic gradients in allele frequency and is scored based on individual accessions rather than populations (Yang et al. 2012).
The large number of candidate SNPs in these regions identified in multiple analyses indicate these two regions may harbor variants that are locally adaptive, with selection altering the frequency of nearby SNPs through genetic hitchhiking (Nielsen et al. 2005). Given the density of SNPs assayed in the present study and the relatively rapid decay of LD in wild barley (Morrell et al. 2005, 2006), the detection of multiple SNPs associated with environmental factors is likely to occur only in regions with suppressed recombination. These two high FST regions are 5 to 7 cM, potentially including hundreds of genes; thus, the patterns observed are likely due to chromosome structural variants that, through the inhibition of genetic exchange between chromosomal rearrangements, have the effect of slowing migration.
The Eastern accessions occur in a region that, on average, is more arid than the region occupied by the Western accessions, where most samples were collected from populations along the Mediterranean coast. The precipitation pattern is reflected in the environmental association analysis. Both of the two putative chromosome rearrangements on 2H and 5H are associated with precipitation variables (Figure 2).
Comparisons among genetic maps can provide confirmation of the presence of chromosomal rearrangements through the inference of alternative mapping order in populations where both mapping parents carry the alternative arrangement (Thomas et al. 2008; Lowry and Willis 2010) or through evidence of suppression of crossover when parents differ for the chromosomal arrangement (Fang et al. 2012). It is possible to identify mapping parents that differ for chromosomal rearrangements based on the presence of variants (SNPs) that are private (or nearly private) to the putative rearrangement (Fang et al. 2012) or in cases where the geographic distribution of the putative rearrangement is well-defined, using parents from geographic regions where the rearrangement is more common. Both putative structural rearrangements in wild barley occur in the eastern portion of the wild range. Barley is a cultigen with multiple origins (Morrell and Clegg 2007), with a greater contribution of eastern wild barley ancestry among Asian landraces (Morrell and Clegg 2007; Morrell et al. 2014). Barley doubled haploid genetic mapping populations compared by Muñoz-Amatriaín et al. (2011) include three populations that could potentially differ for the rearrangement: the Japanese malting barley cultivar Haruna Nijo by wild barley OUH602 (Sato et al. 2009); Haruna Nijo by food barley cultivar Akashinriki (Sato et al. 2011); and the Oregon Wolfe Barley (OWB) population (Costa et al. 2001; Stein et al. 2007; Szűcs et al. 2009).
Examining the genomic regions on 2H and 5H with highest allele frequency differentiation and levels of LD among wild barley accessions, we identify an average of 24 SNPs on 2H and 17 SNPs on 5H segregating among mapping parents in these genetic maps. The two Haruna Nijo maps include a single inferred crossover on both 2H and 5H, whereas the OWB maps include a larger number of crossovers on 2H and two crossovers on 5H. Crossover number in these genetic maps does not differ dramatically from that observed in the same genomic regions in mapping populations for Morex × Barke or Morex × Steptoe. Morex, Barke, and Steptoe are western cultivated barleys and are likely to bear the standard arrangement. Given the uncertainty of the presence of the putative rearrangements in mapping parents and limited marker density within the putative rearranged genomic regions, the present genetic maps neither convincingly refute nor support the presence of structural rearrangements.
We also noted that the two putative chromosome rearrangements are not differentiated by large numbers of private SNPs (Figure 3) as observed at the largest inversion (Inv1n) in teosinte, the wild progenitor of maize (Fang et al. 2012). This may be attributable in the discovery of SNPs primarily among western cultivars, which are unlikely to carry the rearrangement. Moreover, the putative translocation on 2H incorporates the centromere and the putative rearrangement on 5H is close to the centromeric region. Therefore, an alternative explanation for the patterns observed in these two genomic regions could be exceptional effects arising from suppressed recombination in the centromeric regions (Mayer et al. 2012).
Useful wild barley alleles
Identification of functional variation that indicates local adaptation can contribute to sustained crop improvement. Crop wild progenitors have a history of adaptation to their local environments that is orders of magnitude older than the time since domestication, so wild populations have been exposed to natural selection for many more generations. Moreover, a domestication bottleneck decreases nucleotide diversity and causes the loss of valuable variants (Eyre-Walker et al. 1998). For these reasons, wild populations are expected to carry many novel nucleotide sequence variants and functional adaptations that are not present in domesticates.
We used several approaches to identify nucleotide polymorphisms potentially involved in local adaptation. SNPs that are outliers in the FST, environmental association, and SPA analyses are potentially linked to loci contributing to local adaptation. The FST comparison assumes that genetic markers with extreme allele frequency differences among populations may contribute to local adaptation (Lewontin and Krakauer 1973), but this method has several limitations. First, the Lewontin and Krakauer approach suffers from a number of assumptions, including that all populations diverged at the same time (Nei and Maruyama 1975). Second, comparison of allele frequency differences with FST requires the prior identification of populations. Environmental association and SPA analyses complement and are consistent with the FST results. The results of both analyses support the conclusion that selection has contributed to the observed allele frequency differentiation likely driven by environmental factors or correlated selection pressures (Coop et al. 2010).
A number of SNPs within previously characterized barley loci are outliers in one or more of the analyses reported here. The SNP (12_30850) from the gene Cbf4 is above the 95th percentile of FST values for SNP frequencies compared between the Eastern and Western populations. Cbf4 contributes to low-temperature tolerance in barley (Francia et al. 2004). From environmental association analysis, one SNP (11_11361) in Cbf4 and another SNP (11_10989) in a cold-regulated gene blt14 (Cattivelli and Bartels 1990; Grossi et al. 1998) are among outliers of PC1, which includes the temperature variables. SPA analysis reveals that SNP 12_30850 in Cbf4 also shows a strong geographic gradient in allele frequency. These results establish the value of geographic analyses as a screen to identify potentially adaptive alleles.
Summary
Two large pericentromeric regions on 2H and 5H make strong contributions to the population structure observed in wild barley. These two genomic regions are putative chromosome structural rearrangements that harbor variants that appear to contribute to environmental adaptation. In particular, the SNPs in these chromosomal regions are shown to be associated with temperature and precipitation variables. It will be important to determine how specific genetic variants within these rearrangements are associated with local adaptation. Nevertheless, the identification of genomic regions associated with environmental adaptation suggests an opportunity for crop improvement.
Supplementary Material
Acknowledgments
The authors thank Thomas Kono for assistance with determination of SNP ancestral state and helpful discussion, and Allison Haaning, Tanja Pyhäjärvi, Jeffrey Ross-Ibarra, and Jeremy Yoder for comments on an earlier version of the manuscript. This work was performed using computing resources at the University of Minnesota Supercomputing Institute. We acknowledge funding from the US Department of Agriculture National Institute for Food and Agriculture USDA NIFA 2011- 68002-30029 (to P.L.M.) and University of Minnesota Doctoral Dissertation Fellowship (to Z.F.) Support was also received from the Lieberman-Okinow Endowment at the University of Minnesota and the USAID-funded Cereals Comparative Genomics Initiative.
Footnotes
Communicating editor: D. Zamir
Literature Cited
- Albrechtsen A., Nielsen F. C., Nielsen R., 2010. Ascertainment biases in SNP chips affect measures of population divergence. Mol. Biol. Evol. 27: 2534–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. H. D., Zohary D., Nevo E., 1978. Outcrossing rates and heterozygosity in natural populations of Hordeum spontaneum Koch in Israel. Heredity 41: 49–62. [Google Scholar]
- Caldwell K. S., Russell J., Langridge P., Powell W., 2006. Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species, Hordeum vulgare. Genetics 172: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattivelli L., Bartels D., 1990. Molecular cloning and characterization of cold-regulated genes in barley. Plant Physiol. 93: 1504–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., 1966. Population structure and human evolution. Proc. R. Soc. Lond. B Biol. Sci. 164: 362–379. [DOI] [PubMed] [Google Scholar]
- Close T. J., Bhat P. R., Lonardi S., Wu Y., Rostoks N., et al. , 2009. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10: 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. I., Galinat W. C., 1984. Potential use of alien germplasm for maize improvement. Crop Sci. 24: 1011–1015. [Google Scholar]
- Coop G., Witonsky D., Di Rienzo A., Pritchard J. K., 2010. Using environmental correlations to identify loci underlying local adaptation. Genetics 185: 1411–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa J. M., Corey A., Hayes P. M., Jobet C., Kleinhofs A., et al. , 2001. Molecular mapping of the Oregon Wolfe Barleys: a phenotypically polymorphic doubled-haploid population. Theor. Appl. Genet. 103: 415–424. [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Gaut R. L., Hilton H., Feldman D. L., Gaut B. S., 1998. Investigation of the bottleneck leading to the domestication of maize. Proc. Natl. Acad. Sci. USA 95: 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K., 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Pyhäjärvi T., Weber A. L., Dawe R. K., Glaubitz J. C., et al. , 2012. Megabase-scale inversion polymorphism in the wild ancestor of maize. Genetics 191: 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Eule-Nashoba A., Powers C., Kono T. Y., Takuno S., et al. , 2013. Comparative analyses identify the contributions of exotic donors to disease resistance in a barley experimental population. G3 (Bethesda) 3: 1945–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetch T. G., Steffenson B. J., Nevo E., 2003. Diversity and sources of multiple disease resistance in Hordeum spontaneum. Plant Dis. 87: 1439–1448. [DOI] [PubMed] [Google Scholar]
- Francia E., Rizza F., Cattivelli L., Stanca A. M., Galiba G., et al. , 2004. Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’(winter) × ‘Tremois’(spring) barley map. Theor. Appl. Genet. 108: 670–680. [DOI] [PubMed] [Google Scholar]
- Gattepaille L. M., Jakobsson M., 2012. Combining markers into haplotypes can improve population structure inference. Genetics 190: 159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman S. J., 1997. RST Calc: a collection of computer programs for calculating estimates of genetic differentiation from microsatellite data and determining their significance. Mol. Ecol. 6: 881–885. [Google Scholar]
- Goudet J., 2005. Hierfstat, a package for R to compute and test hierarchical F‐statistics. Mol. Ecol. Notes 5: 184–186. [Google Scholar]
- Grossi M., Giorni E., Rizza F., Stanca A. M., Cattivelli L., 1998. Wild and cultivated barleys show differences in the expression pattern of a cold-regulated gene family under different light and temperature conditions. Plant Mol. Biol. 38: 1061–1069. [DOI] [PubMed] [Google Scholar]
- Haasl R. J., Payseur B. A., 2010. Multi-locus inference of population structure: a comparison between single nucleotide polymorphisms and microsatellites. Heredity 106: 158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D. R., 1990. Vavilov’s concept of centres of origin of cultivated plants: its genesis and its influence on the study of agricultural origins. Biol. J. Linn. Soc. Lond. 39: 7–16. [Google Scholar]
- Hijmans R. J., Guarino L., Cruz M., Rojas E., 2001. Computer tools for spatial analysis of plant genetic resources data: 1. DIVA-GIS. Plant Genet. Resour. Newsl. 127: 15–19. [Google Scholar]
- Hijmans R. J., Cameron S. E., Parra J. L., Jones P. G., Jarvis A., 2005. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25: 1965–1978. [Google Scholar]
- Hill W. G., Robertson A., 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38: 226–231. [DOI] [PubMed] [Google Scholar]
- Huynh L. Y., Maney D. L., Thomas J. W., 2011. Chromosome-wide linkage disequilibrium caused by an inversion polymorphism in the white-throated sparrow (Zonotrichia albicollis). Heredity 106: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob S. S., Rodder D., Engler J. O., Shaaf S., Ozkan H., et al. , 2014. Evolutionary history of wild barley (Hordeum vulgare subsp. spontaneum) analyzed using multilocus sequence data and paleodistribution modeling. Genome Biol. Evol. 6: 685–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M., Rosenberg N. A., 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806. [DOI] [PubMed] [Google Scholar]
- Jones H., Leigh F. J., Mackay I., Bower M. A., Smith L. M. J., et al. , 2008. Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol. Biol. Evol. 25: 2211–2219. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M., Barton N., 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T., Linde-Laursen I., 1988. Spontaneous chromosomal rearrangements in cultivated and wild barleys. Theor. Appl. Genet. 75: 237–243. [Google Scholar]
- Kono T. Y., Seth K., Poland J. A., Morrell P. L., 2014. SNPMeta: SNP annotation and SNP metadata collection without a reference genome. Mol Ecol Resour 14: 419–425. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R. C., Krakauer J., 1973. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics 74: 175–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Z., Sjakste T. G., Röder M. S., Ganal M. W., 2003. Development and genetic mapping of 127 new microsatellite markers in barley. Theor. Appl. Genet. 107: 1021–1027. [DOI] [PubMed] [Google Scholar]
- Lin J. Z., Brown A. H., Clegg M. T., 2001. Heterogeneous geographic patterns of nucleotide sequence diversity between two alcohol dehydrogenase genes in wild barley (Hordeum vulgare subspecies spontaneum). Proc. Natl. Acad. Sci. USA 98: 531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q., Rabanal F. A., Meng D., Huber C. D., Farlow A., et al. , 2013. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45: 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B., Willis J. H., 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8: e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews D. E., Carollo V. L., Lazo G. R., Anderson O. D., 2003. GrainGenes, the genome database for small-grain crops. Nucleic Acids Res. 31: 183–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K. F., Waugh R., Brown J. W., Schulman A., Langridge P., et al. , 2012. A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell P. L., Clegg M. T., 2007. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc. Natl. Acad. Sci. USA 104: 3289–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell P. L., Lundy K. E., Clegg M. T., 2003. Distinct geographic patterns of genetic diversity are maintained in wild barley (Hordeum vulgare ssp. spontaneum) despite migration. Proc. Natl. Acad. Sci. USA 100: 10812–10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell P. L., Toleno D. M., Lundy K. E., Clegg M. T., 2005. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proc. Natl. Acad. Sci. USA 102: 2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell P. L., Toleno D. M., Lundy K. E., Clegg M. T., 2006. Estimating the contribution of mutation, recombination and gene conversion in the generation of haplotypic diversity. Genetics 173: 1705–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell P. L., Gonzales A. M., Meyer K. K. T., Clegg M. T., 2014. Resequencing data indicates a modest effect of domestication on diversity in barley: a cultigen with multiple origins. J. Hered. 105: 253–264. [DOI] [PubMed] [Google Scholar]
- Muñoz-Amatriaín M., Moscou M. J., Bhat P. R., 2011. An improved consensus linkage map of barley based on flow-sorted chromosomes and single nucleotide polymorphism markers. Plant Genome 4: 238–249. [Google Scholar]
- Nei M., Maruyama T., 1975. Lewontin-Krakauer test for neutral genes. Genetics 80: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo E., Beiles A., Zohary D., 1986. Genetic resources of wild barley in the Near East: structure, evolution and application in breeding. Biol. J. Linn. Soc. Lond. 27: 355–380. [Google Scholar]
- Nielsen R., Williamson S., Kim Y., Hubisz M. J., Clark A. G., et al. , 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15: 1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Kimura M., 1973. The model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a genetic population. Genet. Res. 22: 201–204. [DOI] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K., 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2012. R: A language and environment for statistical computing, version 2.15.1. R Foundation for Statistical Computing, Vienna. http://www.r-project.org.
- Ramage R. T., Suneson C. A., 1961. Translocation-gene linkages on barley chromosome 7. Crop Sci. 1: 319–320. [Google Scholar]
- Ramsay L., Macaulay M., Degli Ivanissevich S., MacLean K., Cardle L., et al. , 2000. A simple sequence repeat-based linkage map of barley. Genetics 156: 1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A., Li L. M., Ward R., Pritchard J. K., 2003. Informativeness of genetic markers for inference of ancestry. Am. J. Hum. Genet. 73: 1402–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J., Morrell P. L., Gaut B. S., 2007. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc. Natl. Acad. Sci. USA 104: 8641–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J., Dawson I. K., Flavell A. J., Steffenson B., Weltzien E., et al. , 2011. Analysis of >1000 single nucleotide polymorphisms in geographically matched samples of landrace and wild barley indicates secondary contact and chromosome‐level differences in diversity around domestication genes. New Phytol. 191: 564–578. [DOI] [PubMed] [Google Scholar]
- Saisho D., Purugganan M. D., 2007. Molecular phylogeography of domesticated barley traces expansion of agriculture in the Old World. Genetics 177: 1765–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Nankaku N., Takeda K., 2009. A high-density transcript linkage map of barley derived from a single population. Heredity 103: 110–117. [DOI] [PubMed] [Google Scholar]
- Sato K., Close T. J., Bhat P., Muñoz-Amatriaín M., Muehlbauer G. J., 2011. Single nucleotide polymorphism mapping and alignment of recombinant chromosome substitution lines in barley. Plant Cell Physiol. 52: 728–737. [DOI] [PubMed] [Google Scholar]
- Scheet P., Stephens M., 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78: 629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalenbach I., Léon J., Pillen K., 2009. Identification and verification of QTLs for agronomic traits using wild barley introgression lines. Theor. Appl. Genet. 118: 483–497. [DOI] [PubMed] [Google Scholar]
- Schoen D. J., Brown A. H. D., 2001. The Conservation of Wild Plant Species in Seed Banks. Bioscience 51: 960–966. [Google Scholar]
- Shin J. H., Blay S., McNeney B., Graham J., 2006. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 16: 1–10. [Google Scholar]
- Shriner D., 2011. Investigating population stratification and admixture using eigenanalysis of dense genotypes. Heredity 107: 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139: 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffenson B. J., Olivera P., Roy J. K., Jin Y., Smith K. P., et al. , 2007. A walk on the wild side: mining wild wheat and barley collections for rust resistance genes. Aust. J. Agric. Res. 58: 532–544. [Google Scholar]
- Stein N., Prasad M., Scholz U., Thiel T., Zhang H., et al. , 2007. A 1,000-loci transcript map of the barley genome: new anchoring points for integrative grass genomics. Theor. Appl. Genet. 114: 823–839. [DOI] [PubMed] [Google Scholar]
- Szpiech Z. A., Jakobsson M., Rosenberg N. A., 2008. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szűcs P., Blake V. C., Bhat P. R., Chao S., Close T. J., et al. , 2009. An integrated resource for barley linkage map and malting quality QTL alignment. Plant Genome 2: 134–140. [Google Scholar]
- Takeda S., Matsuoka M., 2008. Genetic approaches to crop improvement: Responding to environmental and population changes. Nat. Rev. Genet. 9: 444–457. [DOI] [PubMed] [Google Scholar]
- Thomas J. W., Cáceres M., Lowman J. J., Morehouse C. B., Short M. E., et al. , 2008. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics 179: 1455–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K., 2003. Libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics 19: 2325–2327. [DOI] [PubMed] [Google Scholar]
- Valdes A. M., Slatkin M., Freimer N. B., 1993. Allele frequencies at microsatellite loci: the stepwise mutation model revisited. Genetics 133: 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R. K., Marcel T. C., Ramsay L., Russell J., Röder M. S., et al. , 2007. A high density barley microsatellite consensus map with 775 SSR loci. Theor. Appl. Genet. 114: 1091–1103. [DOI] [PubMed] [Google Scholar]
- Volis S., Mendlinger S., Orlovsky N., 2000. Variability in phenotypic traits in core and peripheral populations of wild barley Hordeum spontaneum Koch. Hereditas 133: 235–247. [DOI] [PubMed] [Google Scholar]
- Volis S., Mendlinger S., Turuspekov Y., Esnazarov U., 2002a Phenotypic and allozyme variation in Mediterranean and desert populations of wild barley, Hordeum spontaneum Koch. Evolution 56: 1403–1415. [DOI] [PubMed] [Google Scholar]
- Volis S., Mendlinger S., Ward D., 2002b Differentiation in populations of Hordeum spontaneum Koch along a gradient of environmental productivity and predictability: plasticity in response to water and nutrient stress. Biol. J. Linn. Soc. Lond. 75: 301–312. [Google Scholar]
- Volis S., Mendlinger S., Ward D., 2002c Adaptive traits of wild barley plants of Mediterranean and desert origin. Oecologia 133: 131–138. [DOI] [PubMed] [Google Scholar]
- Volis S., Verhoeven K. J., Mendlinger S., Ward D., 2004. Phenotypic selection and regulation of reproduction in different environments in wild barley. J. Evol. Biol. 17: 1121–1131. [DOI] [PubMed] [Google Scholar]
- von Korff M., Wang H., Leon J., Pillen K., 2005. AB-QTL analysis in spring barley. I. Detection of resistance genes against powdery mildew, leaf rust and scald introgressed from wild barley. Theor. Appl. Genet. 111: 583–590. [DOI] [PubMed] [Google Scholar]
- von Korff M., Wang H., Leon J., Pillen K., 2006. AB-QTL analysis in spring barley: II. Detection of favourable exotic alleles for agronomic traits introgressed from wild barley (H. vulgare ssp spontaneum). Theor. Appl. Genet. 112: 1221–1231. [DOI] [PubMed] [Google Scholar]
- von Korff M., Wang H., Leon J., Pillen K., 2008. AB-QTL analysis in spring barley: III. Identification of exotic alleles for the improvement of malting quality in spring barley (H. vulgare ssp spontaneum). Mol. Breed. 21: 81–93. [Google Scholar]
- Wang C., Zöllner S., Rosenberg N. A., 2012. A quantitative comparison of the similarity between genes and geography in worldwide human populations. PLoS Genet. 8: e1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. S., Cockerham C. C., 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Wright S. I., Bi I. V., Schroeder S. G., Yamasaki M., Doebley J. F., et al. , 2005. The effects of artificial selection on the maize genome. Science 308: 1310–1314. [DOI] [PubMed] [Google Scholar]
- Wright M. H., Tung C. W., Zhao K., Reynolds A., McCouch S. R., et al. , 2010. ALCHEMY: a reliable method for automated SNP genotype calling for small batch sizes and highly homozygous populations. Bioinformatics 26: 2952–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R. C., 1998. Estimating hierarchical F-statistics. Evolution 58: 950–956. [DOI] [PubMed] [Google Scholar]
- Yang W. Y., Novembre J., Eskin E., Halperin E., 2012. A model-based approach for analysis of spatial structure in genetic data. Nat. Genet. 44: 725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S. J., Gyenis L., Bossolini E., Hayes P. M., Matus I., et al. , 2006. Validation of quantitative trait loci for multiple disease resistance in barley using advanced backcross lines developed with a wild barley. Crop Sci. 46: 1179–1186. [Google Scholar]
- Yun S. J., Gyenis L., Hayes P. M., Matus I., Smith K. P., et al. , 2005. Quantitative trait loci for multiple disease resistance in wild barley. Crop Sci. 45: 2563–2572. [Google Scholar]
- Zohary D., Hopf M., 2000. Domestication of plants in the Old World: the origin and spread of cultivated plants in West Asia, Europe, and the Nile Valley, Oxford University Press, Oxford, UK. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.