Abstract
Using animal models of alcoholism, previous studies suggest that neuropeptide Y (NPY) may be implicated in alcohol preference and consumption due to its role in the modulation of feeding and anxiety. Quantitative trait loci (QTL) analysis previously identified an interval on rat chromosome 4 that is highly associated with alcohol preference and consumption using an F2 population derived from inbred alcohol-preferring (iP) and -nonpreferring (iNP) rats. NPY mapped to the peak of this QTL region and was prioritized as a candidate gene for alcohol-seeking behavior in the iP and iNP rats. In order to identify a potential mechanism for reduced NPY protein levels documented in the iP rat, genetic and molecular components that influence NPY expression were analyzed between iP and iNP rats. Comparing the iP rat to the iNP rat, quantitative real-time polymerase chain reaction detected significantly decreased levels of NPY mRNA expression in the iP rat in the six brain regions tested: nucleus accumbens, frontal cortex, amygdala, hippocampus, caudate-putamen, and hypothalamus. In addition, the functional significance of three previously identified polymorphisms was assessed using in vitro expression analysis. The polymorphism defined by microsatellite marker D4Mit7 in iP rats reduced luciferase reporter gene expression in SK-N-SH neuroblastoma cells. These results suggest that differential expression of the NPY gene resulting from the D4mit7 marker polymorphism may contribute to reduced levels of NPY in discrete brain regions in the iP rats.
Keywords: neuropeptide Y, alcohol preference, selective breeding, mRNA expression, polymorphism, quantitative trait locus
Alcoholism is a complex disorder influenced by the interaction between multiple genes and the environment, exhibiting a heritability that ranges from 50 to 60% in both men and women (Heath et al., 1997). Thus far, however, only the protective effects of the alcohol metabolizing enzymes have been consistently replicated (Foroud and Li, 1999; Thomasson et al., 1993). To identify genetic factors that influence alcoholism, the alcohol-preferring (P) and -nonpreferring (NP) lines were developed from a randomly bred closed colony of Wistar rats through bidirectional selective breeding on the basis of alcohol consumption and preference (Li et al., 1991). In this model, P rats display the characteristics that are considered necessary for an animal model of alcoholism (Cicero, 1979). Subsequently, inbred P (iP) and NP (iNP) strains have been established that maintain highly discordant alcohol consumption scores.
Previous studies suggest that neuropeptide Y (NPY) is implicated in the modulation of alcohol consumption in P and NP rats (Cowen et al., 2004; Thiele and Badia-Elder, 2003; Pandey et al., 2003). P rats display lower levels of NPY immunoreactivity in various regions of the brain, including the central nucleus of the amygdala, hippocampus and the frontal cortex and higher levels in the paraventricular hypothalamic nucleus and arcuate nucleus of the hypothalamus (Ehlers et al., 1998; Hwang et al., 1999). Furthermore, decreased levels of NPY are associated with increased anxiety in P rats (Colombo, 1997; Stewart et al., 1993), and i.c.v. infusion of NPY has been shown to reduce ethanol intake in the P rat (Gilpin et al., 2003). NPY is localized to an interval that is highly associated with alcohol preference and consumption, mapping to a quantitative trait locus with a lod score of 9.2 on rat chromosome 4, using an F2 population bred from iP and iNP rats (Carr et al., 1998; Bice et al., 1998).
In addition, mice deficient in NPY exhibit increased alcohol consumption compared with wild-type mice, and an over-expression of NPY decreases ethanol consumption in transgenic mice (Thiele et al., 1998). In humans, a polymorphism in NPY (Leu7Pro) was significantly associated with dependence in European American and Finnish alcoholics, both exhibiting an increased frequency of the Pro7 allele compared with controls (Lappalainen et al., 2002; Zhu et al., 2003). Therefore, these studies have established NPY as an excellent candidate gene for alcoholism and alcohol preference, documenting an inverse relationship between NPY levels and alcohol consumption.
While a relationship between NPY and alcohol consumption has been established, the molecular and physiological mechanism responsible for this relationship remains to be elucidated. Because NPY is likely to function in memory, stress, hypertension, feeding and emotion (Heilig and Thorsell, 2002), this study was conducted to better understand the genetic and molecular components that influence NPY expression in iP and iNP rats. Thus, to further characterize the relationship between NPY and alcohol preference in P and NP rats, mRNA expression was evaluated in several brain regions between alcohol-naive iP and iNP rats, and transient transfection assays were performed on three previously identified polymorphisms (Bice et al., 1998) to determine their functional significance.
EXPERIMENTAL PROCEDURES
Dissection of brain regions
Five iP and four iNP alcohol-naive adult male rats were killed, and the entire brain was removed and dissected using the coordinates of Paxinos and Watson (Paxinos and Watson, 1998) to produce six subregional tissue samples: 1) nucleus accumbens, 2) frontal cortex, 3) amygdala, 4) hippocampus, 5) caudate-putamen, and 6) hypothalamus. These subregions were selected because they have been implicated in the mesocorticolimbic dopamine system, reciprocally interacting with the VTA to regulate alcohol-drinking behavior (McBride and Li, 1998). The nucleus accumbens and caudate putamen are dissected from a 2 mm section generated by a coronal cut at 2 mm anterior to the optic chiasm (Bregma 1.70 mm) and a coronal cut at the optic chiasm (Bregma −0.26 mm). The nucleus accumbens is dissected bilaterally by cutting below the rhinal fissure and trimming off the olfactory tubercles and cortical tissue at the ventral and ventrolateral borders of the slice. From the remaining tissue the caudate putamen (bilateral) is dissected below the corpus callosum leaving the septum hanging in the center. The hypothalamus is dissected from the tissue posterior to the optic chiasm by making an incision 2 mm to both the right and the left of the midline and a cut in front of the mammillary bodies as the caudal limit. The amygdala is dissected by a cut at the lateral borders of the lateral hypothalamus (Bregma −2.12 mm) and ventral of the rhinal fissure, with cortical tissue then removed at the lateral edges of the dissected slice. The caudal border of the amygdalar dissection is the rostroventral border of the CA3 subfield of Ammon’s horn (−4.16 mm). The entire hippocampus is dissected from the remaining brain by a midline incision between the hemispheres and rolling the hippocampus out of the cerebral cortex. All of the experiments utilized in this study conformed to local and international guidelines on the ethical use of animals in order to minimize the number of animals used and their suffering.
Isolation of RNA
The microdissected tissues were snap frozen on dry ice and stored at −80 °C until RNA isolation. RNA was isolated according to the RNeasy Midi manufacturer’s protocol (Qiagen, Valencia, CA, USA), and the isolated RNA was resuspended in diethyl-pyrocarbonate-treated water, treated with DNase I, and stored at −80 °C.
Quantitative real-time RT-PCR (polymerase chain reaction)
Using the ABI PRISM 7700 Sequence Detection System (PE Biosystems, Wellesley, MA, USA), the relative mRNA expression levels of NPY in the iP and iNP rats were determined in the nucleus accumbens, frontal cortex, amygdala, hippocampus, caudate-putamen, and hypothalamus by quantitative real-time RT-PCR (qRT-PCR). cDNA was generated from the six selected brain regions of each of the five iP and four iNP rats. cDNA template, generated from 50 ng total RNA, was added to each PCR reaction that contained 0.1 μM forward and reverse primers and SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Each reaction was performed in triplicate. The integrated software package accompanying the ABI PRISM 7700 was utilized to select the NPY-1F and NPY-1R primers for qRT-PCR (Table 1). The specificity of these primers was confirmed using agarose gel analysis. Each sample was amplified for 40 cycles, and the cycle threshold (Ct) was determined for each cDNA template. The Ct refers to the cycle number at which the fluorescence of the amplified product reached an arbitrary threshold that was within the exponential phase of amplification. Relative values of expression were determined for each sample using the standard curve method, and these values were normalized to the Ct values of GAPDH, a standard “housekeeping” control gene, using glyceraldehyde phosphate dehydrogenase (GAPDH)-F and GAPDH-R primers (Table 1) for PCR amplification. t-Tests were performed to detect significant differences in expression between the iP and iNP samples.
Table 1.
List of oligos
| Real-time PCR | |
| NPY-1F | 5′-agatactactccgctctgcga-3′ |
| NPY-1R | 5′-ggcattttctgtgctttctct-3′ |
| GAPDH-F | 5′-cagtcaaggctgagaatggga-3′ |
| GAPDH-R | 5′-gggatctcgctcctggaag-3′ |
| Vector construction | |
| Luc-1F | 5′-cggatccaactttgacttccaacag-3′ |
| Luc-1R | 5′-cggatccatcaactctggaaaaac-3′ |
| Luc-2F | 5′-cggatccaagtactcccttagagactg-3′ |
| Luc-2R | 5′-cggatcctaaaacacaagaggcaaa-3′ |
| Luc-3F | 5′-ctctagaatgaaacttgctctcctgactt-3′ |
| Luc-3R | 5′-atctagatagtcacaccaggtgttcagtc-3′ |
| Luc-4R | 5′-gacgatagtcatgccccgcg-3′ |
Luciferase reporter constructs
Reporter constructs were created to analyze the effects of three previously identified polymorphisms located in the 2nd intron, 3rd intron, and the 3′UTR (Bice et al., 1998; Larhammar et al., 1987) (Fig. 2). First, to amplify the regions encompassing these polymorphisms, three primer pairs were designed: Luc-1F and Luc-1R for the second intron, Luc-2F and Luc-2R for the third intron, and Luc-3F and Luc-3R for the 3′UTR (Table 1). Using iP and iNP genomic DNA, six fragments were amplified and ligated into the pCR2.1-TOPO vector. Sequence analysis was then performed to confirm that there were no errors. The resulting TOPO constructs were digested with either BamHI or XbaI, depending on the desired site of insertion of each fragment into the pGL-3 Promoter vector (Promega, Madison, WI, USA). These sites were determined based on the location of each fragment in the NPY gene. The resulting fragments were gel-isolated and ligated into a pGL-3 Promoter vector that was either digested with BamHI or XbaI. Thus, the fragments encompassing the polymorphisms in the 2nd and 3rd introns were ligated into the pGL-3 Promoter vector in a site following the SV40 late poly(A) signal, while the 3′UTR fragments were ligated between the luciferase gene and the SV40 late poly(A) signal (Fig. 3). To determine fragment orientation, each fragment was amplified using either the fragment specific forward or reverse primer paired with Luc-4R, a primer specific to the pGL-3 Promoter vector. The constructs containing the correct orientation of each fragment were isolated using the EndoFree Plasmid Maxi Kit (Qiagen).
Fig. 2.
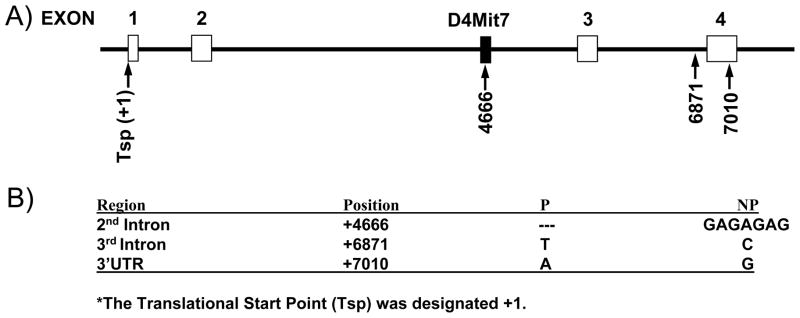
Polymorphisms identified in the NPY gene. Sequence analysis was previously performed on the NPY gene and identified three polymorphisms in the 2nd intron, 3rd intron and 3′UTR (Bice et al., 1998). (A) The diagram depicts the NPY gene. NPY’s four exons are represented with white boxes, while a black line depicts the intronic regions. The microsatellite marker D4Mit7 is represented with a black box. The three polymorphisms that have been identified in the NPY gene are labeled with arrows and their position relative to the translational start point (Tsp) that was designated +1. (B) Three polymorphisms are defined by their regional location in the NPY gene, their position relative to the Tsp (+1), and the sequence difference detected between iP and iNP rats.
Fig. 3.
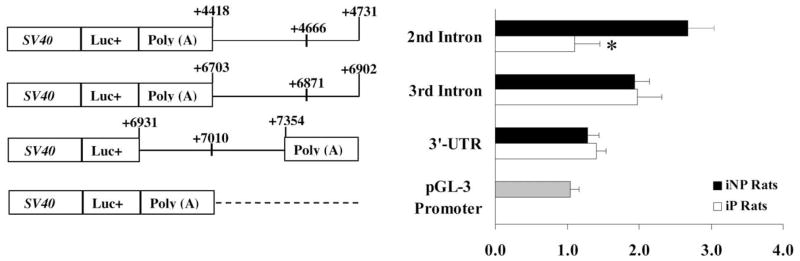
Functional importance of the NPY polymorphisms identified in the 2nd intron, 3rd intron, and 3′UTR. The Luc constructs were transiently transfected into SK-N-SH cells, and the resulting effect on luciferase expression was subsequently determined. SV40, luc+, and poly(A) denote the SV40 promoter, the luciferase gene, and the SV40 late poly(A) signal, respectively. The polymorphisms, located at +4666, +6871, and +7010, are noted in addition to the relative insertion site of each fragment into the pGL-3 Promoter vector. The activity of each construct was normalized to the internal control plasmid, pRL-CMV, and was expressed as fold change compared with the activity of the pGL-3 Promoter vector, which was designated as 1. The bars and fold change show the mean±S.E.M. of the results from seven independent transfection experiments performed in triplicate, using two different plasmid preparations. The significance of differences in the resulting mean values within and between multiple constructs was analyzed using ANOVA. An asterisk denotes a significant reduction in luc expression.
Transient transfection and luciferase assays
Human neuroblastoma SK-N-SH cells were cultured in Eagle’s Minimal Essential Medium containing 7.5% NaHCO3, 2 mM Glut-max, 0.1 mM non-essential amino acids, 1 mM pyruvate, 10% FBS (Invitrogen, Carlsbad, CA, USA) and were maintained at 37°C in a humidified 5% CO2 incubator. Twenty-four hours before transfection, 4.0×104 cells were plated into each well of a 24 well plate; 0.5 μg of each pGL-3 luciferase test plasmid was transfected per well using Tfx 50 reagent (Promega); 2.5 ng of CMV renilla vector (pRL-CMV) was cotransfected with each pGL-3-luciferase test plasmid to serve as an internal control for transfection efficiency. Cells were incubated for 48 h, washed, and harvested using passive lysis buffer. Cell extracts were assayed for firefly and Renilla luciferase activities in a TD-20/20 Luminometer, using the Dual-Luciferase Reporter Assay System (Promega). Luciferase assays were performed three to six times in triplicate, using plasmids that were independently purified at least twice.
RESULTS
NPY exhibits reduced mRNA expression in alcohol-naive iP rats compared with iNP rats in multiple brain regions
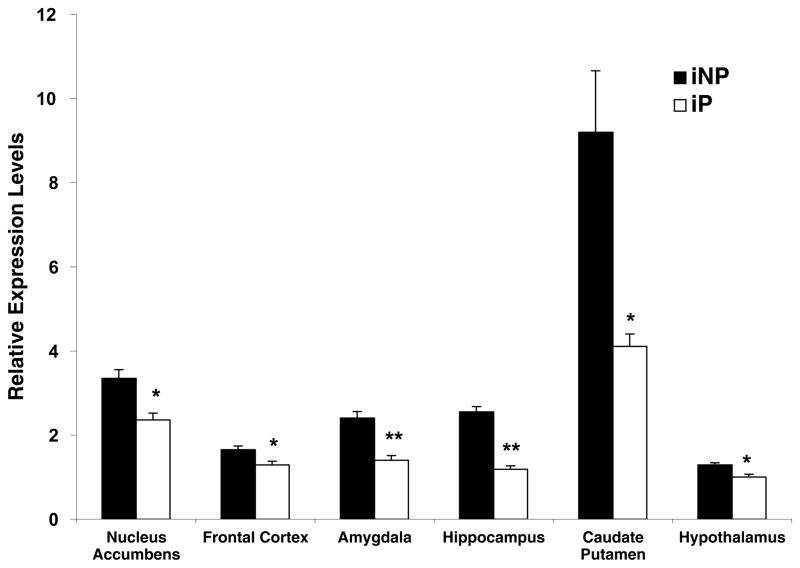
To assess the endogenous difference in NPY mRNA expression between iP and iNP rats, discrete brain regions dissected from alcohol-naive iP and iNP rats were analyzed using qRT-PCR. NPY mRNA expression was observed at significantly decreased levels in the nucleus accumbens (P=0.006), the frontal cortex (P=0.04), the amygdala (P=0.0004), the hippocampus (P<0.00001), the caudate putamen (P=0.02), and the hypothalamus (P=0.02) in the iP rat (Fig. 1). The greatest difference in expression comparing iNP to iP was in the hippocampus and caudate putamen (2.2-fold difference), and the smallest difference in expression was in the frontal cortex and hypothalamus (1.3-fold difference; Fig. 1).
Fig. 1.
NPY mRNA expression is decreased in alcohol-naive P rats compared with NP rats. qRT-PCR analysis was utilized to compare the relative levels of NPY mRNA between alcohol-naive iP and iNP rats in the nucleus accumbens, the frontal cortex, the amygdala, the hippocampus, the caudate putamen, and the hypothalamus. All values were determined using the standard curve method and compared with mRNA expression in the iP hypothalamus, which was arbitrarily designated 1. The graph depicts the mean±S.E.M. of the results from five independent iP and four independent iNP experiments performed in triplicate, using separate preparations of cDNA. Significant differences in regional NPY mRNA expression between iP and iNP rats was determined using the Student’s t-test. * P<0.05; ** P<0.0005.
Microsatellite marker D4Mit7 polymorphism reduced luciferase activity in vitro using SK-N-SH cells
The microsatellite marker D4Mit7 is located in the 2nd intron of NPY. To determine the probable effects of the previously identified polymorphisms on NPY gene expression, PCR was employed to generate DNA fragments containing the polymorphisms in the 2nd intron (D4Mit7), 3rd intron and 3′UTR. The fragments containing the 2nd and 3rd intron polymorphisms were cloned into the pGL-3 Promoter luciferase vector downstream the SV40 late poly(A) signal, while the 3′UTR fragments were ligated between the luciferase gene and the SV40 late poly(A) signal (Fig. 2). All constructs were transiently transfected into SK-N-SH cells, which constitutively express the endogenous NPY gene. Luciferase expression was expressed in fold-change compared with the pGL-3 Promoter-luciferase (luc) construct (Fig. 3). The iP-(2nd intron)-luc construct defined by the D4Mit7 microsatellite marker exhibited a significant decrease in expression in SK-N-SH cells compared with the iNP-(2nd intron)-luc construct (P<0.05). The 3rd intron-luc and the 3′UTR-luc vectors did not significantly alter luciferase activity in SK-N-SH cells.
DISCUSSION
In this study, alcohol-naive iP rats exhibited a reduced level of NPY mRNA expression compared with iNP rats in the nucleus accumbens, frontal cortex, amygdala, hippocampus, caudate putamen, and hypothalamus. Three polymorphisms in the NPY gene were assessed for their functional significance in modulating NPY expression. In vitro expression analysis using neuroblastoma SK-N-SH cells yielded a significant decrease in expression of the iP-(2nd intron)-luc construct compared with the iNP-(2nd intron)-luc construct (P<0.05). These results suggest that an endogenous decrease in NPY expression may be influencing the phenotypic decrease in NPY protein expression that has been previously observed in various brain regions when comparing P and NP rats.
The reduced level of NPY mRNA expression observed in alcohol-naive iP rats compared with iNP rats corroborated the previous evidence of a reduction in NPY protein levels in central nucleus of the amygdala, hippocampus, and the frontal cortex (Ehlers et al., 1998; Hwang et al., 1999). In contrast, iP animals exhibited significantly lower levels of NPY mRNA expression in the hypothalamus in the present study, whereas, in a previous study, higher levels of NPY immunoreactivity were detected in the paraventricular hypothalamic nucleus of P rats than NP rats (Hwang et al., 1999). Lack of correlation between mRNA and protein levels may have resulted from procedural differences between the two studies. In our study, the entire hypothalamus was dissected, whereas a very specific region of the hypothalamus was quantified in the immunohistochemistry studies. Lower levels of NPY mRNA have been documented in high alcohol drinking rats compared with the low alcohol drinking rats (Hwang et al., 1999). No difference in NPY mRNA expression was observed between NP rats compared with P rats (Caberlotto et al., 2001). Together, these data suggest that the NPY expression in the hypothalamus may not play a defining role in the modulation of alcohol preference, whereas its expression in the amygdala, a region implicated in anxiety and the reinforcing properties of alcohol may be influential in the regulation of alcohol-seeking behavior (Koob et al., 1998; Cowen et al., 2004).
In vitro expression analysis revealed a significant decrease in luciferase expression in iP-(2nd intron)-luc construct compared with the iNP-(2nd intron)-luc, providing a potential mechanism for the reduced mRNA and protein expression. Furthermore, the functional insignificance of the 3′UTR polymorphism, a primary site for the regulation of mRNA stability (Nair and Menon, 2000; Wang and Kiledjian, 2000; Loflin and Lever, 2001), suggests that NPY expression is regulated at the transcriptional level. Although neuroblastoma cells do not fully reflect regulatory mechanisms underlying NPY expression in neuronal cells, they show that the D4Mit7 polymorphism is functional in a cell line that expresses NPY.
The reduced expression exhibited by one variant of the D4Mit7 polymorphism is consistent with the lower NPY mRNA and protein levels detected in discrete brain regions of the iP strain compared with the iNP strain. The polymorphism defined by the marker D4Mit7 is localized to a GC rich region, possibly involved in transcriptional regulation. Intronic polymorphisms have been associated with regulation of gene expression (Fiskerstrand et al., 1999; Arnold et al., 2000; Bream et al., 2000; Agarwal et al., 2000). Thus, polymorphic regions, previously used as genetic markers, may function in regulating gene expression (Katsuki et al., 1996). It is postulated that these alternating purine-pyrimidine tracts form Z-DNA (left-handed helices) or cruciform regions that influence gene expression, and some CA repeat sequences do exhibit moderate enhancer activities (Hamada et al., 1984). However, the difference in expression associated with the D4Mit7 polymorphism may not fully account for the observed expression difference in NPY detected between iP and iNP rats. Therefore, additional studies will be necessary to define the functional significance of additional polymorphisms discovered in the NPY gene. It is also possible that sequencing additional 5′region will identify other functional polymorphisms since regulatory elements can be located many kilobases upstream in the gene.
The inverse relationship between levels of NPY in various regions of the brain and alcohol consumption has been documented in previous literature (Badia-Elder et al., 2001; Thiele et al., 1998). However, the molecular mechanism of NPY remains to be elucidated even in the P and NP rats where the difference in NPY expression appears to be well defined (Ehlers et al., 1998; Hwang et al., 1999). This study has attempted to further characterize NPY expression in order to ultimately distinguish NPY expression as a primary source of the decrease in NPY protein expression observed in various brain regions between P and NP rats.
In conclusion, the effects of NPY levels on alcohol preference in the P and NP rats have been established in previous studies (Badia-Elder et al., 2001). This study provided evidence of decreased mRNA expression in several brain regions, including the nucleus accumbens, frontal cortex, amygdala, hippocampus, caudate putamen, and hypothalamus. The previously identified D4Mit7 marker polymorphism appeared functional, while the 3rd intron and 3′UTR polymorphisms produced no change in luc expression. All in all, these results further implicated the regulation of NPY gene expression as a likely source for the difference in NPY levels detected between iP and iNP rats.
Acknowledgments
U.S. Public Service grants AA10707 and AA07611.
Abbreviations
- Ct
cycle threshold
- GAPDH
glyceraldehyde phosphate dehydrogenase
- iNP
inbred alcohol-nonpreferring
- iP
inbred alcohol-preferring
- NP
alcohol-nonpreferring
- NPY
neuropeptide Y
- P
alcohol-preferring
- PCR
polymerase chain reaction
- qRT-PCR
quantitative real-time RT-PCR
References
- Agarwal AK, Giacchetti G, Lavery G, Nikkila H, Palermo M, Ricketts M, McTernan C, Bianchi G, Manunta P, Strazzullo P, Mantero F, White PC, Stewart PM. CA-Repeat polymorphism in intron 1 of HSD11B2: effects on gene expression and salt sensitivity. Hypertension. 2000;36:187–194. doi: 10.1161/01.hyp.36.2.187. [DOI] [PubMed] [Google Scholar]
- Arnold R, Maueler W, Bassili G, Lutz M, Burke L, Epplen TJ, Renkawitz R. The insulator protein CTCF represses transcription on binding to the (gt)(22)(ga)(15) microsatellite in intron 2 of the HLA-DRB1(*)0401 gene. Gene. 2000;253:209–214. doi: 10.1016/s0378-1119(00)00271-7. [DOI] [PubMed] [Google Scholar]
- Badia-Elder NE, Stewart RB, Powrozek TA, Roy KF, Murphy JM, Li TK. Effect of neuropeptide Y (NPY) on oral ethanol intake in Wistar, alcohol-preferring (P), and -nonpreferring (NP) rats. Alcohol Clin Exp Res. 2001;25:386–390. [PubMed] [Google Scholar]
- Bice P, Foroud T, Bo R, Castelluccio P, Lumeng L, Li TK, Carr LG. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm Genome. 1998;9:949–955. doi: 10.1007/s003359900905. [DOI] [PubMed] [Google Scholar]
- Bream JH, Carrington M, O’Toole S, Dean M, Gerrard B, Shin HD, Kosack D, Modi W, Young HA, Smith MW. Polymorphisms of the human IFNG gene noncoding regions. Immunogenetics. 2000;51:50–58. doi: 10.1007/s002510050008. [DOI] [PubMed] [Google Scholar]
- Caberlotto L, Thorsell A, Rimondini R, Sommer W, Hyytia P, Heilig M. Differential expression of NPY and its receptors in alcohol-preferring AA and alcohol-avoiding ANA rats. Alcohol Clin Exp Res. 2001;25:1564–1569. [PubMed] [Google Scholar]
- Carr LG, Foroud T, Bice P, Gobbett T, Ivashina J, Edenberg H, Lumeng L, Li TK. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin Exp Res. 1998;22:884–887. [PubMed] [Google Scholar]
- Cicero TJ. A critique of animal analogues of alcoholism. In: Majchrowicz E, Noble EP, editors. Biochemistry and pharmacology of ethanol. Vol. 2. New York: Plenum Press; 1979. pp. 533–560. [Google Scholar]
- Colombo G. ESBRA-Nordmann 1996 Award Lecture: ethanol drinking behaviour in Sardinian alcohol-preferring rats. Alcohol Alcohol. 1997;32:443–453. doi: 10.1093/oxfordjournals.alcalc.a008279. [DOI] [PubMed] [Google Scholar]
- Cowen MS, Chen F, Lawrence AJ. Neuropeptides: implications for alcoholism. J Neurochem. 2004;89:273–285. doi: 10.1111/j.1471-4159.2004.02394.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Li TK, Lumeng L, Hwang BH, Somes C, Jimenez P, Mathe AA. Neuropeptide Y levels in ethanol-naive alcohol-preferring and nonpreferring rats and in Wistar rats after ethanol exposure. Alcohol Clin Exp Res. 1998;22:1778–1782. [PubMed] [Google Scholar]
- Fiskerstrand CE, Lovejoy E, Gerrard L, Quinn JP. An intronic domain within the rat preprotachykinin-A gene containing a CCCT repetitive motif acts as an enhancer in differentiating embryonic stem cells. Neurosci Lett. 1999;263:141–144. doi: 10.1016/s0304-3940(99)00127-5. [DOI] [PubMed] [Google Scholar]
- Foroud T, Li TK. Genetics of alcoholism: a review of recent studies in human and animal models. Am J Addict. 1999;8:261–278. doi: 10.1080/105504999305677. [DOI] [PubMed] [Google Scholar]
- Gilpin NW, Stewart RB, Murphy JM, Li TK, Badia-Elder NE. Neuropeptide Y reduces oral ethanol intake in alcohol-preferring (P) rats following a period of imposed ethanol abstinence. Alcohol Clin Exp Res. 2003;27:787–794. doi: 10.1097/01.ALC.0000065723.93234.1D. [DOI] [PubMed] [Google Scholar]
- Hamada H, Seidman M, Howard BH, Gorman CM. Enhanced gene expression by the poly(dT-dG). poly (dC-dA) sequence. Mol Cell Biol. 1984;4:2622–2630. doi: 10.1128/mcb.4.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PAF, Dinwiddie SH, Slutske WS, Statham DJ, Dunne MP, Whitfield J, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Heilig M, Thorsell A Brain neuropeptide Y (NPY) in stress and alcohol dependence. Rev Neurosci. 2002;13:85–94. doi: 10.1515/revneuro.2002.13.1.85. [DOI] [PubMed] [Google Scholar]
- Hwang BH, Zhang JK, Ehlers CL, Lumeng L, Li TK. Innate differences of neuropeptide Y (NPY) in hypothalamic nuclei and central nucleus of the amygdala between selectively bred rats with high and low alcohol preference. Alcohol Clin Exp Res. 1999;23:1023–1030. [PubMed] [Google Scholar]
- Katsuki S, Kato J, Nakajima M, Inui N, Sasaki K, Kohgo Y, Niitsu Y. Analysis of CA repeats in first intron of class I ADH gene in Long-Evans Cinnamon rats developing fatal intoxication after ethanol intake. Alcohol Clin Exp Res. 1996;20:33A–35A. doi: 10.1111/j.1530-0277.1996.tb01724.x. [DOI] [PubMed] [Google Scholar]
- Koob GF, Roberts AJ, Schulteis G, Parsons LH, Heyser CJ, Hyytia P, Merlo-Pich E, Weiss F. Neurocircuitry targets in ethanol reward and dependence. Alcohol Clin Exp Res. 1998;22:3–9. [PubMed] [Google Scholar]
- Larhammar D, Ericsson A, Persson H. Structure and expression of the rat neuropeptide Y gene. Proc Natl Acad Sci USA. 1987;84:2068–2072. doi: 10.1073/pnas.84.7.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen J, Kranzler HR, Malison R, Price LH, Van Dyck C, Rosenheck RA, Cramer J, Southwick S, Charney D, Krystal J, Gelernter J. A functional neuropeptide Y Leu7Pro polymorphism associated with alcohol dependence in a large population sample from the United States. Arch Gen Psychiatry. 2002;59:825–831. doi: 10.1001/archpsyc.59.9.825. [DOI] [PubMed] [Google Scholar]
- Li T-K, Lumeng L, Doolittle DP, Carr LG. Molecular associations of alcohol-seeking behavior in rat lines selectively bred for high and low voluntary ethanol drinking. Alcohol Alcohol. 1991;1:121–124. [PubMed] [Google Scholar]
- Loflin P, Lever JE. HuR binds a cyclic nucleotide-dependent, stabilizing domain in the 3′untranslated region of Na(+)/glucose cotransporter (SGLT1) mRNA. FEBS Lett. 2001;509:267–271. doi: 10.1016/s0014-5793(01)03176-3. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Li TK. Animal models of alcoholism: neurobiology of high alcohol-drinking behavior in rodents. Crit Rev Neurobiol. 1998;12:339–369. doi: 10.1615/critrevneurobiol.v12.i4.40. [DOI] [PubMed] [Google Scholar]
- Nair AK, Menon KM. Regulatory role of the 3′untranslated region of luteinizing hormone receptor: effect on mRNA stability. FEBS Lett. 2000;471:39–44. doi: 10.1016/s0014-5793(00)01365-x. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Carr LG, Heilig M, Ilveskoski E, Thiele TE. Neuropeptide y and alcoholism: genetic, molecular, and pharmacological evidence. Alcohol Clin Exp Res. 2003;27:149–154. doi: 10.1097/01.ALC.0000052706.21367.0E. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotoxic coordinates. 4. San Diego, CA: Academic Press; 1998. [Google Scholar]
- Stewart RB, Gatto GJ, Lumeng L, Li T-K, Murphy JM. Comparison of alcohol-preferring (P) and nonpreferring (NP) rats on tests of anxiety and for the anxiolytic effects of ethanol. Alcohol. 1993;10:1–10. doi: 10.1016/0741-8329(93)90046-q. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Marsh DJ, Ste Marie L, Bernstein IL, Palmiter RD. Ethanol consumption and resistance are inversely related to neuropeptide Y levels. Nature. 1998;396:366–369. doi: 10.1038/24614. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Badia-Elder NE. A role for neuropeptide Y in alcohol intake control: evidence from human and animal research. Physiol Behav. 2003;79:95–101. doi: 10.1016/s0031-9384(03)00109-4. [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Crabb DW, Edenberg HJ, Li TK. Alcohol and aldehyde dehydrogenase polymorphisms and alcoholism. Behav Genet. 1993;23:131–136. doi: 10.1007/BF01067417. [DOI] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol Cell Biol. 2000;20:6334–6341. doi: 10.1128/mcb.20.17.6334-6341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Pollak L, Mottagui-Tabar S, Wahlestedt C, Taubman J, Virkkunen M, Goldman D, Heilig M. NPY Leu7Pro and alcohol dependence in Finnish and Swedish populations. Alcohol Clin Exp Res. 2003;27:19–24. doi: 10.1097/01.ALC.0000050642.62233.44. [DOI] [PubMed] [Google Scholar]