Abstract
A change in the timing or rate of developmental events throughout ontogeny is referred to as heterochrony, and it is a major evolutionary process in plants and animals. We investigated the genetic basis for natural variation in the timing of vegetative phase change in the tree Eucalyptus globulus, which undergoes a dramatic change in vegetative morphology during the juvenile-to-adult transition. Quantitative trait loci analysis in an outcross F2 family derived from crosses between individuals from a coastal population of E. globulus with precocious vegetative phase change and individuals from populations in which vegetative phase change occurs several years later implicated the microRNA EglMIR156.5 as a potential contributor to this heterochronic difference. Additional evidence for the involvement of EglMIR156.5 was provided by its differential expression in trees with early and late phase change. Our findings suggest that changes in the expression of miR156 underlie natural variation in vegetative phase change in E. globulus, and may also explain interspecific differences in the timing of this developmental transition.
Keywords: QTL, microRNA, eucalypts, heterochrony, adaptation
Transition to the reproductive state is a key event in animal and plant development, and it is often accompanied by changes in somatic or vegetative traits. These changes may be rapid, synchronized, and dramatic, as seen in the well-known examples of animal metamorphosis and plant heteroblasty, or they may be more subtle, gradual, and unsynchronized. The timing of these transitions is usually under strong genetic control (Gould 1977; Guerrant 1988; MacKinney and MacNamara 1991) and can be subject to evolutionary change in a phenomenon referred to as heterochrony (MacKinney and MacNamara 1991). It has long been argued that markedly different morphologies in organisms of equivalent age may arise from relatively simple genetic changes in genes that control developmental timing, and that this may provide a means of rapid adaptive evolution (Gould 1977; Guerrant 1988; MacKinney and MacNamara 1991). Evidence for heterochronic evolution in plants in response to recent climate change is already emerging (Franks and Weis 2008).
Although heteroblasty is evident in many woody plant genera including Hedera, Pinus, and Acacia (Climent et al. 2006; Zotz et al. 2011), the evolutionary significance of heteroblasty is clearly exemplified in trees of the genus Eucalyptus (Barber 1965; Potts and Wiltshire 1997). Many Eucalyptus species are strongly heteroblastic, with a striking and abrupt change from the juvenile to the adult vegetative phase. Phase change in eucalypts is usually most obvious in leaf morphology and orientation but can also include changes in leaf anatomy, physiology, chemical composition, and resistance to pests and diseases, as well as changes in stem shape, bark type, and the anatomy and composition of wood (Potts and Wiltshire 1997; James and Bell 2001; Lawrence et al. 2003; Goodger et al. 2007; Jaya et al. 2010). This variation suggests that the different leaf phases in heteroblastic eucalypts may confer distinct advantages in specific biotic or abiotic environments (Jordan et al. 2000; James and Bell 2001). In eucalypts, the timing of vegetative phase change is under strong genetic control and may vary markedly both within and between species (Jordan et al. 1999; Hamilton et al. 2011). Further, while reproduction is normally associated with the adult foliage type, at least 30 species from diverse eucalypt lineages are reproductive while bearing typical juvenile foliage in their canopy, and many of these never develop adult foliage (Potts and Wiltshire 1997). This process is likely to reflect heterochronic speciation. For example, it is thought that the rare Tasmanian endemic E. risdonii has recently speciated from the heteroblastic E. tenuiramis by retaining juvenile foliage for life (Wiltshire et al. 1998). Similar heterochronic differentiation separates other closely related species throughout the eucalypt phylogeny, suggesting this process has occurred independently numerous times in eucalypt evolution (Potts and Wiltshire 1997).
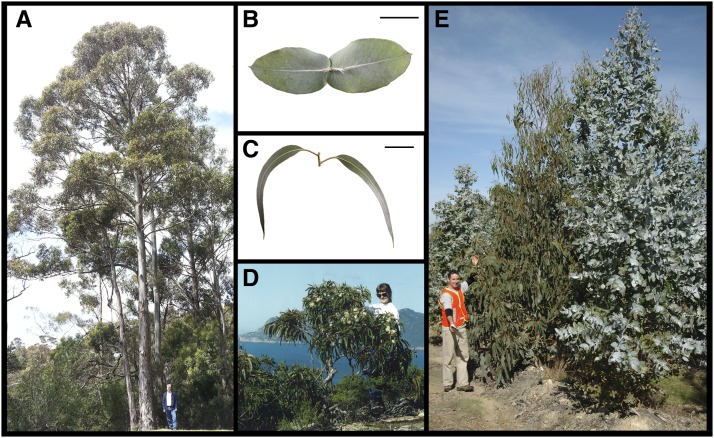
The forest tree Eucalyptus globulus is one of the best known heteroblastic plants and undergoes a dramatic vegetative phase change (Figure 1) (Hamilton et al. 2011; Zotz et al. 2011). E. globulus also provides a particularly striking example of heterochrony, with the occurrence of precocious reproductive and vegetative phase change in several dwarf coastal cliff-top populations (Figure 1) that are thought to have evolved in response to drought and salt stress and/or high winds (Dutkowski and Potts 1999; Jordan et al. 2000; Foster et al. 2007). Among these populations, the most extreme case of precocious reproductive and vegetative phase change occurs in the Lighthouse population at Wilsons Promontory in southeastern Victoria (Dutkowski and Potts 1999; Jordan et al. 2000). In common garden trials, most progeny from the cliff-top trees produce adult foliage and many have flower buds by the second year of growth, whereas progenies from most other populations, including adjacent disjunct populations, are taller and remain vegetatively and reproductively juvenile for 5 or more years (Jordan et al. 2000). However, despite much study of this ecotypic differentiation, the molecular basis of this variation is yet to be explored.
Figure 1.
Heteroblastic and heterochronic variation in Eucalyptus globulus. E. globulus is usually a tall forest tree (A). However, on exposed granitic cliff-tops, E. globulus grows as a precocious ecotype that typically reaches less than 4 m and is often multi-stemmed (D). E. globulus undergoes a dramatic change in vegetative morphology from juvenile (B) to adult (C) foliage (scale bars indicate 5 cm). There is broad genetic-based variation in the timing of this transition (E) from precocious (left) to late (right) in ontogeny in E. globulus.
Although heteroblasty has been of interest to researchers for more than a century (Goebel 1889), it is only in the past 10 years that advances in molecular genetics have given insight into the underlying molecular mechanisms. Studies in Arabidopsis and maize have identified two miRNAs that act sequentially in a pathway that is central to regulation of vegetative phase change (Wu and Poethig 2006; Chuck et al. 2007; Wu et al. 2009). MiR156 promotes juvenility, is strongly expressed in seedlings, and its expression decreases during development, whereas miR172 is negatively regulated by miR156 and has the opposite role and expression pattern. MiR156 is nearly identical in sequence to miR157 and both target the same members of the SQUAMOSA PROMOTER BINDING-LIKE (SPL) gene family (Rhoades et al. 2002; Poethig 2009), while miR172 targets members of the APETALA2 (AP2) family (Poethig 2009; Wu et al. 2009). Recent evidence suggests that the same miRNAs are involved in phase change in woody plants, including E. globulus, implying that they may regulate vegetative phase change in all flowering plants (Wang et al. 2011). However, although various studies have identified natural and induced mutants in which changes to these miRNAs or their targets affect phase change (Xie et al. 2005; Chuck et al. 2007; Usami et al. 2009; Wu et al. 2009), a link between polymorphism in genes involved in this regulatory network and adaptation in natural populations has not been demonstrated. Here, we aim to elucidate the molecular genetic basis of adaptive differentiation between E. globulus ecotypes with precocious vegetative phase change and those in which the timing of vegetative phase change is more characteristic of the species norm. We present evidence to suggest that genetic variation affecting the miRNA regulatory network plays a major role in ecotypic differentiation, highlighting the evolutionary importance of this regulatory network and heterochronic differentiation as an adaptive solution in plants.
Materials and Methods
QTL study
Genetic material and trial design:
Linkage mapping and quantitative trait loci (QTL) analyses were conducted in an inter-provenance outcross F2 family (Lighthouse F2). This family was constructed from crossing two unrelated F1 individuals (the parents) in 2002, each derived from crossing individuals (the grandparents) with precocious [Wilsons Promontory Lighthouse (LH)] and “normal” [from either King Island in Bass Strait (KI) or Taranna in the southeast of Tasmania (TA)] vegetative phase change in 1995, thus producing the following pedigree for the F2 family: 614LH/KI440//615LH/TA423. The F2 family was planted at two sites, Boyer and Geeveston, approximately 50 km apart in southern Tasmania (Supporting Information, Table S1). Seed was sown in April 2006, with seedlings raised in glasshouse conditions before being planted in a randomized block design including various control families at Geeveston in November 2006, and at Boyer between May and July 2007. QTL analyses were conducted on a total of 467 F2 individuals, 158 from Boyer and 309 from Geeveston.
Assessment and analysis of phenotypic traits:
Vegetative phase change was scored in both trials at intervals of approximately 6 months, from trial planting until all trees had produced adult vegetation (∼4.5 years of age). For the purpose of scoring, foliage was classified as juvenile, intermediate (bearing characteristics of both juvenile and adult foliage), or adult, as depicted in Figure S1. The timing of phase change was quantified by counting the number of nodes (cotyledons = node 0) at which transitions between these foliage types occurred, with the traits termed “last juvenile node” and “number of intermediate nodes.” While measuring phase change traits, the height to each transition was also recorded to enable direct comparisons with previous studies (Jordan et al. 1999, 2000).
Flowering was quantified in terms of time and node that plants reached reproductive maturity, as well as the timing of flowering each year once reproductive maturity had been reached. These traits were scored in the flowering seasons beginning in 2009 and 2010. “Flowering precocity” represented a simple presence/absence of flowers at the end of the flowering year at each site and “node to first flower” was the node at which the first flower bud occurred. The proportions of flowers open, and yet to open, were recorded at least fortnightly during each flowering year. This was then converted to a peak flowering time (“anthesis time”) following the method of Jones et al. (2011).
Tree height was measured in June 2010. Tree shape was calculated as tree height divided by the maximum canopy width. Tree shape was calculated using measurements made at 23 months of age in both field trials. Height measurements and internode counts were used to calculate mean internode lengths for juvenile and intermediate vegetative phases. The Pearson correlation coefficients were calculated between traits using PROC CORR of SAS.
Linkage mapping and QTL analysis:
QTL analyses were performed using the linkage map described by Hudson et al. (2012). Briefly, all linkage maps were constructed using JoinMap 4.0 (Van Ooijen 2006). Individual parental maps were built using 503 F2 individuals before constructing a sex-averaged consensus map. The consensus map contained 50 microsatellite (SSR) and 1010 DArT markers. To reduce computational demands, a subset of these markers (391) was used for QTL analyses. Specifically, most 3:1 segregating DArT markers were removed from the consensus map while retaining all SSR markers and an even distribution of DArT markers segregating in a 1:1 ratio at approximately 2- to 5-cM intervals. Linkage group numbering corresponds to the 11 main scaffolds of the reference E. grandis genome sequence (v1.1 www.phytozome.net) (Myburg et al. 2014).
Putative eucalypt homologs of genes involved in vegetative and reproductive phase change in Arabidopsis were also mapped in this study (Wu and Poethig 2006; Wu et al. 2009; Chen et al. 2010; Wang et al. 2011). These included miR156, miR157, and miR172 loci, the genes targeted by these miRNAs, and genes involved in flowering (Table S2). As miR156, miR157, and miR172 precursor loci are not yet annotated in the E. grandis genome (Myburg et al. 2014), these were identified by performing a BLAST search with the mature miR156, miR157, and miR172 sequences to identify the stem loop sequence on Phytozome (http://www.phytozome.com/eucalyptus.php). Putative homologs of genes targeted by miR156/157 and miR172 were identified using the annotations on ORCAE (http://bioinformatics.psb.ugent.be/orcae/overview/Eugra) and Eucgenie (http://www.eucgenie.org/), respectively. Putative homologs of key flowering genes were identified by Vining et al. (unpublished data). All homologs were then placed on the linkage map based on extrapolation using the position of their closest flanking DArT markers in the E. grandis genome sequence (annotated at http://eucgenie.bi.up.ac.za/) using the “neighbors” approach (Cone et al. 2002). The average distance between each homolog genes and its flanking markers was 1845 kb.
QTL analyses were performed with MapQTL 6.0 (Van Ooijen 2009) using 467 individuals of the Lighthouse F2. Phenotypic data were site-adjusted (site mean of zero; performed in SAS 9.2 using PROC STANDARD) and combined across sites for QTL analysis. Permutation tests were run in MapQTL 6.0 to determine LOD significance thresholds at genome-wide and chromosome-wide levels for all traits (1000 permutations) (Churchill and Doerge 1994). Putative QTL were declared at two significance thresholds: significant (genome-wide type I error α = 0.05) and suggestive (chromosome-wide type I error rate α = 0.05). Following the detection of putative QTL in interval mapping, markers linked to QTL that exceeded the suggestive (α = 0.05) threshold were chosen as cofactors in restricted multiple-QTL model (rMQM) mapping. For each trait, rMQM analyses were performed using an iterative approach until no further QTL were detected, selected cofactor markers were the closest marker to each QTL, and QTL positions were stable. The regression algorithm implemented in MapQTL 6.0 was used in interval and rMQM mapping.
Population genomics analysis
An outlier marker analysis was conducted to detect potentially adaptive loci that differentiated the precocious Lighthouse population and the nearest normal population (Tidal River, approximately 14 km NW). The Lighthouse population is likely to have evolved from the adjacent Tidal River population (Foster et al. 2007) and was chosen to minimize differentiation due to stochastic processes, such as neutral drift, which generally increases with greater geographical/genetic separation. Thirty-two individuals from the Lighthouse population and 30 individuals from the Tidal River population were genotyped using DArTseq marker technology (DArT Pty. Ltd. Canberra, ACT) (Sansaloni et al. 2011). For each DArTseq marker, the average read depth was 9× and an absolute minimum sequence read depth of 3× in any individual/marker was used. BayeScan outlier analysis was conducted on 14,708 binary DArTseq markers following the removal of markers with the following: minor allele frequencies ≤2 out of 62 individuals (2875 markers) (Foll and Gaggiotti 2008; Roesti et al. 2012); more than 20% missing data in any one population; and quality scores (Q) <2. Outlier detection was performed using BayeScan v2.1 (Foll and Gaggiotti 2008; Foll et al. 2010; Fischer et al. 2011). Default BayeScan parameters were applied with a prior odds value of 10. Outlier markers were defined as those having a log10 (PO) value ≥0. Although this corresponds to a relatively weak level of outlier significance [a log10 (PO) ≥0<0.5 value is considered “barely worth mentioning” according to Jeffrey’s Bayes factor interpretation], markers at this significance threshold were mapped to compare their position to detected QTL.
To estimate the position of outlier markers on the Lighthouse F2 linkage map, BLAST searches were first performed (using the sequence supplied with each marker; all 69 bp in length) to identify the homologous position(s) of outlier markers in the E. grandis genome (http://bioinformatics.psb.ugent.be/webtools/bogas/). Linkage map positions were then estimated for each of these positions following the method of Cone et al. (2002).
Expression analysis
Ten plants of precocious phase change ecotype (from four open-pollinated families of the Lighthouse provenance) and nine plants of normal phase change ecotype (six plants from three open-pollinated families of Tidal River and three plants from a Taranna family related to a grandparent of the F2) were grown in a randomized design in a glasshouse under natural light. After 8 months, juvenile leaf material at node 10 of each plant (i.e., 9–10 biological replicates) was harvested, frozen in liquid nitrogen, and stored at −70°. Plants were grown to monitor phase change phenotype; the normal ecotype remained juvenile after 2 years of growth (average 44 nodes), whereas all precocious ecotype plants had undergone phase transition by this stage.
Leaf material was crushed in liquid nitrogen in a mortar and pestle before nucleic acid extraction. RNA was isolated using the SV Total RNA Isolation System (Promega, Sydney, NSW, Australia) with the following modifications: 1% PEG (MW 20,000) was added to the lysis buffer, the optional 70° 3-min incubation step was included, and the DNase incubation was increased to 30 min. Genomic DNA was extracted using a CTAB method (McKinnon et al. 2004). Quality and quantity of nucleic acids were estimated using NanoDrop 8000 Spectrophotometer (Thermo Scientific). The Tetro cDNA Synthesis Kit (Bioline, London, UK) was used for cDNA synthesis, using 500 ng RNA in a volume of 20 µL. A negative control (omitting reverse-transcriptase enzyme) was included to confirm there was no genomic DNA contamination of RNA samples. Thirty microliters of RNase-free water was used to dilute cDNA for use in quantitative real-time PCR (qRT-PCR) or regular PCR. Primers for qRT-PCR (Table S3) were designed to target four miR156 precursor loci (EgrMIR156.1, EgrMIR156.4, EgrMIR156.5, EgrMIR156.10) identified by the BLAST searches of E. grandis described above using Primer3 v.0.4.0 (Rozen and Skaletsky 2000). Primers spanned the stem loop structure of each gene. Primers for EglSPL3, EglSPL9, and the housekeeping gene EglEIF4 were those used by Wang et al. (2011) and primer sequences for another housekeeping gene, EglACT2, were provided by the same authors (M.Y. Park personal communication) (Table S3). Primers were first tested on E. globulus gDNA and cDNA using regular PCR (conditions as in Jones et al. 2011) using the annealing temperatures in Table S3. The resulting PCR products were cleaned and sequenced at the Australian Genome Research Facility (Brisbane, Queensland, Australia) to confirm the identity of the amplified product. Once confirmed in E. globulus, loci were renamed with the “Egl” prefix (Table S3). Sequences were deposited in GenBank, with accession numbers KJ948420 to KJ948423. Conditions used for qRT-PCR reactions were as in Jones et al. (2011). Each reaction was performed in duplicate and the Cq values of these two technical replicates were averaged. For the housekeeping genes EglEIF4 and EglACT2, an unpaired two-tailed t test between mean Cq values for precocious and normal phase change phenotypes (n = 10 and n = 9 biological replicates, respectively; see description of genetic material above) was performed in GraphPad Prism Version 6.01 for Windows (GraphPad Software, La Jolla, CA), applying Welch’s correction. These housekeeping genes did not differ in expression between the ecotypes (P = 0.65 for EglEIF4 and P = 0.79 for EglACT2); therefore, EglEIF4 was used for subsequent normalization of genes of interest. Reactions were normalized as in Hecht et al. (2011) and the t test between mean values for precocious and normal phase change phenotypes was conducted as described above.
Sequence analysis of EglMIR156.5
To investigate sequence variation in the EglMIR156.5 region, a 1310-bp region encompassing the 87-bp MIR156.5 stem loop and constituting part of the gene was sequenced in five Wilsons Promontory Lighthouse and five Tidal River individuals. Four primer pairs (Table S3) amplifying overlapping fragments were designed with PerlPrimer (Marshall 2004) and Primer3web (Koressaar and Remm 2007; Untergrasser et al. 2012). The template used for primer design was a consensus E. globulus sequence generated through the mapping of Illumina short-sequence reads (NCBI SRA library accession numbers SRX116786 and SRX059820; 54.8 G bases in total) to a 10-kb segment of the E. grandis BRASUZ1 genome sequence (version 1.1) that contained EgrMIR156.5 (scaffold 3 from 50,817,705 to 50,827,704 bp). The short sequence reads were mapped using default settings in BWA-0.6.1 (Li and Durbin 2009) with the consensus sequence (average basepair read coverage of 23.5) being called with SAM tools (Li et al. 2009). PCR conditions were as in Jones et al. (2011) using the annealing temperatures in Table S3. PCR products were cleaned and sequenced at Macrogen (Seoul, South Korea) and aligned using Sequencher version 4.8 (Gene Codes Corporation, Ann Arbor, MI; http://www.genecodes.com). Sequences were deposited in GenBank, with accession numbers KJ933702 to KJ933711.
Results
QTL analyses
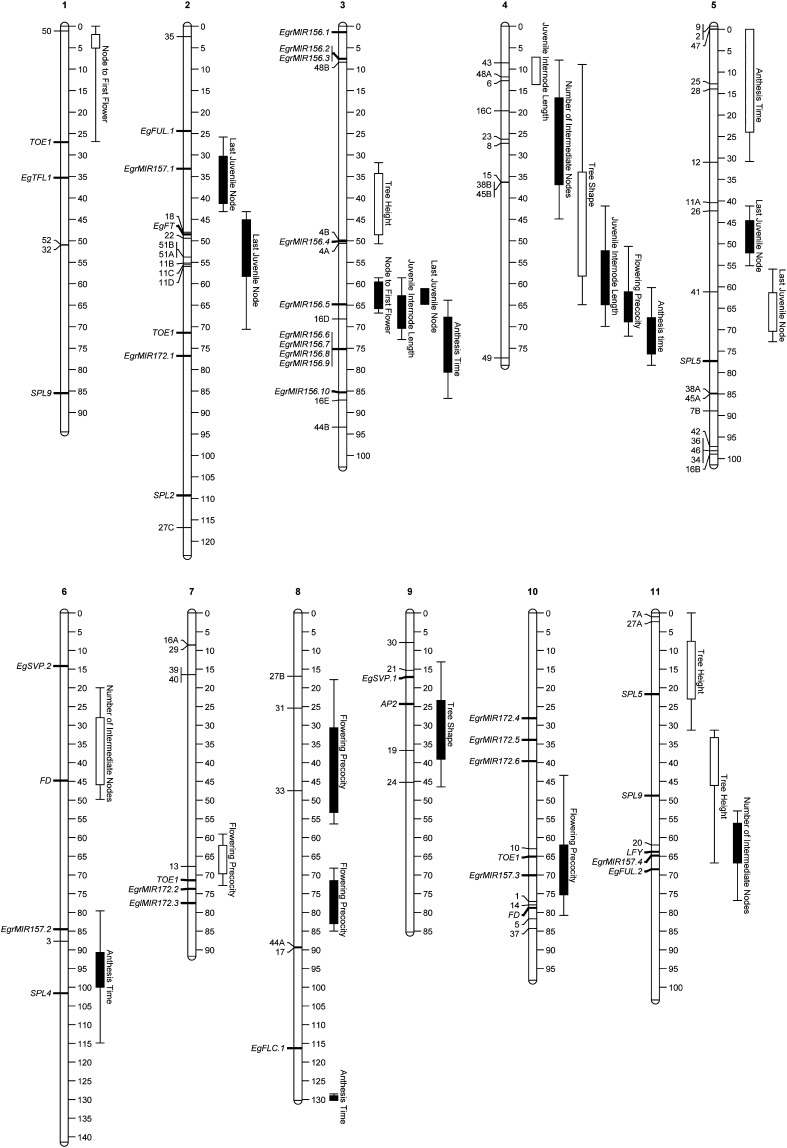
To investigate the genetic basis of phenotypic differences between the precocious and normal ecotypes of E. globulus, we first conducted QTL analyses using a large outcross F2 pedigree (n = 467) designed specifically to maximize segregation of ecotypic differences. In addition to traits related to vegetative phase change (last juvenile node and number of intermediate nodes), QTL analyses were also conducted for other traits that were likely to differentiate the ecotypes, namely: node to first flower; flowering precocity; anthesis time; tree height; tree shape; and juvenile internode length. In total, 28 QTL were identified exceeding the chromosome-wide significance level (α < 0.05), of which two to five were detected for each trait (Table 1). QTL were located on all 11 linkage groups (Figure 2). In line with the strong correlations between many of the traits analyzed (Table S4), several QTL were co-located (i.e., their 2-LOD support intervals overlapped) (Figure 2). Of the 28 QTL detected, nine were bi-parentally inherited, i.e., segregated in both F1 parents crossed to produce the F2 (Table 1). We define these QTL as “ecotypic QTL” because their bi-parental inheritance indicated these were likely to be associated with population-level differentiation between the precocious and normal ecotypes. These nine ecotypic QTL were located on five chromosomes (with three co-located on LG3) and were associated with vegetative and reproductive phase change, juvenile internode length, and anthesis time.
Table 1. QTL detected in the Lighthouse Eucalyptus globulus F2 family using rMQM mapping.
| Trait | QTL | |||||
|---|---|---|---|---|---|---|
| LG | cMa | Adj. markerb | LOD | PVEc | SEGd | |
| Vegetative phase change traits | ||||||
| Last juvenile node | 2 | 32.4 | ePt-641876 | 4.4** | 1.6 | F |
| 2 | 48.7 | ePt-568767 | 4.7** | 1.7 | F | |
| 3 | 62.1 | ePt-639243 | 103** | 62.8 | Ecotypic | |
| 5 | 48.6 | ePt-641489 | 4.4** | 1.6 | M | |
| 5 | 65.1 | ePt-571521 | 4.0* | 1.4 | Ecotypic | |
| Number of intermediate nodes | 4 | 27.9 | Es54 | 5.4** | 3.8 | Ecotypic |
| 6 | 34.8 | Embra627 | 3.9* | 2.5 | F | |
| 11 | 62.1 | ePt-575083 | 9.1** | 6.5 | Ecotypic | |
| Flowering traits | ||||||
| Node to first | 1 | 3.1 | Embra11 | 3.3* | 3.7 | M |
| flower | 3 | 61.0 | ePt-639927 | 15.9** | 20.2 | Ecotypic |
| Flowering | 4 | 66.9 | ePt-568492 | 9.4** | 5.2 | F |
| precocity | 7 | 65.3 | ePt-504063 | 4.3* | 2.4 | F |
| 8 | 44.6 | ePt-638446 | 5.4** | 3.1 | M | |
| 8 | 78.7 | ePt-640315 | 8.2** | 4.6 | Ecotypic | |
| 10 | 70.9 | ePt-572657 | 4.7** | 2.6 | M | |
| Anthesis time | 3 | 73.0 | ePt-570139 | 4.7** | 2.1 | F |
| 4 | 72.4 | Embra36 | 5.2** | 2.4 | Ecotypic | |
| 5 | 0.9 | Embra618 | 3.4* | 1.5 | M | |
| 6 | 94.7 | ePt-504481 | 5.2** | 2.4 | F | |
| 8 | 129.5 | Es76 | 8.0** | 3.8 | Ecotypic | |
| Tree height and shape traits | ||||||
| Tree height | 3 | 42.4 | ePt-571733 | 3.2* | 2.7 | F |
| 11 | 15.2 | ePt-570063 | 3.6* | 3.1 | F | |
| 11 | 39.5 | Eg99 | 3.5* | 3.0 | F | |
| Tree shape | 4 | 54.2 | ePt-564417 | 3.4* | 3.0 | F |
| 9 | 30.2 | ePt-505052 | 4.4** | 3.9 | M | |
| Juvenile | 3 | 66.8 | ePt-640855 | 15.6** | 10.4 | Ecotypic |
| internode | 4 | 10.9 | ePt-600106 | 3.8* | 2.5 | M |
| length | 4 | 59.6 | ePt-570676 | 5.8** | 3.8 | F |
QTL LOD peak position.
Adjacent marker to QTL LOD peak.
The percent variation explained for each QTL.
Segregation of the QTL effect (M = male; F = female; Ecotypic = segregation from both parents or bi-parental). LOD significance: * = chromosome-wide α ≤ 0.05 and ** = genome-wide α ≤ 0.05.
Figure 2.
The location of QTL, candidate genes, and outlier markers on the Lighthouse F2 linkage map in Eucalyptus globulus. Particularly notable is the co-location between the major QTL for last juvenile node and node to first flower on LG3 with EgrMIR156.5. Bars and lines indicate one-LOD and two-LOD QTL confidence intervals, respectively. QTL exceeding genome-wide (α ≤ 0.05) and chromosome-wide (α ≤ 0.05) significance thresholds are indicated by filled and empty bars, respectively. Significant outlier markers (Table S6) and candidate genes (Table S2) are shown on the left of linkage groups. Outlier markers are designated by a number followed by a letter (A–E) if they were present in multiple copies in the E. grandis genome sequence.
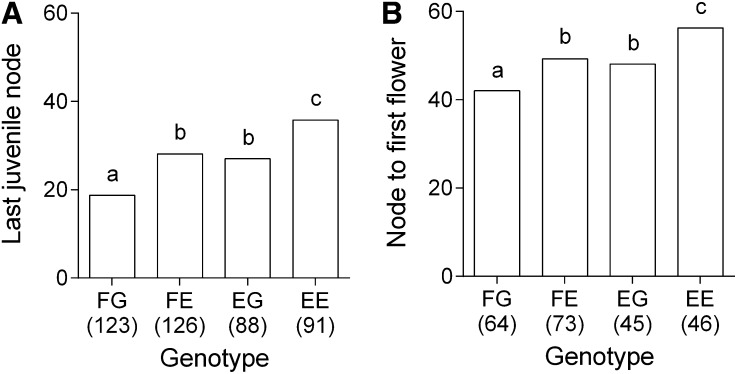
Whereas most QTL were of relatively small effect [82% individually accounted for less than 5% of the phenotypic variation explained (PVE)], major effect QTL were detected for last juvenile node (62.8% PVE) and, to a lesser extent, for node to first flower (20.2% PVE). These QTL co-located with two other QTL for anthesis time and juvenile internode length on linkage group 3 (LG3) (Figure 2) and the results point to the effect of multiple loci in this region. The QTL for last juvenile node and node to first flower possibly represent the effects of a single pleiotropic locus because their peaks were close to each other (Table 1), their inheritance was bi-parental, their allelic effects were both additive (Figure 3), they segregated in coupling, and these traits were strongly positively correlated in the mapping family (r = 0.64; P < 0.0001). As would be expected, individuals inheriting alleles from both Lighthouse grandparents at these major QTL on LG3 underwent reproductive and vegetative phase change significantly earlier than heterozygotes or individuals homozygous for normal phase change alleles (Figure 3), indicating this locus strongly influences the timing of both transitions. However, the QTL for anthesis time and juvenile internode length were most likely controlled by different loci. This is supported by greater distances between peak positions for these QTL. Furthermore, the allelic effects of the QTL for juvenile internode length were dominant, and the QTL for anthesis time segregated from the female parent only, in contrast to the inheritance and mode of QTL effects at the co-located major effect QTL in this region (Table 1).
Figure 3.
Co-dominant and bi-parental inheritance of the major QTL for vegetative and reproductive phase change in Eucalyptus globulus. Phenotypic means for last juvenile node and node to first flower are shown for each genotype class of Embra1656 (a microsatellite marker less than 3.4 cM from the QTL peak on LG3 for either trait). Genotype classes indicate the inheritance of alleles from grandparental ecotypes: FG = homozygous precocious (i.e., contains two precocious grandparental alleles, inherited from Lighthouse 614LH and 615LH grandparents); FE = heterozygous genotype (615LH and KI440); EG = heterozygous genotype (614LH and TA423); and EE = homozygous normal (KI440 and TA423). The sample sizes of genotype classes are shown in parentheses. Letters above bars indicate significant differences (within traits; adjusted Tukey α = 0.05) between genotype classes.
Identification of candidate genes
In view of the central role of the miR156 pathway in regulation of vegetative phase change and flowering (Poethig 2009; Wu et al. 2009), we surveyed the entire E. grandis genome (v1.1 www.phytozome.net) for genes related to this pathway (miR156, miR157, and miR172 precursors, the target of these miRNAs, and flowering genes) as potential candidates underlying the major effect QTL on LG3. In Arabidopsis, there are eight miR156 precursors (MIR156A–MIR156H), four miR157 precursors (MIR157A–MIR157D), and five miR172 precursors (MIR172A–MIR172E) (Xie et al. 2005). These genes are not yet annotated in the E. grandis genome; our BLAST searches identified 10 miR156, four miR157, and six putative miR172 precursor loci in the main scaffolds of the E. grandis genome sequence (Table S2). The positions of these candidate genes were estimated on the Lighthouse F2 linkage map (Figure 2) and these loci were distributed among 10 of the 11 linkage groups (Figure 2). Interestingly, all of the miR156 precursors (EgrMIR156.1–EgrMIR156.10) occurred on LG3, of which one (EgrMIR156.5) co-located with the major QTL for last juvenile node (Figure 2). Specifically, this gene was located within the region defined by markers spanning the 2-LOD confidence interval of the last juvenile node QTL, which corresponds to ∼2.5 Mbp of the E. grandis genome sequence. We also searched this region of the E. grandis genome for other potential candidate genes. None of the annotated genes in this region (scaffold 3 from 48,300,000 to 50,840,000 bp, v1.1 www.phytozome.net, accessed February 18, 2014) had gene ontology related to, or an Arabidopsis homolog involved in, flowering or vegetative phase change (Table S5), providing evidence that the EgrMIR156.5 locus is the best candidate gene in this region.
Expression and sequence analyses
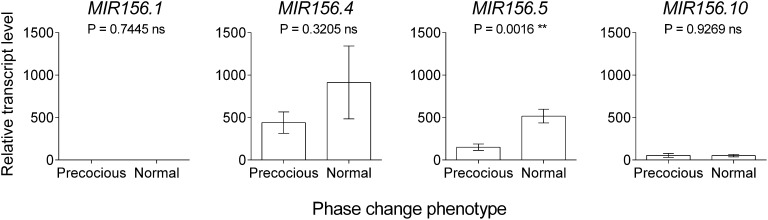
Positional evidence from the QTL analysis implicated EgrMIR156.5 as the potential cause of the difference in the timing of phase change between the precocious and the normal populations used in this study. To test this hypothesis, we used qRT-PCR to determine the abundance of the EglMIR156.5 precursor in juvenile leaves of these ecotypes. The expression of EglMIR156.5 was significantly (P = 0.0016) higher in juvenile leaves at node 10 of plants of normal phase change phenotype than in juvenile leaves at the same node on plants with the precocious phase change phenotype (Figure 4). This finding is consistent with the conserved role of miR156 suppressing vegetative phase change and provides further evidence that this locus underlies the observed difference in timing of vegetative phase change. In addition, transcript levels of the stem loop region of three other miR156 precursor loci (EglMIR156.1, EglMIR156.4, and EglMIR156.10) were monitored in the same tissue. The expression of EglMIR156.4 followed a similar trend to that of EglMIR156.5, but the difference between ecotypes was not significant (Figure 4). EglMIR156.1 and EglMIR156.10 were not or were poorly expressed in both ecotypes (Figure 4). To determine if the difference in expression of EglMIR156.5 between ecotypes had an effect on genes downstream in the pathway, we analyzed the expression of the eucalypt homologs of SPL3 and SPL9, genes that are inhibited by mature miR156 and promote vegetative phase change and flowering when mature miR156 levels are low (Chen et al. 2010). As expected, EglSPL3 levels were higher in precocious compared with normal ecotypes, although this difference was not significant (Figure S2). EglSPL9 was not expressed in either ecotype (Figure S2).
Figure 4.
Expression of four miR156 precursors in precocious and normal phase change ecotypes of Eucalyptus globulus. The qRT-PCR analyses of miR156 precursors were performed using node 10 juvenile leaves. Expression was normalized as the percentage of expression above EglEIF4 expression. Shown are the mean and SE of the expression in 10 plants of precocious phase change phenotype (representing four families from the Wilsons Promontory population) and nine plants of normal phase change phenotype (representing four families from Tidal River and Taranna provenances). *Significant difference between mean values of EglMIR156.5 expression for precocious and normal phase change phenotypes in the t test; ** = α ≤ 0.01. Expression of EglMIR156.1 was so low that it is not visible here.
Sequence surrounding the EglMIR156.5 stem loop (up to 1310 bp) was obtained for five Lighthouse and five Tidal River wild trees. No fixed polymorphic differences were detected between populations. The sequence(s) responsible for the differential expression of this gene may therefore reside outside the immediate stem-loop region.
Population genomics analysis
To further explore the genetic basis of ecotypic differentiation, a genome scan was performed using 14,708 DArTseq markers to identify “outlier markers” undergoing differential selection between the precocious Lighthouse population (n = 32) and the nearest normal population, Tidal River (n = 30). The position of outlier markers on the Lighthouse F2 linkage map was estimated to enable comparison with the position of QTL and candidate genes, providing population-level positional support for some of the QTL identified as contributing to ecotypic differentiation. The mean population FST value between Lighthouse and Tidal River populations was 0.25, consistent with isolation and drift in the small Lighthouse population (Jones et al. 2013). In total, 59 outlier markers (0.4%) were identified at the log10 (PO) ≥0 significance threshold. Only markers with fixed, or near-fixed, differences in allele frequency between populations were detected as outliers. According to Jeffrey’s Bayes factor interpretation, 16 outlier markers were significant at “strong” and “substantial” thresholds, respectively, with 27 markers significant at “the barely worth mentioning” threshold. For the 59 outlier markers, the maximum q-value at which all markers were significant was 0.23, meaning that 23% (or 14) of the outlier markers identified may be false positives.
At a minimum of 50 out of 69 bp (72%) sequence match, 52 of the 59 outlier markers returned BLAST matches to the 11 main scaffolds of the E. grandis genome. Ten markers returned multiple equal-quality sequence alignments [allowing for a maximum of two single nucleotide polymorphisms (SNPs) between alignments] (Table S6). Thus, the 52 outlier markers were mapped to 68 genomic positions, distributed across all 11 chromosomes (Figure 2). Importantly, outlier markers provided positional support for six of the nine ecotypic QTL (Figure 2). The co-location between ecotypic QTL and outlier markers occurred more commonly than expected by chance (χ2df1 = 4.4; P = 0.035). These results provide independent validation of genomic regions identified in the QTL analysis, suggesting that many of the polymorphic loci with significant phenotypic effects in our F2 family also differentiate the natural populations and are subject to selection. No significant outliers were located within the confidence intervals of the major-effect QTL for node to first flower and last juvenile node on LG3. However, a marker with nearly fixed differences in allele frequencies between the Lighthouse and Tidal River populations (fixed in Lighthouse and fixed except for two plants in Tidal River) occurred ∼0.5 and 2.5 cM from the major-effect QTL peak for node to first flower and last juvenile node on LG3, respectively, supporting the identification of these QTL as key contributors to differentiation of the precocious and normal ecotypes.
Discussion
Identifying the genetic basis of natural variation in developmental trajectories and understanding the evolutionary potential of this variation are major challenges of evolutionary biology. The demonstration that miRNAs regulate the timing of vegetative phase change not only in model annual plants but also across a wide variety of plant taxa including woody perennials (Huijser and Schmid 2011; Wang et al. 2011) now provides the possibility to explore the molecular basis of heterochronic evolution in natural populations. In this study we examined the molecular basis of heterochronic differentiation between naturally occurring ecotypes of the forest tree E. globulus with precocious vegetative phase change and those in which the timing of vegetative phase change is more characteristic of the species norm. Our results provide evidence suggesting that genetic variation affecting the miRNA regulatory network plays a major role in differentiating these ecotypes.
Polymorphism affecting the miRNA regulatory network underlies intraspecific variation in the timing of vegetative phase change in E. globulus
While functional genomic studies have provided detailed information regarding the genetic basis of a number of traits in model organisms, much less is known regarding how polymorphism at these loci contributes to adaptation in natural populations. Consistent with the role of miR156 as a “master regulator” of vegetative phase change in plants (Poethig 2010), our QTL and expression analyses provide independent evidence that genetic variation affecting this regulatory network underlies differentiation in the timing of vegetative phase change between the E. globulus ecotypes examined. Specifically, the detection of an ecotypic QTL for vegetative phase change (last juvenile node) of extremely large effect occurring adjacent to EgrMIR156.5 implicates this locus as a positional candidate. The detection of a QTL for reproductive phase change at essentially the same genomic location (node to first flower) (Figure 2) provides strong additional support for this candidate locus, because the genes targeted by miR156/157 are some of the only loci known to influence both vegetative phase change and flower initiation (Huijser and Schmid 2011). Further support for the role of the miR156 regulatory network is provided by the genetic-based difference in expression of EglMIR156.5 between ecotypes with marked differences in the timing of vegetative phase change. In Arabidopsis, miR156 is highly expressed during early shoot development, suppressing vegetative phase change, and declines rapidly during the juvenile-to-adult vegetative transition (Yang et al. 2011). Therefore, our finding of significantly reduced expression of EglMIR156.5 in juvenile leaves of E. globulus plants with precocious phase change, relative to those with normal phase change, is consistent with genetic variation affecting the miRNA regulatory network contributing to heterochronic differentiation.
Recent evidence from Arabidopsis suggests that only two of the eight miR156 precursor genes are likely to make a significant contribution to the overall level and developmental regulation of the mature miR156 (Yang et al. 2011, 2013; Yu et al. 2013). Similarly, of the four miR156 precursor loci we analyzed in E. globulus, only two, EglMIR156.4 and EglMIR156.5, were expressed and only the latter varied significantly in expression between precocious and normal ecotypes. Because the relative contribution of individual miR156 precursor loci to the mature pool of miR156 and how this varies in different taxa are not well-understood, it is quite possible that in E. globulus ecotypic variation in the expression of EglMIR156.5 alone could result in substantially reduced miRNA156 levels, and therefore precocious phase change.
Assuming that variation in the expression of EglMIR156.5 underlies the major effect QTL differentiating precocious and normal phase change ecotypes, the lack of fixed sequence differences between these ecotypes directly in or around the stem-loop region suggests that the difference in expression of EglMIR156.5 may be due to a mutation in parts of the gene we have not sequenced or in a cis-regulatory element near this locus. In other systems, causative mutations at loci with major phenotypic effects on developmental traits have been shown to lie outside the coding unit of the underlying gene, as in the case of the Corngrass1 (Chuck et al. 2007) and teosinte branched1 loci, which are both important in maize domestication (Clark et al. 2006; Studer et al. 2011). In the case of the severe Corngrass1 mutant, which fixes plants in the juvenile stage, overexpression of the miR156b/c locus is caused by a retro-transposon insertion close to the transcription initiation site (Chuck et al. 2007). In the case of the transcription factor teosinte branched1, the causative mutations are also transposable element insertions but lie much further upstream, approximately 60–70 kb from the gene itself (Studer et al. 2011; Zhou et al. 2011).
Alternatively, it is formally possible that the QTL may represent the effect of a locus near but distinct from EglMIR156.5, which may act in trans to regulate expression of EglMIR156.5 specifically or of multiple miR156 precursor loci. While our expression analysis of the miR156 precursor loci was not exhaustive, the fact that EglMIR156.5 was the only locus to display differential expression between precocious and normal phase change ecotypes is consistent with the regulation of this locus alone. Another possibility is that a distinct mutation in the QTL region could affect EglMIR156.5 expression through local alteration in chromatin structure. Clearly, without the identification of an obvious causal polymorphism, the evidence that the major effect QTL reflects a mutation in the EglMIR156.5 gene or in a cis-regulatory element near this locus is inconclusive. Nonetheless, it remains the most plausible explanation from the available evidence. Future expression analysis of loci surrounding EglMIR156.5 and additional miR156 precursor loci, as well as detailed sequence analysis of the wider genomic region surrounding EglMIR156.5, should shed more detail on the exact nature of the genetic differentiation between the ecotypes examined.
Evolutionary implications
Heterochrony has been widely implicated in the adaptive evolution of a diverse group of woody perennial genera (Li and Johnston 2000; Climent et al. 2006), including Eucalyptus (Barber 1965; Potts and Wiltshire 1997; Wiltshire et al. 1998), and numerous studies have argued that the timing of vegetative phase change, in particular, has adaptive significance in eucalypts (Wiltshire et al. 1991; Jordan et al. 2000; Hamilton et al. 2011). In E. globulus, it has been argued that the retention of juvenile foliage may be advantageous in mesic environments, whereas early transition to the more xeromorphic adult may confer an advantage in environments where water is limited (Jordan et al. 2000). The main selective forces in the cliff-top environment inhabited by the precocious ecotype are likely to be exposure to strong winds, salt spray, and drought (King 1999; James and Bell 2001). Plants can have similar physiological responses to drought and salt stress because both limit water uptake (Munns 2002), whereas wind exposure causes both drought and mechanical stress. Therefore, early phase change may have provided trees with a selective advantage by reducing susceptibility to desiccation and wind damage earlier in development. Outlier markers were located within three of the five QTL for last juvenile node, consistent with differential selection acting on the timing of phase change between the different ecotypes.
The outlier analysis identified many genomic regions under differential selection between the ecotypes. Many of these outliers were located within QTL confidence intervals, pointing to the possibility that these traits are also under differential selection between the ecotypes. Additionally, many outlier loci did not co-locate with QTL. Some of these outliers are likely to reflect variation in phenotypic traits that were not measured in this study, but are under differential selection between the ecotypes. Taken together, these results indicate that ecotypic divergence has involved various distinct traits and many genomic regions. Given that linkage disequilibrium (LD) probably decays rapidly in E. globulus, as in most tree species (Thavamanikumar et al. 2013), it is likely that many more markers than we used (14,708 markers) would have been necessary to saturate the genome for exhaustive outlier marker detection. Our low marker coverage probably explains why we did not find an outlier marker near EglMIR156.5. However, the low marker coverage also means that we have most likely underestimated the number of genomic regions involved in the differentiation of our ecotypes.
While it has long been argued that heterochronic evolution can produce marked changes in morphology through few genetic changes (Gould 1977; Guerrant 1988; MacKinney and MacNamara 1991), to date there has been little direct proof for this. Our findings address this long-standing issue. The major QTL for phase change had an exceptionally large effect (62.8% PVE) when it is considered that individual QTL generally account for less than 5% of variation in most quantitative traits in forest trees, crop plants, and humans (Visscher 2008; Buckler et al. 2009; Neale and Kremer 2011), as was the case for the majority of the QTL detected in this study. This implies that the underlying gene or genes is likely to have profound effects on the timing of vegetative phase change. The segregation of QTL effects from both F1 parents indicates that alleles at this locus differentiated both grandparents with the precocious ecotype from those with normal phase change. Such loci with major effect have also been found to underlie variation in developmental traits in salamander [Pisum sativum (Wiltshire et al. 1994); Ambystoma sp. (Voss and Smith 2005); and Zea mays (Chuck et al. 2007)], collectively supporting the potential for rapid heterochronic evolution with few genetic changes.
While our results suggest ecotypic differentiation is a result of genome-wide adaptation, and most differences involve QTL of small effect (consistent with the bulk of plant and animal studies of ecotypic differentiation or early speciation reported to date), it is possible that the major effect QTL linked to the microRNA gene was the primary mutation that allowed the colonization and persistence of E. globulus in an ecologically extreme coastal habitat. Under this scenario, following the establishment of E. globulus with the precocious mutation in the cliff-top habitat (e.g., Wilsons Promontory), limited gene flow with subsequent selection may have led to a gradual build-up of genome-wide adaptation to produce the contemporary pattern of genomic differentiation that distinguishes the precocious Wilsons Promontory population from the adjacent “normal” Tidal River population. By focusing on a natural system where variation in the timing of vegetative phase change appears to be an important adaptation, we highlight the potential evolutionary importance of the miR156 regulatory network in heterochronic evolution in flowering plants.
Supplementary Material
Acknowledgments
We thank Norske Skog and Forestry Tasmania for providing land for the field trials. Paul Tilyard, Sascha Wise, Stephen Ridge, and James Marthick are gratefully acknowledged for providing technical assistance. Robert Wiltshire is acknowledged for discussion of this study and helping with photos. Funding for this project was provided by the Australian Research Council (DP0770506 and DP110101621).
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Barber H. N., 1965. Selection in natural populations. Heredity 20: 551–572. [Google Scholar]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J., et al. , 2009. The genetic architecture of maize flowering time. Science 325: 714–718. [DOI] [PubMed] [Google Scholar]
- Chen X. B., Zhang Z. L., Liu D. M., Zhang K., Li A. L., et al. , 2010. SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J. Integr. Plant Biol. 52: 946–951. [DOI] [PubMed] [Google Scholar]
- Chuck G., Cigan A. M., Saeteurn K., Hake S., 2007. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39: 544–549. [DOI] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. M., Wagler T. N., Quijada P., Doebley J., 2006. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 38: 594–597. [DOI] [PubMed] [Google Scholar]
- Climent J., Chambel M. R., Lopez R., Mutke S., Alia R., et al. , 2006. Population divergence for heteroblasty in the Canary Island pine (Pinus canariensis, Pinaceae). Am. J. Bot. 93: 840–848. [DOI] [PubMed] [Google Scholar]
- Cone K. C., McMullen M. D., Bi I. V., Davis G. L., Yim Y.-S., et al. , 2002. Genetic, physical, and informatics resources for maize. On the road to an integrated map. Plant Physiol. 130: 1598–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkowski G. W., Potts B. M., 1999. Geographic patterns of genetic variation in Eucalyptus globulus ssp. globulus and a revised racial classification. Aust. J. Bot. 47: 237–263. [Google Scholar]
- Fischer M. C., Foll M., Excoffier L., Heckel G., 2011. Enhanced AFLP genome scans detect local adaptation in high-altitude populations of a small rodent (Microtus arvalis). Mol. Ecol. 20: 1450–1462. [DOI] [PubMed] [Google Scholar]
- Foll M., Gaggiotti O., 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A bayesian perspective. Genetics 180: 977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M., Fischer M. C., Heckel G., Excoffier L., 2010. Estimating population structure from AFLP amplification intensity. Mol. Ecol. 19: 4638–4647. [DOI] [PubMed] [Google Scholar]
- Foster S. A., McKinnon G. E., Steane D. A., Potts B. M., Vaillancourt R. E., 2007. Parallel evolution of dwarf ecotypes in the forest tree Eucalyptus globulus. New Phytol. 175: 370–380. [DOI] [PubMed] [Google Scholar]
- Franks S. J., Weis A. E., 2008. A change in climate causes rapid evolution of multiple life-history traits and their interactions in an annual plant. J. Evol. Biol. 21: 1321–1334. [DOI] [PubMed] [Google Scholar]
- Goebel K., 1889. Über die Jugendzustände der Pflanzen. Flora 72: 1–45. [Google Scholar]
- Goodger J. Q. D., Choo T. Y. S., Woodrow I. E., 2007. Ontogenetic and temporal trajectories of chemical defence in a cyanogenic eucalypt. Oecologia 153: 799–808. [DOI] [PubMed] [Google Scholar]
- Gould S. J., 1977. Ontogeny and phylogeny, Harvard University Press, Cambridge, MA. [Google Scholar]
- Guerrant E., 1988. Heterochrony in plants: the intersection of evolution ecology and ontogeny, pp. 111–133 in Heterochrony in evolution, edited by McKinney M. E. Plenum Press, New York. [Google Scholar]
- Hamilton M. G., Tilyard P. A., Williams D. R., Vaillancourt R. E., Wardlaw T. J., et al. , 2011. The genetic variation in the timing of heteroblastic transition in Eucalyptus globulus is stable across environments. Aust. J. Bot. 59: 170–175. [Google Scholar]
- Hecht V., Laurie R. E., Vander Schoor J. K., Ridge S., Knowles C. L., et al. , 2011. The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23: 147–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C. J., Kullan A. R. K., Freeman J. S., Faria D. A., Grattapaglia D., et al. , 2012. High synteny and colinearity among Eucalyptus genomes revealed by high-density comparative genetic mapping. Tree Genet. Genomes 8: 339–352. [Google Scholar]
- Huijser P., Schmid M., 2011. The control of developmental phase transitions in plants. Development 138: 4117–4129. [DOI] [PubMed] [Google Scholar]
- James S. A., Bell D. T., 2001. Leaf morphological and anatomical characteristics of heteroblastic Eucalyptus globulus ssp globulus (Myrtaceae). Aust. J. Bot. 49: 259–269. [Google Scholar]
- Jaya E., Kubien D. S., Jameson P. E., Clemens J., 2010. Vegetative phase change and photosynthesis in Eucalyptus occidentalis: architectural simplification prolongs juvenile traits. Tree Physiol. 30: 393–403. [DOI] [PubMed] [Google Scholar]
- Jones R. C., Vaillancourt R. E., Gore P. L., Potts B. M., 2011. Genetic control of flowering time in Eucalyptus globulus ssp. globulus. Tree Genet. Genomes 7: 1209–1218. [Google Scholar]
- Jones R. C., Steane D. A., Lavery M., Vaillancourt R. E., Potts B. M., 2013. Multiple evolutionary processes drive the patterns of genetic differentiation in a forest tree species complex. Ecol. Evol. 3: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan G., Potts B. M., Wiltshire R., 1999. Strong, independent quantitative genetic control of vegetative phase change and first flowering in Eucalyptus globulus ssp. globulus. Heredity 83: 179–187. [DOI] [PubMed] [Google Scholar]
- Jordan G. J., Potts B. M., Chalmers P., Wiltshire R. J. E., 2000. Quantitative genetic evidence that the timing of vegetative phase change in Eucalyptus globulus ssp globulus is an adaptive trait. Aust. J. Bot. 48: 561–567. [Google Scholar]
- King D. A., 1999. Juvenile foliage and the scaling of tree proportions, with emphasis on Eucalyptus. Ecology 80: 1944–1954. [Google Scholar]
- Koressaar T., Remm M., 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23: 1289–1291. [DOI] [PubMed] [Google Scholar]
- Lawrence R., Potts B. M., Whitham T. G., 2003. Relative importance of plant ontogeny, host genetic variation, and leaf age for a common herbivore. Ecology 84: 1171–1178. [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Johnston M. O., 2000. Heterochrony in plant evolutionary studies through the twentieth century. Bot. Rev. 66: 57–88. [Google Scholar]
- MacKinney M. L., MacNamara K. J., 1991. Heterochrony: the evolution of ontogeny, Plenum Press. [Google Scholar]
- Marshall O. J., 2004. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 20: 2471–2472. [DOI] [PubMed] [Google Scholar]
- McKinnon G. E., Vaillancourt R. E., Steane D. A., Potts B. M., 2004. The rare silver gum, Eucalyptus cordata, is leaving its trace in the organellar gene pool of Eucalyptus globulus. Mol. Ecol. 13: 3751–3762. [DOI] [PubMed] [Google Scholar]
- Munns R., 2002. Comparative physiology of salt and water stress. Plant Cell Environ. 25: 239–250. [DOI] [PubMed] [Google Scholar]
- Myburg, A. A., D. Grattapaglia, G. A. Tuskan, U. Hellsten, R. D. Hayes, et al., 2014 Genome of Eucalyptus grandis—a global tree for fiber and energy. Nature 510: 356–362. [DOI] [PubMed] [Google Scholar]
- Neale D. B., Kremer A., 2011. Forest tree genomics: growing resources and applications. Nat. Rev. Genet. 12: 111–122. [DOI] [PubMed] [Google Scholar]
- Poethig R. S., 2009. Small RNAs and developmental timing in plants. Curr. Opin. Genet. Dev. 19: 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R. S., 2010. The past, present, and future of vegetative phase change. Plant Physiol. 154: 541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts B. M., Wiltshire R. J. E., 1997. Eucalypt genetics and genecology, pp. 56–91 in Eucalypt ecology: individuals to ecosystems, edited by Williams J., Woinarski J., Cambridge University Press, Cambridge, UK. [Google Scholar]
- Rhoades M. W., Reinhart B. J., Lim L. P., Burge C. B., Bartel B., et al. , 2002. Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Roesti M., Salzburger W., Berner D., 2012. Uninformative polymorphisms bias genome scans for signatures of selection. BMC Evol. Biol. 12: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., 2000. Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by Krawetz S., Misener S. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Sansaloni, C. P., C. D. Petroli, D. Jaccoud, J. Carling, F. Detering et al., 2011 Diversity Arrays Technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus, pp. 54 in BMC Proc.
- Thavamanikumar S., Southerton S. G., Bossinger G., Thumma B. R., 2013. Dissection of complex traits in forest trees—opportunities for marker-assisted selection. Tree Genet. Genomes 9: 627–639. [Google Scholar]
- Studer A., Zhao Q., Ross-Ibarra J., Doebley J., 2011. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43: 1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergrasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., et al. , 2012. Primer3 - new capabilities and interfaces. Nucleic Acids Res. 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami T., Horiguchi G., Yano S., Tsukaya H., 2009. The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136: 955–964. [DOI] [PubMed] [Google Scholar]
- Van Ooijen J., 2006. JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations, Kyazma BV, Wageningen, Netherlands. [Google Scholar]
- Van Ooijen J., 2009. MapQTL 6.0, software for the mapping of quantitative trait loci in experimental populations of diploid species, Kyazma, Wageningen, Netherlands. [Google Scholar]
- Visscher P. M., 2008. Sizing up human height variation. Nat. Genet. 40: 489–490. [DOI] [PubMed] [Google Scholar]
- Voss S. R., Smith J. J., 2005. Evolution of salamander life cycles: A major-effect quantitative trait locus contributes to discrete and continuous variation for metamorphic timing. Genetics 170: 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. W., Park M. Y., Wang L. J., Koo Y. J., Chen X. Y., et al. , 2011. MiRNA control of vegetative phase change in trees. PLoS Genet. 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire R. J. E., Murfet I. C., Reid J. B., 1994. The genetic control of heterochrony - evidence from developmental mutants of Pisum sativum L. J. Evol. Biol. 7: 447–465. [Google Scholar]
- Wiltshire R. J. E., Potts B. M., Reid J. B., 1991. A paedomorphocline in Eucalyptus - natural variation in the E. risdonii / E. tenuiramis complex. Aust. J. Bot. 39: 545–566. [Google Scholar]
- Wiltshire R. J. E., Potts B. M., Reid J. B., 1998. The genetic control of reproductive and vegetative phase change in the Eucalyptus risdonii/E. tenuiramis complex. Aust. J. Bot. 46: 45–63. [Google Scholar]
- Wu G., Poethig R. S., 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M. Y., Conway S. R., Wang J.-W., Weigel D., et al. , 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. X., Allen E., Wilken A., Carrington J. C., 2005. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Conway S. R., Poethig R. S., 2011. Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development 138: 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xu M., Koo Y., He J., Poethig R. S., 2013. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Cao L., Zhou C.-M., Zhang T.-Q., Lian H., et al. , 2013. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2: e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Zhang J., Yan J., Song R., 2011. Two transposable element insertions are causative mutations for the major domestication gene teosinte branched 1 in modern maize. Cell Res. 21: 1267–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotz G., Wilhelm K., Becker A., 2011. Heteroblasty—a review. Bot. Rev. 77: 109–151. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.