Abstract
Living fossils are lineages that have retained plesiomorphic traits through long time periods. It is expected that such lineages have both originated and diversified long ago. Such expectations have recently been challenged in some textbook examples of living fossils, notably in extant cycads and coelacanths. Using a phylogenetic approach, we tested the patterns of the origin and diversification of liphistiid spiders, a clade of spiders considered to be living fossils due to their retention of arachnid plesiomorphies and their exclusive grouping in Mesothelae, an ancient clade sister to all modern spiders. Facilitated by original sampling throughout their Asian range, we here provide the phylogenetic framework necessary for reconstructing liphistiid biogeographic history. All phylogenetic analyses support the monophyly of Liphistiidae and of eight genera. As the fossil evidence supports a Carboniferous Euramerican origin of Mesothelae, our dating analyses postulate a long eastward over-land dispersal towards the Asian origin of Liphistiidae during the Palaeogene (39–58 Ma). Contrary to expectations, diversification within extant liphistiid genera is relatively recent, in the Neogene and Late Palaeogene (4–24 Ma). While no over-water dispersal events are needed to explain their evolutionary history, the history of liphistiid spiders has the potential to play prominently in vicariant biogeographic studies.
Keywords: living fossils, genetic diversity, plesiomorphies, vicariance, dispersal, ancestral areas
1. Introduction
Living fossils are those lineages that were once more diverse than today, or that have survived over long evolutionary timescales with little morphological change [1]. Textbook examples include plants such as the Gingko tree and the cycads, and animals such as coelacanth fishes and horseshoe crabs. These living fossils are hypothesized to exhibit ancient roots and early diversification patterns [2,3]. However, such assertions are rarely tested within a time-calibrated phylogenetic framework. The latest dating analyses of extant cycads and coelacanths have discovered that although these lineages are ancient, their extant species are a surprisingly recent radiation [1,4]. Within arachnids, liphistiid spiders are widely recognized as ‘living fossils' [5] as they are the only surviving group of the suborder Mesothelae that have retained spider plesiomorphies such as abdominal tergites and spinnerets located in the middle of the abdominal venter (figure 1a,b) [6–9]. Because of this plesiomorphic morphology, the family and genera of living liphistiid spiders native only to parts of Southeast and East Asia are assumed to be ancient and exhibit early diversification patterns. This assumption, however, remains untested.
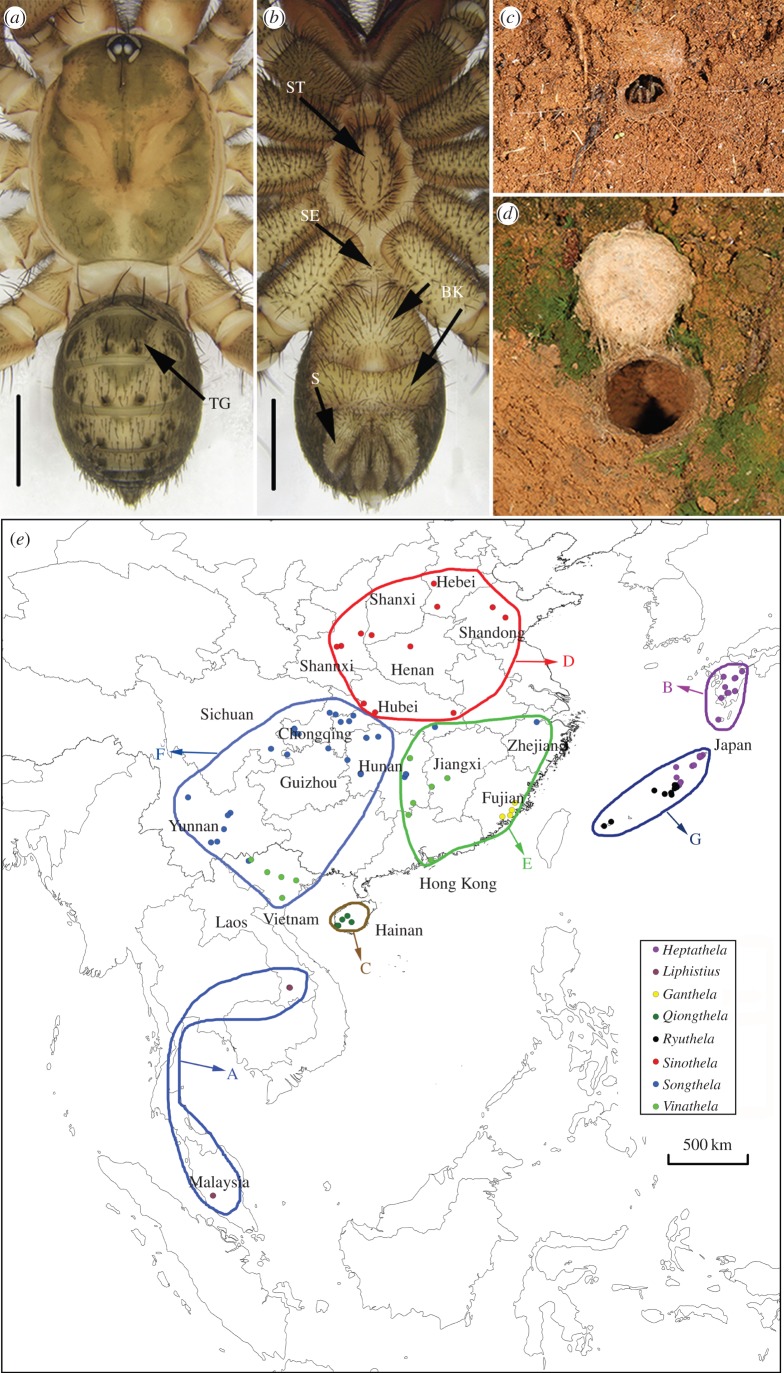
Figure 1.
Phenotypical plesiomorphies of liphistiid spiders, and a map with the sampling localities. (a) Dorsal view of Ryuthela nishihirai; (b) ventral view of R. nishihirai; (c) trapdoor of Liphistius with radiating signal lines; (d) heptatheline trapdoor (note absence of signal lines). BK, book lungs; S, spinnerets; SE, sternite, ST, sternum; TG, tergite. Scale bars, 2 mm; (e) map with the sampling localities. Colours, corresponding with those in figure 3, represent eight liphistiid genera. Letters denote areas for biogeographic reconstruction: A: Southeast Asia; B: Kyushu; C: Hainan; D: North China (north of the Yangtze River); E: East China (Fujian, Hubei, Hunan, Hong Kong, Jiangxi and Zhejiang); F: West China and North Vietnam (Chongqing, Guizhou, Hubei (Enshi, Jianshi and Lichuan), Hunan (Cili, Fenghuang and Zhangjiajie), Sichuan and Yunnan); G: Ryukyu archipelago. (Online version in colour.)
The family Liphistiidae consists of 89 extant species-level taxa currently grouped in three genera, and displays an interesting geographical distribution confined to Southeast and East Asia [10]. The group is a species-poor, but seemingly ancient lineage of relatively large, dispersal-limited, unusually long-lived (5–18 years), ground-dwelling spiders that build trapdoor burrows (figure 1c,d) [8,9,11]. Since their discovery [12], classical authors [13] divide Liphistiidae into two subfamilies, Liphistiinae with a single genus, Liphistius and 50 species, and Heptathelinae with 39 species within two genera, Heptathela and Ryuthela [10]. Liphistiines are geographically separated from heptathelines, Liphistius occurring in Southeast Asia, and Heptathela and Ryuthela confined to East Asia (figure 1e).
As the sister clade to all other known spiders, Mesothelae should play prominently in any attempts at reconstructing the arachnid tree of life and testing alternative biogeographic hypotheses. Surprisingly, however, the research on Mesothelae and Liphistiidae has thus far been devoid of comprehensive phylogenies. Although liphistiid exemplars are routinely used in family level or higher spider phylogenetic research, these works either use solely morphological data [7–9,14–17], or where molecular data are used, employ a single or at the most two representatives of the family [18–21] except for a recent study reporting the molecular phylogeny for the genus Ryuthela [22]. To date, no molecular phylogeny has been available for the family and for other genera.
Representing an ancient lineage with a geographically restricted distribution and very likely limited colonization ability, the biogeography of liphistiids is of great interest. It is, therefore, surprising how little we know about the origin of Mesothelae (the basal-most branch of all spiders) and Liphistiidae (the only Mesothelae family with extant species), and how geological and biological processes in the Earth's history might have affected them. The extant Mesothelae are only distributed in East and Southeast Asia, from where no Palaeozoic arachnids are known, and the only fossil species is the Late Carboniferous Palaeothele montceauensis [23], known from the part of Euramerica that corresponds to today's France [23,24]. At that time in the Earth's history (295 Ma), those regions where Mesothelae are found today were on landmasses isolated from Pangea [25–32]. Recent authors [13,33] hypothesized that Mesothelae originated in Euramerica prior to its integration with Pangea, and that during the Late Triassic the ancestor of Liphistiidae colonized East and Southeast Asia from Euramerica over one of several possible landmasses. The occurrence of Mesothelae on the islands of southern Japan (Kyushu and Ryukyu Archipelago) was explained both through vicariant origin in the Tertiary when the Japanese island arc separated from mainland Asia [33], or alternatively, as a consequence of dispersal events over-land bridges from east China during the Pleistocene [9]. Considering the acute absence of a liphistiid phylogeny, none of the above biogeographic hypotheses have been empirically tested.
Here we use molecular data from our original sampling to test the monophyly of Liphistiidae and its genera. We then devise a time-calibrated phylogeny to elucidate the origin of the stem group Mesothelae and its major clades. We use these data to examine the biogeographic history of these spiders, and, given their sedentary lifestyle and long evolutionary history, propose that the lineage is an ideal model for reconstructing past events. Finally, we test the assumption that present-day liphistiids exhibit not only early origin but also diversification patterns, as would be expected from their labelling as living fossils.
2. Material and methods
The greater detailed methods are described in the electronic supplementary material.
(a). Material and phylogenetic inference
Our study primarily focused on heptathelines (the liphistiids of East Asia), and we added two species of liphistiines from Laos and Malaysia (figure 1e). Our ingroup sampling thus had a total of 124 liphistiid specimens from 105 localities (figure 1e). We sampled mygalomorph outgroups of the families Atypidae and Ctenizidae and augmented original data with sequences from GenBank (electronic supplementary material, table S1). We performed parsimony [34], maximum-likelihood (ML) [35] and Bayesian (BI) [36] phylogenetic analyses using nucleotide data from five genes (CO1, 16S, 28S, H3, ITS2) (electronic supplementary material, table S2) and selecting the best fit DNA substitution model for each of the four partitions [37].
(b). Calibrations and divergence dating
To estimate divergence times within Liphistiidae, we devised a matrix whose taxon sample maximized fossil calibration points. Analyses conducted using only fossil-based time constraints yielded extremely low estimates for mtDNA substitution rates, more than twice lower than any value reported in the literature [38–41]. Therefore, we conducted additional analyses with normal distribution on the mtDNA rate prior based on information available in the literature. We linked the mitochondrial genes (CO1 and 16S) in a single clock, assigning the substitution rate parameter a normal prior with a mean 0.017 and standard deviation 0.0045, as recently estimated for other spider lineages [42]. For the nuclear genes (28S, H3 and ITS2) and to speed-up analyses, we restricted the parameter space by setting diffuse, uniform priors to the starting value of 0.00115, with the minimum and maximum bounds set to 0.0001 and 0.0115. These values are based on the universal substitution rate proposed for arthropod mtDNA [43], and were selected under the assumption that the nuclear genes are about one order of magnitude slower than mitochondrial (starting value) and generally no nuclear protein coding gene will show higher rates than the mitochondrial genes (upper bound) [44].
Based on the jModeltest results, we used the substitution models GTR + I + G for CO1, 16S, 28S and ITS2 and TN93 + I + G for H3, and base frequencies were set to estimate except for H3 and ITS2 which were set to equal. A starting tree including the time constraints was generated with starttree (http://bodegaphylo.wikispot.org/starttree_program) and incorporated to the analysis by modifying the xml file. All analyses were run assuming a lognormal relaxed clock for each gene partition and a Yule tree prior. We specified an exponential prior for the yule.birthRate with a mean of 0.05, estimated using the pyule tool (available at https://github.com/joaks1/pyule). For each analysis, three independent chains were run for 50 million generations. Chain convergence and correct mixing of each MCMC chain was assessed with TRACER v. 1.6 [45]. Ten per cent of the first generation of each chain was removed as burnin and the remaining values were combined into a single file with the help of LogCombiner [46] and consensus trees were obtained with TreeAnnotator [46].
(c). Biogeographical analyses
To infer biogeographical events, we defined seven discrete geographical areas (figure 1e) and reconstructed ancestral distributions on a simplified matrix using three alternative methods in RASP v. 3.0 [47,48]: (i) statistical dispersal–vicariance analysis (S-DIVA; [47]), with a maximum of three areas per node; (ii) BI binary MCMC analysis (BBM; [47,49]), with 5000 cycles using 10 chains, sampling every 100 cycles, allowing for a maximum of three areas, giving a ‘custom’ root distribution, using the JC + G substitution model; and (iii) dispersal–extinction–cladogenesis analysis (DEC; [50]), setting the maximum areas to three [48], with an unconstrained model allowing for equal rates of dispersal (1.0) between areas at any time.
3. Results
(a). Phylogenetic reconstruction
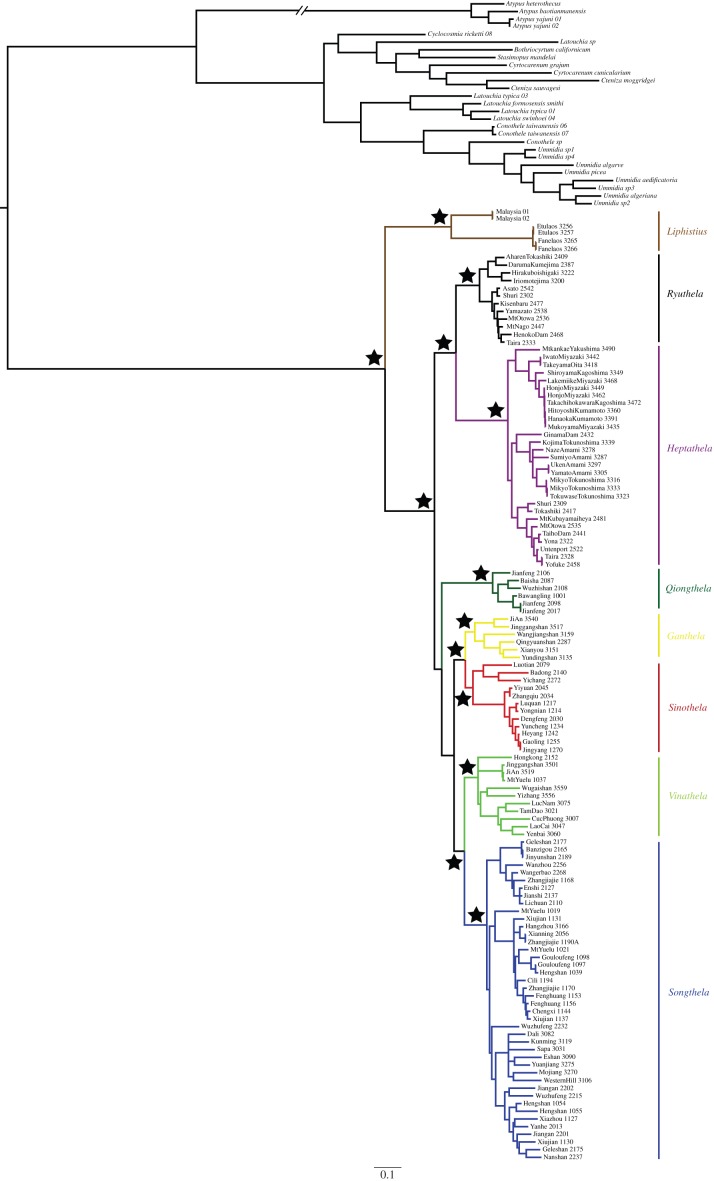
All analyses, BI, parsimony and ML under the four partition schemes consistently recover Liphistiidae monophyly with high support values (posterior probability, hereafter PP, ML bootstrap support, BS and parsimony jackknife, JK, all reaching 100%), and all analyses converged on similar within-family topologies (figure 2). The monophyly of Liphistius, in our taxonomic sample represented by exemplars from Malaysia and Laos, is well supported (PP = 1, JK = 100 and BS = 100) and this clade was always sister to heptathelines, a clade uniting all remaining liphistiids (figure 2). The samples from Japan fit into two reciprocally monophyletic and highly supported clades (figure 2), one including all species of Ryuthela from Ryukyu Island and southern part of Okinawa, and the other clade consisting of some but not all Heptathela (those from Ryukyu Island and Kyushu; figure 2). The fact that all other Heptathela species, those from the Asian mainland, fall into other clades makes Heptathela paraphyletic. In an accompanying paper [51], we revise the genus level taxonomy based on the phylogenetic evidence (this paper) and morphological and natural history diagnoses. Here, we refer to those taxonomic decisions by treating Liphistius Schiödte, 1849 and Ryuthela Haupt, 1983 as valid genera, and Heptathela sensu lato as six genera (nomenclature formalized in [51]): Ganthela, Heptathela sensu stricto, Qiongthela, Sinothela Haupt, 2003, Songthela Ono, 2000, Vinathela Ono, 2000.
Figure 2.
Summary phylogenetic results. The topology is from the partitioned BI analysis of the full matrix. Black stars indicate solid branch supports, i.e. above the threshold in more than half of the 12 analyses using BI inference, ML and parsimony (thresholds of posterior probabilities > 0.95, boostrap > 70%, jackknife > 70%). (Online version in colour.)
(b). Divergence dating and biogeographical reconstructions
Results of the analyses with and without the informed prior in the mtDNA substitution rate resulted in markedly different time frames of diversification for liphistiids (electronic supplementary material, table S3 and figure S1; figure 3a). Informing the substitution rate resulted in a compression of the liphistiid clade towards the present, yielding dates about one-third younger than the fossil, unconstrained analysis (electronic supplementary material, table S3). The estimated rate of substitution (ucld.mean) of the mtDNA under the unconstrained fossil analysis yielded very low values, about twice lower than those reported in the literature [38–41] (electronic supplementary material, table S4). In addition, some of the parameter values reported for the unconstrained fossil analysis (coefficient of variation of the 28S and ITS2) were higher than those recommended in the literature (greater than 1) (electronic supplementary material, table S4). On the other hand, increasing the time range of the maximum bound had little to no effect on the results (electronic supplementary material, tables S3 and S4, column rates and rates (20%)).
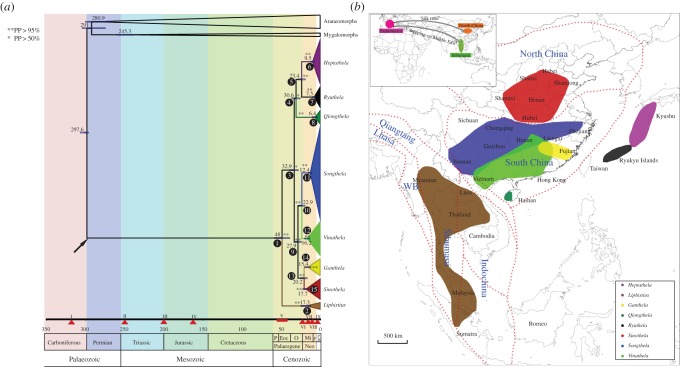
Figure 3.
Divergence time estimates and geographical distribution of liphistiid genera. (a) Chronogram from BEAST with inferred node ages in million years and 95% confidence intervals (bars), estimated using fossil calibrations and informed priors on the mtDNA substitution rates. The arrow indicates the fossil age of Palaeothele montceauensis but does not imply direct ancestry. Black numbered circles represent the major nodes matching those in electronic supplementary material, figure S2. Triangles with Roman numbers mark relevant geological events that may have affected the dispersal routes and diversification of mesothelid and liphistiid lineages. I, Pangea started to form; II, Pangea started to break-up and Sibumasu terrane contacted with Eurasia; III, Sibumasu terrane accreted to the Indochina, South China and North China plates; IV, North China plate connected to Eurasia; V, India collided with Eurasia; VI, the Yangtze River formed; VII, Japanese islands separated from East Asia; VIII, Taiwan formed; IX, Hainan Island separated from the mainland. (b) General distribution maps of the eight genera delimited in this study, and the demarcation of main continental plates and terranes that formed Southeast and East Asia (in part from [52]). Inset map shows the two most plausible of the three mutually exclusive hypotheses explaining the eastward dispersal routes of liphistiid ancestors (simplified on today's land masses, but see text for details). WB, West Burma. (Online version in colour.)
The time-calibrated phylogenetic analyses based on fossil (exponential prior with 95% of the posterior density falls within the range 10% older than the minimum bound) and on substitution rates estimated the basal-most spider divergence between the Mesothelae (treated as a stem group) and the Opisthothelae (represented by Mygalomorphae and Araneomorphae) at 297.6 Ma (295–306 Ma) (figure 3a). Given the estimated age of the extant mygalomorphs and araneomorphs at 245.3 Ma (240–259 Ma) and 280.8 (262–297 Ma), respectively, and a long, 250 million year Mesothelae branch, the Palaeogene origin of the family Liphistiidae estimated at 48 Ma (39–58 Ma) is relatively recent. Heptathelinae origin is estimated at 32.9 Ma (28–39 Ma). The origins of all liphistiid genera fall into the Late Palaeogene and Neogene (4–36 Ma). Within Heptathelinae, the clade uniting eastern island Oligocene taxa Heptathela (9.5; 7–12 Ma), Qiongthela (6.4; 4–10 Ma) and Ryuthela (7.5; 5–10 Ma) originated at 30.6 Ma (25–36 Ma). The clade of a comparable Oligocene age (27.9; 23–33 Ma) unites the mainland genera Ganthela (15.4; 11–20 Ma), Sinothela (17.7; 14–22 Ma), Songthela (12.4; 10–15 Ma) and Vinathela (16.2; 13–20 Ma). The extant diversity of the Heptathelinae genera can be traced to the Miocene (approx. 4–22 Ma), while Liphistius may have diversified slightly earlier (17.3; 11–24 Ma).
Ancestral area reconstructions indicate a number of likely dispersal and vicariant events (electronic supplementary material, figure S2 and table S5). None of postulated dispersal events in the liphistiid biogeographic history, however, need to invoke any over-water dispersal. The ancestral area for liphistiids is as likely to be Southeast as East Asia, which since that geological time (approx. 48 Ma) continued to be a single land mass. The heptatheline ancestors occupied the areas corresponding to today's Hainan, East China or Ryukyu archipelago, at the time (approx. 33 Ma) all part of continental Asia. Likewise, the ancestors of today's island taxa (Heptathela s.s., Ryuthela, Qiongthela) occupied either Hainan or Ryukyu archipelago before they were islands (approx. 31 Ma). These three genera diversified vicariantly, before or when their ancestral areas became continental islands, i.e. when Kyushu separated from the Asian continent with the opening of the Japanese sea approximately 15 Ma [53–55], and when Ryukyu archipelago was gradually isolated from Asia approximately 10–5.3 Ma [56–61]. On the mainland, the separation of the lineages Sinothela and Ganthela (approx. 20; 16–25 Ma) corresponds well to the timing of the Yangtze River origin approximately 23 Ma [55,62]. This suggests that their vicariant origin on each side of the river is still mirrored in today's distributions. Similarly, Vinathela and Songthela have never in their 23 Ma (19–28 Ma) history left the areas E and F (figure 1e) over Yangtze to the north or over the ocean to the south and east, and neither has Ganthela in approximately 15 Ma (15–20 Ma). Similarly, the approximately 6 Ma (3–10 Ma) evolution of Qiongthela invokes no over-water dispersals from Hainan.
4. Discussion
Bristowe [5] referred to Liphistiidae as a ‘family of living fossil spiders' [5] because they ‘retained many characters from their ancestors and resemble fossil spiders of the Carboniferous period’. Our extensive fieldwork over their present range (figure 1e) discovered numerous new localities and species and, albeit emphasizing heptathelines, secured a taxon sample needed for the first comprehensive species-level phylogeny of Liphistiidae. Our phylogenetic (figures 2 and 3a) and biogeographic (electronic supplementary material, figure S2) analyses enabled rigorous testing of the monophyly of the family, its two subfamilies, its genera and species groups and the timing of the major divergences and the ancestral ranges. Interesting strong evolutionary and biogeographic patterns emerge. While extant liphistiids indeed possess arachnid plesiomorphies (figure 1a,b) not shared with any other spiders, and the Mesothelae lineage traces back as far as the Carboniferous, the origin of Liphistiidae and its diversification are surprisingly much more recent than once thought, repeating the patterns recently detected also in other textbook living fossils [1,4]. We trace the origin of the family in Southeast or East Asia to the Late Palaeogene and Eocene (39–58 Ma), and the origins of all eight genera to the Late Oligocene and Neogene (figure 3a; electronic supplementary material, figure S1 and table S3). However, liphistiids do belong to a very long evolutionary branch that connects them with the only mesothelean fossil known from Eurasia (figure 3a), and consequently, some of their phenotypic traits resemble the hypothetically ancestral spiders. In the current absence of any additional fossil data, this long phylogenetic branch suggests that mesothelean evolution must account for a very slow eastward move with possibly numerous extinction events. Below, we discuss three possible scenarios and suggest that the eventual discovery of new mesothelean fossils may support one over the other, depending on its location. Finally, we align liphistiid natural history, in particular their sedentary lifestyle (figure 1c,d), to the emerging phylogenetic and biogeographic patterns to reveal that they apparently lack over-water dispersal ability, and this makes them ideal biogeographic models.
(a). Phylogeny and dating analysis
We provided the first phylogenetic hypothesis for the family Liphistiidae, with a particular focus on the genera and species groups within Heptathelinae. We used exemplars obtained through our several years of sampling throughout their range, obtained data on five mitochondrial and nuclear markers, and analysed them along with numerous mygalomorph outgroups using three phylogenetic approaches (BI inference, ML and parsimony) under four partition schemes. The results (figures 2 and 3a) have converged on the following general patterns: (i) the monophyly of the family Liphistiidae is well supported; (ii) the genus Liphistius is the sister group to Heptathelinae, the latter containing all other liphistiids; (iii) within Heptathelinae, Ryuthela is monophyletic but the genus ‘Heptathela’ s.l. as currently known is paraphyletic.
Liphistiid monophyly has never been questioned, and the clade has been recovered in all morphological and molecular analyses that focused on higher level phylogenetics [7–9,14,15,17–20,63]. However, these studies only included a small subset of liphistiid diversity, typically one Liphistius and/or one Heptathela [7,8,18–20]. Our study is the first to test and confirm liphistiid monophyly with a comprehensive taxon sampling and using molecular data.
Two distinct clades can be identified within Liphistiidae: Liphistiinae and Heptathelinae. Liphistiinae contains only Liphistius whose monophyly has not been disputed [9,13]. Our dating analysis estimates the crown group Liphistius to be approximately 17 Ma, but its stem may be as old as approximately 48 Ma. Its sister lineage, Heptathelinae, which contains all other liphistiids from China, Japan and Vietnam, originated during the Eocene/Oligocene. Our results unequivocally confirm the monophyly of Ryuthela in agreement with morphological [9,13] and preliminary molecular datasets [22]. However, all phylogenetic evidence refutes the monophyly of ‘Heptathela’ s.l., contradicting most recent taxonomic literature [13,64].
The monophyly of each of the mainland Chinese genera Songthela, Vinathela, Sinothela, and in part, Ganthela is well supported in the phylogeny, and also by the dating analysis and morphological diagnostics [51]. The exception is the suboptimal clade support for Ganthela in the parsimony analyses, and poorly resolved relationships within Songthela in most analyses. The apparent conflict in the data may be due to inadequate taxonomic sampling in south and southwest China. The position of the genus Qiongthela from Hainan remains unresolved. With relatively low support, the phylogenetic analyses place it as sister to all mainland taxa (figure 2), but the dating analysis places it as sister to the doublet Ryuthela and Heptathela s.s. the remaining two island genera (figure 3a; electronic supplementary material, table S5). More sampling and data are needed to resolve this topological instability for the three genera that may be crucial for the understanding of liphistiid island biogeographic patterns.
(b). Historical biogeography
Mesothelid spiders are sedentary, dispersal-limited spiders whose evolutionary history dates back to the very origin of spiders in the Late Carboniferous. They have therefore already attracted considerable biogeographical interest [9,33,64–66], but our view is that they should play a more prominent role in future reconstructions of biogeographical histories. As all prior hypotheses have been devoid of molecular data, the here presented phylogenetic framework and the inferred chronogram (figures 2 and 3a) shed new light on the origin and biogeography of the group. Interestingly, the reconstructed ancestral areas (electronic supplementary material, figure S2) and dispersal versus vicariant events (electronic supplementary material, table S5) do not require any explanations of over-water dispersal in liphistiid biogeographic history. Apparently, effective water barriers include not only bodies of ocean, but also major rivers, notably Yangtze.
The available fossil evidence supports the ‘Euramerican origin hypothesis' for Mesothelae (figure 3b inset; [13,33]). Our various dating analyses agree that the spider origin, and thus the basal-most split between Mesothelae and Opisthothele was in the Late Carboniferous and Early Permian, approximately 295–320 Ma (figure 3a; electronic supplementary material figure S1 and table S3). During the Late Carboniferous and Permian, the Laurentia and the Baltica plates together formed the equatorially situated Euramerica [28,28,31,67]. It was there that the only known Mesothelae fossil lived around 295 Ma [23,24]. By contrast, the regions inhabited today by extant liphistiids (Southeast and East Asia) correspond to isolated microplates of that time, and are not known to contain any Palaeozoic arachnid fossils. Therefore, all evidence points to the origin of Mesothelae in Euramerica.
The long evolutionary history from the Mesothelae origin in Euramerica to modern time is estimated at about 298 Ma. Prior to our dating analysis, the node Liphistiidae was traditionally viewed as ‘primitive’ and ancient, but in fact our dating analysis places its origin at ‘only’ approximately 48 Ma, and therefore, recovers an extraordinarily long (approx. 250 Ma) branch between the time of the mesothelean fossil (295 Ma) and the origin of Liphistiidae. Even the alternative analysis that only used fossil data unconstrained for mtDNA substitution rates uncovered a relatively recent origin of Liphistiidae at approximately 124 Ma, implying a minimum estimate of the mesothelean branch of 177 Ma. This means that liphistiid ancestors must have arrived east over Laurasia (or Gondwana, see below) over a long geological time.
The available palaeogeographical and palaeoclimatological data [28,29,31,52,67–71] allow for three possibilities of such a long journey over land from Laurasia to Asia (figure 3b; see electronic supplementary material): (i) the liphistiid ancestors may have taken a southern route along the northern margins of Gondwana, to which Euramerica connected during the Late Carboniferous [33], then drifted north towards Southeast Eurasia on Sibumasu, i.e. eastern Burma, western Thailand, northwestern peninsular Malaysia and a part of Sumatra [68–70,72–74]. We name this scenario the ‘Out of Gondwana’ hypothesis; (ii) they may have taken a route along the southern margin of Laurasia by stepping onto the Cimmerian continent strip [33]. This is the ‘Stepping-on Middle East’ hypothesis (figure 3b, inset); and finally (iii) they may have travelled through northern Laurasia and on to the North China plate after it had connected to Laurasia during the Mid-Jurassic. We name this the ‘Silk road’ hypothesis (figure 3b, inset). Wherever the over-land dispersal took the liphistiid ancestors during this long evolutionary time, they (along with any branches they may have formed) must have repeatedly gone extinct along the way. Finding new fossil Mesothelae representatives from the Late Carboniferous to Eocene is therefore expected, but depending on the route taken, these fossils could be on either of the continental masses of Gondwana, Central and North Asia, Sibumasu or China. See electronic supplementary material for discussion on plausibility of these hypotheses and their predictions.
Liphistiidae diverged in the Palaeogene and diversified during the Neogene (figure 3a; electronic supplementary material figure S1 and table S3). The estimated diversification times and ancestral area reconstructions (figure 3a; electronic supplementary material figures S1 and S2, tables S3 and S5) suggest that the origins of Heptathela s.s. Qiongthela, and Ryuthela, the three genera exclusively inhabiting the southwestern Japanese islands and Hainan Island, predate the break-up of the islands form the mainland estimated at 15 Ma for Japan and approximately 2–2.5 Ma for Hainan [53,55,56,75–77]. Our results thus support Schwendinger's hypothesis [33], and refute the explanation of liphistiids dispersing onto Japanese islands over land bridges during the Pleistocene [9]. Likewise, the occurrence of Qiongthela on Hainan Island requires no ad hoc over-water dispersal events.
We can now also explain the absence of liphistiids from Taiwan. The island namely formed de novo about 9 Ma during the collision of the Philippine Sea plate with the Eurasian plate [78]. In accordance with their sedentary terrestrial life history and an over 298 Ma long evolutionary history of slow, over-land dispersal, the best explanation at hand is that liphistiids have never set their tarsi on Taiwan, not even during relatively short-lived land bridges that formed between the southern Japanese islands and the Asian mainland during the Pleistocene glacial cycles [79–82].
5. Conclusion
Our results combined with the fossil record indicate that Mesothelae originated in Euramerica in the Carboniferous and Permian boundary, and suggest that the period that followed was marked by a long eastward over-land dispersal of liphistiid ancestors towards the origin of Liphistiidae in Southeast or East Asia. The notable pattern seen is an up to 250-Ma-long lag between the stem of Mesothelae and the origin of extant Liphistiidae, combined with the occurrence of likely extinction events during the evolutionary history of the family. Since their origin in the Palaeogene Asia is relatively recent, we argue that liphistiids are modern spiders that happened to have retained certain characters considered plesiomorphic within spiders. As such, their labelling as living fossils is accurate from the phenotypical stance, however, their diversification is much more recent than expected.
Organismal dispersal abilities profoundly affect their diversification patterns [83] and biogeographic work benefits from identifying such lineages that due to their ancient origins and limited dispersal abilities retain their genetic imprint and biogeographic signal [84]. The family Liphistiidae should join other prominent litter or soil-dwelling spiders [40,85–88] and be considered an excellent model for biogeographic research on a par with selected few phylogenetically old terrestrial arthropod lineages, such as harvestmen [89], scorpions [90], centipedes [91] and velvet worms [84]. Both the reconstructed biogeographic history of Liphistiidae and their natural history namely suggest that these spiders are very poor and slow dispersers that, rather than ever move over-water or over short-lived land bridges, prefer to maintain their restricted ranges, or ride vicariantly on drifting continents.
Supplementary Material
Acknowledgements
We thank numerous people for local assistance, help and advice: Gary Ades, Paul Crow, Yorkie Wong and Zoie Wong in Hong Kong, Xianjin Peng, Luyu Wang, Bo Wu, Chengqiong Wu, Tingbang Yang and Zizhong Yang in China, Chu Thi Thao and Neuyen Thi Dinh in Vietnam, Zoltán Korsós, Mamoru Toda and Bo Wu in Japan. We thank Ingi Agnarsson, Danwei Huang, Wei Shong Hwang, Nina Vidergar and Matjaž Gregorič for kind help and/or advice on molecular resources and data analyses, Peter Jäger for providing information on collection locality in Laos, Gonzalo Giribet for commenting on an early draft, and Reta Mehta as well as two anonymous reviewers for their insightful comments. Finally, we thank the staff of the Centre for Behavioural Ecology and Evolution (Hubei University) and the Behavioural Ecology and Sociobiology Lab (National University of Singapore) for support, in particular, Zhanqi Chen, Seok Ping Goh, Xiaoguo Jiao, Hongze Li, Jie Liu, Yu Peng, Xiaoyan Wang, Chen Xu, Long Yu and Zengtao Zhang.
Data accessibility
DNA sequences can be accessed via GenBank (KP229800-KP230400); phylogenetic matrices can be accessed via Dryad: http://dx.doi.org/10.5061/dryad.b8d6m; and all other datasets supporting this study are made available as part of the electronic supplementary material.
Funding statement
This work was supported in part by the NSFC (grant no. 31272324) and the Singapore Ministry of Education (AcRF Tier 1 grant no. R-154-000-591-112) grants to D.L., and by the Slovenian Research Agency grant nos. P1-10236 and MU-PROM/12-001 to M.K.
Authors' contributions
D.L., M.K. and X.X. designed this study; D.L., M.K., F.X.L., H.O., D.S.P., Y.N-R., X.X., X.X. and Z.S.Z. conducted fieldwork; and X.X., R.-C.C., M.A.A., D.L. and M.K. performed data analyses. All the authors contributed to the writing, revising of the manuscript and gave final approval for publication.
References
- 1.Nagalingum NS, Marshall CR, Quental TB, Rai HS, Little DP, Mathews S. 2011. Recent synchronous radiation of a living fossil. Science 334, 796–799. ( 10.1126/science.1209926) [DOI] [PubMed] [Google Scholar]
- 2.Norstog K, Nicholls TJ. 1997. The biology of the cycads. Ithaca, NY: Comstock Pub. Associates. [Google Scholar]
- 3.Watson J, Cusack HA. 2005. Cycadales of the English Wealden. Palaeontogr. Soc. Monogr. 622, 1–189. [Google Scholar]
- 4.Inoue JG, Miya M, Venkatesh B, Nishida M. 2005. The mitochondrial genome of Indonesian coelacanth Latimeria menadoensis (Sarcopterygii: Coelacanthiformes) and divergence time estimation between the two coelacanths . Gene 349, 227–235. ( 10.1016/j.gene.2005.01.008) [DOI] [PubMed] [Google Scholar]
- 5.Bristowe WS. 1975. A family of living fossil spiders. Endeavour 34, 115–117. ( 10.1016/0160-9327(75)90130-1) [DOI] [Google Scholar]
- 6.Pocock RI. 1892. XXXVIII. - Liphistius and its bearing upon the classification of spiders. Ann. Mag. Nat Hist. Ser. 6 10, 306–314. ( 10.1080/00222939208677416) [DOI] [Google Scholar]
- 7.Platnick NI, Gertsch WJ. 1976. The suborders of spiders: a cladistic analysis (Arachnida, Araneae). Am. Mus. Nov. 2607, 1–15. [Google Scholar]
- 8.Coddington JA, Levi HW. 1991. Systematics and evolution of spiders (Araneae). Annu. Rev. Ecol. Syst. 22, 565–592. ( 10.1146/annurev.es.22.110191.003025) [DOI] [Google Scholar]
- 9.Haupt J. 2003. The Mesothelaea—monograph of an exceptional group of spiders (Araneae: Mesothelae) (morphology, behaviour, ecology, taxonomy, distribution and phylogeny). Zoologica 154, 1–102. [Google Scholar]
- 10.World Spider Catalog. 2015. World spider catalog. version 16 Bern, Switzerland: Natural History Museum; See http://wsc.nmbe.ch (accessed 13 April 2015). [Google Scholar]
- 11.Bristowe WS. 1976. A contribution to the knowledge of liphistiid spiders. J. Zool. Lond. 178, 1–6. ( 10.1111/j.1469-7998.1976.tb02260.x) [DOI] [Google Scholar]
- 12.Schiödte JC. 1849. Om en afigende sloegt af spindlernes orden. Naturhistorisk Tidsskr. 2, 617–624. [Google Scholar]
- 13.Schwendinger PJ, Ono H. 2011. On two Heptathela species from southern Vietnam, with a discussion of copulatory organs and systematics of the Liphistiidae (Araneae: Mesothelae). Rev. Suisse Zool. 118, 599–637. [Google Scholar]
- 14.Haupt J. 1983. Vergleichende Morphologie der Genitalorgane und Phylogenie der liphistiomorphen Webspinnen (Araneae: Mesothelae). I. Revision der bisher bekannten Arten. Z. Zool. Syst. Evol. Forsch. 21, 275–293. ( 10.1111/j.1439-0469.1983.tb00296.x) [DOI] [Google Scholar]
- 15.Haupt J. 1984. Comportement sexuel, morphologie génitale et phylogenèse des araignées liphistiomorphes. Rev. Arachnol. 5, 161–168. [Google Scholar]
- 16.Platnick N, Goloboff PA. 1985. On the monophyly of the spider suborder Mesothelae (Arachnida: Araneae). New York Entomol. Soc. 93, 1265–1270. [Google Scholar]
- 17.Kraus O, Kraus M. 1993. Divergent transformation of chelicerae and original arrangement of eyes in spiders (Arachnida, Araneae). Mem. Queensl. Mus. 33, 579–584. [Google Scholar]
- 18.Agnarsson I, Coddington JA, Kuntner M. 2013. Systematics—progress in the study of spider diversity and evolution. In Spider research in the 21st century (ed. Penney D.), pp. 58–111. Manchester, UK: Siri Scientific Press. [Google Scholar]
- 19.Bond JE, Garrison NL, Hamilton CA, Godwin RL, Hedin M, Agnarsson I. 2014. Phylogenomics resolves a spider backbone phylogeny and rejects a prevailing paradigm for orb web evolution. Curr. Biol. 24, 1765–1771. ( 10.1016/j.cub.2014.06.034) [DOI] [PubMed] [Google Scholar]
- 20.Fernández R, Hormiga G, Giribet G. 2014. Phylogenomic analysis of spiders reveals nonmonophyly of orb weavers. Curr. Biol. 24, 1772–1777. ( 10.1016/j.cub.2014.06.035) [DOI] [PubMed] [Google Scholar]
- 21.Sharma PP, Kaluziak ST, Pérez-Porro AR, González VL, Hormiga G, Wheeler WC, Giribet G. 2014. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol. Biol. Evol. 31, 2963–2984. ( 10.1093/molbev/msu235) [DOI] [PubMed] [Google Scholar]
- 22.Tanikawa A. 2013. Phylogeny and genetic variation in the spiders of the genus Ryuthela (Araneae: Liphistiidae). Acta Arachnol. 62, 41–49. ( 10.2476/asjaa.62.41) [DOI] [Google Scholar]
- 23.Selden PA. 1996. Fossil mesothele spiders. Nature 379, 498–499. ( 10.1038/379498b0) [DOI] [Google Scholar]
- 24.Selden PA. 1996. First fossil mesothele spider, from the Carboniferous of France. Rev. Suisse Zool. 2, 585–596. [Google Scholar]
- 25.McLoughlin S. 2001. The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Aust. J. Bot. 49, 271–300. ( 10.1071/BT00023) [DOI] [Google Scholar]
- 26.Scotese CR. 2002. Plate tectonic maps and continental drift animations. PALEOMAP Project. See www.scotese.com/earth.htm (accessed 11 October 2014).
- 27.Rogers JJW, Santosh M. 2003. Supercontinents in earth history. Gondwana Res. 6, 375 ( 10.1016/S1342-937X(05)70993-X) [DOI] [Google Scholar]
- 28.Metcalfe I. 2002. Permian tectonic framework and palaeogeography of SE Asia. J. Asian Earth Sci. 20, 551–566. ( 10.1016/S1367-9120(02)00022-6) [DOI] [Google Scholar]
- 29.Metcalfe I. 2011. Tectonic framework and Phanerozoic evolution of Sundaland. Gondwana Res. 19, 3–21. ( 10.1016/j.gr.2010.02.016) [DOI] [Google Scholar]
- 30.Seton M, et al. 2012. Global continental and ocean basin reconstructions since 200 Ma. Earth Sci. Rev. 113, 212–270. ( 10.1016/j.earscirev.2012.03.002) [DOI] [Google Scholar]
- 31.Metcalfe I. 2013. Gondwana dispersion and Asian accretion: tectonic and palaeogeographic evolution of eastern Tethys. J. Asian Earth Sci. 66, 1–33. ( 10.1016/j.jseaes.2012.12.020) [DOI] [Google Scholar]
- 32.Torsvik TH, Cocks LRM. 2013. Gondwana from top to base in space and time. Gondwana Res. 24, 999–1030. ( 10.1016/j.gr.2013.06.012) [DOI] [Google Scholar]
- 33.Schwendinger PJ. 2009. Liphistius thaleri, a new mesothelid spider species from southern Thailand (Araneae, Liphistiidae). Contrib. Nat. Hist. 12, 1253–1268. [Google Scholar]
- 34.Goloboff PA, Farris JS, Nixon KC. 2008. TNT, a free program for phylogenetic analysis. Cladistics 24, 774–786. ( 10.1111/j.1096-0031.2008.00217.x) [DOI] [Google Scholar]
- 35.Zwickl DJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, The University of Texas at Austin. [Google Scholar]
- 36.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 ( 10.1038/nmeth.2109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bond J, Hedin M, Ramirez M, Opell B. 2001. Deep molecular divergence in the absence of morphological and ecological change in the Californian coastal dune endemic trapdoor spider Aptostichus simus. Mol. Evol. 10, 899–910. ( 10.1046/j.1365-294X.2001.01233.x) [DOI] [PubMed] [Google Scholar]
- 39.Macías-Hernández NE, Oromí P, Arnedo MA. 2010. Integrative taxonomy uncovers hidden species diversity in woodlouse hunter spiders (Araneae, Dysderidae) endemic to the Macaronesian archipelagos. Syst. Biod. 8, 531–553. ( 10.1080/14772000.2010.535865) [DOI] [Google Scholar]
- 40.Bidegaray-Batista L, Arnedo MA. 2011. Gone with the plate: the opening of the Western Mediterranean basin drove the diversification of ground-dweller spiders. BMC Evol. Biol. 11, 317 ( 10.1186/1471-2148-11-317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Planas E, Ribera C. 2014. Uncovering overlooked island diversity: colonization and diversification of the medically important spider genus Loxosceles (Arachnida: Sicariidae) on the Canary Islands. J. Biogeogr. 41, 1255–1266. ( 10.1111/jbi.12321) [DOI] [Google Scholar]
- 42.Bidegaray-Batista L, Ferrández MÁ, Arnedo MA. 2014. Winter is coming: Miocene and Quaternary climatic shifts shaped the diversification of Western-Mediterranean Harpactocrates (Araneae, Dysderidae) spiders. Cladistics 30, 428–446. ( 10.1111/cla.12054) [DOI] [PubMed] [Google Scholar]
- 43.Brower AVZ. 1994. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl Acad. Sci. USA 91, 6491–6495. ( 10.1073/pnas.91.14.6491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Opatova V, Arnedo MA. 2014. From Gondwana to Europe: inferring the origins of Mediterranean Macrothele spiders (Araneae, Hexathelidae) and the limits of the family Hexathelidae. Invert. Syst. 28, 361–374. [Google Scholar]
- 45.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 ( 10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Y, Harris AJ, He XJ. 2010. S-DIVA (Statistical Dispersal-Vicariance Analysis): a tool for inferring biogeographic histories. Mol. Phylogenet. Evol. 56, 848–850. ( 10.1016/j.ympev.2010.04.011) [DOI] [PubMed] [Google Scholar]
- 48.Yu Y, Harris AJ, He XJ. 2014. RASP (reconstruct ancestral state in phylogenies) 3.0. See http://mnh.scu.edu.cn/soft/blog/RASP.
- 49.Nylander JAA, Olsson U, Alstrom P, Sanmartìn I. 2008. Accounting for phylogenetic uncertainty in biogeography: a Bayesian approach to dispersal–vicariance analysis of the thrushes (Aves: Turdus). Syst. Biol. 57, 257–268. ( 10.1080/10635150802044003) [DOI] [PubMed] [Google Scholar]
- 50.Ree RH, Smith SA. 2008. Maximum likelihood inference of geographic range evolution by dispersal local extinction and cladogenesis. Syst. Biol. 57, 4–14. ( 10.1080/10635150701883881) [DOI] [PubMed] [Google Scholar]
- 51.Xu X, Liu FX, Chen J, Ono H, Li D, Kuntner M. 2015. A genus level taxonomic revision of primitively segmented spiders (Mesothelae: Liphistiidae). Zookeys 488, 121–151. ( 10.3897/zookeys.488.8726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Metcalfe I. 1998. Palaeozoic and Mesozoic geological evolution of the SE Asian region: multidisciplinary constraints and implications for biogeography. In Biogeography and geological evolution of SE Asia (eds Hall R, Holloway JD.), pp. 25–41. Leiden, The Netherlands: Backhuys Publishers. [Google Scholar]
- 53.Otofuji Y, Matsuda T, Nohda S. 1985. Opening mode of the Japan Sea inferred from paleomagnetism of the Japan arc. Nature 317, 603–604. ( 10.1038/317603a0) [DOI] [Google Scholar]
- 54.Otofuji Y, Itaya T, Matsuda T. 1991. Rapid rotation of Southwest Japan: paleomagnetism and K-Ar ages of Miocene volcanic rocks of southwest Japan . Geophys. J. Intern. 105, 397–405. ( 10.1111/j.1365-246X.1991.tb06721.x) [DOI] [Google Scholar]
- 55.Yin A. 2010. Cenozoic tectonic evolution of Asia: a preliminary synthesis. Tectonophysics 488, 293–325. ( 10.1016/j.tecto.2009.06.002) [DOI] [Google Scholar]
- 56.Kimura M. 2000. Paleogeography of the Ryukyu Islands. Tropics 10, 5–24. ( 10.3759/tropics.10.5) [DOI] [Google Scholar]
- 57.Kimura M. 2002. The formation age of the Ryukyu Arc and migration of biota to the arc. Naha, Okinawa: Okinawa Times Co. [Google Scholar]
- 58.Kimura M. 2003. Land connections between Eurasian continent and Japanese islands—related to human migration. Mig. Diff. 4, 14–33. [Google Scholar]
- 59.Ota H. 1998. Geographic patterns of endemism and speciation in amphibians and reptiles of the Ryukyu Archipelago, Japan, with special reference to their paleogeographical implications. Res. Popul. Ecol. 40, 189–204. ( 10.1007/BF02763404) [DOI] [Google Scholar]
- 60.Ota H. 2000. The current geographic faunal patterns of reptiles and amphibians of the Ryukyu Archipelago and adjacent regions. Tropics 10, 51–62. ( 10.3759/tropics.10.51) [DOI] [Google Scholar]
- 61.Otsuka H, Takahashi A. 2000. Pleistocene vertebrate faunas in the Ryukyu Islands: their migration and extinction. Tropics 10, 25–40. ( 10.3759/tropics.10.25) [DOI] [Google Scholar]
- 62.Zheng HB, Clift PD, Wang P, Tada R, Jia JT, He MY, Jourdan F. 2013. Pre-Miocene birth of the Yantze River. Proc. Natl Acad. Sci. USA 110, 7556–7561. ( 10.1073/pnas.1216241110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bond JE, Hendrixson BE, Hamilton CA, Hedin M. 2012. A reconsideration of the classification of the spider infraorder Mygalomorphae (Arachnida: Araneae) based on three nuclear genes and morphology. PLoS ONE 7, e38753 ( 10.1371/journal.pone.0038753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ono H. 2000. Zoogeographic and taxonomic notes on spiders of the subfamily Heptathelinae (Araneae, Mesothelae, Liphistiidae). Mem. Natl Sci. Mus. 33, 145–151. [Google Scholar]
- 65.Paik K. 1953. A study on the geographical distribution of Heptathela kimurai Kishida. Acta Arachnol. 13, 53–68. ( 10.2476/asjaa.13.63) [DOI] [Google Scholar]
- 66.Haupt J. 2003. Zoogeography in southern Japan as revealed by ground-living arachnids. Rev. Suisse Zool. 110, 133–139. [Google Scholar]
- 67.Cox CB, Moore PD. 2005. Biogeography: an ecological and evolutionary approach, 7th edn Malden, MA: Blackwell Publishing. [Google Scholar]
- 68.Ali JR, Aitchison JC. 2005. Greater India. Earth Sci. Rev. 72, 169–188. ( 10.1016/j.earscirev.2005.07.005) [DOI] [Google Scholar]
- 69.Ali JR, Aitchison JC. 2006. Positioning Paleogene Eurasia problem: solution for 60–50 Ma and broader tectonic implications. Earth Planet. Sci. Lett. 251, 148–155. ( 10.1016/j.epsl.2006.09.003) [DOI] [Google Scholar]
- 70.Ali JR, Aitchison JC. 2008. Gondwana to Asia: plate tectonics, paleogeography and the biological connectivity of the Indian sub-continent from the Middle Jurassic through latest Eocene (166–35 Ma). Earth Sci. Rev. 88, 145–166. ( 10.1016/j.earscirev.2008.01.007) [DOI] [Google Scholar]
- 71.Torsvik TH, Cocks LRM. 2004. Earth geography from 400 to 250 Ma: a palaeomagnetic, faunal and facies review. J. Geol. Soc. Lond. 161, 555–572. ( 10.1144/0016-764903-098) [DOI] [Google Scholar]
- 72.Golonka J, Krobicki M, Pajak J, Giang NV, Zuchiewicz W. 2006. Phanerozoic palaeogeography of Southeast Asia. Geolines 20, 40–43. [Google Scholar]
- 73.Golonka J. 2007. Phanerozoic paleoenvironment and paleolithofacies maps. Mesozoic. Geologia 33, 211–264. [Google Scholar]
- 74.Clouse RM, Giribet G. 2010. When Thailand was an island—the phylogeny and biogeography of mite harvestmen (Opiliones, Cyphophthalmi, Stylocellidae) in Southeast Asia. J. Biogeogr. 37, 1114–1130. ( 10.1111/j.1365-2699.2010.02274.x) [DOI] [Google Scholar]
- 75.Zeng ZX, Zeng XZ. 1989. Physical geography of Hainan Island. Beijing, China: Science Press. [Google Scholar]
- 76.Zhao HT, Wang LR, Yuan JY. 2007. Origin and time of Qiongzhou Strait. Marine Geol. Quat. Geol. 27, 33–40. [Google Scholar]
- 77.Huang Y, Guo X, Ho SY, Shi H, Li J, Li J, Cai B, Wang Y. 2013. Diversification and demography of the oriental garden lizard (Calotes versicolor) on Hainan Island and the adjacent mainland. PLoS ONE 8, e64754 ( 10.1371/journal.pone.0064754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sibuet JC, Hsu SK. 2004. How was Taiwan created? Tectonophysics 379, 159–181. ( 10.1016/j.tecto.2003.10.022) [DOI] [Google Scholar]
- 79.Wageman JM, Hilde TMC, Emery KO. 1970. Structural framework of East China Sea and Yellow Sea. Am. Assoc. Pet. Geol. Bull. 54, 1611–1643. [Google Scholar]
- 80.Ye XQ. 1982. On the formation and development of the geology and geomorphology of Taiwan . J. Central China Teachers College 1982, 83–89. [Google Scholar]
- 81.Juan VCC. 1986. Thermal-tectonic evolution of the Yellow sea and East China Sea—Implication for transformation of continental to oceanic crust and marginal basin formation. Tectonophysics 125, 231–244. ( 10.1016/0040-1951(86)90016-8) [DOI] [Google Scholar]
- 82.Teng LS. 1992. Geotectonic evolution of Tertiary continental margin basins of Taiwan. Petrol. Geol. Taiwan 27, 1–19. [Google Scholar]
- 83.Agnarsson I, Cheng RC, Kuntner M. 2014. A multi-clade test supports the intermediate dispersal model of biogeography. PLoS ONE 9, e86780 ( 10.1371/journal.pone.0086780) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murienne J, Daniels SR, Buckley TR, Mayer G, Giribet G. 2014. A living fossil tale of Pangaean biogeography. Proc. R. Soc. B 281, 20132648 ( 10.1098/rspb.2013.2648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hendrixson B, Bond J. 2007. Molecular phylogeny and biogeography of an ancient Holarctic lineage of mygalomorph spiders (Araneae: Antrodiaetidae: Antrodiaetus). Mol. Phylogenet. Evol. 42, 738–755. ( 10.1016/j.ympev.2006.09.010) [DOI] [PubMed] [Google Scholar]
- 86.Hedin M, Starrett J, Hayashi C. 2013. Crossing the uncrossable: novel trans-valley biogeographic patterns revealed in the genetic history of low-dispersal mygalomorph spiders (Antrodiaetidae, Antrodiaetus) from California. Mol. Ecol. 22, 508–526. ( 10.1111/mec.12130) [DOI] [PubMed] [Google Scholar]
- 87.Opatova V, Bond JE, Arnedo MA. 2013. Ancient origins of the Mediterranean trap-door spiders of the family Ctenizidae (Araneae, Mygalomorphae). Mol. Phylogenet. Evol. 69, 1135–1145. ( 10.1016/j.ympev.2013.08.002) [DOI] [PubMed] [Google Scholar]
- 88.Wood HM, Matzke NJ, Gillespie RG, Griswold CE. 2013. Treating fossils as terminal taxa in divergence time estimation reveals ancient vicariance patterns in the palpimanoid spiders. Syst. Biol. 62, 264–284. ( 10.1093/sysbio/sys092) [DOI] [PubMed] [Google Scholar]
- 89.Giribet G, Sharma PP, Benavides LR, Boyer SL, Clouse RM, de Bivort BL, Kawauchi GY, Murienne J, Schwendinger PJ. 2012. Evolutionary and biogeographic history of the harvestman suborder Cyphophthalmi (Arachnida, Opiliones)—an ancient and global group of arachnids. Biol. J. Linn. Soc. 105, 92–130. ( 10.1111/j.1095-8312.2011.01774.x) [DOI] [Google Scholar]
- 90.Bryson RW, Jr, Savary WE, Prendini L. 2013. Biogeography of scorpions in the Pseudouroctonus minimus complex (Vaejovidae) from south-western North America: implications of ecological specialization for pre-Quaternary diversification. J. Biogeogr. 40, 1850–1860. [Google Scholar]
- 91.Giribet G, Edgecombe GD. 2012. Reevaluating the arthropod tree of life. Annu. Rev. Entomol. 57, 167–186. ( 10.1146/annurev-ento-120710-100659) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences can be accessed via GenBank (KP229800-KP230400); phylogenetic matrices can be accessed via Dryad: http://dx.doi.org/10.5061/dryad.b8d6m; and all other datasets supporting this study are made available as part of the electronic supplementary material.