Abstract
The transfer of genes between populations is increasingly important in a world where pollinators are declining, plant and animal populations are increasingly fragmented and climate change is forcing shifts in distribution. The distances that pollen can be transported by small insects are impressive, as is the extensive gene flow between their own populations. We compared the relative ease by which small insects introduce genetic markers into their own and host-plant populations. Gene flow via seeds and pollen between populations of an Asian fig species were evaluated using cpDNA and nuclear DNA markers, and between-population gene flow of its pollinator fig wasp was determined using microsatellites. This insect is the tree's only pollinator locally, and only reproduces in its figs. The plant's pollen-to-seed dispersal ratio was 9.183–9.437, smaller than that recorded for other Ficus. The relative effectiveness of the pollinator at introducing markers into its own populations was higher than the rate it introduced markers into the plant's populations (ratio = 14 : 1), but given the demographic differences between plant and pollinator, pollen transfer effectiveness is remarkably high. Resource availability affects the dispersal of fig wasps, and host-plant flowering phenology here and in other plant–pollinator systems may strongly influence relative gene flow rates.
Keywords: Agaonidae, Ficus, gene flow, insect dispersal, seed dispersal, Slatkin's paradox
1. Introduction
Dispersal between populations plays a vital role in shaping the genetic structure of flowering plant populations. As a cohesive force that unites individual plant species into real evolutionary units [1], dispersal is of great interest amid rising concerns about the persistence of populations within increasingly fragmented landscapes. Gene flow is usually achieved via dispersal of seeds and pollen [2], but dispersal of pollen is almost always more significant than gene flow mediated by movements of seeds [3], except at small spatial scales, e.g. [4]. In addition to reducing overall among-population differentiation, dispersal of pollen between populations can also introduce new genes, and thereby rescue declining populations by reducing inbreeding depression and promoting offspring fitness [5]. Maintenance of inter-population pollen transfer should, therefore, be considered when drafting long-term management strategies for plants in fragmented habitats or facing declines in pollinators [6].
Insects are the sole pollen vectors of many flowering plants, especially in tropical and subtropical regions [7]. The foraging behaviour of the insects that visit their flowers determines which species can act as pollinators, how much pollen they collect and how far the pollen can be transferred [6,8]. Dispersal kernels of insects, and pollen flow mediated by them, have traditionally been expected to be left skewed, with most individuals dispersing over short distances and gene flow between populations being the result of rare long-distance dispersal events. Direct observations of insect movements are difficult, especially if they are small, and impractical for recording rare long-distance dispersal [9], but molecular markers have made the detection of these rare events much easier. Average pollination distances of hundreds of metres are reported [10], and are particularly long among some tropical trees [11,12], where paternity analysis has detected examples of pollen flow between trees growing tens or even hundreds of kilometres apart [8,13].
The distances that pollinators travel is only one aspect of inter-population pollen transfer. The quantities of pollen that they collect and subsequently deposit on appropriate flowers are equally important [14], and the latter may vary according to how far an insect has dispersed. Insects generally acquire and deposit pollen passively during sequences of visits to flowers. In general, longer times between floral visits, or more intervening floral visits, will result in fewer pollen grains being deposited, due to grooming behaviour and abrasion [15]. Insects that have dispersed longer distances may also be weaker, less active and less likely to deposit the pollen they carry. Consequently, insects that have travelled further are likely to deposit less pollen than more locally dispersing individuals.
Insect dispersal also contributes to gene flow between their own populations. Realized gene flow among populations of small insects is often high, and in contradiction to the apparently localized movements of individual insects [16]. This apparent contradiction (Slatkin's paradox) may have been resolved because there is increasing evidence that small flying insects can disperse over large distances [8,9,17,18]. Much of this evidence is based on analysis of the pollen that the insects are carrying, and in the same way as transportation of pollen between populations does not necessarily ensure seed set, so the fecundity of insects after they have dispersed long distances may be reduced [19]. In the case of pollinating insects, any declines in their ability to reproduce after dispersal need not necessarily be proportionate to changes in their ability to pollinate, so assessments of pollen flow between plant populations do not necessarily reflect the extent of gene flow between populations of their pollinators.
Identification of plant offspring that result from between-population pollination events allows the extent and direction of gene flow between populations to be estimated using Bayesian approaches (e.g. [20]), but the likelihood that pollen grains carried between populations will result in the addition of new genes into plant populations has not been estimated quantitatively. This is because we do not know how many insects entered focal populations, how much of the appropriate pollen they carried and how much they deposited on appropriate stigmas. Also, most plants are pollinated by more than one insect species, each of which will have differing relative contributions to pollen transfer that are probably to vary in space and time.
Here, we combine information derived from between-population gene flow in a plant and in its host-specific unique pollinator to determine the relative effectiveness of gene flow in the two species. Our verbal definition of pollinator effectiveness for dispersing insects moving between populations is the ratio of genetic markers introduced and becoming established in a pollen vector's population compared with the markers that it introduces and that become established in host-plant populations via the pollen it carried. Estimates of pollen-mediated gene flow between populations of fig trees can be obtained by comparing bi-parentally and uni-parentally inherited markers (reflecting pollen and seed inheritance, respectively) [21], and gene flow among their pollinators can be estimated using bi-parentally inherited markers [22]. In combination, these allow the relative effectiveness of gene flow in fig trees and fig wasps to be estimated quantitatively. Because of their strongly contrasting generation times, we hypothesize that pollinators disperse their own genes far more readily than plant genes and that the relative effectiveness of gene flow should be much smaller than 1.
To test the above hypothesis, we assessed pollinator effectiveness in a fig species (Ficus, Moraceae). Each fig species is exclusively pollinated by one or a small number of species of host-plant-specific fig wasps (Agaonidae) that enter the trees' globular inflorescences (figs) in order to lay their eggs [23]. Pollinating fig wasps are short-lived, weak-flying insects, but paternity analyses and population structuring of their host populations suggest that whereas some species disperse locally [24], others disperse across much longer distances [8,13,25], initially using fast-flowing air to transport them passively in whichever direction it is moving [26,27].
In this study, the focal plant species is an Asian fig, Ficus pumila. Firstly, we estimate pollen flow between populations by comparing its genetic structure based on cpDNA and nuclear DNA markers. Then we estimate gene flow of its pollinating fig wasp Wiebesia pumilae using nuclear microsatellites. Finally, we calculate the relative effectiveness of the pollinator at introducing genes into its own populations and those of its host plant.
2. Material and methods
(a). Study system
Ficus pumila L. is a functionally dioecious creeping fig tree that grows on trees and walls. It is widely distributed in subtropical China. The large, pear-shaped figs contain thousands of tiny female flowers. Figs of female individuals produce only seeds, whereas figs on male plants support development of fig wasp offspring [28]. Foundress females of the pollinator fig wasp W. pumilae Hill enter the figs to lay their eggs, but cannot reproduce if they enter a female fig. Their wings are removed on entry into the figs and once they enter a fig they do not re-emerge. Usually several females enter each receptive fig. Female F. pumila produce one crop of figs each year, pollinated in spring and early summer. Male trees generally produce two crops a year with a spring/early summer maturing crop that releases the fig wasps that pollinate female trees, and a second crop that matures in summer/autumn [29]. The male figs that release adult fig wasps in late spring contain large numbers of dehiscent male flowers that release pollen that covers the fig wasps before they emerge. Conversely, adult fig wasps released from their natal figs in late summer disperse at a time when there are no receptive female figs to enter, and their natal male figs produce no pollen. Using microsatellites, moderate levels of genetic diversity and low between-population differentiation have been recorded in F. pumila populations growing in fragmented landscapes, suggesting moderate to high gene flow among populations, including those located on different islands [30].
Ficus pumila supports three closely related and largely allopatrically distributed Wiebesia pollinators in China [28]. Unlike many fig wasps, Wiebesia species are passive pollinators that do not actively collect and disperse pollen. Based on the fine-scale spatial genetic structure of a F. pumila population, Wang et al. [31] inferred that its pollen is dispersed further than its seeds, and is routinely carried further than 1 km. Wiebesia pumilae (Wiebesia sp. 2 of Chen et al. [28]) is the only pollinator of F. pumila in South China. A single W. pumilae female that enters a female fig of F. pumila results in the production on average of 1000 seeds [32]. If she enters a male fig she can produce around 500 offspring [32], but most figs are entered by several foundresses (up to 10 or more), and competition for oviposition sites together with interference between females reduces the numbers of eggs that each female can lay.
(b). Collections of Ficus pumila and its pollinating wasps
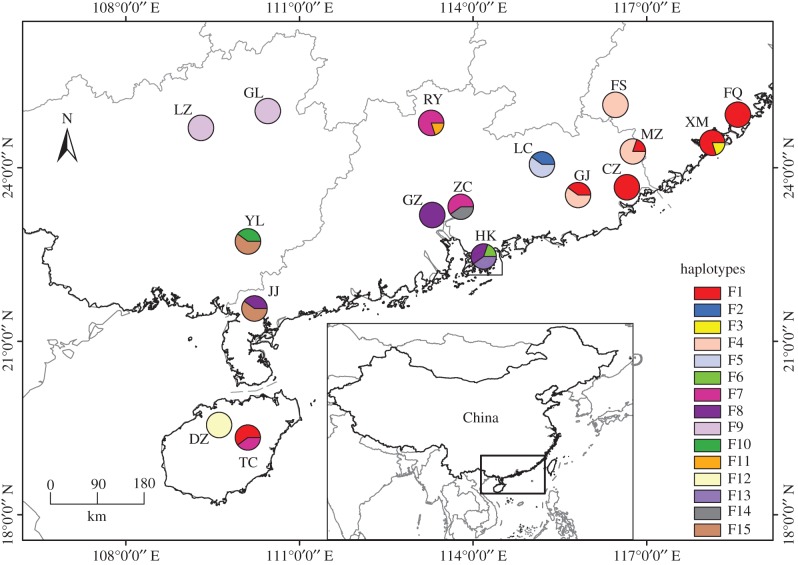
Although its three associated fig wasps are mostly distributed allopatrically, there are some areas of overlap, so we focused our study in South China, where only W. pumilae is present [28]. A total of 17 populations, separated by up to 1100 km, were sampled (figure 1). Between seven and 27 plant individuals were sampled in each population, with each plant separated by at least 30 m to avoid repeat-sampling of the same individuals. About five healthy leaves were collected from each plant and dried using silica gel. Fig wasps were collected from male trees by placing mature figs that did not have exit holes into netting bags and letting the adult fig wasps emerge naturally. The fig wasps were stored in absolute ethanol at 4°C.
Figure 1.
Locations of Ficus pumila sample sites in South China and the distribution of its cpDNA haplotypes. Populations names are abbreviated to two letters, and haplotypes are represented by different colours. (Online version in colour.)
(c). Analyses of microsatellites and cpDNA sequencing in Ficus pumila
Total genomic DNA of F. pumila was extracted from about 30 mg of leaves dried in silica gel, using a Plant Genomic DNA Kit (Tiangen, Beijing, China). Eight nuclear microsatellite loci (FP9, FP38, FP102, FP134, FP213, FP540, FP556 and FP601) were genotyped using fluorescently labelled PCR primers as described by Zhang et al. [33]. The amplification products were mixed into two groups (group 1: FP9, FP134, FP213, FP556; group 2: FP38, FP102, FP540, FP601), and each mixture was scanned on an ABI 3730 Automated DNA Sequencer (Applied Biosystems, Foster City, CA, USA). Allele sizes were scored using PEAKSCANNER (Applied Biosystems).
For chloroplast DNA of F. pumila, three noncoding regions, trnS-trnG [34], atpF-atpH [35] and trnC-ycf6 [36] were amplified in a volume of 50 μl, which included approximately 60 ng of genomic DNA, 0.2 mM dNTPs, 0.2 uM of each primer, 1 × PCR buffer, 2 mM Mg2+ and 0.4 U of DNA Taq polymerase (Sangon), under the following conditions: 5 min denaturation at 94°C; 35 cycles of 45 s at 94°C, 45 s at 58°C, 1 min at 72°C; and a final extension of 72°C for 8 min. We also amplified the three cpDNA fragments of F. sarmentosa var. henryi (the most closely related species in the study region) and two out-group species, F. pubigera and F. erecta. PCR products were cleaned and sequenced in both directions on an ABI 3730 DNA Sequence Analyzer.
(d). Microsatellite analyses of Wiebesia pumilae
Genomic DNA of the pollinating wasps was isolated from whole bodies of single females using the modified method of Sambrook et al. [37]. Genotyping was carried out using 10 microsatellite primers developed previously [38] with 5′-labelled with fluorescent dye on the forward primer. The PCR amplification was performed in a volume of 10μl. The amplification products were combined into three mixtures (mixture 1: WP447 (6-FAM), WP294 (ROX) and WP076 (6-FMA); mixture 2: WP403 (ROX), WP554 (TAMRA), WP399 (HEX) and WP231 (6-FAM); mixture 3: WP522 (6-FAM), WP439 (HEX) and WP004 (6-FAM)), and each mixture was scanned on an ABI 3730 Automated DNA Sequencer. Allele sizes were scored using PEAKSCANNER.
(e). Analyses of genetic structure
For nSSRs of the plant and its pollinator, tests for deviation from Hardy–Weinberg equilibrium (HWE) were performed with genepop v. 4.0 [39] using exact tests followed by sequential Bonferroni corrections [40]. Linkage disequilibrium (LD) among loci per population was conducted using FSTAT v. 2.9.3 [41]. Genetic diversity was estimated using the following parameters: mean number of alleles per locus (NA), allelic richness per locus (AR, correcting for sample size to the minimal sample size), observed (HO) and unbiased expected heterozygosities (HE). These analyses were performed using FSTAT and TFPGA [42]. Population genetic differentiation FST(n) [43] was evaluated based on all loci using FSTAT. Isolation-by-distance patterns in F. pumila and its pollinator were tested by using Mantel tests with the R package ‘vegan’ [44].
For cpDNA of F. pumila, sequences (GenBank accession numbers: KJ576907–KJ576923) were aligned using Clustal w, implemented in MEGA v. 4.0 [45]. DnaSP [46] was used to count the number of haplotypes. Population differentiation was estimated by calculating FST(c) with 1000 permutations in Arlequin v. 3.11 [47]. The phylogenetic tree was constructed by the maximum-likelihood approach using PHYML v. 3.0 [48]. The appropriate nucleotide substitution model (TPMuf + I) was chosen by JMODELTEST v. 2.1.5 [49] based on AIC criterion. Node support was estimated with 100 bootstrap replicates.
A Bayesian approach to infer population structure of F. pumila was performed in structure 2.3.1 [50]. We ran the admixture model with correlated frequencies, and 10 independent runs for each K (from 1 to 10) were performed with 100 000 MCMC repetitions and a burn-in of 10 000. We used LnP(D), the posterior probability of the data for a given K, to identify the most probable number of clusters using ΔK values [51]. After the best K was chosen, all individuals were assigned to the K populations probabilistically by using a burn-in of 300 000 and 1 000 000 MCMC repetitions.
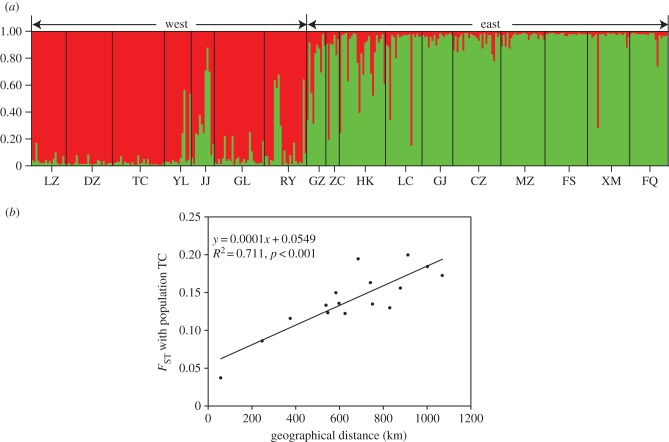
The structure analysis divides individuals into at least two clusters, even if all individuals belong to a single panmictic population. F. pumila populations showed latitudinal and longitudinal gradients in genetic composition, which might be the output of contact of two genetic clusters or caused by dispersal, given the neutral markers used in this study. To infer the potential cause and the most likely direction of dispersal [52], we tested the relationship between genetic and spatial distances to the most southern (population TC), most northern (population FS), most western (population LZ) and most eastern population (FQ) using a linear regression in R [53].
(f). Estimation of pollinator effectiveness
We defined pollinator effectiveness (PE) using the following equation
| 2.1 |
where Nmp is pollen gene flow (number of pollen grains per generation) of the plant, Nmi is gene flow (number of individuals per generation) of the pollinating insect, Lgp is generation length (years) of the plant and Lgi is the generation length (years) of the pollinating insect. Generation lengths (to reaching maturity) of F. pumila and W. pumilae average about 10 and 0.5 years, respectively (R Zhao and XY Chen 2010, unpublished data). However, fig wasps of the summer generation can themselves reproduce, but do not pollinate female figs. That means that the insect spreads its genes twice a year, but only spreads the plant genes once a year. Thus, we applied a value of 1 per year instead of 0.5 years per generation in this specific case.
To estimate pollinator effectiveness, we have to obtain gene flow of the pollinating insect (Nmi) and pollen-mediated gene flow (Nmp). Under the assumptions of Wright's [22] infinite island model of population structure, we can estimate Nmi from the fixation of alleles among populations of the pollinating wasp.
For parentally inherited markers, such as nuclear DNA allozymes or microsatellites, fixation index and gene flow in plant species have the following relationship [22]
where Nms and Nmp are seed and pollen gene flow, respectively.
In most angiosperms, Nms can be estimated using maternally inherited markers, such as cpDNA markers. For dioecious plants with a 1 : 1 breeding sex ratio, the relationship between cpDNA genetic differentiation (FST(c)) and seed gene flow can be expressed as: FST(c) = 1/(Nms + 1) [54]. Based on the above equations, pollen-mediated gene flow can then be estimated using
| 2.2 |
Owing to their extreme polymorphism, genetic differentiation estimates based on microsatellites are generally underestimates [55], and produce overestimates of gene flow. However, F. pumila and W. pumilae both have moderate genetic variation and display similar FST values, so biases in estimations of gene flow should be low. The estimated gene flow values were also slightly lower than those obtained using a private allele approach [56] in Genepop, which again suggests that any biases were weak.
To check whether pollinator effectiveness PE was related to distance, we estimated pairwise PE based on pairwise differentiation between populations, and tested its relationship with spatial distance.
We also estimated the pollen-to-seed dispersal ratio in F. pumila. Assuming a low rate of seed migration, for dioecious plants with a 1 : 1 sex ratio, the pollen-to-seed dispersal ratio (r) can then be estimated by Ennos' [21] method
| 2.3 |
3. Results
Diagnostic loci confirmed that all the fig wasps in the study populations were W. pumilae (=Wiebesia sp. 2). In total, 331 F. pumila and 316 W. pumilae were genotyped using microsatellite loci. In F. pumila, deviation from HWE was found at two loci (FP9 in populations RY and LC; FP134 in populations TC, CZ, MZ and FS). No LD was observed. In W. pumilae, four loci were found to deviate from HWE (WP447 in XM; WP294 in FQ; WP076 in LZ; WP399 in DZ, RY, LC, GJ). No LD was detected among W. pumilae populations.
The mean number of alleles (NA) across all eight loci in populations of F. pumila ranged from 3.6 to 7.0 with a mean of 5.4. Allelic richness (A) was lowest in population FS (3.1) and highest in population JJ (5.2). Mean observed heterozygosity (HO) ranged from 0.50 to 0.80, with an average of 0.63. The expected heterozogosity per population (HE) was between 0.55 and 0.72, with an average of 0.66 (table 1). A total of 15 chloroplast haplotypes were found in the 17 populations of F. pumila, with the Hong Kong population having the most haplotypes (figure 1). The ML tree indicated that F. pumila haplotypes were clustered together as a sister clade to F. sarmentosa var. henryi (electronic supplementary material, figure S1), suggesting no cytoplasm transfer from other local Ficus species.
Table 1.
Sampling information and genetic diversity of populations of Ficus pumila and its specific pollinating wasp Wiebesia pumilae. # loci, number of loci; n, sample size; NA, number of alleles per locus; A, allelic richness; HO, observed heterozygosity; HE, expected heterozygosity; FST(n), nuclear DNA microsatellite-based fixation index; # hap., number of haplotypes; FST(c), cpDNA haplotype-based fixation index. Numerals in parentheses are ranges of values except those of FST(n). Means are presented ± s.d.
| Ficus pumila | Wiebesia pumilae | |
|---|---|---|
| nDNA SSRs | ||
| # loci | 8 | 10 |
| n | 19 ± 6 (7–27) | 19 ± 8 (8–30) |
| NA | 5.4 ± 0.9 (3.6–7.0) | 6.4 ± 1.5 (2.8–7.9) |
| A | 4.3 ± 0.5 (3.1–5.2) | 5.2 ± 0.8 (2.8–5.9) |
| HO | 0.63 ± 0.08 (0.50–0.79) | 0.67 ± 0.06 (0.58–0.76) |
| HE | 0.66 ± 0.04 (0.55–0.72) | 0.72 ± 0.07 (0.49–0.80) |
| FST(n) | 0.123 (95% CI: 0.099–0.151) | 0.059 (95% CI: 0.048–0.071) |
| cpDNA | ||
| # hap. | 15 | / |
| FST(c) | 0.750 ** | / |
**p < 0.001.
In populations of W. pumilae, NA was between 2.8 and 7.9 with an average of 6.4. HO and HE ranged from 0.58 to 0.76 and 0.49 to 0.80, respectively. Allelic richness was lowest in population GZ (2.8), and highest in LZ (5.9) (table 1).
Mantel tests revealed a pattern of isolation-by-distance in populations of F. pumila (r = 0.527, p < 0.001) (figure 2), but not in its pollinator (r = 0.152, p = 0.149). The structure analysis indicated a gradient in genetic composition of F. pumila populations (figure 3a). A significant positive relationship between genetic and spatial distances was found to the most southern (r2 = 0.711, p < 0.001; figure 3b), northern (r2 = 0.371, p = 0.007), western (r2 = 0.581, p < 0.001) and eastern (r2 = 0.349, p = 0.009) populations, suggesting that dispersal other than secondary contact of two genetic clusters played a critical role in shaping genetic structure of these populations of F. pumila. The coefficient of determination for the relationship between genetic and spatial distances was highest to the most southern population TC, and southern populations were located in the west of the studied region, hinting that a most likely dispersal pattern was first from Hainan Island (populations TC and DZ) to the mainland and then from the west to the east.
Figure 2.
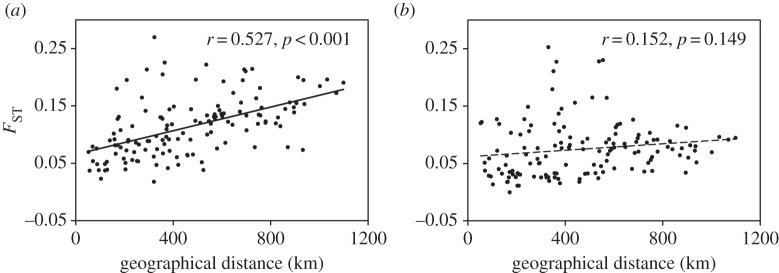
The relationships between genetic differentiation and geographical distance in South China populations of (a) Ficus pumila and (b) Wiebesia pumilae.
Figure 3.
(a) Genetic clusters of individuals from 17 Ficus pumila populations assigned by STRUCTURE. The red columns indicate the western group, and the green columns the eastern group. The populations (left–right) are arranged from west to east. (b) A linear regression between geographical distances from the most southern population of Ficus pumila (TC) and the genetic differences of these populations from population TC.
Based on nuclear variation, the populations of F. pumila were moderately differentiated, with a fixation index (FST(n)) of 0.123 (95% CI: 0.099–0.151) and a calculated gene flow (Nm) of 1.783 individuals per generation, which was smaller than that estimate based on the frequencies of private alleles (3.282). Large differentiation in cpDNA was observed among populations (FST(c) = 0.750, p < 0.001). Based on differentiation between cpDNA and nuclear DNA variation, we obtained values for the pollen-to-seed dispersal ratio (r) of 9.183 and 9.437 when FST(n) was estimated by FST and RST, respectively.
Low levels of genetic differentiation were found among populations of the pollinator (FST(n) = 0.059, 95% CI: 0.048–0.071). Gene flow between populations (Nmi) was estimated to be 3.987 individuals per generation. This value was slightly lower than that estimated from private alleles (4.688).
Pollen-mediated gene flow (Nmp) between populations was estimated at 2.898 pollen grains per generation. From equation (2.1), inter-population pollinator effectiveness was calculated to be 0.0727, meaning that for every 13.8 pollinating wasps from outside populations that successfully introduced markers into its own populations, one marker was introduced into populations of F. pumila, via the pollen that it carried. PE was 0.0959 and 0.0989 within the eastern and western population clusters, respectively, much larger than that between the two clusters (0.0205). A slight but non-significant decline in PE was present as spatial distances between populations increased (electronic supplementary material, figure S2).
4. Discussion
(a). Dispersal in Ficus pumila and its pollinating wasps
Pollinating fig wasps play an important role in transferring their hosts' genes. However, the wasps are weak fliers and their long-distance dispersal depends on their ability to use the wind. Most dioecious fig trees are understory species and remain below the canopy, where wind speed is very slow [57]. Thus, strong genetic structure was expected in dioecious fig trees and their pollinating wasps [57], as has been found in another dioecious creeper in China [24]. However, F. pumila is a creeper that can approach the forest canopy, or cover rocks or abandoned walls. This will allow its pollinating wasps to more easily make use of the wind to disperse over long distances. Genetic differentiation is low among South Chinese W. pumilae populations separated by up to 1100 km, confirming that the wasps disperse widely between populations. Genetic differentiation of the host F. pumila was also not large over this wide range. Further north, F. pumila is pollinated by a different Wiebesia species, which displays similarly extensive dispersal between populations [30]. Clearly, both of these pollinators disperse the pollen of F. pumila over wide areas.
Our result is consistent with those from monoecious figs, most of which are canopy trees or forest–canopy hemi-epiphytes. For example, the pollinator of monoecious F. racemosa showed limited genetic structure across a 1600 km expanse of continental southeast Asia [58]. A weaker dispersal ability has been inferred among the pollinating wasps associated with some dioecious figs, based on their rates of recovery after local extinctions. In 1998, an El Niño event resulted in an absence of figs on the trees and the consequent local extinction of pollinators of fig trees at Lambir Hills National Park, Sarawak, Malaysia, Borneo. Several fig wasp species had recolonized within 1 year, but recovery of pollinators associated with monoecious species was more rapid [59]. Elsewhere, a relatively continuous distribution of high-density populations may be responsible for the dioecious understory species F. hirta having extensive pollen dispersal across its range, as shown by its populations’ weak genetic differentiation [60].
Extreme events such as droughts, hurricanes and harsh winters can lead to the local extinction of fig wasp populations, while at the same time leaving host-plant populations intact [14,57,59,61]. Similar extreme events, especially if repeated, would disengage the genetic structuring of the pollinator populations from those of their host plants. If the wasps can disperse to long distances, such events reduce the genetic structuring of pollinator populations, relative to those of their hosts. Alternatively, strong genetic structure will be observed in the fig wasp populations due to bottlenecks or founder effects resulting from a small number of colonizers. Dramatic environmental events are not infrequent in South China and most years there are typhoons that could cause large fluctuations in the sizes of W. pumilae populations. High inter-population dispersal of W. pumilae is evident because its populations are less differentiated (FST = 0.059) than those of its host (FST(n) = 0.123).
Movements of pollinators, in combination with seed dispersal, determine gene flow between the plants they visit. Microsatellites are often assumed to overestimate gene flow [55], but our estimates based on genetic differentiation in F. pumila populations were lower than estimates using private alleles, suggesting that they are not inflated. The fruit bats and birds that eat ripe figs of F. pumila [62,63] are capable of dispersing fig seeds over long distances [64]. Our estimates of the relative contribution of pollen and seeds to gene flow in F. pumila (9.183–9.437) is less than half of that recorded for another dioecious fig tree, F. hirta [17]. They are also lower than those recorded for most other plants, where a median value of 17 was reported by Petit et al. [3]. Nevertheless, the pollen-to-seed dispersal ratio shows that the nuclear genome is less structured than the cytoplasmic genomes, as was indicated previously by a study of the plant's fine-scale spatial genetic structure, which concluded that seed dispersal in an area elsewhere in the plant's range was mainly within a radius of 1 km [31].
(b). Pollinator effectiveness
The extensive dispersal displayed by Wiebesia species is achieved despite the limitations imposed by their short adult lifespans and low flight speeds [9]. Long-distance dispersal events may be a feature of many such small insects, not just fig wasps [18,65,66] and provide a likely explanation for ‘Slatkin's paradox’, that direct observations of insect dispersal underestimate their potential to generate gene flow [8,17]. In the case of fig wasps, where they are the sole dispersers of their host's pollen, gene flow among the insect and plant populations is intimately linked.
Genetic studies of plant populations can provide estimates of the proportion of seeds or seedlings sired by pollen originating from outside focal populations, but give no indication of how many pollinators were responsible for moving the pollen. Partially consistent with our initial hypothesis, our comparison of the relative abilities of a fig wasp to introduce markers that become established in its own and into its host-plant's populations showed that markers are introduced more readily into the insect's populations. For every 14 insects that dispersed between populations and successfully introduced genetic markers into their own populations, one pollen grain successfully introduced markers into the plant's populations. Pollen is haploid, whereas eggs that result in female offspring are diploid, which should favour the introduction of pollinator markers. No significant relationship was found between pairwise pollinator effectiveness and spatial distance between populations as a whole or within each of the two population clusters, indicating that inter-population pollinator effectiveness was not influenced by the distances between populations. Fig wasps can use fast-flowing winds for long-distance dispersal, and variation in wind speed and direction may make variation in the distances the wasps are carried insignificant.
Although W. pumilae introduces markers into its own populations at a higher rate than it transfers markers into populations of its host, its pollinator effectiveness can nonetheless be seen as being remarkably high, given the differences in demography between the fig tree and its pollinator. As in most plant species, the vast majority of seeds produced by F. pumila, including those sired by pollen from other populations, must fail to become established plants [67]. By contrast, female fig wasps that have successfully entered a male fig have a much better chance of producing adult offspring that can themselves reproduce.
Factors that might be responsible for a lowered relative effectiveness of introducing markers into the pollinator's own populations include a greater likelihood that those W. pumilae that have dispersed long distances will enter female, rather than male figs. Between-population pollen flow only takes place in late spring because there is only one crop of female figs each year. Gene flow between its pollinator populations will be mainly in late summer, because very few receptive male figs are produced in spring. Any factors that favour more long-distance dispersal in late summer rather than spring will therefore favour gene flow between plant populations. Wind speeds in the region do not differ consistently between these two seasons, so ease of dispersal is unlikely to be responsible. The ‘selfish’ fruiting phenology of F. pumila provides a more likely explanation, because it results in fig wasps that emerge from figs in spring having to leave their natal male trees and make themselves liable to undertake long-distance dispersal. This is because those individuals that emerge from figs in spring find themselves on male trees where few if any receptive figs are present, so their only chance for reproduction is if they disperse in search of figs on other trees. Given that the reproductive success of the male plants depends on the fig wasps entering figs on female trees, this is clearly advantageous for the male plants. By contrast, pollinators that emerge from figs in autumn will often find receptive figs on their natal male trees and dispersal from these trees will be unnecessary. There are no female figs to pollinate at this time, so fig wasp populations are increased on their natal trees, ready to emerge the following spring, which is again to the tree's advantage, but reduces the likelihood that the fig wasps will undertake long-distance flights. This effect may be further increased because those fig wasps that do disperse and successfully reach a fig on a non-natal male tree may be late-arrivals and face greater competition for oviposition sites from more locally-dispersed individuals. Those fig wasps that have dispersed long distances are also likely to be weaker than others, and capable of laying fewer eggs, even in figs where there is no competition for oviposition sites. Pollination is achieved when the insects walk around the inside of a fig, whereas egg laying involves energetically demanding repeated probing down the styles of each flower where an egg is laid. Consequently, the rigours of long-distance flight are likely to impact more on oviposition rates than pollination rates.
Slatkin's paradox reflects a surprising extent of gene flow among populations of small insects, given their apparently poor dispersal abilities. Our results have generated a somewhat contradictory paradox, namely that the extent of dispersal evident from a small insect's movement of plant markers was not reflected to the expected extent in the dispersal of its own genes. We have suggested that manipulation of the pollinators' dispersal behaviour by their host plant is largely responsible for this apparent anomaly in our study species, but comparative studies of pollination effectiveness in other systems are required before any general conclusions can be reached. Nonetheless, our study emphasizes that caution is required when using plant population structure to infer the behaviour of their pollen vectors.
Supplementary Material
Supplementary Material
Acknowledgements
The authors thank Jaco Greeff, Xin-Sheng Hu, Da-Yong Zhang and Yan Chen for constructive suggestions and discussions on the paper. Anonymous referees significantly enhanced the final version of the manuscript.
Data accessibility
The data used in this paper can be accessed via Dryad: doi:10.5061/dryad.g50b0.
Funding statement
This work was funded by the National Natural Sciences Foundation of China (31270416), the Doctoral Program of the Ministry of Education of China (20110076110013) and the Fundamental Research Funds for the Central Universities (78220028) to X.Y.C. M.L. was also supported by the Graduate School of East China Normal University (CX2011003 and XRZZ2011023).
Authors' contributions
X.-Y.C. designed the study. M.L., F.-E.P. and J.Z. conducted the experiments. M.L., S.C. and X.-Y.C. performed analyses and wrote the manuscript. All authors approved the manuscript.
References
- 1.Ellstrand NC. 2014. Is gene flow the most important evolutionary force in plants? Am. J. Bot. 101, 737–753. ( 10.3732/ajb.1400024) [DOI] [PubMed] [Google Scholar]
- 2.Hu X-S, He F. 2006. Seed and pollen flow in expanding a species' range. J. Theor. Biol. 240, 662–672. ( 10.1016/j.jtbi.2005.11.002) [DOI] [PubMed] [Google Scholar]
- 3.Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG. 2005. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 14, 689–701. ( 10.1111/j.1365-294X.2004.02410.x) [DOI] [PubMed] [Google Scholar]
- 4.Chen X-Y, Fan X-X, Hu X-S. 2008. Roles of seed and pollen dispersal in natural regeneration of Castanopsis fargesii (Fagaceae): implications for forest management. Forest Ecol. Manag. 256, 1143–1150. ( 10.1016/j.foreco.2008.06.014) [DOI] [Google Scholar]
- 5.Walisch TJ, Colling G, Poncelet M, Matthies D. 2012. Effects of inbreeding and interpopulation crosses on performance and plasticity of two generations of offspring of a declining grassland plant. Am. J. Bot. 99, 1300–1313. ( 10.3732/ajb.1100479) [DOI] [PubMed] [Google Scholar]
- 6.Hadley AS, Betts MG. 2012. The effects of landscape fragmentation on pollination dynamics: absence of evidence not evidence of absence. Biol. Rev. 87, 526–544. ( 10.1111/j.1469-185X.2011.00205.x) [DOI] [PubMed] [Google Scholar]
- 7.Scopece G, Cozzolino S, Johnson SD, Schiestl FP. 2010. Pollination efficiency and the evolution of specialized deceptive pollination systems. Am. Nat. 175, 98–105. ( 10.1086/648555) [DOI] [PubMed] [Google Scholar]
- 8.Ahmed S, Compton SG, Butlin RK, Gilmartin PM. 2009. Wind-borne insects mediate directional pollen transfer between desert fig trees 160 kilometers apart. Proc. Natl Acad. Sci. USA 106, 20 342–20 347. ( 10.1073/pnas.0902213106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Compton SG. 2002. Sailing with the wind: dispersal by small flying insects. In Dispersal ecology (eds Bullock JM, Kenward RE, Hails RS.), pp. 113–133. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 10.Ashley MV. 2010. Plant parentage, pollination, and dispersal: how DNA microsatellites have altered the landscape. Crit. Rev. Plant Sci. 29, 148–161. ( 10.1080/07352689.2010.481167) [DOI] [Google Scholar]
- 11.Kremer A, et al. 2012. Long-distance gene flow and adaptation of forest trees to rapid climate change. Ecol. Lett. 15, 378–392. ( 10.1111/j.1461-0248.2012.01746.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manoel RO, Alves PF, Dourado CL, Gaino APSC, Freitas MLM, Moraes MLT, Sebbenn AM. 2012. Contemporary pollen flow, mating patterns and effective population size inferred from paternity analysis in a small fragmented population of the Neotropical tree Copaifera langsdorffii Desf. (Leguminosae-Caesalpinioideae). Conserv. Genet. 13, 613–623. ( 10.1007/s10592-011-0311-0) [DOI] [Google Scholar]
- 13.Nason JD, Herre EA, Hamrick JL. 1998. The breeding structure of a tropical keystone plant resource. Nature 391, 685–687. ( 10.1038/35607) [DOI] [Google Scholar]
- 14.Bronstein JL, Hossaert-McKey M. 1996. Variation in reproductive success within a subtropical fig/pollinator mutualism. J. Biogeogr. 23, 433–446. ( 10.1111/j.1365-2699.1996.tb00005.x) [DOI] [Google Scholar]
- 15.Muchhala N, Thomson JD. 2012. Interspecific competition in pollination systems: costs to male fitness via pollen misplacement. Funct. Ecol. 26, 476–482. ( 10.1111/j.1365-2435.2011.01950.x) [DOI] [Google Scholar]
- 16.Jones A. 2010. Reconciling field observations of dispersal with estimates of gene flow. Mol. Ecol. 19, 4379–4382. ( 10.1111/j.1365-294X.2010.04778.x) [DOI] [PubMed] [Google Scholar]
- 17.Yu H, Nason JD, Ge X, Zeng J. 2010. Slatkin's paradox: when direct observation and realized gene flow disagree. A case study in Ficus. Mol. Ecol. 19, 4441–4453. ( 10.1111/j.1365-294X.2010.04777.x) [DOI] [PubMed] [Google Scholar]
- 18.McLaren GF, Reid S, Colhoun KM. 2010. Long-distance movement of New Zealand flower thrips (Thrips obscuratus Crawford) (Thysanoptera: Thripidae) into Central Otago orchards. N Z Entomol. 33, 5–13. ( 10.1080/00779962.2010.9722185) [DOI] [Google Scholar]
- 19.Gibbs M, Breuker CJ, Van Dyck H. 2010. Flight during oviposition reduces maternal egg provisioning and influences offspring development in Pararge aegeria (L.). Physiol. Entomol. 35, 29–39. ( 10.1111/j.1365-3032.2009.00706.x) [DOI] [Google Scholar]
- 20.Beerli P, Palczewski M. 2010. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 185, 313–326. ( 10.1534/genetics.109.112532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ennos RA. 1994. Estimating the relative rates of pollen and seed migration among plants populations. Heredity 72, 250–259. ( 10.1038/hdy.1994.35) [DOI] [Google Scholar]
- 22.Wright S. 1951. The genetical structure of populations. Ann. Eugen. 15, 323–354. ( 10.1111/j.1469-1809.1949.tb02451.x) [DOI] [PubMed] [Google Scholar]
- 23.Weiblen GD. 2002. How to be a fig wasp. Annu. Rev. Entom. 47, 299–330. ( 10.1146/annurev.ento.47.091201.145213) [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Jiang Z-X, Compton SG, Liu M, Chen X-Y. 2011. Genetic diversity and differentiation of the extremely dwarf Ficus tikoua in Southwestern China. Biochem. Syst. Ecol. 39, 441–448. ( 10.1016/j.bse.2011.06.006) [DOI] [Google Scholar]
- 25.Zavodna M, Arens P, Van Dijk PJ, Partomihardjo T, Vosman B, Van Damme JMM. 2005. Pollinating fig wasps: genetic consequences of island recolonization. J. Evol. Biol. 18, 1234–1243. ( 10.1111/j.1420-9101.2005.00937.x) [DOI] [PubMed] [Google Scholar]
- 26.Ware AB, Compton SG. 1994. Dispersal of adult female fig wasps: 2. Movements between trees. Entomol. Exp. Appl. 73, 231–238. ( 10.1111/j.1570-7458.1994.tb01860.x) [DOI] [Google Scholar]
- 27.Compton SG, Ellwood MDF, Davis AJ, Welch K. 2000. The flight heights of chalcid wasps (Hymenoptera, Chalcidoidea) in a lowland Bornean rain forest: fig wasps are the high fliers. Biotropica 32, 515–522. ( 10.1111/j.1744-7429.2000.tb00497.x) [DOI] [Google Scholar]
- 28.Chen Y, Compton SG, Liu M, Chen X-Y. 2012. Fig trees at the northern limit of their range: the distributions of cryptic pollinators indicate multiple glacial refugia. Mol. Ecol. 21, 1687–1701. ( 10.1111/j.1365-294X.2012.05491.x) [DOI] [PubMed] [Google Scholar]
- 29.Liu M, Zhao R, Chen Y, Zhang J, Compton SG, Chen X-Y. 2014. Competitive exclusion among fig wasps achieved via entrainment of host plant flowering phenology. PLoS ONE 9, e97783 ( 10.1371/journal.pone.0097783) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Shi MM, Ai B, Gu JM, Chen XY. 2008. Genetic variation in island and mainland populations of Ficus pumila (Moraceae) in eastern Zhejiang of China. Symbiosis 45, 37–44. [Google Scholar]
- 31.Wang R, Ai B, Gao B-Q, Yu S, Li Y-Y, Chen X-Y. 2009. Spatial genetic structure and restricted gene flow in a functionally dioecious fig, Ficus pumila L. pumila (Moraceae). Popul. Ecol. 51, 307–315. ( 10.1007/s10144-008-0126-0) [DOI] [Google Scholar]
- 32.Chen Y, Li H-Q, Ma W-L. 2003. Egg-laying and pollinating behaviour of Blastophaga pumilae. Acta Entom. Sini. 46, 35–39. [Google Scholar]
- 33.Zhang J, Jiang K, Shi YS, Liu M, Chen XY. 2011. Development and polymorphism of microsatellite primers in Ficus pumila L. (Moraceae). Am. J. Bot. 98, e170–e172. ( 10.3732/ajb.1000340) [DOI] [PubMed] [Google Scholar]
- 34.Hamilton MB. 1999. Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol. Ecol. 8, 521–523. [PubMed] [Google Scholar]
- 35.Lahaye R, et al. 2008. DNA barcoding the floras of biodiversity hotspots. Proc. Natl Acad. Sci. USA 105, 2923–2928. ( 10.1073/pnas.0709936105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw J, et al. 2005. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 92, 142–166. ( 10.3732/ajb.92.1.142) [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 38.Liu M, Wang X-Y, Cui M-Y, Chen Y. 2009. Fifteen polymorphic microsatellite loci in Wiebesia pumilae (Hill) (Agaonidae). Conserv. Genet. Resour. 1, 189–191. ( 10.1007/s12686-009-9046-3) [DOI] [Google Scholar]
- 39.Rousset F. 2008. GENEPOP'007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. ( 10.1111/j.1471-8286.2007.01931.x) [DOI] [PubMed] [Google Scholar]
- 40.Rice WR. 1989. Analyzing tables of statistical tests. Evolution 43, 223–225. ( 10.2307/2409177) [DOI] [PubMed] [Google Scholar]
- 41.Goudet J. 1995. FSTAT (version 1.2): a computer program to calculate F-statistics. J. Hered. 86, 485–486. [Google Scholar]
- 42.Miller MP. 1997. Tools for population genetic analyses (TFPGA) v1.3: a windows program for the analysis of allozyme and molecular genetic data. Flagstaff, AZ: Department of Biological Sciences, Northern Arizona University. [Google Scholar]
- 43.Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. ( 10.2307/2408641) [DOI] [PubMed] [Google Scholar]
- 44.Oksanen J, et al. 2013. vegan: community ecology package version 2.0–9. http://vegan.r-forge.r-project.org/ .
- 45.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. ( 10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- 46.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. ( 10.1093/bioinformatics/btp187) [DOI] [PubMed] [Google Scholar]
- 47.Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50. [PMC free article] [PubMed] [Google Scholar]
- 48.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 49.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 ( 10.1038/nmeth.2109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. ( 10.1111/j.1365-294X.2005.02553.x) [DOI] [PubMed] [Google Scholar]
- 52.Liu M, Zhang J, Chen Y, Compton SG, Chen X-Y. 2013. Contrasting genetic responses to population fragmentation in a coevolving fig and fig wasp across a mainland–island archipelago. Mol. Ecol. 22, 4384–4396. ( 10.1111/mec.12406) [DOI] [PubMed] [Google Scholar]
- 53.R Development Core Team. 2014. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 54.Hamilton MB, Miller JR. 2002. Comparing relative rates of pollen and seed gene flow in the island model using nuclear and organelle measures of population structure. Genetics 162, 1897–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hedrick PW. 2005. A standardized genetic differentiation measure. Evolution 59, 1633–1638. ( 10.1111/j.0014-3820.2005.tb01814.x) [DOI] [PubMed] [Google Scholar]
- 56.Slatkin M. 1985. Rare alleles as indicators of gene flow. Evolution 39, 53–65. ( 10.2307/2408516) [DOI] [PubMed] [Google Scholar]
- 57.Alvarez N, et al. 2010. Phylogeography and historical biogeography of obligate specific mutualisms. In The biogeography of host–parasite interactions (eds Morand S, Krasnov BR.), pp. 31–39. Oxford, UK: Oxford University Press. [Google Scholar]
- 58.Kobmoo N, Hossaert-Mckey M, Rasplus JY, Kjellberg F. 2010. Ficus racemosa is pollinated by a single population of a single agaonid wasp species in continental South-East Asia. Mol. Ecol. 19, 2700–2712. ( 10.1111/j.1365-294X.2010.04654.x) [DOI] [PubMed] [Google Scholar]
- 59.Harrison RD. 2000. Repercussions of El Niño: drought causes extinction and the breakdown of mutualism in Borneo. Proc. R. Soc. Lond. B 267, 911–915. ( 10.1098/rspb.2000.1089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu H, Nason JD. 2013. Nuclear and chloroplast DNA phylogeography of Ficus hirta: obligate pollination mutualism and constraints on range expansion in response to climate change. New Phytol. 197, 276–289. ( 10.1111/j.1469-8137.2012.04383.x) [DOI] [PubMed] [Google Scholar]
- 61.Joseph KJ. 1958. Recherches sur les chalcidiens Blastophaga psenes L. et Philotrypesis caricae L. du figuier (Ficus carica L.). Ann. Sci. Nat. Zool. 13, 187–260. [Google Scholar]
- 62.Shanahan M, So S, Gompton SG, Gorlett R. 2001. Fig-eating by vertebrate frugivores: a global review. Biol. Rev. 76, 529–572. ( 10.1017/S1464793101005760) [DOI] [PubMed] [Google Scholar]
- 63.Corlett RT. 2006. Figs (Ficus, Moraceae) in urban Hong Kong, South China. Biotropica 38, 116–121. ( 10.1111/j.1744-7429.2006.00109.x) [DOI] [Google Scholar]
- 64.Shilton LA, Altringham JD, Compton SG, Whittaker RJ. 1999. Old World fruit bats can be long-distance seed dispersers through extended retention of viable seeds in the gut. Proc. R. Soc. Lond. B 266, 219–223. ( 10.1098/rspb.1999.0625) [DOI] [Google Scholar]
- 65.Francuski L, Ludoški J, Milankov V. 2013. Phenotypic diversity and landscape genetics of Eristalis tenax in a spatially heterogeneous environment, Durmitor Mountain (Montenegro). Ann. Zool. Fenn. 50, 262–278. ( 10.5735/085.050.0502) [DOI] [Google Scholar]
- 66.Zayed A, Packer L. 2007. The population genetics of a solitary oligolectic sweat bee, Lasioglossum (Sphecodogastra) oenotherae (Hymenoptera: Halictidae). Heredity 99, 397–405. ( 10.1038/sj.hdy.6801013) [DOI] [PubMed] [Google Scholar]
- 67.Crawley MJ. 1990. The population dynamics of plants. Phil. Trans. R. Soc. Lond. B 330, 125–140. ( 10.1098/rstb.1990.0187) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this paper can be accessed via Dryad: doi:10.5061/dryad.g50b0.