Abstract
We report a method for generating Drosophila germline mutants effectively via injection of the complex of the purified Cas9 protein, tracrRNA, and gene-specific crRNAs, which may reduce delayed mutations because of the transient activity of the Cas9 protein, combined with the simple mutation detection in GO founders by the T7E1 assay.
Keywords: Cas9 protein, RGEN, Drosophila
In the past few years, new technologies to knockout the genes of interests have been developed, including zinc-finger nucleases (ZFNs) (Kim et al. 2011), transcription activator-Like effector nucleases (TALENs) (Kim et al. 2013), and RNA-guided engineered nucleases (RGENs) derived from the clustered regularly interspaced short palindromic repeat RNA/CRISPR-associated (CRISPR/Cas) system (Gaj et al. 2013). Among them, RGENs are the latest gene knockout tools that were originally identified as an acquired immunity-like system in bacteria to protect the host from invading viruses or plasmids (Wiedenheft et al. 2012; Mali et al. 2013).
RGENs consist of three components: Cas9 endonuclease, CRISPR RNA (crRNA) that complementarily binds to the target site of the genomic DNA, and trans-activating CRISPR RNA (tracrRNA). This Cas9/tracrRNA/crRNA tripartite complex or its modified version, the bipartite complex consisting of Cas9/chimeric RNA (also called single-guided RNA or sgRNA), can cleave chromosomal DNA in a targeted manner and induce mutations efficiently in the vicinity of target sites in model organisms such as mouse, zebrafish, and C. elegans or in cell lines via the error-prone nonhomologous end-joining (NHEJ) DNA repair system (Cho et al. 2013a,b; Mali et al. 2013; Wang et al. 2013; Sung et al. 2014). RGENs were also successfully applied to Drosophila melanogaster, another major genetic model organism, but with differences in detailed methodologies; Cas9 was delivered as DNA plasmid (Gratz et al. 2013), mRNA (Bassett et al. 2013; Yu et al. 2013), or transgenic expression under a germ-cell–specific promoter (Kondo and Ueda 2013; Ren et al. 2013). Similarly, sgRNA was injected as DNA (Gratz et al. 2013a; Ren et al. 2013), RNA (Bassett et al. 2013; Yu et al. 2013), and transgenic expression under a universal promoter (Kondo and Ueda 2013). These approaches showed various efficiencies in terms of germ-line transmission rates and the percentages of mutants in F1 embryos (Gratz et al. 2013b; Bassett and Liu 2014; Beumer and Carroll 2014).
Here, we used an alternative approach in which the in vitro pre-formed ribonucleoprotein complex of the purified Cas9 protein, in vitro transcribed tracrRNA, and target gene–specific crRNAs was injected into the Drosophila embryos to knockout the gene of interest and validated the mutation-inducing efficiency based on the T7 endonuclease I (T7E1) assay in G0 founders and F1 mutants. The mutational sequences were later confirmed by direct sequencing. Our results indicate that our CRISPR/tracrRNA/crRNA complex injection induced loss-of-function mutations efficiently with low toxicity. The combined usage of the purified Cas9 protein and the T7E1 assay can help to validate candidate crRNAs for injection to choose efficient crRNAs in vitro and in vivo. In addition, the pre-formed ribonucleoprotein complex consisting of the purified Cas9 protein/tracrRNA/crRNA shown to act immediately after injection and degrade rapidly may alleviate delayed mutation events, helping reduce off-target effects and mosaicism by RGENs.
Results and Discussion
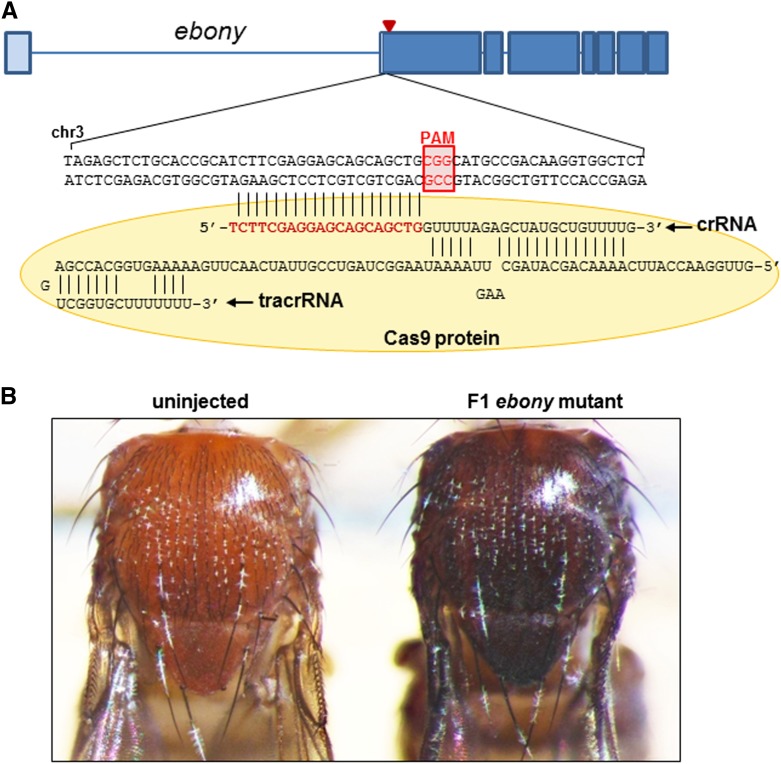
To test whether the tripartite ribonucleoprotein (RNP) complex of purified Cas9 protein, tracrRNA, and gene-specific crRNA (“RGEN-RNP” below) can generate mutations in the target sites of Drosophila genome, we first chose the ebony (e) gene in the third chromosome dictating the body color (Figure 1B). We designed two e-specific crRNAs that can bind to the second exon upstream of the start codon (Figure 1A). Syncytial blastoderm-stage w1118 embryos were microinjected with each complex of Cas9/RNAs at different concentrations, grown to adulthood, and individually crossed to D/e, Ser, TM3 to see if mutations were generated and transmitted through the germline cells into the F1 generation (see Supporting Information, Figure S1A for the cross scheme). It was found that the complex with only one of the two tested crRNAs gave rise to the F1 progeny with the e mutant phenotype (Figure 1B and Table 1). With the injection of crRNA(330 ng)-containing RGEN-RNP, 26 “G0 founder” flies, defined as adult flies that yielded at least one F1 mutant, out of 191 flies screened produced the F1 mutants (germline transmission rate, defined as the percentage of G0 founder/G0 adults screened, 26/191 = 13.61%) (Table 1), indicating that this method is capable of inducing loss-of-function mutations efficiently at target sites. Such rate decreased significantly at a higher concentration of crRNA (660 ng crRNA-containing complex, 2/77 = 2.6%) (Table 1), suggesting that injected doses of the RGEN-RNP are critical for the efficient induction of mutations. The percentages of the F1 mutants in individual F1 offspring clutches from G0 founders varied from 1.8% to 25% with the 330 ng crRNA-containing RGEN-RNP (Table 1).
Figure 1.
Generating novel ebony mutants using Cas9 protein/TracrRNA/crRNA complex. (A) Genomic structure of the ebony gene. The target nucleotide sequence upstream of the start codon (red triangle) neighboring the PAM sequence is complementary to the 5′ end of 20 bp of crRNA (shown in red). TracrRNA and Cas9 protein (shown as yellow circle) form the ribonucleoprotein complex along with gene-specific crRNA. (B) Compared to the brown body color of the wild-type adults (uninjected; left), the ebony F1 mutant (right) shows dark black body color, the phenotype due to the disrupted ebony gene by the injection of Cas9 protein/TracrRNA/ebony–specific crRNA complex.
Table 1. Efficiency of germline-transmitted mutations.
| crRNA (ng) | Number of Injected Embryos | Number of G0 Adults Screened | Number of Mosaic G0 Adults | Number of G0 Founder | Germline Transmission Rate (%) | Percentage of Mutants in F1 Offspring (%) | |
|---|---|---|---|---|---|---|---|
| Female | Male | ||||||
| e (660) | 400 | 42 | 35 | 0 | 2 | 2.6% (2/77) | 1.9–3.5 |
| e (330) | 900 | 110 | 81 | 0 | 26 | 13.61% (26/191) | 1.8–25 |
| sn (660) | 400 | 64 | 53 | 3 | 5 | 7.81% (5/64) | 1.7–3.5 |
| sn (330) | 400 | 20 | 16 | 3 | 4 | 20% (4/20) | 2.4–8.4 |
| sn (130) | 600 | 82 | 74 | 1 | 0 | 0 | 0 |
G0 founders yielded at least one F1 mutant, the germline transmission rate is defined as the percentage of G0 founders/G0 adults screened, and the percentage of mutants in F1 offspring means the percentage of the F1 mutant/F1 adults in each clutch.
To detect mutations and evaluate the efficiency induced by the RGEN-RNP on injection, we used the T7E1 assay that can recognize and cleave mismatched base pairs of wild-type and mutation-harboring DNA (Kim et al. 2009; see supplementary Materials and Methods in File S1 for details). Injection of the RGEN-RNP with 330 ng of the e-specific crRNA produced cleaved bands with various intensities in individual G0 founders by the T7E1 assay (Figure 2A). Such bands are thought to reflect different amounts of mismatched DNAs mostly due to the somatic mutations in injected individuals. Germline-transmitted F1 mutants were also confirmed by similar cleaved patterns but with more distinct bands in the T7E1 assay, presumably because they are heterozygous for mutations (Figure 2B). These results show that the simple and straightforward T7E1 assay can be used to detect mutations induced by RGEN-RNPs in the G0 and F1 generations of Drosophila. This assay can be also useful to decide on the best-working crRNA among multiple candidate crRNAs when tested in G0 mosaic individuals.
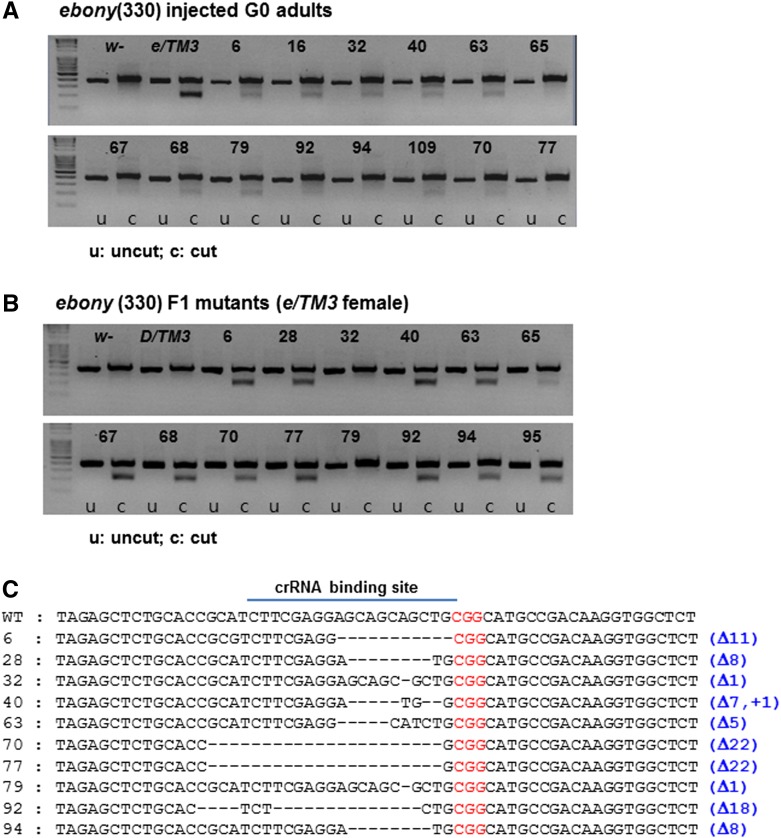
Figure 2.
Identification of ebony mutations by T7E1 assay and direct sequencing. (A) DNA-mismatched mutations induced by the injection of Cas9/RNA complex containing 330 ng of e crRNA were detected in G0 adult flies by the T7E1 assay. Each sample consists of two lanes as a pair in the agarose gel: the left lane is “uncut (u),” meaning no treatment, whereas the right is “cut (c),” digested by the T7E1 endonuclease. The numbers on top of each sample represent independently injected individual adult flies. The “cut” lanes show additional lower DNA bands in addition to ∼400 bp of uncut PCR-amplified DNA because mismatched DNA induced by the Cas9/RNA complex were digested by the T7E1. Diverse intensities of lower bands presumably reflect different degrees of mosaic mutations. w− is the negative control for the T7E1 assay, whereas e/TM3, heterozygous for ebony, is the positive control. (B) The T7E1 assay of individual F1 mutant flies. The format of the gel is similar to (A). The same numbers of samples in (A) and (B) indicate the relationship of parents and their offspring. Both w− and D/TM3 are negative controls. (C) Direct sequencing of the target site in individual F1 mutants revealed various deletion mutations. The numbers on the left indicate the individuals in (B) from which the DNA sequences were derived. Although rare, the deletion and insertion occurred simultaneously during nonhomologous end-joining DNA repair process (in the case of number 40). CGG in red denotes the PAM sequence.
To identify the nature of mutations induced by RGEN-RNP injection, we subcloned and sequenced the target genomic site of the ebony gene in the F1 mutant individuals previously confirmed by the T7E1 assay. A series of small deletion mutations, ranging from 1 base pair (bp) to 22 bp, was identified. Most of them cause the early stop of translation because of out-of-frame mutations and are expected to generate a nonfunctional protein (Figure 2C).
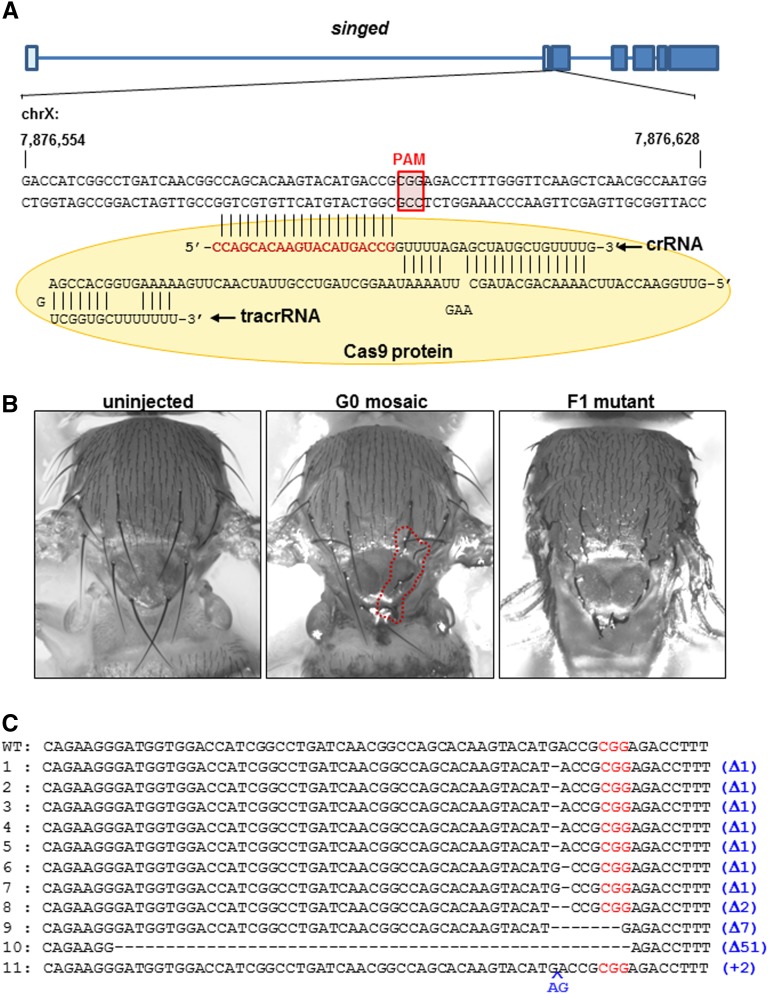
We further confirmed our RGEN-RNP–mediated gene knockout method for another gene, singed (sn), which is an X-chromosome gene that determines bristle formation (Figure 3, A and B). Injection of RGEN-RNP produced partially gnarled bristles even in the G0 adults as mosaic patterns, suggesting the induction of somatic mutations by sn-specific RGEN-RNP (Figure 2B and Table 1). Interestingly, one of the three mosaic G0 adult flies was the female fly, indicating the biallelic mutations in X chromosomes, suggesting high activity of the injected RGEN-RNP. When injected G0 adult female flies were individually crossed with wild-type males (Figure S1B) and the gnarled bristle phenotype was examined in F1 males, it was found that 20% (4/20, 330-ng crRNA injection) and 7.81% (5/64, 660-ng crRNA injection) of the G0 adult females carried the loss-of-function mutations in the sn gene of germline cells (Table 1). Direct sequencing encompassing the sn target site of F1 males confirmed insertion/deletion (indel) mutations ranging from 51 bp deletion to 2 bp insertion (Figure 3C). Some of these mutations were also confirmed by the T7E1 assay, showing the cleaved patterns (Figure S2).
Figure 3.
Identification of novel singed mutations. (A) Genomic structure of the singed gene. Similar to the ebony gene in Figure 1A, the target nucleotide sequence neighboring the PAM sequence is complementary to the 5′ end of 20 bp of crRNA (shown in red). TracrRNA and Cas9 protein (shown in yellow circle) singed-specific crRNA form the ribonucleoprotein complex. (B) Compared to the straight bristles of the wild-type (uninjected; left) the singed F1 mutants show the gnarled bristles (right), the phenotype due to the disruption of the singed gene by the injection of Cas9 protein/TracrRNA/singed–specific crRNA complex. Interestingly, this injection leads to gnarled bristles in some G0 founders in a mosaic fashion (middle, red circle), indicating somatic mutations induced by the Cas9/RNA complex. (C) Direct sequencing of the target site in individual F1 mutants reveals various insertion/deletion mutations. The numbers on the left side indicate individual F1 mutants. Deletions were the prevailing mutations while insertion was identified in one individual (blue in number 11). CGG in red denotes the PAM sequence.
Taken together, our results demonstrated that not only the RGEN-RNP-mediated (consisting of purified Cas9 protein, tracrRNA, and gene-specific crRNA) gene knockout method efficiently induces loss-of-function mutations for the Drosophila genes but also an optimal dose of the complex is required for its maximal activity because their knockdown efficiency appears to be dose-dependent (Table 1).
In using the CRISPR/Cas9 system to generate Drosophila knockouts, our study adopted two methodologies different from previous approaches: we used the purified Cas9 protein instead of cas9 DNA plasmid (Gratz et al. 2013a), in vitro transcribed cas9 RNA (Bassett et al. 2013; Yu et al. 2013), or germline-specific nos-cas9 transgenic fly (Kondo and Ueda 2013; Ren et al. 2013), and we the combined tracrRNA and gene-specific crRNA, which were prepared separately instead of using single-guided (or chimeric) RNA (sgRNA) (Jinek et al. 2012).
The ribonucleoprotein complex consisting of purified Cas9 protein and sgRNA was successfully applied for generating gene knockouts in C. elegans (Cho et al. 2013) and recently in mice and zebrafish (Sung et al. 2014), but it has not been tested in Drosophila thus far. The germline transmission rate (the percentage of G0 founders that yields at least a single F1 mutant) in our study (ranging from 2.6% to 20%) is relatively lower than that reported in other species using Cas9 protein/sgRNA complex (more than 20%) (Sung et al. 2014) and other Drosophila studies using TALEN RNA for trh gene (17% to 39%) (Kondo et al. 2014), cas9 RNA/sgRNA injection for yellow or w gene (12.4% to 68%) (Bassett et al. 2013, Yu et al. 2013), and germline-specific nos-cas9 transgenic expression for w, neuropeptides, and miRNAs (88.2% to 100%) (Ren et al. 2013; Kondo and Ueda 2013). In contrast, our germline transmission rate is higher than that using plasmids for cas9 and sgRNA (∼5%) (Gratz et al. 2013a). The difference in these rates may originate from the different efficiency of sgRNA vs. tracrRNA/crRNA or the distinctive nature of the target genes tested in different studies (e.g., some genes are more targetable than others). The inherent difference of Drosophila from other species in genome editing processes may also contribute to such differences. Of note, it has been reported that the tracrRNA/crRNA combination similar to the one used in our study is at least as efficient as sgRNA and it may even contribute to the reduction of off-target effects (Cho et al. 2014), which is one of the major concerns of the Cas9/RNA system.
Despite the relatively low efficiency of the RGEN-RNP injection in our study compared to the germline-specific nos-cas9 transgenic system or cas9 RNA injection, using the purified Cas9 protein pre-formed with tracrRNA/crRNA (or sgRNA) instead of cas9 DNA or RNA has a few advantages over other methods. First, the transient activity of the RGEN-RNP system can help alleviate delayed mutation events on delivery. Continuous expression of cas9 DNA in germ cells (Kondo and Ueda 2013; Ren et al. 2013) or its fairly stable RNA injection (Bassett et al. 2013; Sung et al. 2014; Yu et al. 2013) into developing embryos may elicit mutational events for a prolonged period of the development. This can increase the chance of unwanted off-target effects as well as on-target mutations and mutational mosaicism in independent cells of embryos, leading to multiple mutant alleles in following generations and potentially complicating further analyses. Contrary to the transgenic expression and RNA injection of cas9, the action of the purified Cas9 protein is expected to be rapid and transient because of its delivery as a form of protein and its short half-life. We have recently shown in human cell lines that the RGEN-RNP system acts immediately on delivery and the purified Cas9 protein was degraded within 24 hr after being applied to cultured cells (S. Kim et al. 2014). In this report, we also validated that the RGEN-RNP system induced off-target mutations quite rarely (up to 13-fold less than Cas9 plasmid did; confirmed by deep sequencing) without compromising on-target mutation efficiency (S. Kim et al. 2014). However, direct comparisons of mutational mosaicism in animal models by the RGEN-RNP and other forms of cas9 remain to be determined.
Another advantage of using the RGEN-RNP system is that designed sgRNAs or crRNAs can be validated for their efficiency for inducing mutations to select the best ones by using the purified Cas9 protein in an in vitro cleavage assay before the actual transfection/injection into cells or embryos is embarked (Sung et al. 2014). Furthermore, such a system can be exploited to genotype the heterozygous and homozygous mutant progenies as an alternative RFLP (restriction fragment length polymorphism) without requiring specific restriction enzymes and recognized sites (J. M. Kim et al. 2014). However, our RGENP-RNP method requires an additional step of preparing recombinant Cas9 protein using a bacterial in vitro translation system (S. Kim et al. 2014), and the relative instability of Cas9 protein compared to DNA or RNA of Cas9 for long-term storage may be practically problematic.
The selection of target gene–specific crRNAs or sgRNAs that work most efficiently in vivo among multiple candidates predicted from bioinformatics is one of the key requirements to generate mutants of animal models using the CRISPR/Cas9 system. Several methods have been developed that allow the detection of mutations in the target DNA induced by genome editing tools in vivo, preferably in the G0 founders. For example, Bassett et al. (2013) and Gratz et al. (2013a) adopted high-resolution melting analysis (HRMA) and the surveyor kit (Transgenomic, Inc), respectively. The T7E1 assay used in this study is similar to the surveyor kit in that DNA mismatches are detected by digestion of a specialized endonuclease, but it is simpler and can detect mutations using genomic DNA from Drosophila G0 founders (Figure 2).
Our current study proposes a strategy to generate Drosophila mutants for a certain gene. First, an investigator designs multiple crRNAs (or sgRNAs) that are predicted to show minimal off-target effects using web-based software such as http://flyrnai.org/crispr/ (Hsu et al. 2013) and http://crispr.mit.edu/ (Ren et al. 2013). The potential off-target sites can be checked at this step (http://www.rgenome.net/cas-offinder/) (Bae et al. 2014). Simultaneously, the purified Cas9 protein is prepared by in vitro translation. Second, the efficiency of multiple crRNAs (or sgRNA) with different doses and ratios with Cas9 protein are verified and the best-working ones are selected by performing the aforementioned in vitro cleavage assay using a RGEN-RNP (mentioned above) and the T7E1 assay after PCR using genomic DNA from G0 individuals as the DNA template. If one prefers the in vivo assay and wants to save time, then the in vitro cleavage assay can be skipped. Next, G0 founders exhibiting cleaved patterns by the T7E1 assay are crossed to identify lines that carry germline-transmitted mutations. Finally, heterozygous and homozygous mutant progenies in the following generations can be genotyped using either RGEN-RNP RFLP or T7E1 assays, and the mutagenesis efficiency can be calculated.
In summary, our method reported in this study is a valid alternative to be considered for generating Drosophila mutants with a reasonable efficiency and as little time and effort as possible, especially when the prevention of delayed mutation events is preferred to reduce the off-target effects and mosaicism.
Supplementary Material
Acknowledgments
This work was supported by NRF-2011-0023507 from MSIP (Korea) and KRIBB initiative program (Korea).
Footnotes
These authors contributed equally to this work.
Communicating editor: H. D. Lipshitz
Literature Cited
- Bae S., Park J., Kim J. S., 2014. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30: 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A., Liu J. L., 2014. CRISPR/Cas9 mediated genome engineering in Drosophila. Methods DOI: 10.1016/j.ymeth.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P., Liu J. L., 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Reports 4: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K. J., Carroll D., 2014. Targeted genome engineering techniques in Drosophila. Methods 68: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Lee J., Carroll D., Kim J. S., 2013a Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195: 1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim J. M., Kim J. S., 2013b Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31: 230–232. [DOI] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim Y., Kweon J., Kim H. S., et al. , 2014. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F., 3rd, 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013a Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Wildonger J., Harrison M. M., O’Connor-Giles K. M., 2013b CRISPR/Cas9-mediated genome engineering and the promise of designer flies on demand. Fly (Austin) 7: 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., et al. , 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Lee H. J., Kim H., Cho S. W., Kim J. S., 2009. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 19: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. M., Kim D., Kim S., Kim J. S., 2014. Genotyping with CRISPR-Cas-derived RNA-guided endonucleases. Nat. Commun. 5: 3157. [DOI] [PubMed] [Google Scholar]
- Kim S., Lee M. J., Kim H., Kang M., Kim J. S., 2011. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat. Methods 8: 7. [DOI] [PubMed] [Google Scholar]
- Kim S., Kim D., Cho S. W., Kim J., Kim J. S., 2014. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. Apr 30 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kweon J., Kim A., Chon J. K., Yoo J. Y., et al. , 2013. A library of TAL effector nucleases spanning the human genome. Nature biotechnology 31: 251–258. [DOI] [PubMed] [Google Scholar]
- Kondo S., Ueda R., 2013. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195: 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Sakuma T., Wada H., Akimoto-Kato A., Yamamoto T., et al. , 2014. TALEN-induced gene knock out in Drosophila. Dev. Growth Differ. 56: 86–91. [DOI] [PubMed] [Google Scholar]
- Mali P., Esvelt K. M., Church G. M., 2013. Cas9 as a versatile tool for engineering biology. Nat. Methods 10: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Sun J., Housden B. E., Hu Y., Roesel C., et al. , 2013. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. USA 110: 19012–19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y. H., Kim J. M., Kim H. T., Lee J., Jeon J., et al. , 2014. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 24: 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., et al. , 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B., Sternberg S. H., Doudna J. A., 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338. [DOI] [PubMed] [Google Scholar]
- Yu Z., Ren M., Wang Z., Zhang B., Rong Y. S., et al. , 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 195: 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.